Significance
The opportunistic pathogen Streptococcus pneumoniae (pneumococcus) participates in horizontal gene transfer through genetic competence and produces antimicrobial peptides called “bacteriocins.” Here, we show that the competence and bacteriocin-related ABC transporters ComAB and BlpAB share the same substrate pool, resulting in bidirectional crosstalk between competence and bacteriocin regulation. We also clarify the role of each transporter in bacteriocin secretion and show that, based on their transporter content, pneumococcal strains can be separated into a majority opportunist group that uses bacteriocins only to support competence and a minority aggressor group that uses bacteriocins in broader contexts. Our findings will impact how bacteriocin regulation and production is modeled in the many other bacterial species that use ComAB/BlpAB-type transporters.
Keywords: pneumococcus, competence, bacteriocin, regulation, ABC transporter
Abstract
The opportunistic pathogen Streptococcus pneumoniae (pneumococcus) uses natural genetic competence to increase its adaptability through horizontal gene transfer. One method of acquiring DNA is through predation of neighboring strains with antimicrobial peptides called “bacteriocins.” Competence and production of the major family of pneumococcal bacteriocins, pneumocins, are regulated by the quorum-sensing systems com and blp, respectively. In the classical paradigm, the ABC transporters ComAB and BlpAB each secretes its own system’s signaling pheromone and in the case of BlpAB also secretes the pneumocins. While ComAB is found in all pneumococci, only 25% of strains encode an intact version of BlpAB [BlpAB(+)] while the rest do not [BlpAB(−)]. Contrary to the classical paradigm, it was previously shown that BlpAB(−) strains can activate blp through ComAB-mediated secretion of the blp pheromone during brief periods of competence. To better understand the full extent of com-blp crosstalk, we examined the contribution of each transporter to competence development and pneumocin secretion. We found that BlpAB(+) strains have a greater capacity for competence activation through BlpAB-mediated secretion of the com pheromone. Similarly, we show that ComAB and BlpAB are promiscuous and both can secrete pneumocins. Consequently, differences in pneumocin secretion between BlpAB(+) and BlpAB(−) strains derive from the regulation and kinetics of transporter expression rather than substrate specificity. We speculate that BlpAB(−) strains (opportunists) use pneumocins mainly in a narrowly tailored role for DNA acquisition and defense during competence while BlpAB(+) strains (aggressors) expand their use for the general inhibition of rival strains.
The opportunistic pathogen, Streptococcus pneumoniae (pneumococcus) can cause serious illnesses such as pneumonia, meningitis, and bacteremia, with the greatest disease burden in the very young and the elderly. The natural niche of pneumococcus is the human nasopharynx, and colonization of this niche is a prerequisite for invasive pneumococcal disease. Pneumococcus colonizes up to 60% of young children (1, 2). As many as half of those who are colonized carry multiple pneumococcal strains (3).
Pneumococcus, a naturally competent bacterium (4), can exploit the large pool of genetic material available to it (1, 2, 5) in the nasopharynx. Natural competence allows pneumococcus to take up new genetic material through horizontal gene transfer and recombination. Multiple studies have documented that recombination occurs with great frequency in pneumococcal lineages that are globally distributed (6), geographically isolated (7), and even confined to a single patient (8). Additionally, to compete with other bacteria found in the nasopharynx, pneumococcus produces small antimicrobial peptides called “bacteriocins.” Pneumocins are the major family of bacteriocins encoded by pneumococcus. The pneumocin locus, blp, is found in all sequenced strains of pneumococcus (9). Pneumocin-producing organisms inhibit sensitive strains and have a fitness advantage in both in vitro biofilms and competitive colonization of the mouse nasopharynx (10, 11).
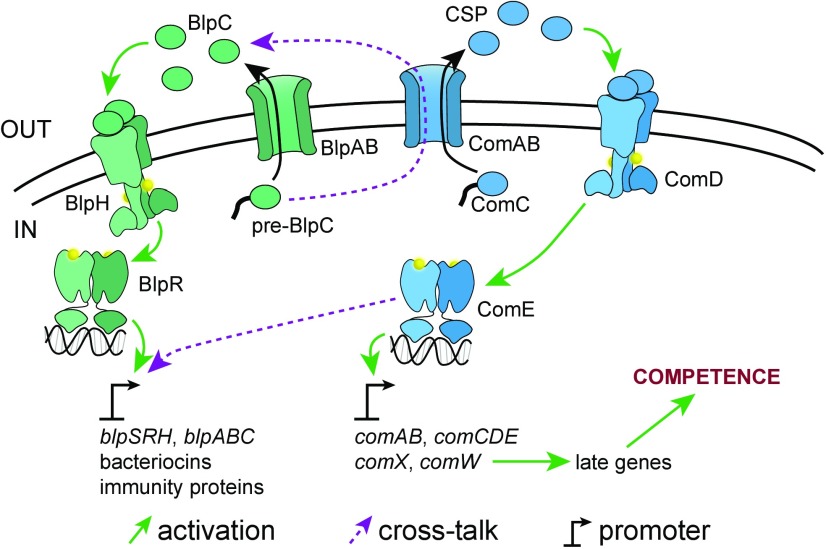
Both competence and pneumocin production in pneumococcus are under strict regulation by two separate but similar systems (Fig. 1). The com system regulates competence. In this system, a peptide prepheromone, ComC, is processed and secreted by a transporter complex ComAB (12, 13). After processing and secretion, the mature pheromone, now called “competence-stimulating peptide” (CSP), accumulates extracellularly. Once a threshold concentration is reached, CSP signals through the ComDE two-component system to up-regulate the set of so-called “early (competence) genes” (14). The early genes include comAB and comCDE, creating a positive feedback loop. Up-regulation of early gene expression starts a regulatory cascade mediated by the alternative sigma factor ComX that ultimately leads to competence development (15). The com system integrates many environmental and physiological signals, such as cell density (16), pH (17), antibiotic stress (18), and protein mistranslation (19). As a result, the propensity for competence activation can differ greatly from one set of conditions to another.
Fig. 1.
com and blp regulation in pneumococcus.
Meanwhile, the blp locus regulates pneumocin production in a manner similar to com and competence (20). In the prototypical case, a small peptide pheromone, BlpC, is processed and secreted by the BlpAB transporter complex. Mature BlpC then signals through the BlpHR two-component system to up-regulate the entire blp locus. Unlike com, the entire blp regulon is directly controlled by BlpR. The up-regulation of the regulatory system forms another positive feedback loop, while the up-regulation of the so-called “bacteriocin immunity region” (BIR) within the blp locus results in the production of a diverse array of pneumocins and their immunity proteins. As with competence, the propensity for blp activation is environment- and context-dependent.
Due to the autoinducing nature of com and blp, activation tends to proceed synchronously among all cells within a population once the pheromone concentration threshold is reached (13). However, different strains can encode different pherotypes of CSP and BlpC along with matched cognate ComD and BlpH receptors (21, 22). In general, each pherotype efficiently activates only its cognate receptor type. Therefore, CSP/BlpC signaling and synchronous com/blp activation is restricted by pherotype. Pherotype diversity among pneumococci may have evolved as a method for cells to privilege clonal or closely related cells which are more likely to have a matched pherotype. Such cells would then share in the benefits of competence activation or be protected from pneumocin-mediated killing while strains with mismatched pherotypes would not.
While com and blp were originally thought to operate independently, two recent studies have shown that com positively influences the regulation of blp (11, 23). This occurs through two mechanisms (Fig. 1, purple dotted arrows): (i) ComE directly up-regulates the transcription of blp genes, and (ii) ComAB processes and secretes BlpC in addition to ComC/CSP. As a result, com activation also induces blp activation. This crosstalk from com to blp allows the coordinated regulation of the two systems. In a biofilm model, pneumocin production resulted in an increase in transformation efficiency (11). The competence regulon itself encodes so-called “fratricide effectors” which target and kill noncompetent pneumococci (24). The role of fratricide seems to be to increase access to extracellular DNA during competence (25, 26). Therefore, pneumococcus may have evolved com-to-blp crosstalk to increase access to genetic material during competence by using pneumocins to augment fratricide.
While the com regulatory system is found intact in nearly all strains of pneumococcus, 75% of sequenced pneumococcal strains contain mutations that disrupt the blpA and/or blpB genes (27). Strains harboring these mutations are predicted to be incapable of making functional BlpAB transporter. These BlpAB(−) strains are dependent on com activation to activate their own blp system (com-dependent blp activation) via the com-to-blp crosstalk described above (11, 23). They can also respond to BlpC pheromone secreted by other strains, since their BlpHR sensory apparatus remains intact. However, it is not clear if BlpAB(−) strains can effect pneumocin-mediated killing. Pneumocins, similar to the pheromones CSP and BlpC, need to be processed and secreted before they can exert their effects. BlpAB is the putative transporter for pneumocins, so the loss of BlpAB would be expected to render cells incapable of secreting active pneumocins. Two previous studies have reported conflicting results on whether BlpAB(−) strains exhibit pneumocin-mediated inhibitory activity (27, 28).
At the heart of both the com and blp systems lie the peptide transporters ComAB and BlpAB, which belong to the superfamily of ATP-binding cassette (ABC) transporters. ComAB and BlpAB are formed from A and B subunits. The B subunits, encoded by comB and blpB, serve unknown functions. The A subunits, encoded by comA and blpA, contain all the known functional domains. Each A subunit contains an N-terminal peptidase domain followed by the channel-forming transmembrane domain and a C-terminal nucleotide-binding domain (NBD) (29). The peptidase domain catalyzes the first step of transport called “processing”: the cleavage of the peptide substrate immediately C-terminal to a conserved double-glycine (GG) motif (sometimes also GA or GS), thereby removing the substrate’s N-terminal signal sequence (29, 30). After processing, the now-mature peptide is secreted out of the cell using energy provided by the ATP-hydrolyzing NBD. While ComAB’s ability to secrete both ComC and BlpC has revealed some degree of promiscuity in its substrate selection process, it is unknown if BlpAB shares this property or just how far it extends.
In this study, we demonstrate that BlpAB can secrete CSP and show the functional significance of this in the 25% of strains that are BlpAB(+). In addition, we demonstrate that ComAB can process and secrete pneumocins, suggesting that BlpAB(−) strains can support pneumocin-mediated inhibition but only under competence-permissive conditions. We also show that temporal regulation of ComAB and BlpAB expression leads to differences in pneumocin transport efficiency in BlpAB(+) and BlpAB(−) strains. Finally, we demonstrate that BlpAB(+) strains are more effective competitors than BlpAB(−) strains in a mouse nasopharyngeal colonization model. These findings suggest that the diversification of the pneumococcal population into BlpAB(+) and BlpAB(−) types occurred to allow for a small pool of costly antagonistic strains that can eliminate competitors when environmental conditions mandate this approach.
Results
Development of a com/blp Dual Luciferase Reporter.
To facilitate the study of com–blp crosstalk, we developed a dual luciferase reporter system to monitor com and blp activation in the same population of cells in real time. For our two luciferases, we chose the NanoLuc luciferase (Nluc; Promega) and the red-emitting Luciola italica luciferase (RFluc; Targeting Systems). We selected these two candidates because their emission spectra have little overlap and can be conveniently separated using optical filters (Fig. 2B), and they use different substrates with no cross-reactivity (31, 32). We also validated that the Nluc substrate furimazine (also known as “Nano-Glo substrate”) was suitable for use in live pneumococcal cultures (SI Appendix, Fig. S1).
Fig. 2.
The dual com/blp luciferase reporter allows simultaneous monitoring of com and blp activation kinetics. (A) Schematic representation of PcomAB-Nluc and PBIR-RFLuc constructs. Bent arrows denote promoters. Hairpins denote transcriptional terminators. (B) Diagram of the emission spectra of Nluc (blue) and RFluc (red) and the optical filter sets used to separate their signals. (C) An R6-derived wild-type dual reporter strain was grown in a 96-well plate in THY broth at pH 7.1. Cells were monitored for growth (right y axes, light shading), com activation (Upper, left y axis, dark shading), and blp activation (Lower, left y axis, dark shading). At t = 60 min, cells were treated with either mock treatment (open circles) or 100 ng/mL CSP (closed squares). Data are plotted as the average ± SD of four wells. (D) An R6-derived wild-type dual reporter strain was grown in a 96-well plate in THY broth at pH 7.4 without treatment. Cells were monitored as in C. Data from two representative wells are shown, one with spontaneous com and blp activation (filled circles), and one without (open circles). RLU, relative luminescence units.
We created a transcriptional fusion of Nluc to the ComE-regulated comAB promoter (PcomAB) and a transcriptional fusion of RFluc to the BlpR-regulated proximal BIR promoter (PBIR) (Fig. 2A). These two constructs were then transformed into wild-type unencapsulated (R6) and encapsulated (D39) strains engineered to encode intact versions of blpAB. In these dual reporters Nluc activity correlates with com activation, and RFluc activity correlates with blp activation (SI Appendix).
We confirmed that the dual reporter successfully recapitulates what has been previously published regarding the response of the com and blp systems to CSP treatment (11, 23, 33, 34) (Fig. 2C and SI Appendix, Fig. S2A). Using it, we are also able to detect spontaneous com and blp activation events when cells are grown under permissive conditions (Fig. 2D and SI Appendix, Fig. S2B). Therefore, the dual reporter provides accurate and convenient simultaneous readouts of com and blp activation in live cells.
BlpAB Processes and Secretes ComC/CSP.
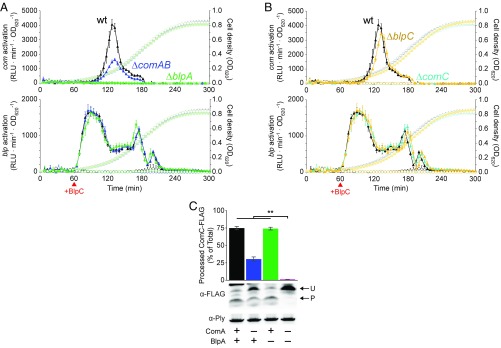
We used the dual luciferase reporter to assay the response of cells to treatment with synthetic BlpC pheromone during growth in THY broth (Todd Hewitt broth + 0.5% yeast extract) at pH 7.1, which is nonpermissive for spontaneous com activation. We noticed that BlpC treatment led to activation of the com system (Fig. 3 A and B). This BlpC-induced com activation was delayed by roughly 30 min compared with activation of the blp system, which responds almost immediately to BlpC. Given this delay, we reasoned that it was unlikely this com activation resulted from direct up-regulation of PcomAB by BlpR. Moreover, BlpC-induced com activation is completely abolished in the ΔblpA and ΔcomC mutants but persists in the ΔcomAB and ΔblpC mutants (Fig. 3 A and B). These data led us to hypothesize that the mechanism underlying the cross-activation was BlpAB-mediated CSP secretion.
Fig. 3.
BlpAB processes and secretes ComC/CSP. (A and B) R6-derived dual reporter strains were grown in 96-well plates in THY broth at pH 7.1. Cells were monitored for growth (right y axes, light shading), com activation (Upper, left y axis, dark shading), and blp activation (Lower, left y axis, dark shading). At t = 60 min, cells were treated with either mock treatment (open circles) or 100 ng/mL BlpC (closed triangles). Data from one representative experiment are shown and are plotted as the average ± SD of four wells. (C) ComC-FLAG processing in wild-type (black), ΔcomAB (blue), ΔblpA (green), and ΔcomABΔblpA (magenta) strains. Cells were grown in THY broth at pH 7.1, induced with 100 ng/mL CSP and 100 ng/mL BlpC, and whole-cell lysates were collected 30 min later for Western blot. Representative blots are presented, showing the unprocessed (U) and processed (P) forms of ComC-FLAG and a pneumolysin (Ply) loading control. The amount of processed ComC-FLAG as a percentage of total (processed and unprocessed) ComC-FLAG is quantified and plotted as the average ± SE of four independent experiments. **P < 0.01, ANOVA with Tukey’s HSD test.
To test our hypothesis, we employed a previously developed peptide-processing assay (35). Using FLAG-tagged peptides, the unprocessed and processed forms can be separated by size using SDS/PAGE and detected via Western blot. FLAG-tagged peptides cannot be secreted by ComAB/BlpAB and are retained within the cytoplasm (35), most likely because the high charge density of the FLAG tag interferes with loading into the transporter channel. Therefore, this assay assesses only the processing step of transport.
We engineered R6 strains expressing C-terminally FLAG-tagged ComC (ComC-FLAG) in place of wild-type ComC from the native comC locus. Using these strains, we assayed for ComC processing in the wild-type, ΔcomAB, ΔblpA, and ΔcomABΔblpA backgrounds 30 min after induction with CSP and BlpC in THY broth (pH 7.1) (Fig. 3C). The double mutant was included as a control to evaluate for the presence of a transporter or transporters other than ComAB or BlpAB that might contribute to ComC processing. The double mutant showed only a negligible amount of ComC processing, suggesting there are no other such transporters. Meanwhile, we confirmed that ComAB can process ComC, consistent with established models of com regulation. We also observed ComC processing in the ΔcomAB strain expressing only BlpAB. These results indicate that BlpAB can complete at least the first step of ComC transport (processing) and when taken together with the com activation kinetics data (Fig. 3 A and B) strongly suggest that BlpAB can carry out the second step (secretion) as well. Therefore, we conclude that BlpAB can transport ComC/CSP and that this mechanism is responsible for the observed BlpC-induced com activation.
Both ComAB and BlpAB Process and Secrete Pneumocins.
Having established that ComAB and BlpAB both secrete CSP and BlpC (11, 23), we sought to determine if the promiscuity of these transporters extended to other substrates as well. The current model of substrate recognition by ComAB/BlpAB posits that the peptidase domain of the transporters interacts with the substrates’ N-terminal signal sequences. In addition to the double-glycine motif, four specific hydrophobic residues in the signal sequence (SI Appendix, Fig. S3A, yellow highlights) are important for this interaction (30). These residues are conserved across all com- and blp-regulated double-glycine peptides found in pneumococcus. Moreover, the residues in the transporter peptidase domain that are thought to participate in substrate recognition (36) are also highly conserved in both ComA and BlpA (SI Appendix, Fig. S3C, yellow highlights). Given this, we hypothesized that ComAB and BlpAB recognize and transport the same pool of substrates, including the pheromones ComC/CSP and BlpC, the pneumocins, and the competence-induced bacteriocins CibAB.
We were particularly interested in testing this hypothesis on the pneumocins for several reasons. First, while long suspected to be BlpAB, the transporter or transporters responsible for pneumocin secretion have never been definitively identified. Second, the question of whether ComAB can secrete the pneumocins is an important unanswered question given the ability of the com system to up-regulate pneumocin expression and the absence of BlpAB in 75% of strains. Since all pneumocins share very similar signal sequences (SI Appendix, Fig. S3A), we reasoned that the details of transport would be similar, if not identical, across the different pneumocins. As such, for use in subsequent experiments we chose a representative pneumocin, BlpIP133 (henceforth referred to as “BlpI”), which has been previously shown to inhibit sensitive strains when expressed with its partner, BlpJ (27).
To assess BlpI secretion, we chose to employ the HiBiT tag detection system (Promega). The HiBiT tag is a short, 11-residue peptide tag (VSGWRLFKKIS) that associates with the inactive LgBiT luciferase fragment with subnanomolar affinity to complement the latter’s luciferase activity (37, 38). This system allows highly sensitive detection of HiBiT-tagged peptides and proteins using a bioluminescence assay (38).
After validating the assay system (SI Appendix, Fig. S4), we used R6 strains expressing BlpI-HiBiT from the native, BlpR-regulated proximal BIR promoter to evaluate BlpI secretion 60 min after BlpC and CSP induction in the wild-type and transporter-deletion backgrounds (Fig. 4A). The amount of BlpI-HiBiT detected in the supernatant of the double mutant was 180-fold less than that of the wild type, confirming that ComAB and BlpAB are the primary contributors to BlpI secretion. We also found that both single mutants secreted significantly more BlpI-HiBiT than the double mutant. In contrast, we detected similar levels of BlpI in the cell-associated fractions of all strains (Fig. 4A, Lower). We corroborated these findings using a BlpI-FLAG processing assay (Fig. 4B), which showed high levels of processing in both single transporter mutants. Therefore, we conclude that both ComAB and BlpAB can process and secrete BlpI and likely the rest of the pneumocins as well.
Fig. 4.
ComAB and BlpAB process and secrete the pneumocin BlpI. (A) BlpI-HiBiT secretion in wild-type (black), ΔcomAB (blue), ΔblpA (green), and ΔcomABΔblpA (magenta) strains. Concentration of BlpI-HiBiT normalized to cell density detected in the supernatant (Upper) and cell-associated (Lower) fractions 60 min after induction with 200 ng/mL CSP and 200 ng/mL BlpC in THY broth (pH 7.1). Data are plotted as the average ± SE of three independent experiments. (B) BlpI-FLAG processing. Cells were grown in THY broth at pH 7.1 and were induced with 100 ng/mL CSP and 100 ng/mL BlpC; whole-cell lysates were collected 30 min later. Representative blots are presented, showing the unprocessed (U) and processed (P) forms of BlpI-FLAG and a pneumolysin (Ply) loading control. The amount of processed BlpI-FLAG as a percentage of total (processed and unprocessed) BlpI-FLAG is quantified and plotted as the average ± SE of four independent experiments. n.s., not significant; ***P < 0.001; ANOVA with Tukey’s HSD test.
Pheromone Secretion by BlpAB Enhances Competence Activation and Allows Competence-Independent blp Activation.
Having determined that ComAB and BlpAB are functionally redundant for secretion of pheromones and pneumocins, we sought to determine what advantages, if any, intact BlpAB confers to pneumococcus. First, we hypothesized that ComC/CSP secretion by BlpAB could drive spontaneous com activation, similar to the way BlpC secretion by ComAB can drive spontaneous blp activation; if true, then we should be able to observe spontaneous com activation following blp activation in a ΔcomAB mutant. To test this, we used our dual reporter to monitor spontaneous com activation in a panel of transporter- and pheromone-deletion mutants grown in THY broth at pH 7.4 in a 96-well plate. Each well was independently assessed for com activation events, defined as when the com activation level of the well rose above an empirically determined threshold value. If a well reached its maximum observed cell density as measured by OD620 before a com activation event occurred, then it was censored. Finally, the event data were fit to Kaplan–Meier estimators and plotted as the cumulative probability of com activation versus cell density.
Under these conditions, we observed within-strain stochasticity in the pattern of com activation; the unencapsulated R6 wild-type strain activated com in only a subset of identically inoculated wells, and there was large variation in the timing of com activation among those wells in which it did occur (Fig. 5A). Contrary to our hypothesis, we did not observe com activation in the ΔcomAB mutant. However, we did observe a defect in com activation in the ΔblpA mutant compared with the wild-type strain. This defect was not caused by an upstream deficiency in blp activation, since blp activation was not observed before com activation in any strain. Due to conflicting reports about the effect of capsule on CSP signaling (39, 40), we tested whether we could reproduce this phenotype in an encapsulated strain. Indeed, we observed the same competence defect in the ΔblpA mutant compared with the wild-type strain in the encapsulated D39 background (Fig. 5B). Therefore, under these conditions the presence of a capsule does not appreciably affect BlpAB-mediated blp-to-com crosstalk.
Fig. 5.
BlpAB enhances spontaneous com activation and drives com-independent blp activation. (A–C) Spontaneous com activation in THY broth and CDM+ medium. R6 (A and C) and D39 (B) dual reporter strains were grown in 96-well plates in THY broth at pH 7.4 (A and B) or in CDM+ medium at pH 7.0 (C). All six (wild-type, pheromone, and transporter deletion) strains are presented in A and C; only the wild-type and transporter deletion strains are presented in B. Each well was continuously monitored for growth and com activation. Data were fit to the Kaplan–Meier estimator. Wells that did not experience a com activation event before cells reached their maximum density were censored (crosses). n = 36 (A and B) or 18 (C) wells per strain, pooled from three independent experiments. ***P < 0.001; **P < 0.01; n.s., not significant; log-rank test with Holm’s correction for multiple comparisons. (D) Cell density at com activation plotted against cell density at blp activation in individual wells from C of the wild-type and ΔblpA strains. Points that fall on the dotted line represent wells in which blp activation occurred at the same time as com activation. Points that fall to the left of the dotted line represent blp activation occurring after com activation. Points that fall to the right of the dotted line (highlighted in red) represent blp activation occurring before com activation.
To assess the contribution of com and blp activation states of the starter cultures used to inoculate media at the beginning of the assay, we repeated the previous experiment using inocula from starter cultures of R6 strains grown in THY broth at pH 6.8, a condition under which neither com nor blp activation occurs. This produced results that closely resemble those obtained using pH 7.4 starter cultures (SI Appendix, Fig. S5A). When analyzing these data, we noticed that two wells of the ΔcomAB mutant activated com (SI Appendix, Fig. S5A, blue line). We reasoned that this must represent blp-dependent com activation. Indeed, the com and blp activation kinetic curves of the two wells show peaks of blp activation that are concurrent with the peaks of com activation (SI Appendix, Fig. S5C). We also observed the same phenomenon in one well containing the D39 ΔcomAB mutant (Fig. 5B, blue line). These data show that BlpAB is sufficient to drive com activation during spontaneous blp activation under certain conditions.
We next tested whether enhanced com activation by BlpAB occurred under other growth conditions. We repeated the spontaneous com activation experiment using CDM+ medium, a more minimal medium than THY broth that is used for maintaining pneumococcal biofilms (41). Unlike in THY broth, we did not observe a com activation defect in the R6 ΔblpA mutant in CDM+ medium compared with the wild type (Fig. 5C).
From the same experiment in CDM+ medium, we were also able to gather data on blp activation using the dual reporter. When analyzing these data, we noticed that a significant number of wells of the wild-type strain activated blp before com (Fig. 5D, Left, red dots). This com-independent blp activation occurred in just over half of the wells (10/18) in the wild-type strain compared with zero wells in the ΔblpA mutant (Fig. 5D, Right). Consistent with previous work (11), these results show that BlpAB can promote and is required for com-independent blp activation.
BlpAB Ensures Efficient Transport of Pneumocins.
Next, we wanted to investigate whether BlpAB(+) strains, which possess both ComAB and BlpAB, enjoy advantages in pneumocin secretion over BlpAB(−) strains, which possess only ComAB. The previous BlpI secretion assay (Fig. 4A) was performed on cells simultaneously induced with saturating concentrations of both CSP and BlpC. These conditions are unlikely to resemble what occurs during natural, spontaneous activation of com and blp.
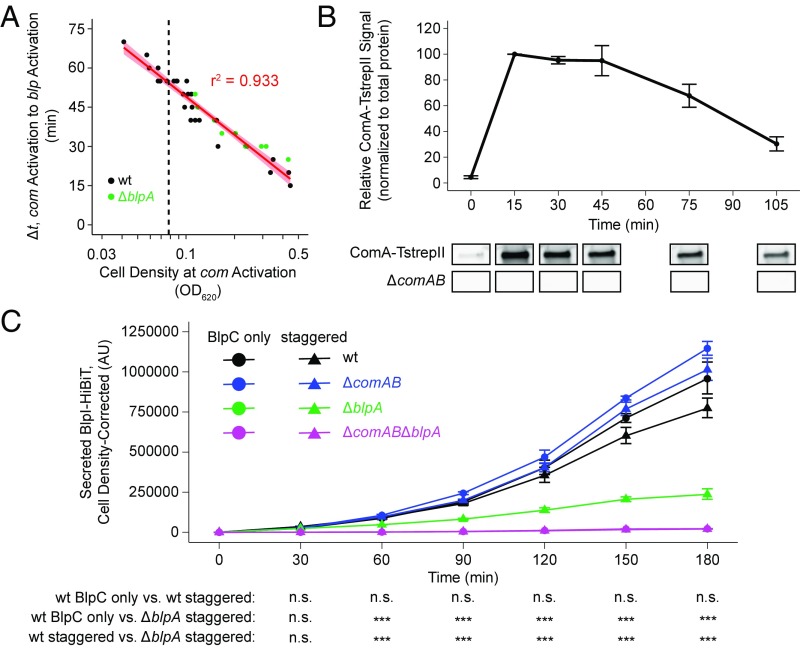
First, we found that the pheromone concentrations required to induce strains to levels and with kinetics similar to spontaneous activation were 10 ng/mL for CSP and 25 ng/mL for BlpC (SI Appendix, Fig. S6 A–C), much lower than the 100–200 ng/mL used in previous processing and secretion assays. Second, in the absence of an exogenous source of BlpC, BlpAB(−) strains can activate blp only in a com-dependent fashion. This com-dependent blp activation also happens in BlpAB(+) strains. In both strain backgrounds, blp activation in this manner occurs after com activation following a cell density-dependent delay (Fig. 6A), the length of which is not affected by the presence of BlpAB (P > 0.05, ANCOVA). Third, we observed that ComA protein levels following CSP induction decrease rapidly after 45 min (Fig. 6B). This is consistent with the rapid shut-off in transcription activity from PcomAB seen with the luciferase reporter (SI Appendix, Fig. S6C). Given these data, we hypothesized that BlpAB(−) strains, which must rely on ComAB for secretion, secrete less pneumocins during com-dependent blp activation than BlpAB(+) strains, which can use both ComAB and BlpAB.
Fig. 6.
BlpAB promotes efficient pneumocin secretion. (A) Correlation of the time delay between spontaneous com and blp activation with cell density at the point of com activation in wild-type (black) and ΔblpA (green) strains in THY broth at pH 7.4. Data were taken from the same experiments as in Fig. 5A and represent all wells in which blp activation was observed. The red line and shading represent the linear regression estimate ± 95% CI. The dashed vertical line is placed at OD620 0.078. (B) Twin-Strep-tagged ComA (ComA-TstrepII) protein levels following treatment with 10 ng/mL CSP at OD620 0.078 in THY broth at pH 7.1. Samples were collected at the indicated time points, and membrane fractions were assayed by quantitative Western blot with anti–Strep-tag II antibody. Representative blots of samples from the strain expressing ComA-TstrepII (Upper) and from a ΔcomAB negative control (Lower) are shown. For quantification, ComA signal was normalized to total protein and expressed relative to the 15-min time point, which was set to a value of 100. Data are plotted as the average ± SE of three independent experiments. (C) BlpI-HiBiT secretion over time in wild-type (black), ΔcomAB (blue), ΔblpA (green), and ΔcomABΔblpA (magenta) strains grown in THY broth at pH 7.1. Beginning at OD620 0.02 (t = −45 min), cells were given two treatments at −45 and 0 min: BlpC-only (mock treatment followed by 25 ng/mL BlpC) and staggered treatment (10 ng/mL CSP followed by 25 ng/mL BlpC). Samples were collected every 30 min and assayed for BlpI-HiBiT in the supernatant fractions. Data were corrected for differences in cell density between samples independently per time point and are plotted as the average ± SE of three independent experiments. Green circles (ΔblpA, BlpC only) are obscured behind the magenta plots. n.s., not significant; ***P < 0.001; ANOVA with Tukey’s HSD test.
To test this hypothesis, we assayed BlpI secretion under conditions similar to spontaneous com and blp activation. The strains used in this experiment were engineered to express pheromone-receptor mismatched CSP2–ComD1 and BlpC6A–BlpHR6 pairs. Therefore, these strains are deficient in autoactivation of both com and blp to ensure that differences in transporter content (and hence pheromone secretion) did not affect activation kinetics. Cells were induced with either BlpC alone (to mimic com-independent blp activation), with CSP and BlpC together (“simultaneous treatment”), or with CSP followed by BlpC 45 min later (“staggered treatment,” to mimic com-dependent blp activation) (SI Appendix, Fig. S6D). Consistent with our hypothesis, we observed a small but statistically significant increase in BlpI secretion from the wild-type strain compared with the ΔblpA mutant in the staggered-treatment group (SI Appendix, Fig. S6E). This difference was not seen in the simultaneous-treatment group. Additionally, when comparing the staggered- with the simultaneous-treatment group, a large decrease in BlpI secretion was seen only in the strains possessing ComAB, and of these two strains the wild-type strain, which possesses both ComAB and BlpAB, suffered the smaller decrease.
The differences seen between the wild-type and ΔblpA strains in the previous experiment were small when assessed at 105 min after CSP treatment. However, given our observation that ComAB levels decrease continuously past 45 min post-CSP treatment, the differences should increase over time. To test this hypothesis, we monitored BlpI-HiBiT secretion over time following the BlpC-only or staggered treatments (Fig. 6C). Consistent with falling ComAB levels, beginning at 60 min post-BlpC treatment (105 min post-CSP treatment) the ΔblpA mutant in the staggered-treatment group showed increasingly large defects in BlpI-HiBiT secretion over time compared with the wild-type strain in the same treatment group. The same was seen when comparing the ΔblpA mutant in the staggered-treatment group with the wild-type strain in the BlpC-only treatment group. Last, in the BlpC-only treatment group, only BlpAB-containing strains secreted BlpI at levels higher than the double transporter mutant, and the wild-type strain did not secrete more than the ΔcomAB mutant (P > 0.05 at all time points). These data show that BlpI secretion through ComAB (but not BlpAB) decreases rapidly with time during com-dependent blp activation and is negligible during com-independent blp activation.
BlpAB(+) Strains Enjoy a Competitive Advantage over BlpAB(−) Strains During Nasopharyngeal Colonization.
We hypothesized that BlpAB(+) strains’ ability to activate blp independently of com and to secrete greater amounts of pneumocins would give them a competitive advantage over BlpAB(−) strains during nasopharyngeal colonization. To test this, we coinoculated mice with either a BlpAB(+) or BlpAB(−) pneumocin-expressing “killer” strain and a Δblp pneumocin-sensitive strain and assessed competitive indices at 4 d postinoculation. The BlpAB(+) killer strain outcompeted the sensitive strain to a greater extent than the BlpAB(−) killer strain did (Fig. 7A). We also assessed how BlpAB(+) and BlpAB(−) strains fare in direct competition. We coinoculated mice with pairs of either BlpC pherotype-matched or mismatched pneumocin-expressing BlpAB(+) and BlpAB(−) strains and assessed competitive indices at 4 d postinoculation. The coinoculated strains had identical BIRs; therefore, either strain could develop immunity to the other’s pneumocins provided it activates blp at the appropriate time. We observed that the BlpAB(+) strain had a competitive advantage over the BlpAB(−) strain but only when the two strains had mismatched BlpC pherotypes (Fig. 7B). All strains used in these competition assays colonize to similar levels when inoculated alone (SI Appendix, Fig. S8), indicating none have intrinsic colonization defects.
Fig. 7.
BlpAB(+) strains have a competitive advantage over BlpAB(−) strains during mouse nasopharyngeal colonization. All pneumococci are mouse-adapted serotype 19A strains. (A) Competition of BlpAB(+) and BlpAB(−) strains against a pneumocin-sensitive strain. BALB/c mice were coinoculated in a 1:1 ratio with either a BlpAB(+) or BlpAB(−) pneumocin-expressing killer strain and a Δblp (whole locus) sensitive strain. Both killer strains encode a P164-type BIR. The cfu counts from the nasopharynx were obtained at 4 d postinoculation and used to calculate competitive indices. n = 10 mice per competition. (B) Direct competition of BlpAB(+) strains against BlpAB(−) strains. Mice were coinoculated as in A with either a BlpCH6A- or BlpCHR6-expressing BlpAB(+) strain and a BlpCHR6-expressing BlpAB(−) strain. All strains encode a P133-type BIR. Competitive indices were calculated as in A. n = 10 mice per competition. Black bars represent medians. ***P < 0.001 (left-hand vs. right-hand group); Mann–Whitney U test.
Discussion
We have presented evidence showing that competence and bacteriocin regulation in pneumococcus are more entwined than previously thought. While it was known that com could send positive inputs to the blp system, we show here that signals can also travel in the opposite direction due to secretion of CSP by BlpAB. While we found this blp-to-com crosstalk could drive com activation following or concurrently with blp activation (SI Appendix, Fig. S5C), the more common effect of the crosstalk was activation of com at higher frequencies and lower cell densities, even in the absence of overt blp activation (Fig. 5 A and B). This highlights the importance of the basal level of transporter expression as the rate-limiting factor for CSP secretion and com activation; the addition of a second transporter (BlpAB) to augment basal CSP secretion by ComAB is sufficient to increase com activation. This phenotype was medium dependent; it was found in THY broth but not CDM+ medium. We postulate that the threshold for com activation in CDM+ medium is lower than in THY broth because of increased rates of CSP secretion, increased CSP stability in the medium, increased responsiveness to CSP, or some combination of these. This allows ComAB alone to secrete enough CSP to activate com efficiently without the help of BlpAB. Therefore, BlpAB’s role in enhancing com activation depends on environmental conditions and is most pronounced when the threshold for com activation is relatively high. We also found that in THY broth com activation suffered the same defect in the absence of BlpC signaling as in the absence of the BlpAB transporter (Fig. 5A; compare ΔblpC with ΔblpA). We speculate that basal levels of BlpC secretion and signaling lead to noise in blp activation during the preactivation period. This noise results in transient elevated expression of BlpAB in a subpopulation of cells that leads to bursts of CSP secretion which over time help push the total extracellular CSP concentration above the threshold required for com activation. Further studies will be needed to confirm this hypothesis and to further elucidate the determinants of transporter-expression dynamics in the preactivation period and their effects on com and blp activation.
We have also shown that the ability of both ComAB and BlpAB to secrete the other system’s pheromone is not a special case but rather is a natural consequence of the transporters’ promiscuity toward substrates. It was previously shown that purified peptidase domain from BlpA could cleave a synthetic CSP-like analog (42). We confirm here that full-length BlpAB can secrete native ComC/CSP in a live-cell context. We have also observed that both transporters can process and secrete the pneumocin BlpI (Fig. 4). These data support the conclusion that ComAB and BlpAB share the same substrate pool. Finally, our data indicate that the signal sequence–peptidase domain interaction is the primary factor that determines whether a peptide is secreted by ComAB/BlpAB. Both transporters tolerate a wide variety of mature peptides for secretion: small and amphipathic (CSP), charged (BlpC), and large and highly hydrophobic (pneumocins).
The promiscuity of ComAB and BlpAB has multiple functional consequences, which are given added importance by the fact that 75% of pneumococcal strains lack a functional BlpAB but nearly all strains produce a functional ComAB. It was previously unclear whether BlpAB(−) strains could secrete pneumocins. We provide evidence here that these strains can in fact secrete pneumocins during com-dependent blp activation. Over short time frames, the amount of pneumocin secreted by BlpAB(−) strains in this manner is comparable to the amount secreted by BlpAB(+) strains during either com-independent or com-dependent blp activation (Fig. 6C and SI Appendix, Fig. S6E). This suggests that BlpAB(−) strains theoretically can effect pneumocin-mediated inhibition during com-dependent blp activation. This would be true for all but a small minority of strains that cannot activate com or produce ComAB due to acquired mutations in com regulatory genes (43). Obtaining direct evidence for this will be a priority for future studies.
Despite the above, BlpAB(+) strains still enjoy multiple advantages in pneumocin secretion over their BlpAB(−) counterparts due to differences in the regulation and kinetics of ComAB and BlpAB expression. First, BlpAB(+) strains can secrete more pneumocins than BlpAB(−) strains during com-dependent blp activation in a time-dependent fashion (Fig. 6C). The transient nature of ComAB expression during com activation limits pneumocin secretion in BlpAB(−) strains to short bursts following com-dependent blp activation. In contrast, blp activation—and therefore BlpAB expression—is not subject to a rapid shut-off mechanism; in broth culture BlpR-regulated promoters remain highly active after initial activation throughout the exponential phase (Fig. 3 A and B). Thus, BlpAB(+) strains can sustain pneumocin secretion for longer periods of time and generate higher extracellular pneumocin concentrations due to secretion through BlpAB. Second, unlike BlpAB(−) strains, BlpAB(+) strains can activate blp independently of com (Fig. 5D and ref. 11) with no penalty to pneumocin secretion capacity despite ComAB being unavailable to contribute to secretion (Fig. 6C).
Consistent with our in vitro findings, BlpAB(+) strains enjoy a competitive advantage over BlpAB(−) strains during nasopharyngeal colonization in mice. During cocolonization, BlpAB(+) strains were better able than BlpAB(−) strains to outcompete a pneumocin-sensitive strain (Fig. 7A). Moreover, a BlpAB(+) strain directly outcompeted its BlpAB(−) counterpart when both expressed the same pneumocins, but only when the two strains could not cross-activate each other’s blp systems (Fig. 7B). These data indicate that, compared with BlpAB(−) strains, BlpAB(+) strains can use pneumocins more effectively in competition during colonization due to more frequent activation of blp and/or secretion of greater amounts of pneumocins. Accordingly, we propose that BlpAB(+) strains leverage these abilities to act as aggressors, wielding pneumocins as weapons for generalized antibacterial competition. In contrast, BlpAB(−) strains act primarily as opportunists, using pneumocins in a limited capacity to augment fratricide and/or inhibit competitors while they are in a potentially vulnerable state during competence.
Linkage analysis indicates the predominant BlpAB-inactivating mutation moves from strain to strain via recombination and that BlpAB(−) strains occasionally revert to a BlpAB(+) genotype in the same manner (27). This suggests that there are costs and benefits to maintaining an intact BlpAB, and BlpAB(+) and BlpAB(−) strains represent dynamic populations that switch their blpAB genotype in response to different selective pressures favoring an aggressor phenotype over opportunist or vice versa. Currently, genomics data do not indicate that invasion is such a selective pressure; in a collection of human isolates from South Africa (27), the distribution of BlpAB(+) strains did not differ between the invasive and colonizing groups (8/21 invasive vs. 11/30 colonizing, P = 1, Fisher’s exact test). Another set of potential selective pressures is that associated with competence development. The increased propensity for competence activation in BlpAB(+) strains (Fig. 5 A and B) would provide them with greater access to DNA for horizontal gene transfer and an improved ability to cope with DNA-damaging stress. On the other hand, competence is a costly, energy-intensive process that interferes with normal cell metabolism and proliferation (44, 45). The stringent regulation of competence and its rapid shut-off once activated likely are strategies to mitigate its negative impacts. Therefore, the increased frequency of competence activation conferred by an intact BlpAB may be detrimental to fitness in certain cases. Finally, BlpAB(+) strains may incur a fitness cost from frequent blp activation. Consistent with this, we observed a dose-dependent, small but reproducible and statistically significant growth defect in THY broth-grown strains following BlpC treatment (SI Appendix, Fig. S9). Validating these and other pressures in physiologic settings to further define how environmental and genetic contexts influence the adaptive value of maintaining an intact BlpAB presents an attractive target for future studies.
Many streptococci are naturally competent, but only members of the Streptococcus mitis (Mitis) group (to which pneumococcus belongs) and the closely related Streptococcus anginosus group (SAG) employ a ComCDE-type system to regulate competence (46). The ability of non-ComAB ABC transporters to secrete ComC/CSP and influence competence development is of direct relevance in other species of Mitis/Anginosus groups. For instance, Sil (Streptococcus invasion locus) is a bacteriocin-encoding locus (47) found in SAG that is structurally and functionally similar to pneumococcal blp. The regulation of Sil is effectively identical to that of blp, with SilE/D/CR/B/A taking the roles of BlpA/B/C/H/R, respectively. Importantly, SilED is the only ComAB/BlpAB-type transporter found in many SAG strains and therefore is the only potential transporter for ComC/CSP. SAG SilE and SilCR/ComC share the same sequence motifs important for substrate recognition as those found in pneumococcal BlpA/ComA and BlpC/ComC, respectively (SI Appendix, Fig. S3 B and D). Therefore, it is likely that SilED can secrete CSP. Given this, competence in SAG may be chiefly regulated by a bacteriocin locus (Sil) through secretion of CSP through a non-ComAB transporter.
Looking beyond streptococci, ComAB/BlpAB-type transporters are widely distributed among Gram-positive and Gram-negative bacteria, where they primarily function to export bacteriocins and in some cases also the signaling peptide that induces expression of said bacteriocins (29, 48–51). With few exceptions, these peptides and transporters share the same conserved sequence motifs found to be important for substrate recognition in their pneumococcal counterparts (Fig. 3 B and D). This implies that most if not all ABC transporters of this family recognize their substrates not only in the same manner but also using the same sequence motifs. Therefore, the promiscuity of pneumococcal ComAB and BlpAB is likely a general feature of this transporter family. Consistent with this, others have shown that in Lactococcus lactis, the lactococcin Q transporter LaqD can secrete the related but distinct bacteriocin lactococcin G (48), and in Enterococcus faecium, the enterocin transporter EnkT recognizes the signal sequences of a number of bacteriocins from other species (52). This transporter promiscuity has wide-ranging implications for the regulation and biosynthesis of bacteriocins in many different bacterial species. Understanding the details of pheromone and bacteriocin secretion by these ABC transporters will provide key insights into the dynamics of interbacterial communication and competition.
Methods
Bacterial Strains and Growth Conditions.
All strains are derived from the R6 strain P654 (35) (referred to as “PSD100” in ref. 35) or the D39 strain P2055 (SI Appendix). For experiments, pneumococcus was grown in either filter-sterilized THY broth or CDM+ medium (41) at 37 °C. Except where noted otherwise, pneumococcal cultures used for all experiments were inoculated from starter cultures grown in THY broth at pH 7.4 to an OD620 of 0.275 and were frozen at −80 °C in 13% glycerol. Transformations were carried out as previously described (53). Unmarked chromosomal mutations were created via Janus or Sweet Janus exchange (54, 55). See SI Appendix for details. All transformants were verified by PCR fragment length analysis and Sanger sequencing. Antibiotics were used at the following concentrations: chloramphenicol, 2 µg/mL; kanamycin, 500 µg/mL; spectinomycin, 200 µg/mL; and streptomycin, 100 µg/mL.
DNA Manipulation.
PCR for downstream Gibson assembly, transformation, or sequencing applications was performed using Phusion polymerase (E0553; New England Biolabs). All other PCR reactions were performed using Taq polymerase (M0273; New England Biolabs). Primers were designed using primer3 (56, 57) and synthesized by IDT. PCR products were purified using silica columns (28106; Qiagen). Gibson assembly was performed using NEBuilder HiFi DNA Assembly master mix (E2621; New England Biolabs). All codon optimization was performed using OPTIMIZER (58).
com/blp Activation Kinetics Assays.
Dual reporter strains were inoculated 1:150 into filter-sterilized THY broth + 25 mM Hepes, 5 µg/mL catalase, 165 µM d-luciferin (88294; Thermo Fisher Scientific), 1:10,000 Nano-Glo substrate (N113B; Promega), or CDM+ medium + 5 µg/mL catalase, 330 µM d-luciferin, and 1:5,000 Nano-Glo substrate. Cultures were aliquoted into a white, clear-bottomed 96-well plate (301012; Porvair), 200 µL (if no treatment was to be added) or 198 µL (if treatment was to be added) per well. The plate was incubated at 37 °C in a BioTek Synergy HTX plate reader. Every 5 min, the plate was shaken (linear shake setting, 587 cycles/min, 5 s), and absorbance (620 nm) and luminescence (bottom-read mode, 0.90-s integration time, gain 150) were read. The Nluc signal was isolated using a 450/50-nm band-pass (BP) filter (7082208; BioTek); The RFluc signal was isolated using a 610-nm long-pass (LP) filter (7092209; BioTek). For experiments in which pheromone treatments were added to the cells, 2 µL sterile medium containing the appropriate pheromone (treatment) or 2 µL sterile medium alone (mock treatment) was added to each well. Luminescence data were used to calculate com/blp activation levels (SI Appendix).
For analysis of spontaneous com and blp activation events, individual wells were assessed for activation events. An activation event was defined as the com/blp activation level exceeding a threshold value for at least two consecutive time points (SI Appendix). Wells without an activation event before cell density reached its maximum observed value were censored at the time point at which maximum cell density was reached. Once generated, activation event data were fit to the Kaplan–Meier estimator using the survfit() function in the R survival package (v2.41-3). The resulting survival curves were compared with the log-rank test using the survdiff() function in the R survival package (v2.41-3) and were adjusted for multiple comparisons with the Holm correction using the p.adjust() function in R 3.4.2.
Peptide Processing Assays.
R6 strains expressing FLAG-tagged peptides were inoculated 1:150 into THY broth + 25 mM Hepes (pH 7.1) and 5 µg/mL catalase and were grown statically at 37 °C. At OD620 0.078, cells were induced with 100 ng/mL CSP1 and 100 ng/mL BlpCR6. Thirty minutes later, a sample of cells, equivalent to OD620 1.175 × 1 mL, was taken from each culture and pelleted by centrifugation at 5,000 × g for 5 min at 4 °C. The pellets were washed with 1 mL PBS and pelleted again using the same method. The washed pellets were then resuspended in 25 µL 1% SDS, 0.1% Triton X-100 and mixed with an equal volume of 4× Laemmli sample buffer (1610747; Bio-Rad) supplemented with 10% (vol/vol) 2-mercaptoethanol. The resulting mixture was boiled at 95 °C for 10 min and then was stored at −20 °C. The samples were later analyzed by peptide Western blot (SI Appendix). Blots were quantified using Image Studio v5.2. Percent peptide processing was calculated by dividing the signal from the processed peptide band by the sum of the signals from the processed and unprocessed peptide bands. Experiments were repeated four times, and differences between groups were evaluated by applying ANOVA and Tukey’s honest significant difference (HSD) test to the percentage of peptide processing values using the aov() and TukeyHSD() functions in R 3.4.2.
BlpI-HiBiT Secretion Assays.
R6 strains expressing BlpI-HiBiT along with a background control strain expressing Strep-tag II–tagged BlpI were inoculated 1:150 into THY broth + 25 mM Hepes (pH 7.1) and 5 µg/mL catalase and were grown statically at 37 °C. These strains expressed a mismatched BlpC6A–BlpHR6 pair to prevent spontaneous blp activation. At OD620 0.078, cells were induced with 200 ng/mL CSP1 and 200 ng/mL BlpCR6. Sixty minutes later, a sample of culture supernatant of each strain was filter sterilized through a 0.22-µm centrifugal filter (8160; Costar) and diluted 10-fold in sterile THY broth + 25 mM Hepes (pH 7.1) and 5 µg/mL catalase. Samples of mock-treated cultures were not diluted. At the same time, a sample of cells of each strain was taken and pelleted by centrifugation at 5,000 × g, for 5 min at 4 °C. The pellet was washed once with 1× culture volume PBS and pelleted again by centrifugation at 5,000 × g for 5 min at 4 °C. The washed pellet was then resuspended in cell wall digest buffer [20 mM MES (pH 6.5), 0.5 M sucrose, 20 mM MgCl2, 50 U/mL mutanolysin (M9901; Sigma), 0.5 mg/mL lysozyme; adapted from ref. 59] and incubated at 37 °C for 30 min. After incubation, the protoplast suspension was diluted fivefold in 0.25% Triton X-100 and incubated at room temperature for 15 min.
For experiments simulating spontaneous activation, cells expressing mismatched CSP2–ComD1 and BlpC6A–BlpHR6 pairs were used. The background control strain was ΔcomC in lieu of a mismatched CSP–ComD pair. For the single-time point assay, cells were induced starting at OD620 0.078 as follows: BlpC-only: mock treatment followed by 25 ng/mL BlpCR6 45 min later; simultaneous treatment: mock treatment followed by 10 ng/mL CSP1 and 25 ng/mL BlpCR6 45 min later; staggered treatment: 10 ng/mL CSP1 followed by 25 ng/mL BlpCR6 45 min later. Supernatant and cell samples were collected 60 min after BlpC induction in all cases and were processed as above. For the multiple-time point assay, cells were induced starting at OD620 0.02 with either the BlpC-only or staggered treatment as above. Supernatant samples were collected just before BlpC treatment (0 min) and every 30 min thereafter for 3 h and were clarified by centrifugation at 2,750 × g for 5 min at 4 °C. All samples after the 0-min time point were diluted 1:10 in sterile THY broth + 25 mM Hepes (pH 7.1) and 5 µg/mL catalase.
Supernatant and cell lysate samples of BlpI-HiBiT–expressing strains along with standards (SI Appendix) were aliquoted into a white 96-well plate (3917; Costar), 50 µL per well, n = 3 wells (single-time point assays) or n = 1 well (multiple-time point assays) per sample/standard. The samples and standards were then assayed for HiBiT tag by adding 50 µL HiBiT Extracellular Detection Reagent (N2421; Promega) to each well. Luminescence was then read using a BioTek Synergy HTX plate reader with the following settings: top read mode (1.00-mm height), 2-s integration time, gain 135. The BlpI-HiBiT concentrations in the samples were calculated using their luminescence values and the appropriate standard curve. Experiments were repeated three times, and differences between groups were evaluated by applying ANOVA and Tukey’s HSD to the log-transformed BlpI-HiBiT concentrations using the aov() and TukeyHSD() functions in R 3.4.2.
ComA Protein Level Kinetics Assay.
R6 strains expressing Twin-Strep–tagged (60) ComA (ComA-TstrepII) (SI Appendix, Fig. S7) or no ComAB (ΔcomAB) and mismatched CSP2–ComD1 and BlpCR6–BlpH6A pairs were inoculated 1:150 into THY broth + 25 mM Hepes (pH 7.1) and 5 µg/mL catalase, grown statically at 37 °C, and induced with 10 ng/mL CSP1 at OD620 0.078. Samples were taken just before induction (0 min) and at 15, 30, 45, 75, and 105 min postinduction. Membrane fractions were isolated using a protocol adapted from ref. 59 and were analyzed by Western blot (SI Appendix). Blots were quantified using Image Studio v5.2. The Strep-tag II signal was normalized to total protein before being expressed as relative to the signal at the 15-min time point.
Mouse Nasopharyngeal Colonization Assays.
Mouse colonization was performed as previously described (27). Briefly, each strain (see SI Appendix for strain details) was grown in THY broth to an OD620 of 0.5. Cells were gently pelleted and resuspended in sterile PBS at one-fifth the original volume. For coinoculations, PBS mixtures were combined in a 1:1 ratio; for single inoculations, an additional equal volume of sterile PBS was added. Ten microliters of dual- or single-strain mixtures were inoculated into the nasopharynx of unanesthetized 5- to 7-wk-old female BALB/c mice (Taconic). Inocula were plated on selective medium after inoculation to ensure that 1:1 ratios were maintained. No mouse developed evidence of sepsis during the experimental period. Mice were killed with CO2 overdose on day 4, and nasopharyngeal colonization was sampled by cannulation of the trachea toward the nasopharynx and administration of 200 µL of sterile PBS. Samples were collected from the nares and used to obtain colony counts (SI Appendix). Mice were housed in a specific pathogen-free facility, and experiments were performed under an approved protocol in compliance with the University of Michigan Institutional Animal Care and Use Committee recommendations. Differences in colonization densities and competitive indices between groups were evaluated by the Mann–Whitney (two groups) and Kruskal–Wallis (more than groups) tests using the wilcox.test() and kruskal.test() functions in R 3.4.2.
Nucleotide Accession Numbers.
The nucleotide sequences of the PcomAB-Nluc and PBIR-RFluc reporters are deposited in GenBank with accession numbers MH304212 and MH304213, respectively.
Supplementary Material
Acknowledgments
We thank Schyler Bennett for assistance in obtaining the ComA-TstrepII protein kinetics data. This work was supported by NIH Grants R01AI101285, T32GM007863, and T32AI007528.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. MH304212 (PcomAB Nluc), MH304213 (PBIR RFluc), and MH304214 (Sweet Janus)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804668115/-/DCSupplemental.
References
- 1.Aniansson G, et al. Nasopharyngeal colonization during the first year of life. J Infect Dis. 1992;165(Suppl 1):S38–S42. doi: 10.1093/infdis/165-supplement_1-s38. [DOI] [PubMed] [Google Scholar]
- 2.Könönen E, Jousimies-Somer H, Bryk A, Kilp T, Kilian M. Establishment of streptococci in the upper respiratory tract: Longitudinal changes in the mouth and nasopharynx up to 2 years of age. J Med Microbiol. 2002;51:723–730. doi: 10.1099/0022-1317-51-9-723. [DOI] [PubMed] [Google Scholar]
- 3.Turner P, et al. Improved detection of nasopharyngeal cocolonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J Clin Microbiol. 2011;49:1784–1789. doi: 10.1128/JCM.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith F. The significance of pneumococcal types. J Hyg (Lond) 1928;27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogaert D, et al. Variability and diversity of nasopharyngeal microbiota in children: A metagenomic analysis. PLoS One. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chewapreecha C, et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet. 2014;46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiller NL, et al. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 2010;6:e1001108. doi: 10.1371/journal.ppat.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogaardt C, van Tonder AJ, Brueggemann AB. Genomic analyses of pneumococci reveal a wide diversity of bacteriocins - including pneumocyclicin, a novel circular bacteriocin. BMC Genomics. 2015;16:554. doi: 10.1186/s12864-015-1729-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawid S, Roche AM, Weiser JN. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun. 2007;75:443–451. doi: 10.1128/IAI.01775-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wholey WY, Kochan TJ, Storck DN, Dawid S. Coordinated bacteriocin expression and competence in Streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog. 2016;12:e1005413. doi: 10.1371/journal.ppat.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hui FM, Morrison DA. Genetic transformation in Streptococcus pneumoniae: Nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J Bacteriol. 1991;173:372–381. doi: 10.1128/jb.173.1.372-381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Håvarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Håvarstein LS, Gaustad P, Nes IF, Morrison DA. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol. 1996;21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 15.Luo P, Li H, Morrison DA. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol Microbiol. 2003;50:623–633. doi: 10.1046/j.1365-2958.2003.03714.x. [DOI] [PubMed] [Google Scholar]
- 16.Tomasz A. Model for the mechanism controlling the expression of competent state in Pneumococcus cultures. J Bacteriol. 1966;91:1050–1061. doi: 10.1128/jb.91.3.1050-1061.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JD, Morrison DA. Modulation of competence for genetic transformation in Streptococcus pneumoniae. J Gen Microbiol. 1987;133:1959–1967. doi: 10.1099/00221287-133-7-1959. [DOI] [PubMed] [Google Scholar]
- 18.Slager J, Kjos M, Attaiech L, Veening JW. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell. 2014;157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 19.Stevens KE, Chang D, Zwack EE, Sebert ME. Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. MBio. 2011;2:e00071-11. doi: 10.1128/mBio.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Saizieu A, et al. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182:4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iannelli F, Oggioni MR, Pozzi G. Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptoccoccus pneumoniae. FEMS Microbiol Lett. 2005;252:321–326. doi: 10.1016/j.femsle.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Pinchas MD, LaCross NC, Dawid S. An electrostatic interaction between BlpC and BlpH dictates pheromone specificity in the control of bacteriocin production and immunity in Streptococcus pneumoniae. J Bacteriol. 2015;197:1236–1248. doi: 10.1128/JB.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kjos M, et al. Expression of Streptococcus pneumoniae bacteriocins is induced by antibiotics via regulatory interplay with the competence system. PLoS Pathog. 2016;12:e1005422. doi: 10.1371/journal.ppat.1005422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claverys JP, Martin B, Håvarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;64:1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- 25.Johnsborg O, Eldholm V, Bjørnstad ML, Håvarstein LS. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol Microbiol. 2008;69:245–253. doi: 10.1111/j.1365-2958.2008.06288.x. [DOI] [PubMed] [Google Scholar]
- 26.Wei H, Håvarstein LS. Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl Environ Microbiol. 2012;78:5897–5905. doi: 10.1128/AEM.01343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Son MR, et al. Conserved mutations in the pneumococcal bacteriocin transporter gene, blpA, result in a complex population consisting of producers and cheaters. MBio. 2011;2:e00179-11. doi: 10.1128/mBio.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lux T, Nuhn M, Hakenbeck R, Reichmann P. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J Bacteriol. 2007;189:7741–7751. doi: 10.1128/JB.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Håvarstein LS, Diep DB, Nes IF. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 30.Kotake Y, Ishii S, Yano T, Katsuoka Y, Hayashi H. Substrate recognition mechanism of the peptidase domain of the quorum-sensing-signal-producing ABC transporter ComA from Streptococcus. Biochemistry. 2008;47:2531–2538. doi: 10.1021/bi702253n. [DOI] [PubMed] [Google Scholar]
- 31.Hall MP, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michelini E, et al. Spectral-resolved gene technology for multiplexed bioluminescence and high-content screening. Anal Chem. 2008;80:260–267. doi: 10.1021/ac7016579. [DOI] [PubMed] [Google Scholar]
- 33.Rimini R, et al. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol Microbiol. 2000;36:1279–1292. doi: 10.1046/j.1365-2958.2000.01931.x. [DOI] [PubMed] [Google Scholar]
- 34.Alloing G, Martin B, Granadel C, Claverys JP. Development of competence in Streptococcus pneumonaie: Pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol Microbiol. 1998;29:75–83. doi: 10.1046/j.1365-2958.1998.00904.x. [DOI] [PubMed] [Google Scholar]
- 35.Kochan TJ, Dawid S. The HtrA protease of Streptococcus pneumoniae controls density-dependent stimulation of the bacteriocin blp locus via disruption of pheromone secretion. J Bacteriol. 2013;195:1561–1572. doi: 10.1128/JB.01964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishii S, et al. Crystal structure of the peptidase domain of Streptococcus ComA, a bifunctional ATP-binding cassette transporter involved in the quorum-sensing pathway. J Biol Chem. 2010;285:10777–10785. doi: 10.1074/jbc.M109.093781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixon AS, et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 38.Schwinn MK, et al. CRISPR-mediated tagging of endogenous proteins with a luminescent peptide. ACS Chem Biol. 2017;13:467–474. doi: 10.1021/acschembio.7b00549. [DOI] [PubMed] [Google Scholar]
- 39.Moreno-Gámez S, et al. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat Commun. 2017;8:854. doi: 10.1038/s41467-017-00903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prudhomme M, Berge M, Martin B, Polard P. Pneumococcal competence coordination relies on a cell-contact sensing mechanism. PLoS Genet. 2016;12:e1006113. doi: 10.1371/journal.pgen.1006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marks LR, Reddinger RM, Hakansson AP. High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae. MBio. 2012;3:e00200-12. doi: 10.1128/mBio.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishii S, et al. High-throughput screening of small molecule inhibitors of the Streptococcus quorum-sensing signal pathway. Sci Rep. 2017;7:4029. doi: 10.1038/s41598-017-03567-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li G, et al. Addiction of hypertransformable pneumococcal isolates to natural transformation for in vivo fitness and virulence. Infect Immun. 2016;84:1887–1901. doi: 10.1128/IAI.00097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaccaria E, Wells JM, van Baarlen P. Metabolic context of the competence-induced checkpoint for cell replication in Streptococcus suis. PLoS One. 2016;11:e0153571. doi: 10.1371/journal.pone.0153571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergé MJ, et al. A programmed cell division delay preserves genome integrity during natural genetic transformation in Streptococcus pneumoniae. Nat Commun. 2017;8:1621. doi: 10.1038/s41467-017-01716-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin B, Quentin Y, Fichant G, Claverys JP. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 2006;14:339–345. doi: 10.1016/j.tim.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 47.Mendonca ML, et al. The sil locus in Streptococcus anginosus group: Interspecies competition and a hotspot of genetic diversity. Front Microbiol. 2017;7:2156. doi: 10.3389/fmicb.2016.02156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishibashi N, Zendo T, Koga S, Shigeri Y, Sonomoto K. Molecular characterization of the genes involved in the secretion and immunity of lactococcin Q, a two-peptide bacteriocin produced by Lactococcus lactis QU 4. Microbiology. 2015;161:2069–2078. doi: 10.1099/mic.0.000157. [DOI] [PubMed] [Google Scholar]
- 49.Ishibashi N, et al. Gene cluster responsible for secretion of and immunity to multiple bacteriocins, the NKR-5-3 enterocins. Appl Environ Microbiol. 2014;80:6647–6655. doi: 10.1128/AEM.02312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birri DJ, Brede DA, Forberg T, Holo H, Nes IF. Molecular and genetic characterization of a novel bacteriocin locus in Enterococcus avium isolates from infants. Appl Environ Microbiol. 2010;76:483–492. doi: 10.1128/AEM.01597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobson AE, Sanozky-Dawes RB, Klaenhammer TR. Identification of an operon and inducing peptide involved in the production of lactacin B by Lactobacillus acidophilus. J Appl Microbiol. 2007;103:1766–1778. doi: 10.1111/j.1365-2672.2007.03417.x. [DOI] [PubMed] [Google Scholar]
- 52.Sushida H, et al. Evaluation of leader peptides that affect the secretory ability of a multiple bacteriocin transporter, EnkT. J Biosci Bioeng. 2018:S1389-1723(17)30929-5. doi: 10.1016/j.jbiosc.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Dawid S, Sebert ME, Weiser JN. Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J Bacteriol. 2009;191:1509–1518. doi: 10.1128/JB.01213-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sung CK, Li H, Claverys JP, Morrison DA. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol. 2001;67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Thompson CM, Lipsitch M. A modified Janus cassette (Sweet Janus) to improve allelic replacement efficiency by high-stringency negative selection in Streptococcus pneumoniae. PLoS One. 2014;9:e100510. doi: 10.1371/journal.pone.0100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Untergasser A, et al. Primer3–New capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 58.Puigbò P, Guzmán E, Romeu A, Garcia-Vallvé S. OPTIMIZER: A web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wayne KJ, et al. Localization and cellular amounts of the WalRKJ (VicRKX) two-component regulatory system proteins in serotype 2 Streptococcus pneumoniae. J Bacteriol. 2010;192:4388–4394. doi: 10.1128/JB.00578-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt TG, et al. Development of the Twin-Strep-tag® and its application for purification of recombinant proteins from cell culture supernatants. Protein Expr Purif. 2013;92:54–61. doi: 10.1016/j.pep.2013.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.