Significance
This work describes the most extensive discovery and functional characterization of small regulatory RNAs (sRNAs) in Mycobacterium tuberculosis to date. We comprehensively define the sRNAs expressed in M. tuberculosis under five host-like stress conditions. This reference dataset comprehensively defines the expression patterns and boundaries of mycobacterial sRNAs. We perform in-depth characterization of one sRNA, mycobacterial regulatory sRNA in iron (MrsI), which is induced in M. tuberculosis in multiple stress conditions. MrsI is critical for the iron-sparing response in mycobacteria by binding directly to mRNAs encoding nonessential iron-containing proteins to repress their expression. Interestingly, MrsI acts in an anticipatory manner, in which its induction by a variety of stresses primes M. tuberculosis to enter an iron-sparing state more rapidly upon iron deprivation.
Keywords: Mycobacterium tuberculosis, small RNA, iron sparing, RNA-Seq, riboregulation
Abstract
One key to the success of Mycobacterium tuberculosis as a pathogen is its ability to reside in the hostile environment of the human macrophage. Bacteria adapt to stress through a variety of mechanisms, including the use of small regulatory RNAs (sRNAs), which posttranscriptionally regulate bacterial gene expression. However, very little is currently known about mycobacterial sRNA-mediated riboregulation. To date, mycobacterial sRNA discovery has been performed primarily in log-phase growth, and no direct interaction between any mycobacterial sRNA and its targets has been validated. Here, we performed large-scale sRNA discovery and expression profiling in M. tuberculosis during exposure to five pathogenically relevant stresses. From these data, we identified a subset of sRNAs that are highly induced in multiple stress conditions. We focused on one of these sRNAs, ncRv11846, here renamed mycobacterial regulatory sRNA in iron (MrsI). We characterized the regulon of MrsI and showed in mycobacteria that it regulates one of its targets, bfrA, through a direct binding interaction. MrsI mediates an iron-sparing response that is required for optimal survival of M. tuberculosis under iron-limiting conditions. However, MrsI is induced by multiple host-like stressors, which appear to trigger MrsI as part of an anticipatory response to impending iron deprivation in the macrophage environment.
The pathogen Mycobacterium tuberculosis survives in macrophages, where it is exposed to an array of stresses, including iron restriction, nutrient limitation, oxidative stress, low pH, and membrane stress (1–5). Bacteria adapt to these stresses through transcriptional and posttranscriptional responses, including the regulatory functions of trans-encoded small regulatory RNAs (sRNAs). Despite the global impact of M. tuberculosis, mycobacteria are part of a broad group of bacterial pathogens about which very little is known in regard to sRNA-mediated regulation. For example, mycobacterial sRNAs have primarily been identified during growth in rich medium, and there have been few efforts to identify sRNAs involved in stress responses (6–12). Additionally, there has been only minimal characterization of the few validated mycobacterial sRNAs. Indeed, just a few putative targets of a single sRNA in M. tuberculosis have been described, and no direct interaction between an sRNA and predicted mRNA target has been validated (13). It is postulated that the rules for sRNA function defined in other bacteria extend to mycobacteria, although they lack key elements of the sRNA machinery, including any obvious homologs of the sRNA chaperone proteins Hfq and ProQ (14, 15).
In other prokaryotes in which sRNAs have been characterized, sRNAs most commonly act by binding to the 5′ end of trans-encoded mRNA targets, thereby repressing translation of the mRNA and often facilitating target mRNA degradation (16). The interaction between sRNAs and their targets is initiated through a short 5–7-nt sequence of perfect complementarity termed the “seed region” and extends to include a longer region with limited complementarity. Many sRNAs are critical for bacterial stress adaptation and pathogenesis, becoming strongly induced during stress exposure to regulate a set of targets (17–20). For example, expression of the iron-sparing sRNAs RyhB and PrrF of enteric bacteria and Pseudomonas aeruginosa, respectively, is highly up-regulated during iron starvation, and these sRNAs repress mRNAs encoding nonessential iron-containing proteins (21, 22).
Given the relative paucity of information on the identity and function of sRNAs in mycobacteria, we used high-throughput methodologies to comprehensively identify sRNAs expressed in M. tuberculosis during five in vivo relevant stress conditions. Interestingly, a subset of sRNAs is highly induced in multiple stresses. One of these, here renamed mycobacterial regulatory sRNA in iron (MrsI) or ncRv11846, is induced during exposure to iron starvation, oxidative stress, and membrane stress. We identify the regulon of MrsI in pathogenic and nonpathogenic species and determine direct targets by experimentally validating its seed region and the complementary binding site in the 5′ UTR of regulated mRNAs. MrsI is critical for normal adaptation to iron deprivation in M. tuberculosis, and its induction in oxidative stress leads to more rapid regulation of the target bfrA upon iron limitation. This supports a model by which M. tuberculosis uses oxidative and membrane stress responses as gateways to enter an anticipatory iron-sparing state.
Results
Discovery of sRNAs in M. tuberculosis Involved in Stress Responses.
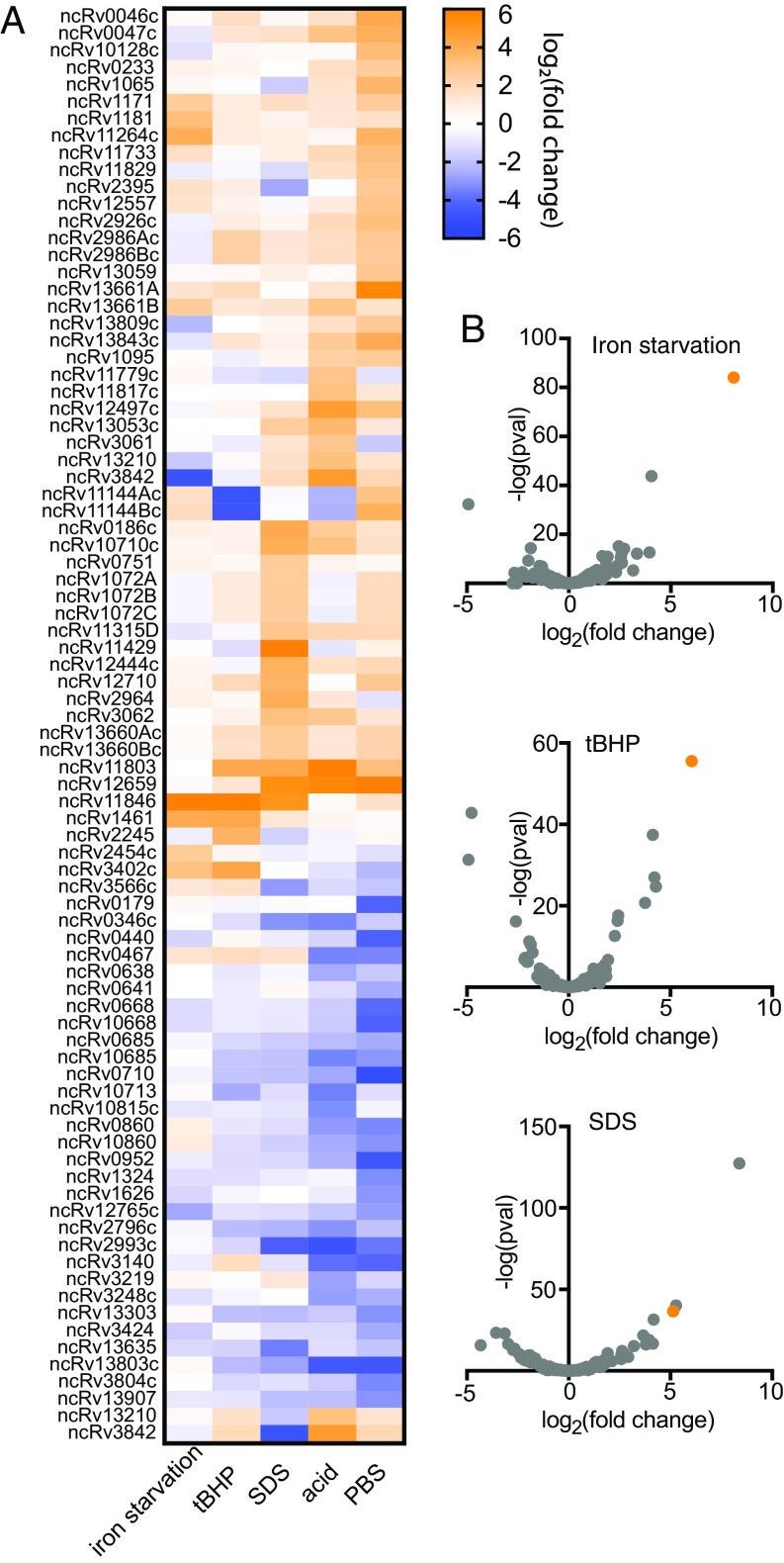
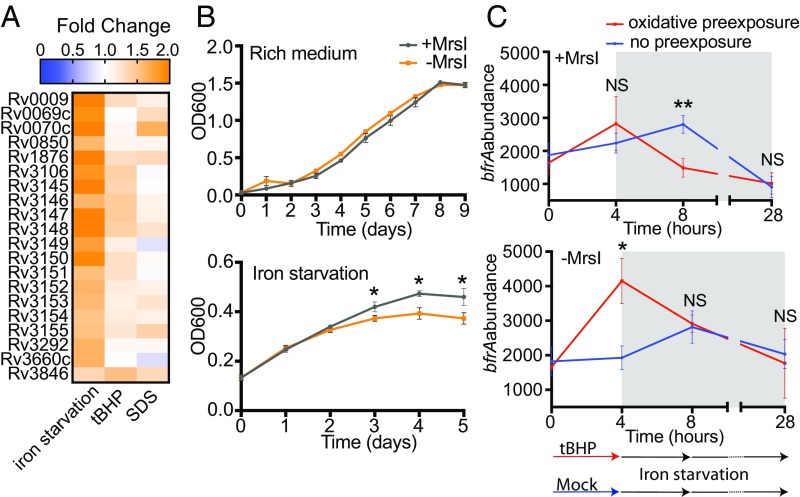
We sought to define the compendium of sRNAs in M. tuberculosis that are constitutively expressed and responsive to stress conditions. Multiple sequencing-based discovery studies in M. tuberculosis have sought to define sRNAs, but only two have used sRNA sequencing (sRNA-seq) approaches, which capture the sRNA boundaries (9, 23). However, these studies searched for only housekeeping sRNAs, and neither provided clues into the physiological roles of these sRNAs. To more systematically define sRNAs in M. tuberculosis and gain insight into their function, we performed sRNA-seq in rich medium and five host-like stress conditions: iron limitation, oxidative stress, membrane stress, acidic pH, and nutrient starvation. We identified sRNAs and their 5′ and 3′ boundaries by using the computational sRNA search tool Bacterial sRNA Finder (BS_Finder) (9, 24). We defined a set of 189 M. tuberculosis candidate sRNAs (Dataset S1), 103 of which had not been identified by previous studies (7, 9, 23). This result highlights the value of performing sRNA discovery under diverse stress conditions. Future work will be required to confirm the features of individual sRNAs by Northern blot analysis and to identify any that may be nonfunctional mRNA degradation products. Importantly, our dataset provides accurate coordinates of sRNA 5′ and 3′ ends as well as extensive differential expression information, and thus serves as a valuable reference source for the field. A total of 82 sRNAs had greater than sixfold differential expression in at least one condition (Fig. 1A and Dataset S1). Most induced sRNAs were specific to a single stress, but a smaller set of sRNAs was induced in multiple stresses, including ncRv11803, ncRv11846, and ncRv12659, which were highly induced in three different stress conditions (Fig. 1A).
Fig. 1.
Differential sRNA-seq in M. tuberculosis. (A) Heat map of the 82 M. tuberculosis sRNAs significantly differentially expressed across three biological replicates (P < 0.05; fold change ≥6) in at least one stress condition. (B) Volcano plots of sRNA differential expression in iron starvation for 24 h (Top), tBHP-mediated oxidative stress for 4 h (Middle), and SDS-mediated membrane stress for 4 h (Bottom). The orange data point in each graph is ncRv11846/MrsI.
The ncRv11846 Homolog in Mycobacterium smegmatis Is an Iron-Sparing sRNA.
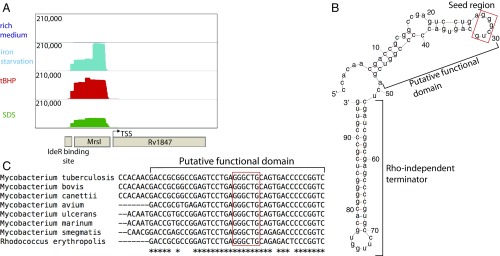
We focused on one of the promiscuously induced sRNAs, ncRv11846, which is the most highly induced sRNA during iron starvation and oxidative stress, and is also highly induced in membrane stress (Fig. 1B, orange data points, and Fig. 2A). ncRv11846 is ∼100 nt and is predicted to be highly structured (Fig. 2B). Transcription of the sRNA starts ∼100 bp upstream of Rv1847, a gene of unknown function that appears to be an independent transcript, given that we have previously identified its distinct transcriptional start site (Fig. 2A) (25). In addition, ncRv11846 has a predicted rho-independent terminator at the 3′ end of the sRNA (Fig. 2B) (26). We used Northern blot analysis to confirm that ncRv11846 is a small independent transcript and is induced in iron starvation and oxidative stress (SI Appendix, Fig. S1). ncRv11846 is highly conserved across Mycobacteriaceae and Nocardiaceae, particularly in a 30-nt stretch preceding the terminator (Fig. 2C). A binding site for the mycobacterial iron-dependent transcription factor IdeR is present near the transcriptional start site of this sRNA in M. tuberculosis and the nonpathogenic relative Mycobacterium smegmatis (27). This is consistent with the increased abundance of the sRNA in growth in iron limiting conditions, suggesting that ncRv11846 might be involved in the bacterial response to iron deprivation.
Fig. 2.
ncRv11846/MrsI is a promiscuously induced, highly structured and conserved sRNA. (A) Read distribution around the ncRv11846/MrsI locus in sRNA-seq data in rich medium and MrsI-inducing stress conditions. The linear scale (shown on the left) shows the number of reads. (B) Predicted minimum free energy secondary structure of ncRv11846/MrsI (mFold). The putative functional domain and rho-independent terminator are marked. The red box denotes the 6-nt seed region. (C) Alignment of the putative functional domain of ncRv11846/MrsI homologs from selected actinobacterial species. The red box denotes the sRNA seed region.
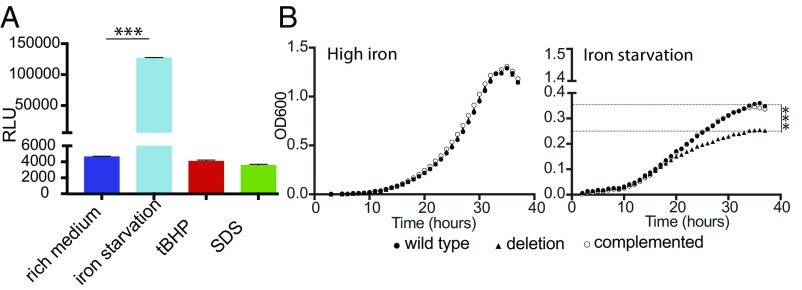
To develop M. smegmatis as a model for dissecting the mechanism of ncRv11846 action, we defined the expression pattern of its homolog in M. smegmatis. This homolog had previously been identified in a screen for sRNAs in M. smegmatis, and both ends were mapped by RACE analysis (10). The promoter of the sRNA in M. smegmatis, defined here as the 200 bp upstream of the 5′ end, was fused to a luciferase reporter to measure induction in each stress condition. Iron deprivation resulted in high levels of induction, but, unlike in M. tuberculosis, neither oxidative stress nor membrane stress induced expression of the sRNA in M. smegmatis (Fig. 3A). This suggests that the core function of the sRNA across mycobacterial species is mediated during iron deprivation and led to it being termed “mycobacterial regulatory sRNA in iron.”
Fig. 3.
ncRv11846/MrsI is induced during iron limitation in M. smegmatis and is involved in adaptation to iron starvation. (A) The promoter of the ncRv11846/MrsI homolog in M. smegmatis was fused to a luciferase reporter, and cells were exposed to stress before measuring luciferase activity (***P < 0.0001, unpaired t test). Error bars represent SD of three replicates. (B) Growth curves of MrsI strains in medium with (Left) and without (Right) iron. Dashed lines indicate the final optical density reached for strains with and without mrsI (***P < 0.0001, unpaired t test).
To investigate the function of MrsI on mycobacterial gene expression and adaptation, we constructed a deletion mutant (∆mrsI) in M. smegmatis. When the cells were grown in iron-rich medium, no growth difference was observed (Fig. 3B, Left). However, in iron-limited medium, M. smegmatis ∆mrsI reached a lower final optical density, whereas MrsI was strongly induced in WT cells (Fig. 3B, Right, and SI Appendix, Fig. S2A). Complementation on an episome restored MrsI levels and growth of the deletion mutant in iron-limiting conditions (Fig. 3B, Right, and SI Appendix, Fig. S2B).
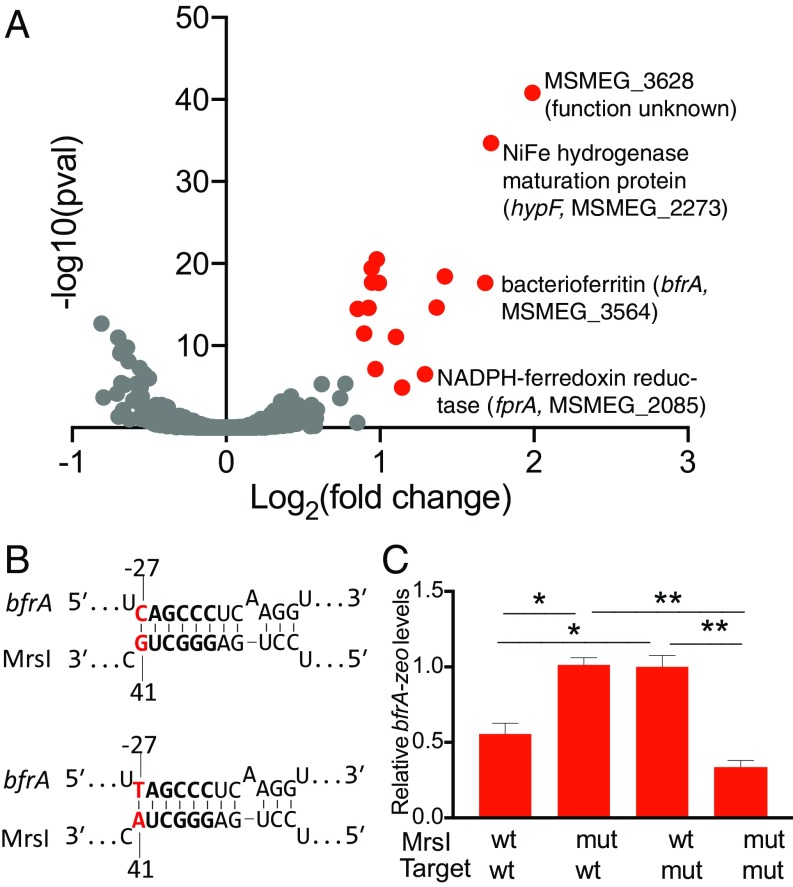
This phenotype is reminiscent of the growth defect observed for deletion of the iron-sparing sRNA RyhB in Escherichia coli and PrrF1 and PrrF2 in Pseudomonas aeruginosa (22, 28). We thus hypothesized that MrsI functions as an iron-sparing sRNA during iron limitation, repressing the expression of nonessential iron-containing proteins to restrict iron for essential functions (28, 29). To test this hypothesis, we used transcriptional profiling to identify genes whose expression is regulated by MrsI during iron deprivation in M. smegmatis (Dataset S2). A total of 20 genes, organized in 12 transcription units, had significantly higher abundance in the mrsI deletion strain compared with the WT and complemented strains, consistent with repression by MrsI (Fig. 4A, red data points, SI Appendix, Fig. S3, and Dataset S2). Of these 12 transcripts, 8 code for nonessential proteins that are predicted to bind iron or be involved with iron metabolism, including the NiFe hydrogenase maturation factor HypF, the bacterioferritin BfrA, and the ferredoxin reductase FprA (SI Appendix, Table S1). We confirmed the higher transcript levels of bfrA and hypF in the mrsI deletion during iron limitation by quantitative RT-PCR (RT-qPCR; SI Appendix, Fig. S4A). Additionally, we performed proteomic analysis on the same strains as mentioned earlier during iron limitation. Gene set enrichment analysis confirmed significant enrichment of the differentially expressed proteins within the gene set of targets identified by transcriptomic analysis (Dataset S3) (30). These results support a model in which MrsI acts as an iron-sparing sRNA in M. smegmatis, whereby it represses the expression of nonessential iron-containing proteins to reserve the dwindling iron stores for essential proteins.
Fig. 4.
MrsI is an iron-sparing sRNA in M. smegmatis and binds directly to the bfrA mRNA. (A) Volcano plot of transcriptomics for WT and ∆mrsI M. smegmatis after 6 h of iron starvation. Red data points indicate genes with elevated levels in the deletion strain compared with WT and complemented strains (fold change >1.5; P < 0.05). (B) Schematic of the WT-WT (Top) and mut-mut (Bottom) binding interaction between MrsI and the target bfrA. The MrsI seed region is in bold, and the bases mutated for the compensatory mutation assay are in red. (C) MrsI regulates bfrA directly. The promoter and 5′ UTR of bfrA were fused to the zeoR gene, and reciprocal mutations were made in the putative interaction sites on MrsI and bfrA-zeoR. Levels of bfrA-zeoR in each strain were measured by RT-qPCR (*P < 0.05 and **P < 0.005, unpaired t test). Error bars represent SD of three replicates.
MrsI Regulates bfrA Through a Direct Interaction.
We hypothesized that MrsI directly regulates transcripts encoding iron-containing proteins. To test this, we first used the sRNA target prediction software programs TargetRNA2 and CopraRNA to agnostically predict direct mRNA interactors of MrsI in M. smegmatis (31, 32). However, of the 115 direct MrsI targets predicted by these tools, none appeared to be regulated in a MrsI-dependent fashion in the expression analysis. We therefore sought to validate our experimentally identified putative targets by manually identifying regions involved in MrsI–mRNA interaction. sRNAs initiate contact with their targets through a short 6–8-nt sequence of perfect complementarity termed the seed region. Because seed regions are usually highly conserved, we focused on the stretch of nucleotides encompassing the 5′ hairpin (Fig. 2C) (33, 34). The 7-nt apical loop of this hairpin was the most promising site, as seed regions are generally single stranded to allow for intermolecular base pairing with target mRNAs (Fig. 2B, red box) (33). Importantly, the 5′ ends of 7 of the 12 differentially expressed transcriptional units have perfect complementarity to 6 nt of the loop (SI Appendix, Fig. S4B and Table S1).
To test if MrsI directly binds to its mRNA targets through the putative 6-nt seed region, we performed reciprocal mutation of mrsI and one candidate target, bfrA (Fig. 4 B and C). The 5′ UTR of bfrA was fused to the exogenous gene zeoR, creating a bfrA-zeoR expression reporter, and mrsI was inducibly expressed from an episomal plasmid. We measured bfrA-zeoR expression during iron limitation by RT-qPCR. We then introduced point mutations into the mrsI seed region and the putative binding site in the bfrA 5′ UTR and assessed the effect on bfrA-zeoR levels. In a strain containing mrsIWTbfrAWT-zeoR, induction of MrsI repressed bfrA-zeoR expression as expected (Fig. 4 B and C). Introduction of a G41A mutation into the putative seed region of mrsI abrogated regulation of the target (Fig. 4C). To determine whether this loss of regulation was caused by true disruption of a seed sequence, we introduced a compensatory C→T mutation to the predicted binding site in the 5′ UTR of bfrA-zeoR (bfrAC→T-zeoR). The bfrAC→T-zeoR mutation restored regulation by mrsIG41A (Fig. 4 B and C), whereas mrsIWT failed to regulate bfrAC→T-zeoR (Fig. 4C). These data demonstrate that MrsI regulates bfrA by direct interaction between a 6-nt seed sequence and a perfectly complementary region in the bfrA 5′ UTR. This represents a validated direct sRNA target in mycobacteria.
MrsI in M. tuberculosis Mediates Extensive Transcriptome Changes During Iron Deprivation.
We next used transcriptional profiling to define the effects of MrsI on the M. tuberculosis transcriptome during iron starvation, oxidative stress, and membrane stress (Fig. 5A and Dataset S4). We used the mycobacterial CRISPR interference system (CRISPRi) to inducibly knock down MrsI expression (35). Knockdown of MrsI resulted in the increased expression of 118 genes, consistent with repression by MrsI (Dataset S4, red). A total of 106 of these genes were differentially expressed during iron deprivation, whereas 5 and 12 genes were differential in oxidative stress and membrane stress, respectively. Thus, although MrsI affects gene expression in three stresses in M. tuberculosis, its effects are most extensive during iron deprivation. We next defined the phenotypic consequences of MrsI-mediated riboregulation in M. tuberculosis during iron starvation. Although no growth defect was observed upon MrsI knockdown in the absence of stress, knockdown of MrsI attenuated growth of M. tuberculosis during iron deprivation, similar to the phenotype observed in M. smegmatis (Fig. 5B).
Fig. 5.
MrsI mediates an anticipatory iron-sparing response in M. tuberculosis. (A) Heat map of genes putatively directly regulated by MrsI in MrsI-inducing stress conditions. (B) Growth curves of M. tuberculosis mrsI knockdown strain with and without CRIPSRi induction in rich medium (Top) and during iron starvation (Bottom; *P < 0.05, unpaired t test). Error bars represent SD of three replicates. (C) Levels of the MrsI target bfrA with and without preexposure to oxidative stress in M. tuberculosis. Three biological replicates of the MrsI knockdown strain were grown with or without oxidative stress for 4 h before iron starvation for 24 h (gray region). Levels of bfrA were measured by using Nanostring. Samples with and without MrsI knockdown (Bottom and Top, respectively) are shown (*P < 0.05 and **P < 0.005, unpaired t test). Error bars represent the SD of three biological replicates.
Two of the genes regulated by MrsI in M. smegmatis, bfrA and fprA, were also regulated in M. tuberculosis during iron deprivation. The MrsI binding sites in both of these transcripts are perfectly conserved between M. smegmatis and M. tuberculosis, suggesting preservation of targeting regions. Additionally, both encode iron-containing proteins consistent with a conserved role for MrsI in iron sparing. To distinguish between direct and indirect effects, we identified other putative direct targets in M. tuberculosis by scanning the 5′ ends of each of the regulated genes for MrsI binding sites. Including bfrA and fprA, 20 genes organized into 9 transcription units contained potential MrsI binding sites in the 5′ UTR. These are therefore putative direct targets (Fig. 5A), and 17 of these genes encode predicted iron-binding proteins. Of the 20 genes, two were differential during oxidative stress and membrane stress. The more robust changes to the transcriptome during MrsI knockdown in iron limitation and the fact that 17 of the 20 putative direct MrsI targets encode nonessential iron-containing proteins support a model in which MrsI functions as an iron-sparing sRNA in M. smegmatis and M. tuberculosis.
MrsI in M. tuberculosis Mediates an Anticipatory Iron Sparing Response.
That MrsI predominantly regulates iron metabolism while it is induced by multiple stresses may be indicative of a stress-adaptation program that integrates multiple signals to anticipate iron starvation and facilitate a more rapid iron-sparing response. We reasoned that preexposure of M. tuberculosis to oxidative stress before iron starvation would lead to more rapid MrsI-mediated repression of its targets. In line with the proposed model, cells preexposed to oxidative stress repressed bfrA expression more quickly under iron limitation than cells not preexposed to oxidative stress (Fig. 5C, Top, time 8 h). This early repression of bfrA is abrogated with MrsI knockdown, confirming that the effect is MrsI-dependent (Fig. 5C, Bottom, time 8 h). Interestingly, with MrsI knockdown, exposure to oxidative stress alone led to increased levels of bfrA, suggesting that this effect is dampened by MrsI in WT cells (Fig. 5C, time 4 h). By 24 h of iron starvation, bfrA repression was the same with or without preexposure to oxidative stress (Fig. 5C, time 28 h). MrsI expression is therefore induced by multiple stresses, which allows for faster regulation of its targets, consistent with a model of an anticipatory response to iron starvation.
Discussion
Although much is known about sRNAs in enteric bacteria, relatively little is understood about these regulators in mycobacteria. Here, we employed high-throughput methodologies to discover M. tuberculosis sRNAs involved in response to five host-like stresses. We generated a reference list of 189 M. tuberculosis sRNAs with well-defined boundaries and profiles of their expression patterns under these relevant conditions (Dataset S1). The results presented here provide the most comprehensive assessment of sRNA-mediated regulation in M. tuberculosis as well as the most detailed functional characterization of any actinobacterial sRNA to date. Importantly, our study also provides a template for conducting similar systematic studies of sRNAs in other species to discover yet-unknown sRNAs and gain valuable insights into their regulatory roles. This will likely prove particularly useful for other important human pathogens for which there is a dearth of information on sRNAs.
We validated a direct interaction between a mycobacterial sRNA and an mRNA target, which provides important insights into target recognition by mycobacterial sRNAs (Fig. 4C). Data from MrsI suggest that the rules for target binding do not precisely mirror those that have been defined in other prokaryotes. Indeed, the established bioinformatic sRNA target prediction tools TargetRNA2 and CopraRNA did not predict that MrsI would directly regulate any of the experimentally identified targets in M. smegmatis. Running TargetRNA2 individually on each of these experimentally identified MrsI targets revealed that only two have favorable binding energies (SI Appendix, Fig. S5). The predicted binding energies for the other MrsI–target pairs are substantially higher (i.e., less favorable) than those of the enterobacterial interactions on which the software is trained, and thus they are omitted from the list of predicted direct targets (SI Appendix, Fig. S5). Our results thus suggest a space for lineage-specific tuning of sRNA target-binding parameters. It is also interesting to note that sRNAs in enterobacteria require RNA chaperone proteins such as Hfq and ProQ to mediate mRNA regulation, despite their comparatively high mRNA affinity (36–38). Although mycobacteria lack a known Hfq or ProQ homolog, the less favorable binding energy of mycobacterial sRNAs suggests that noncanonical RNA chaperones exist in this lineage. Interestingly, studies in Bacillus subtilis have revealed a set of three noncanonical sRNA chaperones involved in the iron-sparing response (39, 40). Further studies will be necessary to identify the sRNA chaperone(s) that mediate MrsI regulation of its mRNA targets and if the binding energies of MrsI–mRNA interactions are representative of all mycobacterial sRNAs.
Although MrsI becomes highly induced in three stresses in M. tuberculosis, its effects on the transcriptome are more extensive during iron starvation (Fig. 5A and Dataset S4). Here, we show that preexposure of M. tuberculosis to oxidative stress results in more rapid MrsI-mediated repression of bfrA during iron starvation (Fig. 5C). This suggests that M. tuberculosis may take advantage of the predictable pattern of its stresses by using oxidative and membrane stresses as warning signals that it has entered a macrophage and will soon become deprived of iron. Therefore, sensing oxidative stress and membrane stress cause M. tuberculosis to enter an anticipatory iron-sparing state, priming MrsI to repress translation of targets such as bfrA. We cannot rule out the possibility that MrsI-mediated repression of these nonessential iron containing proteins could be adaptive in oxidative stress. However, studies in other bacteria suggest that this response alone could be detrimental in oxidative stress, in which iron sequestration by iron-storage proteins has been shown to prevent Fenton reactions from occurring.
Anticipatory responses to predictable sequences of environmental conditions have become increasingly recognized and highlight the intricate adaptation mechanisms bacteria have acquired during coevolution with hosts. For example, an anticipatory metabolic response was recently described for M. tuberculosis, in which exposure to hypoxia induces a metabolic remodeling of cell surface glycolipids and prepares the cell to reinitiate peptidoglycan biosynthesis upon exit from hypoxia (41). Additionally, predictive regulation has been described in E. coli, which resides in the human gut (42, 43). E. coli encounters lactose during gut transit at an earlier point than maltose, and exposure to lactose causes preinduction of a set of maltose utilization operons, preemptively preparing the cells to use maltose as a carbon source (42). Further studies will be necessary to determine the prevalence and functional importance of this anticipatory regulation for pathogenesis of M. tuberculosis and other human-associated bacterial species.
Materials and Methods
Bacterial Strains and Growth Conditions.
All M. tuberculosis strains used are derivatives of H37Rv, and all M. smegmatis strains are derivatives of mc2155. Strains are listed in SI Appendix, Table S2. Bacterial plasmids used in this study are listed in SI Appendix, Table S3, and details of their construction are in SI Appendix, Supplemental Materials and Methods. Oligonucleotides used are listed in SI Appendix, Table S4. M. tuberculosis and M. smegmatis strains were recovered from frozen stocks in Middlebrook 7H9 medium at 37 °C. Kanamycin (20 µg/mL), hygromycin B (50 µg/mL), and anhydrotetrayclcine (ATc; 100–200 ng/mL) were added when appropriate. For iron-starvation experiments, cells were grown in minimal medium with or without 50 µM FeCl3 as described previously (44). The iron-starvation medium, which has no added iron, was additionally treated with Chelex-100 to remove residual iron. For oxidative stress and membrane stress, cells were grown in 7H9 supplemented with tert-Butyl hydroperoxide (tBHP) or SDS, respectively. For nutrient starvation, cells were grown in PBS solution with 0.05% vol/vol tyloxapol. Details of growth conditions for sRNA-seq and transcriptomics are provided in SI Appendix, Supplemental Materials and Methods.
RNA Extraction, RT-qPCR, sRNA-Seq, and Total RNA-Seq.
RNA extraction and RT-qPCR were performed as described previously (35). RNA for sRNA-seq was extracted from M. tuberculosis grown in three biological replicates, and sRNA-seq libraries were prepared as described previously (9). Details of treatment of cells for transcriptomic analysis are provided in SI Appendix, Supplemental Materials and Methods. RNA for total RNA-seq was prepared similarly from two biological replicates by using the KAPA RNA Hyperprep kit (KAPA Biosystems) following the manufacturer’s instructions. For sRNA-seq and total RNA-seq in M. tuberculosis, oxidative stress was induced with 1 mM tBHP and membrane stress was induced with 0.5% SDS. Details of RNA-seq data analysis are provided in SI Appendix, Supplemental Materials and Methods.
Luciferase Assays.
M. smegmatis cells were grown to midlog phase (OD600 0.2–0.8) before exposure to 0.06 mM tBHP (oxidative stress) or 0.02% SDS (membrane stress) for 2 h or growth in medium without iron for 24 h (i.e., iron starvation), and luciferase production was measured by using the RenillaGlo Luciferase Assay kit (Promega) following the manufacturer’s instructions.
Proteomics.
Three replicates of ∆mrsI, complemented, and WT M. smegmatis were expanded to midlog phase in minimal medium supplemented with iron and hygromycin B. Cells were washed once with iron starvation medium and grown in the same medium with hygromycin B and ATc for 10 h. Proteomic analysis was performed by using quantitative LC-MS/MS. Details of proteomic sample preparation and analysis are provided in SI Appendix, Supplemental Materials and Methods. Gene set enrichment analysis (30) was performed by using the Wilcoxon test. The gene set used comprised the genes identified by transcriptomic analysis as regulated by MrsI in M. smegmatis (SI Appendix, Table S1).
Anticipatory Response Experiments.
Three biological replicates of M. tuberculosis were grown in 7H9 medium with or without 24 h of ATc-mediated induction of CRISPRi to midlog phase before transfer to 7H9 medium with or without 1 mM tBHP for 4 h. Samples with CRISPRi induced were supplemented with ATc. Cells were pelleted, washed once with iron-starvation medium, and then transferred to iron-starvation medium with or without ATc for 24 h. RNA was harvested before transfer into each medium and after 4 and 24 h of iron starvation for gene expression analysis by Nanostring (Nanostring Technologies). Details of Nanostring methodology and analysis are provided in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Eric Rubin and all members of the Rubin laboratory for insightful discussions and suggestions and G. Marcela Rodriguez for advice with iron starvation procedures. This work was supported by National Institutes of Health Grants U19 AI107774 and R15 GM102755 (to B.T.), National Science Foundation Fellowship DGE1144152 (to E.R.G.), and EMBO Fellowship ALTF 617-2015 (to T.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All sequencing data have been deposited in the Sequence Read Archive, NCBI, http://www.ncbi.nlm.nih.gov/sra (accession nos. SRP142345, SRP142511, and SRP142541).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718003115/-/DCSupplemental.
References
- 1.Stallings CL, Glickman MS. Is Mycobacterium tuberculosis stressed out? A critical assessment of the genetic evidence. Microbes Infect. 2010;12:1091–1101. doi: 10.1016/j.micinf.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu PT, Stenger S, Tang DH, Modlin RL. Cutting edge: Vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J Immunol. 2007;179:2060–2063. doi: 10.4049/jimmunol.179.4.2060. [DOI] [PubMed] [Google Scholar]
- 3.Purdy GE, Niederweis M, Russell DG. Decreased outer membrane permeability protects mycobacteria from killing by ubiquitin-derived peptides. Mol Microbiol. 2009;73:844–857. doi: 10.1111/j.1365-2958.2009.06801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 5.Saiga H, et al. Lipocalin 2-dependent inhibition of mycobacterial growth in alveolar epithelium. J Immunol. 2008;181:8521–8527. doi: 10.4049/jimmunol.181.12.8521. [DOI] [PubMed] [Google Scholar]
- 6.Li S-K, et al. Identification of small RNAs in Mycobacterium smegmatis using heterologous Hfq. RNA. 2013;19:74–84. doi: 10.1261/rna.034116.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnvig KB, et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnvig KB, Young DB. Identification of small RNAs in Mycobacterium tuberculosis. Mol Microbiol. 2009;73:397–408. doi: 10.1111/j.1365-2958.2009.06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeJesus MA, et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. MBio. 2017;8:e02133-16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai C-H, et al. Identification of novel sRNAs in mycobacterial species. PLoS One. 2013;8:e79411. doi: 10.1371/journal.pone.0079411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miotto P, et al. Genome-wide discovery of small RNAs in Mycobacterium tuberculosis. PLoS One. 2012;7:e51950. doi: 10.1371/journal.pone.0051950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelly S, Bishai WR, Lamichhane G. A screen for non-coding RNA in Mycobacterium tuberculosis reveals a cAMP-responsive RNA that is expressed during infection. Gene. 2012;500:85–92. doi: 10.1016/j.gene.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solans L, et al. The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004183. doi: 10.1371/journal.ppat.1004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haning K, Cho SH, Contreras LM. Small RNAs in mycobacteria: An unfolding story. Front Cell Infect Microbiol. 2014;4:96. doi: 10.3389/fcimb.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smirnov A, et al. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci USA. 2016;113:11591–11596. doi: 10.1073/pnas.1609981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: Expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner EG, Romby P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv Genet. 2015;90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Hébrard M, et al. sRNAs and the virulence of Salmonella enterica serovar Typhimurium. RNA Biol. 2012;9:437–445. doi: 10.4161/rna.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chabelskaya S, Gaillot O, Felden B. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010;6:e1000927. doi: 10.1371/journal.ppat.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 21.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinhart AA, et al. The prrF-encoded small regulatory RNAs are required for iron homeostasis and virulence of Pseudomonas aeruginosa. Infect Immun. 2015;83:863–875. doi: 10.1128/IAI.02707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, et al. An automated approach for global identification of sRNA-encoding regions in RNA-seq data from Mycobacterium tuberculosis. Acta Biochim Biophys Sin (Shanghai) 2016;48:544–553. doi: 10.1093/abbs/gmw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gómez-Lozano M, Marvig RL, Molin S, Long KS. Identification of bacterial small RNAs by RNA sequencing. Methods Mol Biol. 2014;1149:433–456. doi: 10.1007/978-1-4939-0473-0_34. [DOI] [PubMed] [Google Scholar]
- 25.Shell SS, et al. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015;11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czyz A, Mooney RA, Iaconi A, Landick R. Mycobacterial RNA polymerase requires a U-tract at intrinsic terminators and is aided by NusG at suboptimal terminators. MBio. 2014;5:e00931-14. doi: 10.1128/mBio.00931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prakash P, Yellaboina S, Ranjan A, Hasnain SE. Computational prediction and experimental verification of novel IdeR binding sites in the upstream sequences of Mycobacterium tuberculosis open reading frames. Bioinformatics. 2005;21:2161–2166. doi: 10.1093/bioinformatics/bti375. [DOI] [PubMed] [Google Scholar]
- 28.Jacques JF, et al. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol Microbiol. 2006;62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- 29.Oglesby-Sherrouse AG, Murphy ER. Iron-responsive bacterial small RNAs: Variations on a theme. Metallomics. 2013;5:276–286. doi: 10.1039/c3mt20224k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kery MB, Feldman M, Livny J, Tjaden B. TargetRNA2: Identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 2014;42:W124–W129. doi: 10.1093/nar/gku317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright PR, et al. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci USA. 2013;110:E3487–E3496. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richter AS, Backofen R. Accessibility and conservation: General features of bacterial small RNA-mRNA interactions? RNA Biol. 2012;9:954–965. doi: 10.4161/rna.20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papenfort K, Podkaminski D, Hinton JCD, Vogel J. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc Natl Acad Sci USA. 2012;109:E757–E764. doi: 10.1073/pnas.1119414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rock JM, et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol. 2017;2:16274. doi: 10.1038/nmicrobiol.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papenfort K, Förstner KU, Cong J-P, Sharma CM, Bassler BL. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci USA. 2015;112:E766–E775. doi: 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng L, et al. A qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. 2015;160:228–240. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smaldone GT, Antelmann H, Gaballa A, Helmann JD. The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases. J Bacteriol. 2012;194:2586–2593. doi: 10.1128/JB.05567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaballa A, et al. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci USA. 2008;105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eoh H, et al. Metabolic anticipation in Mycobacterium tuberculosis. Nat Microbiol. 2017;2:17084. doi: 10.1038/nmicrobiol.2017.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell A, et al. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 43.Tagkopoulos I, Liu Y-C, Tavazoie S. Predictive behavior within microbial genetic networks. Science. 2008;320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurthkoti K, et al. The capacity of Mycobacterium tuberculosisto survive iron starvation might enable it to persist in iron-deprived microenvironments of human granulomas. MBio. 2017;8:e01092–17. doi: 10.1128/mBio.01092-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.