Significance
p53 research has primarily addressed its cell-autonomous tumor-suppressive features. Yet, recent evidence suggests that, in normal tissues, p53 can also exert cell nonautonomous tumor suppressor activities. Cancer-associated fibroblasts (CAFs) enhance the malignant features of adjacent cancer cells. Surprisingly, we found that the conversion of normal fibroblasts to CAFs entails nonmutational alteration of their p53, such that it now becomes cancer supportive rather than cancer inhibitory. This is achieved through reprogramming the transcriptional output of the CAF p53, rendering it a positive regulator of cancer-promoting genes and secreted proteins. Overall, our study highlights a function of nonmutated p53 in the tumor microenvironment and suggests that molecules that can “re-educate” the renegade p53 may have therapeutic value.
Keywords: p53, fibroblasts, matrix metalloproteinases, tumor microenvironment, lung cancer
Abstract
Within the tumor microenvironment, cancer cells coexist with noncancerous adjacent cells that constitute the tumor microenvironment and impact tumor growth through diverse mechanisms. In particular, cancer-associated fibroblasts (CAFs) promote tumor progression in multiple ways. Earlier studies have revealed that in normal fibroblasts (NFs), p53 plays a cell nonautonomous tumor-suppressive role to restrict tumor growth. We now wished to investigate the role of p53 in CAFs. Remarkably, we found that the transcriptional program supported by p53 is altered substantially in CAFs relative to NFs. In agreement, the p53-dependent secretome is also altered in CAFs. This transcriptional rewiring renders p53 a significant contributor to the distinct intrinsic features of CAFs, as well as promotes tumor cell migration and invasion in culture. Concordantly, the ability of CAFs to promote tumor growth in mice is greatly compromised by depletion of their endogenous p53. Furthermore, cocultivation of NFs with cancer cells renders their p53-dependent transcriptome partially more similar to that of CAFs. Our findings raise the intriguing possibility that tumor progression may entail a nonmutational conversion (“education”) of stromal p53, from tumor suppressive to tumor supportive.
The p53 tumor suppressor acts as a major barrier against cancer development. Wild-type (WT) p53 helps maintain genome integrity and cellular homeostasis by regulating the expression of a plethora of genes involved in the regulation of cell cycle, apoptosis, stem cell differentiation, senescence, DNA repair, and metabolism (1–5). Cancer-associated mutations in the TP53 gene, resulting in production of mutant p53 proteins, can lead not only to loss of its tumor-suppressive functions but often also to gain of tumor-promoting activities, associated with altered p53-dependent transcriptional programs (6). Of note, alterations in the regulatory networks that impinge on p53 may cause genetically WT p53 to adopt features that partly resemble those of bona fide mutant p53 (4, 7, 8). This might convert WTp53 from tumor suppressive to potentially tumor supportive. So far, p53 research has focused primarily on its cell-autonomous functions. However, p53 also possesses cell nonautonomous tumor-suppressive functions (9, 10).
Fibroblasts are a major component of the tumor stroma and play important roles in disease progression and metastasis (11, 12). Cancer-associated fibroblasts (CAFs) differ from their normal counterparts in a variety of structural and functional aspects, and emerge, at least in part, through continuous “education” of the stroma by cancer cells (11, 12). Interestingly, suppression of p53 activity in normal fibroblasts (NFs) promotes acquisition of a “CAF phenotype” (13). Moreover, p53 overexpression in NFs can reduce tumor growth and enhance apoptosis of adjacent tumor cells (14). Mechanistically, inactivation of p53 in NFs augments the expression of proteins such as SDF1/CXCL12 (15, 16) and TSPAN12 (17), which might enhance tumor invasion and malignancy. p53 also modulates macrophage functions in a cell nonautonomous manner, thereby promoting an antitumoral microenvironment (9). CAFs probably harbor very few, if any, genetic modifications and instead are shaped mainly by epigenetic alterations (18–20).
We set out to determine whether nonmutational alterations in fibroblast p53 might contribute to the conversion of NFs into CAFs. We found that CAF p53 indeed differs functionally from NF p53. In particular, CAF p53 contributes to an altered transcriptional program, modifying the CAF secretome and promoting cell-autonomous and nonautonomous distinctive CAF features. Moreover, p53 facilitates a “CAF-like” transcriptional response in NFs cocultivated with cancer cells. We thus propose that altered p53 functionality in cancer-associated stromal cells may actively contribute to a tumor-supportive microenvironment.
Results
p53 Regulates Cell Autonomous CAF-Specific Properties.
While the cell-autonomous and nonautonomous tumor-suppressive functions of p53 in NFs have been studied in detail (9, 15, 16), its impact on the properties of CAFs is less well established. To address this issue, we employed paired NF and CAF cultures derived from the resected lung of the same patient (patient 4731; poorly differentiated adeno-squamous lung carcinoma). Analysis of α-smooth muscle actin (ACTA2) protein and mRNA confirmed that, as expected, the CAFs expressed higher levels of ACTA2 than the corresponding NFs (SI Appendix, Fig. S1 A and B). Both culture types displayed comparable levels of vimentin mRNA (SI Appendix, Fig. S1C). Next, these low-passage cultures were immortalized with human telomerase reverse transcriptase (hTERT) and stably transduced with GFP and subsequently with either p53 shRNA or control LacZ shRNA (SI Appendix, Fig. S1 D and E).
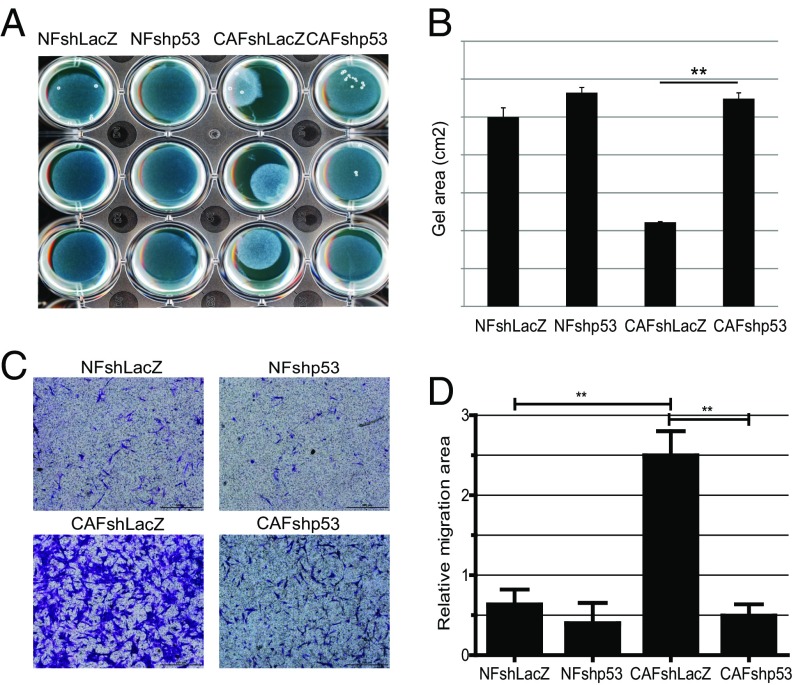
In the tumor microenvironment, CAFs display an activated fibroblast phenotype and can physically remodel the extracellular matrix (ECM) (11). In culture, this is reflected by an increased ability of CAFs to contract a collagen gel in which they are embedded (21). As expected, the immortalized CAFs (CAFshLacZ) displayed markedly greater contractile activity than the corresponding NFs (NFshLacZ; Fig. 1 A and B). Remarkably, silencing of p53 in the CAFs (CAFshp53) strongly compromised this activity (Fig. 1 A and B), implicating p53 as a key contributor to a distinctive CAF feature.
Fig. 1.
p53 controls the cell-autonomous functions in CAFs. (A) hTERT-immortalized NFs and CAFs from patient 4731, stably expressing either p53 shRNA or control LacZ shRNA, were subjected to a collagen gel contraction assay in triplicate and imaged after 24 h. (B) Quantification of the collagen gel area, analyzed as in A. Values represent averages ± SEMs from three independent experiments. (C) Cells as in A were grown in trans-well inserts. The lower chamber was loaded with medium containing EGF (10 ng/mL). Sixteen hours later, cells that had migrated across the membrane were stained with crystal violet and photographed. (Scale bars, 500 μm.) (D) Average migration ± SEM from three independent experiments performed as in C; quantification is described in SI Appendix. **P < 0.01.
CAFs also display increased migration (11, 22). We therefore compared the different immortalized fibroblast populations in a trans-well migration assay, with EGF as a chemoattractant. As expected, the CAFs migrated more avidly than their matched NFs (Fig. 1 C and D; compare CAFshLacZ with NFshLacZ). p53 silencing markedly attenuated this enhanced migration (compare CAFshp53 with CAFshLacZ). Furthermore, partial depletion of p53 diminished the collagen contraction activity and migratory capacity of nonimmortalized CAFs derived from lung tumors of additional patients (SI Appendix, Fig. S2); perhaps not surprisingly, the scope of the effect and the degree of divergence between CAFs and NFs varied among individual patients. Collectively, these observations imply that p53 promotes inherent, cell-autonomous, distinctive characteristics of CAFs.
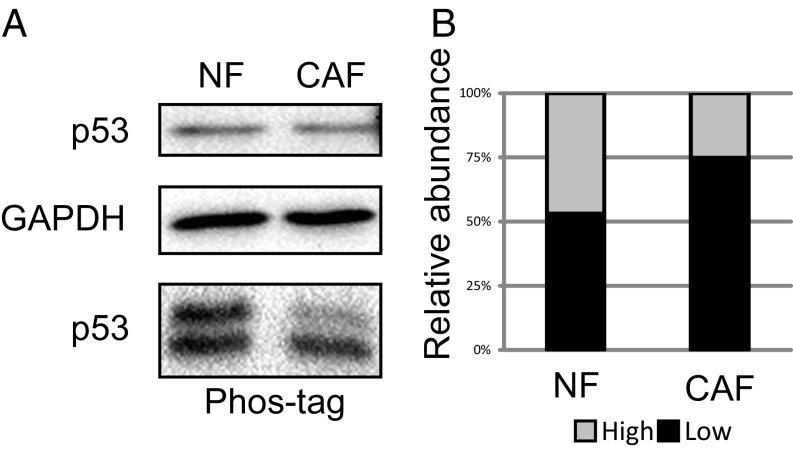
RNA sequencing did not detect any TP53 coding-region mutations in our CAFs. The cellular functions of p53 are largely controlled by posttranslational modifications, including multiple phosphorylation events (23). Interestingly, relative to NFs, the CAFs displayed a selective reduction in phosphorylated forms of p53 (Fig. 2 and SI Appendix, Fig. S3). In some instances, reduced p53 phosphorylation has been linked to altered protein conformation, detectable by specific monoclonal antibodies (7, 24). In agreement, immunoprecipitation analysis with the PAb1620 and PAb240 monoclonal antibodies, recognizing the WT conformation and a mutant conformation of p53, respectively, confirmed that the PAb240-to-PAb1620 ratio was elevated, albeit mildly, in CAF p53 relative to NF p53 (SI Appendix, Fig. S4), suggesting that a small fraction of the CAF p53 had adopted a mutant-like conformation. Together, these observations suggest that the endogenous WT p53 undergoes chemical and structural alterations during the conversion of NFs to CAFs, which may contribute to the distinctive biological features of the CAFs.
Fig. 2.
CAF p53 is hypophosphorylated. (A) Extracts from immortalized NFs and CAFs (patient 4731) were subjected to either standard SDS/PAGE (Top) or 30 µM Phos-tag SDS/PAGE (Bottom), followed by Western blot analysis with the indicated antibodies. (B) The relative abundance of each band in the Phos-tag gel, denoted by its position in the autoradiogram.
p53 Regulates the Expression of CAF-Specific Genes.
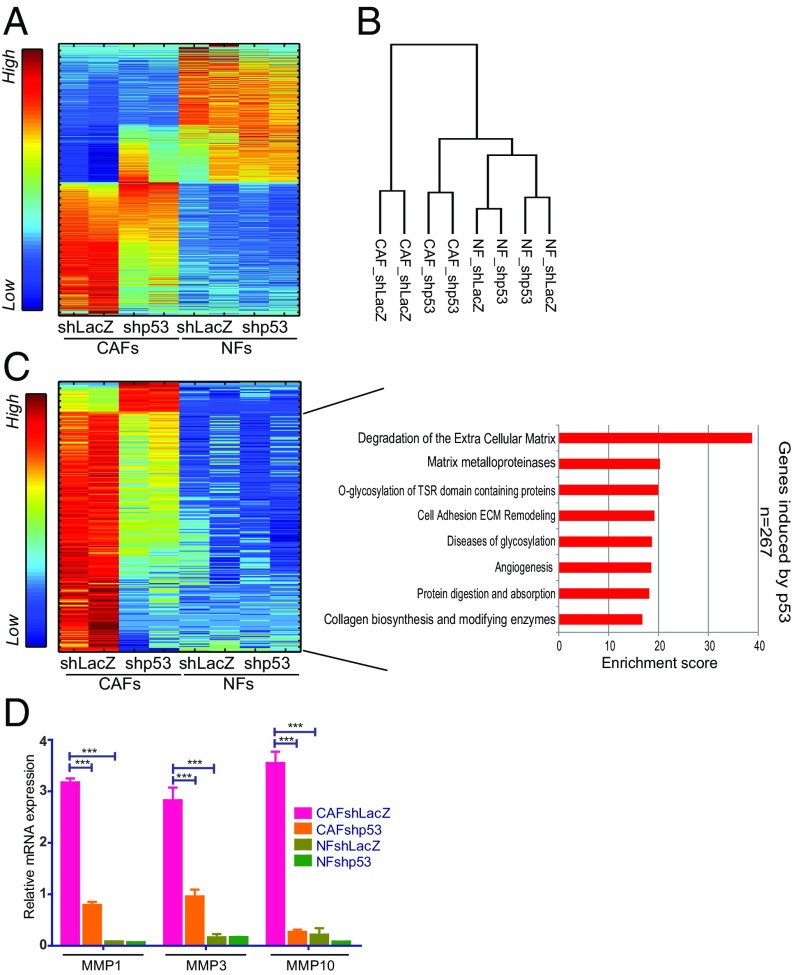
Earlier studies have uncovered differential gene expression patterns between CAFs and NFs in various tumor types (21, 25–27). We therefore performed global transcriptome analysis (RNA-seq) of immortalized NFs and CAFs, with or without stable p53 knockdown. As expected, the expression profile of CAFs differed substantially from that of NFs, with 1,662 genes differentially expressed between the two cell types (Fig. 3A and Dataset S1). Comparison of these differentially expressed genes with a published lung cancer CAF-associated gene signature (21) revealed substantial overlap (SI Appendix, Fig. S5A), authenticating the representative nature of this NF–CAF pair.
Fig. 3.
CAF p53 regulates genes associated with ECM remodeling. (A) SPIN-ordered expression matrix of genes differentially expressed between immortalized NFs and CAFs (patient 4731) (fold-change >1.5 and adjusted P value <0.05; 1,662 genes). Colors indicate relative expression after standardizing each gene (Left bar). (B) Dendrogram showing hierarchical clustering of data from transcriptome analysis of duplicate control and p53-depleted CAF and NF samples (average linkage, Pearson correlation). (C) p53-dependent genes in CAFs (fold-change >1.5 between shLacZ and shp53 in both replicates; 300 genes) were extracted from the set of genes more abundantly expressed in CAFs compared with NFs. Expression levels in both CAFs and NFs were visualized by SPIN-ordered expression matrix; colors indicate relative expression after standardizing each gene (Left bar). Pathway enrichment analysis (GeneAnalytics) for genes expressed preferentially and activated by p53 in CAFs is shown on the Right. (D) Quantification of MMP1, MMP3, and MMP10 mRNA from immortalized fibroblasts (patient 4731) by qRT-PCR. Values (mean ± SEM) were derived from five independent experiments. ***P ≤ 0.001 using one-way ANOVA and Tukey post hoc test. SPIN, sorting points into neighborhood; TSR, thrombospondin type I repeat.
Remarkably, hierarchical clustering of the overall gene expression patterns indicated that p53 silencing in the CAFs rendered their transcriptome more similar to that of NFs (Fig. 3B), further implying that the p53 of CAFs contributes to their “CAFness.” In agreement, while p53 knockdown decreased the expression of canonical p53 target genes in both NFs and CAFs (see SI Appendix, Fig. S5B, and p21 panels in SI Appendix, Fig. S6 for primary NF and CAF cultures from additional patients), p53 silencing in CAFs also reduced the expression of numerous CAF-specific genes, defined by their transcripts being more abundant in control CAFs (CAFs shLacZ; Fig. 3A) than in matched control NFs (NFs shLacZ).
We next focused on the cluster of 300 genes expressed more abundantly in CAFs than in NFs, whose expression in CAFs was p53 regulated (Fig. 3C and Dataset S1). A total of 267 of those genes were positively regulated by CAF p53. Silencing of CAF p53 rendered the expression of those genes more similar to NFs [Fig. 3C, compare CAFs shp53 (columns 3–4) with NFs shLacZ (columns 5–6)]. Notably, GeneAnalytics analysis of those genes revealed significant enrichment for functional pathways related to ECM degradation, matrix metalloproteinases (MMPs), and collagen-related enzymes (Fig. 3C, Right). In particular, MMP1, MMP3, and MMP10 mRNA were up-regulated in CAFs in a partially p53-dependent manner, as further validated by qRT-PCR (Fig. 3D). Importantly, p53-dependent expression of these genes was also observed in primary lung CAFs from additional patients (SI Appendix, Fig. S6). Of note, MMPs are frequently up-regulated in cancer, promoting tumor cell migration, invasion, and metastasis (28).
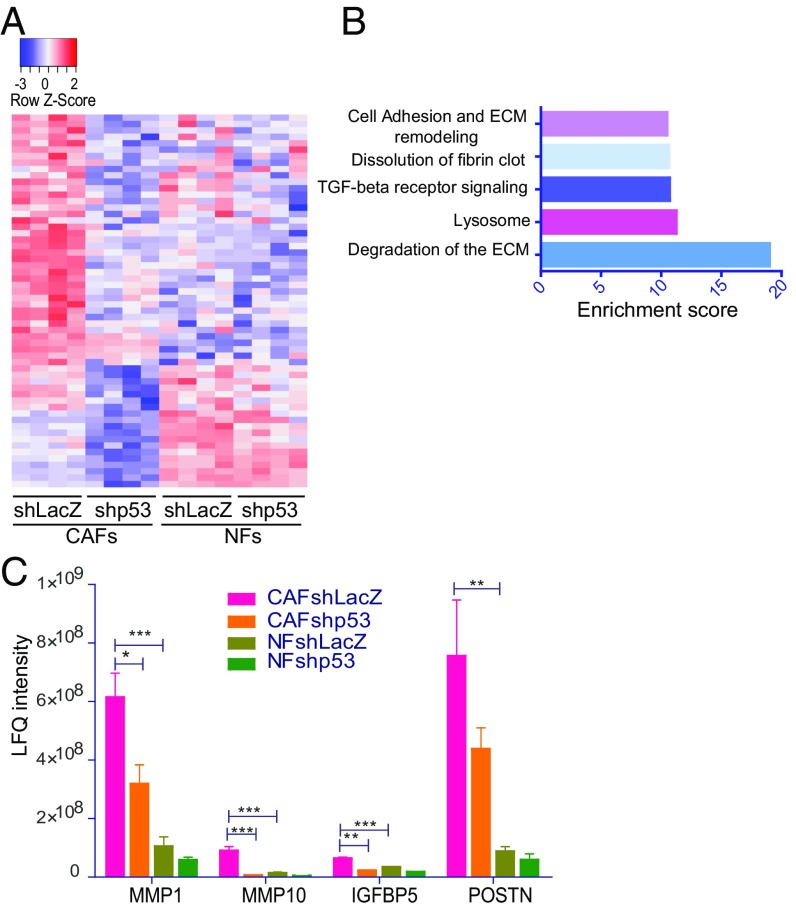
p53 Affects the CAF Secretome.
Proteins secreted by CAFs can modulate the properties of surrounding cancer cells (29). Therefore, we collected conditioned medium (CM) from the various fibroblasts and subjected it to liquid chromatography–MS–based protein analysis. Notably, clustering of the secretome data revealed that the secretion of numerous proteins was elevated in CAFs relative to NFs in a p53-dependent manner (Fig. 4A). The top 58 secreted proteins regulated specifically by p53 in CAFs (Fig. 4A and Dataset S1) were enriched for pathways involved in ECM degradation (Fig. 4B), consistent with the transcriptome analysis. Fig. 4C illustrates the p53-dependent regulation of representative secreted proteins (MMP1, MMP10, IGFBP5, and POSTN). Overall, our data indicate that CAF p53 undergoes alterations that may expand its target gene repertoire to include many CAFness genes, supporting tumor progression.
Fig. 4.
p53 regulates the CAF secretome. (A) Conditioned media of immortalized fibroblasts (patient 4731) were subjected to LC-MS analysis. The heatmap was generated using differentially secreted proteins between CAFshLacZ and CAFshp53 (fold-change >1.5 and adjusted P value <0.05 in four repeats; total = 58 proteins). Colors indicate relative expression (Top bar). (B) Pathway enrichment analysis (GeneAnalytics) for secreted proteins expressed differentially between CAFshLacZ and shp53. (C) Relative abundance of the indicated proteins in CM from immortalized CAFs and NFs, with and without p53 silencing, deduced from the LC-MS analysis. Bars represent normalized LFQ intensities. Values (mean ± SEM) were derived from four independent experiments. *P ≤ 0.05, **P < 0.01, ***P ≤ 0.001 using one-way ANOVA and Tukey post hoc test. LC-MS, liquid chromatography-MS; LFQ, label-free quantification.
CAF p53 Promotes Cancer Cell Migration and Invasion.
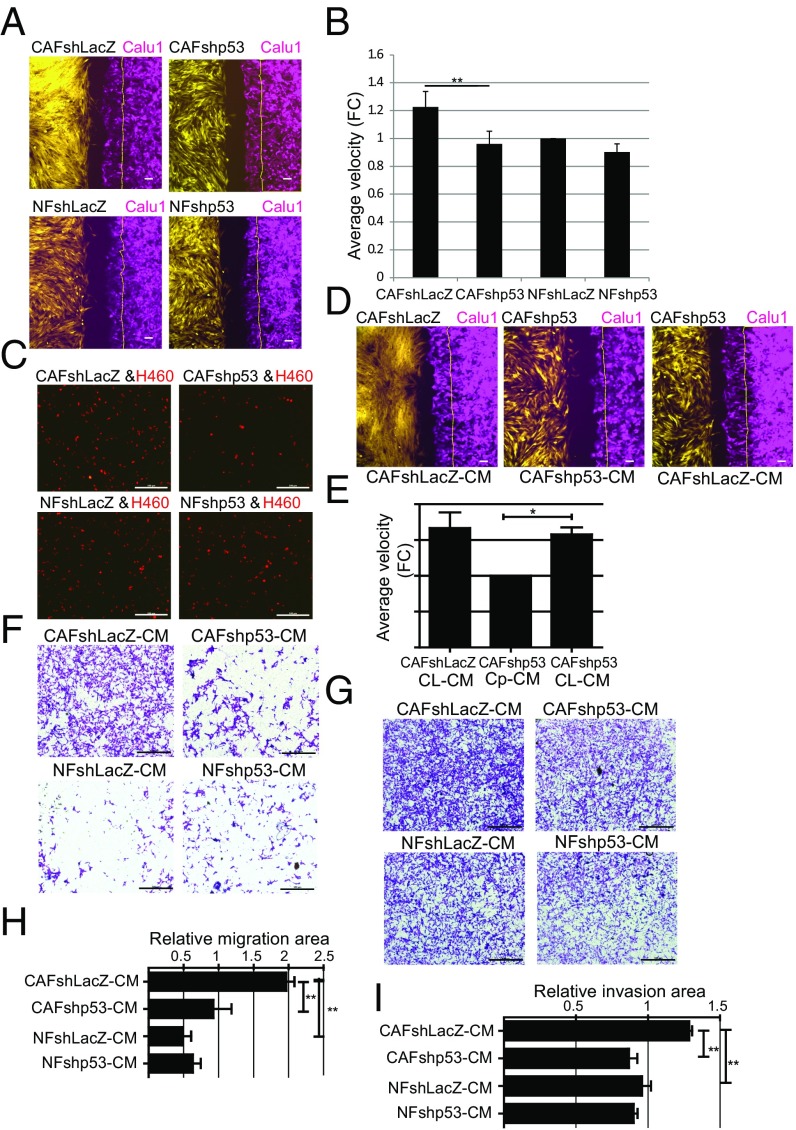
CAFs can promote tumor cell migration and invasion (29, 30). To determine whether CAF p53 can exert cell nonautonomous effects on adjacent cancer cells, we performed live cell imaging of gap closure, with GFP-labeled immortalized fibroblasts seeded on one side of the gap and mCherry-labeled Calu1 lung cancer cells on the other side. As seen in Fig. 5 A and B and Movies S1–S4, the cancer cells indeed migrated faster toward control CAFs (shLacZ) than toward control NFs. Notably, knockdown of CAF p53 reduced cancer cell migration, rendering it comparable to migration toward NFs. Likewise, when mCherry-labeled H460 lung cancer cells were cocultured with GFP-expressing NFs or CAFs, p53 silencing attenuated the ability of CAFs to promote cancer cell migration (Fig. 5C). Hence, the p53 of CAFs contributes to their ability to promote migration of adjacent cancer cells.
Fig. 5.
CAFs promote p53-dependent cancer cell migration and invasion. (A) GFP-expressing immortalized fibroblasts from patient 4731 (yellow) and mCherry-expressing Calu1 cells (magenta) were seeded in 12-well plates containing ibidi culture inserts. The next day, inserts were removed and cells were allowed to migrate. Shown are images snap-captured at 6 h. The thin yellow line on the right indicates the position of the Calu1 front at time = 0. (Scale bars, 100 μm.) (B) Average velocity of Calu1 migration toward the indicated fibroblasts, relative to migration of tumor cells alone, based on 6-h measurements. Values are means ± SEMs of three independent experiments, **P < 0.01. (C) mCherry-labeled H460 cells were seeded together with the indicated immortalized fibroblasts and subjected to a trans-well migration assay toward EGF. mCherry-positive cells that had migrated across the membrane were photographed 16 h later. (Scale bars, 500 μm.) (D) Live cell migration analysis as in A, except that cultures were supplemented with CM from the indicated fibroblasts (listed below the panel) after removal of the insert. (Scale bars, 100 μm.) (E) Average velocity of Calu1 cell migration toward the fibroblasts as assayed in D, relative to the CAFshp53-CM samples. CL-CM, CAFshLacZ-CM; Cp-CM, CAFshp53-CM. Values are means ± SEMs of three independent experiments, *P < 0.05. (F) Calu1 cells were grown in trans-well inserts, and the lower chamber was loaded with CM from the different indicated fibroblasts. Sixteen hours later, migrated cells were stained with crystal violet and photographed. (Scale bars, 500 μm.) (G) Calu1 cells were grown in Matrigel-coated trans-well inserts, and the lower chamber was loaded with CM from the different indicated fibroblasts. Twenty hours later, cells that had invaded across the membrane were stained with crystal violet and photographed. (Scale bars, 500 μm.) (H) Average migration ± SEMs from three independent experiments performed as in F; quantification is described in SI Appendix. **P < 0.01 using one-way ANOVA and Tukey post hoc test. (I) Average invasion ± SEMs from three independent experiments performed as in G, quantified as in H. **P < 0.01 using one-way ANOVA and Tukey post hoc test. FC, fold-change.
Our RNA-seq and secretome data indicated that, in addition to MMPs, CAF p53 promotes the expression of numerous other secreted factors (Fig. 4C, SI Appendix, Fig. S7A, and Dataset S1). Such factors may mediate, at least in part, the p53-dependent effects of the CAFs on the cancer cells. Indeed, CM from p53-proficient control CAF cultures (CAFshLacZ) partly restored the defective migration of Calu1 cancer cells toward p53-depleted CAFs (Fig. 5 D and E and Movies S5–S7). Likewise, in a trans-well assay, CM from CAFs, but not NFs, stimulated in a p53-dependent manner the migration of Calu1 cells (Fig. 5 F and H). Furthermore, CM from p53-depleted CAFs was less potent than control CAF CM in promoting the invasion of lung cancer cells through a Matrigel layer (Fig. 5 G and I). Thus, the cell nonautonomous, promigratory, and proinvasive effects of CAFs on adjacent cancer cells are facilitated by p53-dependent up-regulation of genes encoding secreted factors.
Cancer Cells Modify the p53-Dependent Transcriptional Program in NFs.
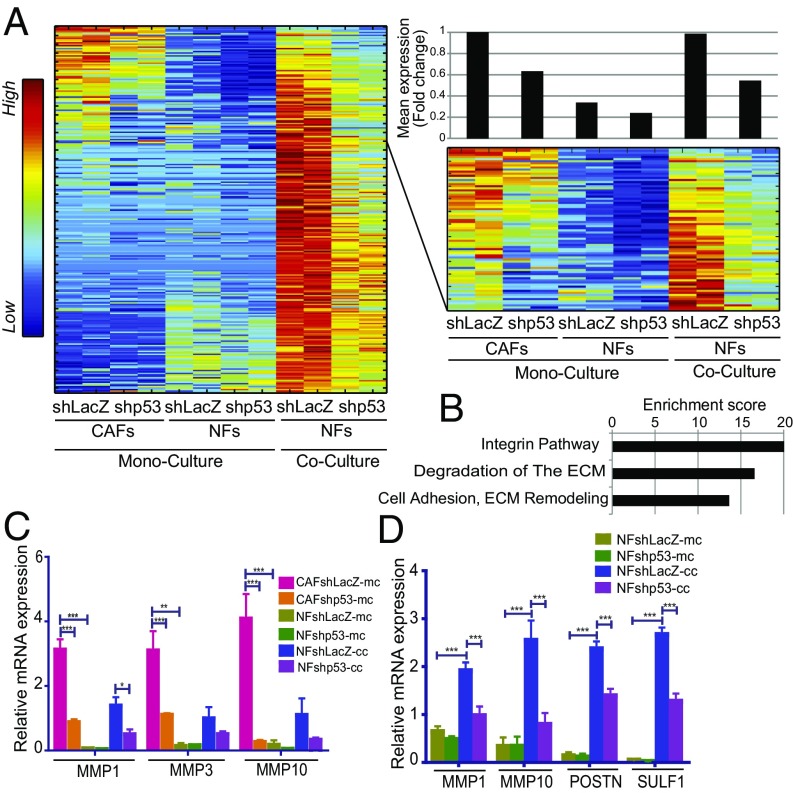
Cancer cells can instruct resident and recruited NFs to gradually acquire features of CAFs, in a process sometimes referred to as “education” (31). To probe a potential involvement of p53 in this process, mCherry-labeled H460 cells were cocultured for 3 d with GFP-expressing control NFs or p53-silenced NFs. Fibroblasts were then separated from cancer cells by FACS sorting (see SI Appendix, Fig. S7B for validation of the purity of each sorted subpopulation), and the purified fibroblasts were subjected to RNA-seq analysis. Cocultivation with cancer cells augmented the expression of 1,193 genes in control NFs, relative to the corresponding monocultured NFs. Importantly, expression of >250 of those genes was partially p53 dependent (Fig. 6A and Dataset S1). Notably, a subset of such genes, whose basal expression in monocultured NFs is relatively low, are constitutively more highly expressed in CAFs in a partially p53-dependent manner (Fig. 6A, Right ). These genes are enriched for processes such as ECM degradation and remodeling, integrin signaling, and cell adhesion (Fig. 6B), reminiscent of the p53-dependent transcription program in CAFs (Fig. 3C). Validation of representative genes by qRT-PCR is shown in Fig. 6C. A similar trend was observed when immortalized NFs were cocultured with another lung cancer cell line, H1299 (Fig. 6D). Thus, p53 contributes to the transcriptional reprogramming of NFs in the continued presence of adjacent cancer cells.
Fig. 6.
Tumor cells promote p53-dependent expression of CAF-associated genes in cocultured NFs. (A) SPIN-ordered expression matrix of genes up-regulated in a p53-dependent manner in immortalized NFs (patient 4731) cocultured with H460 cells, isolated by FACS sorting before RNA extraction. The upper portion of the heatmap is enlarged on the right side of the panel, to highlight the subset of genes also expressed highly in monocultured CAFs. Top Right shows mean fold-change relative to control (column 1) in each pair of replicate samples, calculated for the 77 genes shown in the enlarged panel. (B) Pathway enrichment analysis (GeneAnalytics) of the 77 genes in the right matrix in A. (C) Expression of MMP1, MMP3, and MMP10 mRNA in immortalized CAFs and NFs, grown as monoculture or cocultured with H460 cells and FACS-sorted before RNA extraction. The monoculture data are the same as in Fig. 3D and are included only for comparison. Values are means ± SEMs from four independent experiments. (D) Expression of MMP1, MMP10, POSTN, and SULF1 mRNA in immortalized NFs (patient 4731), grown as monoculture or cocultured with H1299 cells, and FACS-sorted before RNA extraction. Values are means ± SEMs from at least three independent experiments. *P ≤ 0.05, **P < 0.01, and ***P ≤ 0.001 using one-way ANOVA and Tukey post hoc test. SPIN, sorting points into neighborhood.
CAF p53 Promotes Tumor Growth In Vivo.
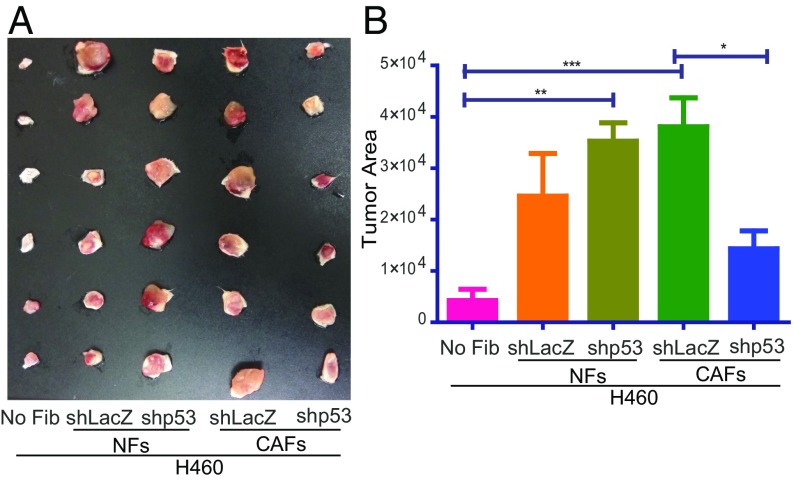
CAFs support tumor growth and cancer progression (11, 21, 29). To investigate the contribution of CAF p53 to these activities, H460 cells were injected s.c. into SCID mice, either alone or in combination with p53-proficient or p53-deficient immortalized NFs and CAFs. Nine days later, tumors were excised and photographed (Fig. 7A); relative tumor size was deduced by quantifying the area occupied by each tumor image (Fig. 7B). In agreement with earlier results (15, 21, 32), control (shLacZ) CAFs showed a trend to promote tumor growth better than control NFs (Fig. 7), while depletion of p53 in the NFs (shp53) showed a trend to render them more tumor supportive. Importantly, depletion of p53 in CAFs greatly reduced their ability to support tumor growth. Essentially similar results were obtained in a second experiment, in which we determined the weights of tumors harvested 12 d postinjection (SI Appendix, Fig. S8). Hence, in agreement with our in vitro observations, CAF p53 also exerts a cancer-promoting effect in vivo. Altogether, our findings reveal a surprising role for p53 in CAFs, and suggest that, during tumor progression, p53 functionality is altered not only in the cancer cells but also in their adjacent stroma.
Fig. 7.
CAF p53 promotes tumor growth in SCID mice. mCherry-expressing H460 cells (5 × 105) were injected, either alone or together, with 1.5 × 106 immortalized NFs or CAFs (patient 4731) stably expressing either shLacZ or shp53, into the right flank of male SCID mice (n = 6 per group; total inoculation volume = 80 µL). (A) Tumors were excised and photographed 9 d later. (B) Relative tumor size was deduced from the photograph by calculating the tumor area with Fiji software (values are means ± SEM from six mice in each group). No Fib, no fibroblasts.
Discussion
In the tumor microenvironment, fibroblasts play major roles in modulating tumor growth and metastasis (11, 32). Remarkably, the impact of stromal fibroblasts on the adjacent cancer cells changes in the course of tumor progression. Initially, resident NFs tend to suppress tumor growth and maintain tissue homeostasis. At that early stage, p53 plays a cell nonautonomous tumor-suppressive role within the fibroblasts, partly by inhibiting their ability to produce and secrete a variety of tumor-promoting factors (15–17, 33). However, continued cross-talk with the cancer cells converts their neighboring fibroblasts into CAFs (11). CAFs originate from a variety of sources (34–36), although local fibroblasts are often the major contributors to the CAF population (37). Upon conversion into CAFs, these cells acquire an ability to support the proliferation and survival of cancer cells and promote their aggressive features, largely through production and secretion of a variety of proteins and other molecules. This can also promote cancer cell resistance to chemotherapy (38, 39).
p53 is a very frequent target for mutational inactivation in cancer (6). Mutations in the TP53 gene, when occurring within cancer cells, do not only ablate p53’s tumor-suppressive functions but may also lead to oncogenic gain-of-function (6). However, while genomic alterations dominate the landscape of cancer cells, the distinctive cancer-promoting features of CAFs are primarily attributed to epigenetic modifications (40, 41) and differential expression of miRNAs (42). Indeed, despite several earlier reports of p53 mutations in the stromal compartment (43), it is now commonly held that CAFs do not harbor p53 mutations (18, 20). However, p53 functionality is often compromised in CAFs (44, 45), and epithelial cells acquire an increased ability to suppress the canonical activity of p53 as they gradually become more transformed and cancerous (45). We now show that not only is the canonical cell nonautonomous tumor-suppressive activity of p53 quenched in CAFs, but CAF p53 is actually rewired to become a significant contributor to the CAFs’ tumor-supportive activities. This entails transcriptional reprogramming, enabling p53 to support the elevated expression of many CAFness genes. Moreover, advanced cancer cells can modulate the transcriptional output of adjacent NFs, partly skewing it toward the program supported by p53 in CAFs. It remains to be determined whether, unlike in CAFs, this requires continuous exposure to cancer cells.
ECM remodeling in the tumor microenvironment is a CAF hallmark (11, 46). MMPs are often overexpressed in both cancer cells and CAFs (47, 48). For example, MMP1 is frequently overexpressed in lung cancer, and is an indicator of poor prognosis (49–51). As shown here, the augmented expression of several MMP genes in lung cancer-derived CAFs is partially p53 dependent, suggesting that the extent of CAF p53 education might affect tumor severity and patient prognosis.
Of note, several CAF p53-dependent genes encode secreted proteins. In addition to MMPs, these also comprise ligands for cancer-relevant surface receptors (e.g., WNT2 and WNT16) (SI Appendix, Fig. S7). Interestingly, another Wnt family member—Wnt7a—drives CAF conversion in breast cancer (52). Such secreted molecules may affect not only cancer cell proliferation, survival, and motility but also response to anticancer agents. For example WNT16, secreted by surrounding CAFs, can contribute to resistance of prostate cancer cells to cytotoxic drugs (53). Hence, the rewired CAF p53 may also render their associated tumors more resistant to chemotherapy.
“Re-education” of tumor-associated stromal cells within the tumor microenvironment has been proposed as a potential therapeutic strategy (30, 34, 54). It is tempting to speculate that agents capable of restoring the normal functionality of CAF p53 may offer an exciting approach toward such a goal.
Materials and Methods
Cell Culture.
All cells were maintained at 37 °C in a 5% (vol:vol) CO2 humidified incubator and regularly tested for mycoplasma (for further details, SI Appendix, Methods).
Establishment of NF and CAF Cultures.
Lung tissue was obtained from patients with newly diagnosed lung cancer from the Klinik Schillerhöhe, immediately after resection. The investigation was approved by the local ethics committee (project numbers 396/2005V and 159/2011BO2) and informed consent was obtained from the patients. Most of the work in the present study employed cells from patient 4731, a 62-y-old woman smoker with a poorly differentiated (G3) adeno-squamous carcinoma, TNM status = pT2apN0M0, with no residual tumor (R0), no lymph node invasion (L0), and no venous infiltration (V0). Some experiments also incorporated cells from patients 8163 and 9585, both with lung adenocarcinoma. CAFs and NFs from the noncancerous margins were isolated and cultured as described previously (14, 19). Altogether, three NF–CAF pairs were employed in the present study (patients 4731, 8163, and 9585). NFs and CAFs from patient 4731 were immortalized as described in SI Appendix, Methods.
Methods for viral infection, collagen gel contraction assay, conditioned media, live cell imaging, trans-well migration and invasion assays, Western blot analysis, immunoprecipitation, RNA isolation, qRT-PCR, FACS analysis, MS, RNA sequencing, in vivo tumor growth, and statistical analysis are described in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Gil Hornung (Israel National Center for Personalized Medicine) for analysis of RNA-seq data, Ayala Sharp for FACS guidance, and Meital Kupervaser and Yishai Levin for MS analysis. This study was supported in part by the Robert Bosch Foundation (Project 11.5.8000.0094), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, Center of Excellence Grant 2084/15 from the Israel Science Foundation, the Innovative Medicines Initiative Joint Undertaking under Grant Agreement 115188 (PREDECT), a research grant from the Comisaroff Family Trust, and the Moross Integrated Cancer Center. M.O. is an incumbent of the Andre Lwoff Chair in Molecular Biology. S.A. is supported by a fellowship from the Planning and Budgeting Committee of the Council for Higher Education (VATAT), Israel. This study is dedicated to the memory of Dr. Heiko van der Kuip.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719076115/-/DCSupplemental.
References
- 1.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Lozano G. p53: Multiple facets of a Rubik’s cube. Annu Rev Cancer Biol. 2017;1:185–201. doi: 10.1146/annurev-cancerbio-050216-121926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Aylon Y, Oren M. The paradox of p53: What, how, and why? Cold Spring Harb Perspect Med. 2016;6:a026328. doi: 10.1101/cshperspect.a026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller PAJ, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 7.Furth N, et al. Down-regulation of LATS kinases alters p53 to promote cell migration. Genes Dev. 2015;29:2325–2330. doi: 10.1101/gad.268185.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trinidad AG, et al. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell. 2013;50:805–817. doi: 10.1016/j.molcel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lujambio A, et al. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2011;473:544. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 12.Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med. 2014;211:1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Procopio M-G, et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat Cell Biol. 2015;17:1193–1204. doi: 10.1038/ncb3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid JO, et al. Cancer cells cue the p53 response of cancer-associated fibroblasts to cisplatin. Cancer Res. 2012;72:5824–5832. doi: 10.1158/0008-5472.CAN-12-1201. [DOI] [PubMed] [Google Scholar]
- 15.Addadi Y, et al. p53 status in stromal fibroblasts modulates tumor growth in an SDF1-dependent manner. Cancer Res. 2010;70:9650–9658. doi: 10.1158/0008-5472.CAN-10-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moskovits N, Kalinkovich A, Bar J, Lapidot T, Oren M. p53 attenuates cancer cell migration and invasion through repression of SDF-1/CXCL12 expression in stromal fibroblasts. Cancer Res. 2006;66:10671–10676. doi: 10.1158/0008-5472.CAN-06-2323. [DOI] [PubMed] [Google Scholar]
- 17.Otomo R, et al. TSPAN12 is a critical factor for cancer-fibroblast cell contact-mediated cancer invasion. Proc Natl Acad Sci USA. 2014;111:18691–18696. doi: 10.1073/pnas.1412062112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosein AN, et al. Breast carcinoma-associated fibroblasts rarely contain p53 mutations or chromosomal aberrations. Cancer Res. 2010;70:5770–5777. doi: 10.1158/0008-5472.CAN-10-0673. [DOI] [PubMed] [Google Scholar]
- 19.Sonnenberg M, et al. Highly variable response to cytotoxic chemotherapy in carcinoma-associated fibroblasts (CAFs) from lung and breast. BMC Cancer. 2008;8:364. doi: 10.1186/1471-2407-8-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu W, et al. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet. 2008;40:650–655. doi: 10.1038/ng.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navab R, et al. Prognostic gene-expression signature of carcinoma-associated fibroblasts in non-small cell lung cancer. Proc Natl Acad Sci USA. 2011;108:7160–7165. doi: 10.1073/pnas.1014506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, et al. Carcinoma-associated fibroblasts lead the invasion of salivary gland adenoid cystic carcinoma cells by creating an invasive track. PLoS One. 2016;11:e0150247. doi: 10.1371/journal.pone.0150247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meek DW, Anderson CW. Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gogna R, Madan E, Kuppusamy P, Pati U. Re-oxygenation causes hypoxic tumor regression through restoration of p53 wild-type conformation and post-translational modifications. Cell Death Dis. 2012;3:e286. doi: 10.1038/cddis.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calon A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 26.Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 27.Herrera M, et al. Functional heterogeneity of cancer-associated fibroblasts from human colon tumors shows specific prognostic gene expression signature. Clin Cancer Res. 2013;19:5914–5926. doi: 10.1158/1078-0432.CCR-13-0694. [DOI] [PubMed] [Google Scholar]
- 28.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: Orchestrating the composition of malignancy. Genes Dev. 2016;30:1002–1019. doi: 10.1101/gad.279737.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell. 2010;17:135–147, and erratum (2010) 17:523. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 32.Olumi AF, et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiaris H, et al. Evidence for nonautonomous effect of p53 tumor suppressor in carcinogenesis. Cancer Res. 2005;65:1627–1630. doi: 10.1158/0008-5472.CAN-04-3791. [DOI] [PubMed] [Google Scholar]
- 34.Quante M, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- 36.Augsten M. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front Oncol. 2014;4:62. doi: 10.3389/fonc.2014.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arina A, et al. Tumor-associated fibroblasts predominantly come from local and not circulating precursors. Proc Natl Acad Sci USA. 2016;113:7551–7556. doi: 10.1073/pnas.1600363113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson TR, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu M, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 41.Bechtel W, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao L, et al. miRNA expression analysis of cancer-associated fibroblasts and normal fibroblasts in breast cancer. Int J Biochem Cell Biol. 2012;44:2051–2059. doi: 10.1016/j.biocel.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Fukino K, Shen L, Patocs A, Mutter GL, Eng C. Genomic instability within tumor stroma and clinicopathological characteristics of sporadic primary invasive breast carcinoma. JAMA. 2007;297:2103–2111. doi: 10.1001/jama.297.19.2103. [DOI] [PubMed] [Google Scholar]
- 44.Hawsawi NM, et al. Breast carcinoma-associated fibroblasts and their counterparts display neoplastic-specific changes. Cancer Res. 2008;68:2717–2725. doi: 10.1158/0008-5472.CAN-08-0192. [DOI] [PubMed] [Google Scholar]
- 45.Bar J, et al. Cancer cells suppress p53 in adjacent fibroblasts. Oncogene. 2009;28:933–936. doi: 10.1038/onc.2008.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27:5287–5297. doi: 10.1200/JCO.2009.23.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauter W, et al. LUCY-Consortium Matrix metalloproteinase 1 (MMP1) is associated with early-onset lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:1127–1135. doi: 10.1158/1055-9965.EPI-07-2840. [DOI] [PubMed] [Google Scholar]
- 49.Li M, et al. Prognostic significance of matrix metalloproteinase-1 levels in peripheral plasma and tumour tissues of lung cancer patients. Lung Cancer. 2010;69:341–347. doi: 10.1016/j.lungcan.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 50.Chen YK, et al. Plasma matrix metalloproteinase 1 improves the detection and survival prediction of esophageal squamous cell carcinoma. Sci Rep. 2016;6:30057. doi: 10.1038/srep30057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin TS, et al. Expression spectra of matrix metalloproteinases in metastatic non-small cell lung cancer. Oncol Rep. 2004;12:717–723. [PubMed] [Google Scholar]
- 52.Avgustinova A, et al. Tumour cell-derived Wnt7a recruits and activates fibroblasts to promote tumour aggressiveness. Nat Commun. 2016;7:10305. doi: 10.1038/ncomms10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012;18:1359–1368. doi: 10.1038/nm.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fridlender ZG, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.