Significance
De novo protein synthesis is critical for memory formation. We found that protein synthesis during acquisition is transiently required for contextual memory formation. We identified one candidate gene, Nrgn (encoding protein neurogranin, Ng) with enhanced translation upon novel-context exposure, and found that experience-dependent translation of Ng in the hippocampus is required for contextual memory formation. Furthermore, fragile-X mental retardation protein interacts with the 3′UTR of the Nrgn mRNA, which is required for activity-dependent translation of Ng in the synaptic compartment and contextual memory formation. Together, these results indicate that experience-dependent and acute translation of Ng in the hippocampus during memory acquisition enables durable context memory encoding.
Keywords: hippocampus, contextual memory, dentate gyrus, ASD, schizophrenia
Abstract
Experience induces de novo protein synthesis in the brain and protein synthesis is required for long-term memory. It is important to define the critical temporal window of protein synthesis and identify newly synthesized proteins required for memory formation. Using a behavioral paradigm that temporally separates the contextual exposure from the association with fear, we found that protein synthesis during the transient window of context exposure is required for contextual memory formation. Among an array of putative activity-dependent translational neuronal targets tested, we identified one candidate, a schizophrenia-associated candidate mRNA, neurogranin (Ng, encoded by the Nrgn gene) responding to novel-context exposure. The Ng mRNA was recruited to the actively translating mRNA pool upon novel-context exposure, and its protein levels were rapidly increased in the hippocampus. By specifically blocking activity-dependent translation of Ng using virus-mediated molecular perturbation, we show that experience-dependent translation of Ng in the hippocampus is required for contextual memory formation. We further interrogated the molecular mechanism underlying the experience-dependent translation of Ng, and found that fragile-X mental retardation protein (FMRP) interacts with the 3′UTR of the Nrgn mRNA and is required for activity-dependent translation of Ng in the synaptic compartment and contextual memory formation. Our results reveal that FMRP-mediated, experience-dependent, rapid enhancement of Ng translation in the hippocampus during the memory acquisition enables durable context memory encoding.
Novel experience induces changes in neuronal protein content and synaptic strength, which eventually leads to memory encoding via long-lasting modifications of the pattern of neural connectivity. It is generally accepted that de novo protein synthesis is required for memory formation (1–3), and multiple windows for protein synthesis exist for different phases of memory formation (4–7). In previous studies, perturbation of protein synthesis for interrupting memory formation was generally initiated before or at the time of the memory acquisition phase, thus leading to the assumption that protein synthesis is required during memory acquisition and immediate consolidation. However, it is unclear how long this first protein synthesis requirement lasts. In some associative learning paradigms, de novo protein synthesis requirement extends 30 min or hours after memory acquisition (8–10), whereas others suggested that only a narrow window was required during memory acquisition (11). Therefore, it is important to describe the temporal window of de novo protein synthesis required for memory encoding for the identification of critical translation targets in memory formation at relevant time points.
The pool of candidate genes undergoing translation in response to neural activity is potentially extensive. For example, preceding evidence suggests that ∼1,000 mRNAs are targets of the activity-dependent translation regulator, the fragile X mental retardation protein (FMRP, encoded by the Fmr1 gene) (12, 13). Many potential FMRP-interacting mRNA targets are involved in pathways important for synaptogenesis, synaptic plasticity, and implicated in neurodevelopmental and psychiatric disorders (12–14). In contrast to this vast landscape of potential targets, the number of bona fide FMRP targets that are functionally validated remains limited, as summarized in Pasciuto and Bagni (15). Studies confirming experience- (not electrical or chemical stimulation) dependent translational regulation (not transcriptionally dependent) of candidate genes are sparse, with the exception of CaMKIIα (16), using light exposure after dark rearing, and Arc (17), using repeated context exposure to induce translation after the initial gene transcription. Although the contribution of these gene targets to learning and memory has been heavily explored, the specific effect of activity-dependent protein-synthesis of any one gene product on synaptic plasticity and learning and memory remains largely unknown.
One attractive candidate that undergoes local translation in an activity-dependent manner is neurogranin (Ng; gene name, Nrgn), a small neuronal protein (78 amino acids) primarily expressed in the somato-dendritic compartment (dendrite, dendritic spine, and cell body) of projection neurons in the cerebral cortex, hippocampus, striatum and some subcortical nuclei, including the amygdala (18–20). Ng belongs to the calpacitin protein family, composed of small, abundant proteins that preferentially bind to the Ca2+-free form of calmodulin (CaM). Ng is the only family member in the postsynaptic compartment in the forebrain (21). It has been hypothesized that Ng regulates the availability of CaM for Ca2+ binding and Ca2+ buffering capacity in neurons (22), thus critically influences Ca2+ or Ca2+/CaM-dependent neuronal processes, such as synaptic plasticity and, consequently, learning and memory (23–25). Nrgn is associated with schizophrenia (26, 27) and a rare genetic disorder with symptoms of intellectual disability, Jacobsen syndrome (28), implying an important role of Ng in cognitive function.
Previous studies have shown that Ng levels change in response to behavioral, environmental, and hormonal stimulation in rodent models, and schizophrenic and aging brains in humans (29–38), with emphasis on regulation at the transcription level. The 3′UTR of the Nrgn mRNA contains a putative dendritic targeting sequence that is potentially important for both translocation and translational control of Ng (39, 40). The Nrgn mRNA is also a potential target of FMRP (12). However, it is unknown whether the expression of Ng is regulated at the translation level by experience.
Here, using a hippocampus-dependent behavioral paradigm (41), specifically separating learning of context from fear-conditioning (42), we report that there is a transient window for de novo protein synthesis during memory acquisition required for contextual memory formation. Through a screening, we identified Ng, which is up-regulated in the hippocampus during this critical window, and this up-regulation is necessary for durable encoding of the memory. We further found that FMRP interacts with the 3′UTR of the Nrgn mRNA, and the FMRP-mediated, novel-context–induced Ng up-regulation is essential for the contextual memory formation.
Results
Contextual Memory Formation Requires Rapid Protein Synthesis.
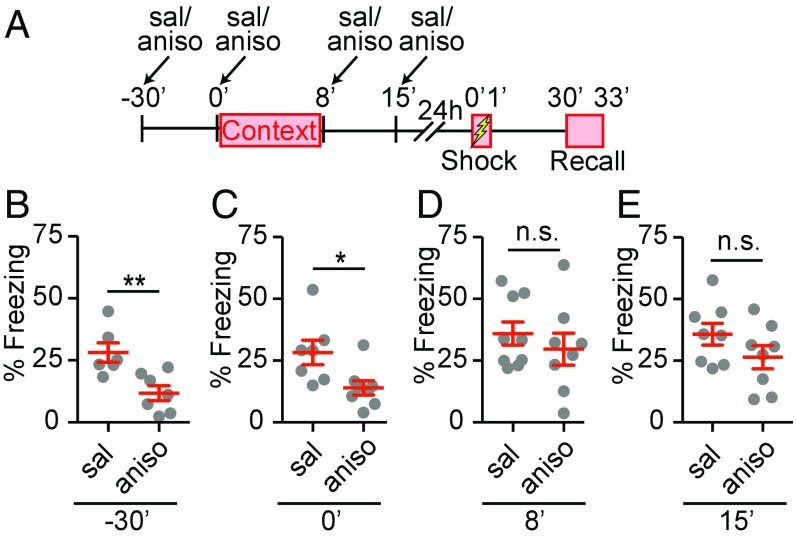
To define the temporal window for de novo protein synthesis required in contextual memory formation, we adapted a contextual memory test in which contextual exposure and associative fear learning, using an electric foot shock, were separated by 24 h (42). Consistent with previous studies, associative learning was established when animals were sufficiently preexposed to the arena in which the shock is later delivered (SI Appendix, Fig. S1), showing it was the sufficient preexposure to the context, rather than other cues caused by handling, which allowed associative learning. This approach separates the contextual memory formation and associative learning, and uses the associative component to interrogate the establishment of contextual memory. Behavioral freezing levels are then used to monitor successful association of foot shock with the context and, hence, determine whether contextual memory was formed or not. Combined with pharmacological manipulation, this procedure enabled us to probe the distinct phases of memory formation with defined temporal control. We intraperitoneally injected the protein synthesis inhibitor anisomycin (150 mg/kg) at four different time points relative to the context exposure (Fig. 1A). Consistent with previous results (42), the intraperitoneal injection of anisomycin 30 min before context exposure (−30 min from the starting point of context exposure) prevented contextual memory formation (Fig. 1B). The intraperitoneal injection immediately before context exposure (0 min from the starting point of context exposure) also prevented contextual memory formation (Fig. 1C), suggesting that protein synthesis during the initial context exposure was required for context memory formation. In contrast, intraperitoneal injection of anisomycin immediately after (8 min after the starting point of context exposure) and 7 min after context exposure (15 min after the starting point of context exposure) did not affect contextual memory formation (Fig. 1 D and E).
Fig. 1.
An immediate and transient phase of protein synthesis is required for contextual memory formation. (A) The schematics for the contextual memory test. (B–E) Quantification of effects of anisomycin, a protein synthesis inhibitor, on percentage of freezing during recall. Saline (sal) or anisomycin (aniso, 150 mg/kg) was intraperitoneally administered 30 min before (A, −30 min, n = 5, sal; n = 7, aniso), immediately before (B, 0 min, n = 7, sal; n = 8, aniso), immediately after (C, 8 min, n = 9, sal; n = 8, aniso), and 7 min after (D, 15 min, n = 8, sal; n = 8, aniso) context exposure. Gray symbols, individual data points; mean ± SEM in red lines. Unpaired Student’s t test, (B) t11 = 3.355, **P = 0.006; (C) t13 = 2.594, *P = 0.02; (D) t15 = 0.80, P = 0.43; (E) t14 = 1.456, P = 0.17; n.s., not significant.
Previous studies reported that anisomycin injected intraperitoneally in rats or mice reaches effective concentrations in the brain cerebrospinal fluid between 15 and 45 min to inhibit protein synthesis for more than 3 h in the hippocampus (43, 44). Considering these observations, our results show that the first phase of protein synthesis requirement ceases shortly after the context exposure, suggesting the first de novo protein synthesis required for context memory formation has a rapid onset, and is transient.
Novel Context Induces Rapid Increase of Ng Protein Levels in the Hippocampus.
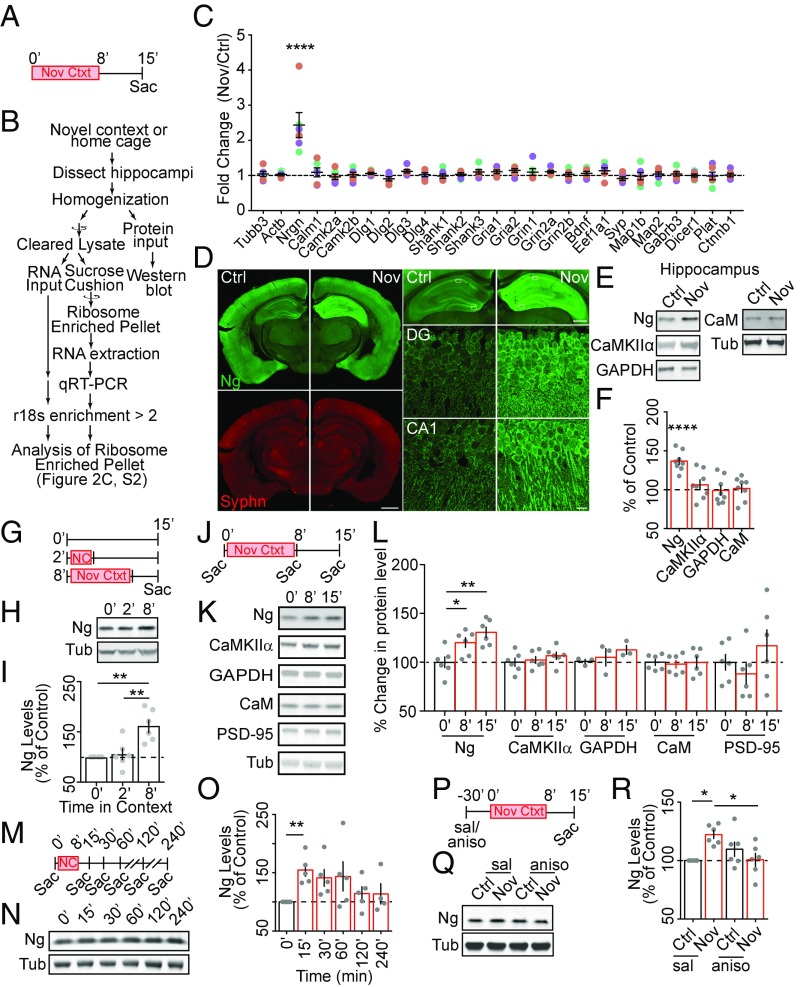
Activation of NMDARs in synaptoneurosomes induces rapid changes in protein synthesis, leading to elevated protein synthesis 15- to 60-min poststimulation (45), a time window coinciding with that required for protein synthesis-dependent memory formation. Many genes are potential targets of activity-dependent translation, which are thought to be involved in memory formation (46, 47). However, little is known about which genes are translationally up-regulated in the hippocampus during the critical time window essential for context memory formation. To identify the potential targets during context exposure, we isolated a polyribosome-enriched mRNA pool (48, 49) from total hippocampal lysate of naïve animals and animals exposed to a novel context (Materials and Methods and Fig. 2 A and B). An array of candidate genes and their gene family members (SI Appendix, Table S1) were tested using qRT-PCR. The selection criteria are: (i) activity-dependent and experience-dependent protein synthesis has been shown, or suggested via functional studies; (ii) putative or proven dendritically targeted mRNA; and (iii) functionally validated FMRP targets relevant to synaptic structure and function (SI Appendix, Dataset S1). We found that, among 28 genes tested, exposure to a novel context induced an increase only of Nrgn mRNAs in the actively translating, polyribosome-enriched mRNA pool (Fig. 2C). This result showed that exposure to novel context induced the selective recruitment of the Nrgn mRNA to the polyribosome-enriched compartment.
Fig. 2.
Ng is up-regulated rapidly following novel-context exposure. (A) The behavioral paradigm for RNA and protein analysis. (B) The workflow for analyzing the ribosome enriched fraction. (C) Quantification of relative levels (Fold-Change Nov/Ctrl) of mRNA candidates in the hippocampal ribosome-enriched fraction from novel-context–exposed (Nov) animals to those from control animals (Ctrl). Each cDNA target was normalized to GAPDH cDNA within each sample, and then the relative fold-change was calculated. Colored symbols, individual data points; colors, different cohorts of animals, n = 7 out of three cohorts; mean ± SEM, in black lines in each data column. One-way ANOVA, F(27, 168) = 8.356, P < 0.0001; followed by Dunnett’s test, compared with Tubb3 ratio as a control, ****P < 0.0001. (D) Representative confocal images of coronal sections obtained from novel-context–exposed (Nov) and control mice (Ctrl). (Left Top) Ng (green) and (Left Bottom) Synaptophysin (red). (Scale bar, 500 μm.) (Right Top) Ng (green) in hippocampus. (Scale bar, 200 μm.) White boxes show area of detail. (Right, Middle, and Bottom) Ng (green) in dentate gyrus and CA1. (Scale bar, 10 μm.) (E and F) Representative Western blot images (E) and quantification (F, n = 8 pairs) of select proteins in hippocampal lysates from control (Ctrl) and novel-context–exposed (Nov) animals. In this and subsequent Western blot quantification graphs, gray symbols, individual data points; mean ± SEM, as the bar graph. One-way ANOVA; F(5, 42) = 10.33, P < 0.0001; (E) F(4, 20) = 0.54, P = 0.70; (F), F(4, 20) = 1.099, P = 0.38; followed by Dunnett’s test, compared with 100%, ****P < 0.0001. (G–I) The behavioral paradigm (G), representative Western blot (H), and quantification (I) of Ng [normalized to tubulin (Tub)] in hippocampal lysate from animals exposed 0 min (n = 6), 2 min (n = 6), and 8 min (n = 6) to the novel context, and collected at 15 min after the initial context exposure. One-way ANOVA, F(2, 15) = 12.43, P = 0.0007; followed by Tukey’s test, compared with the no exposure control, **P < 0.01. (J–L) The behavioral paradigm (J), representative Western blot (K), and quantification (L) of select proteins (normalized to Tub) in hippocampal lysate from animals collected before (0 min, n = 6), immediately after the initial exposure (8 min, n = 6), and 15 min after the initial exposure (15 min, n = 6). One-way ANOVA, Ng, F(2, 15) = 8.37, P = 0.003; CaMKIIα, F(2, 15) = 0.756, P = 0.49; GAPDH (three sets of samples were used), F(2, 6) = 1.014, P = 0.42; CaM, F(2, 15) = 0.06, P = 0.94; PSD-95, F(2, 15) = 1.41, P = 0.27; followed by Dunnett’s test, compared with the no exposure control (0 min), *P < 0.05, **P < 0.01. (M–O) The behavioral paradigm (M), representative Western blot (N), and quantification (O) of Ng in hippocampal lysate from animals collected at different time points relative to the initial context exposure (n = 5 for 0, 15, 30, 60, 120 min; n = 4 for 240 min). One-way ANOVA, F(5, 23) = 3.31, P = 0.02; followed by Dunnett’s test, compared with the no exposure control (0 min), *P < 0.05, **P < 0.01. (P–R) The behavioral paradigm (P), representative Western blot images (Q), and quantification (R) of Ng in hippocampal lysate from control (Ctrl, black bar) and novel-context–exposed (Nov, red bar) animals treated with either saline (sal) or anisomycin (aniso); n = 6 in each group. Two-way ANOVA, interaction F(1, 20) = 12.13, P = 0.002; followed by Tukey’s test. *P < 0.05.
Given the fact that immediate early genes (IEGs) respond to novel-context exposure and enhance their transcription and translation (50, 51), we tested several IEGs as a positive control, including Egr1, Fos, Npas4, Arc, and Homer1, both in the polyribosome-enriched fraction and in the total RNA input. Egr1, Arc, and Homer1 were neither significantly changed in the polyribosome-enriched fraction nor in the total RNA input (SI Appendix, Fig. S2 A and B). Consistent with previous findings (52), we found that Fos and Npas4 were significantly up-regulated in both the polyribosome-enriched fraction and in the total RNA input. Thus, transcription of Fos and Npas4 responds to context exposure rapidly and significantly. We next tested whether the increase of Nrgn mRNAs in the polyribosome-enriched compartment was due to the increase of total Nrgn mRNAs. In contrast to Fos and Npas4, Nrgn mRNA levels in the total RNA input was not increased by novel-context exposure (SI Appendix, Fig. S2B), which indicated that the enhancement of the Nrgn mRNAs in the polyribosomal fraction was due to the recruitment of existing Nrgn mRNAs into the fraction rather than enhanced transcription of Nrgn.
The enhanced Nrgn mRNA in the polyribosome fraction implicates an increase in translation. We therefore analyzed the influence of context exposure on protein levels using immunofluorescence staining and Western blotting. Consistent with the specific increase of the Nrgn mRNA in the hippocampal polyribosome pool, the increased Ng protein levels were detected in the hippocampal CA1, CA3, and dentate gyrus (DG) regions in response to novel-context exposure using immunostaining with an Ng-specific antibody (Fig. 2D). Ng protein levels, but not CamKIIα, GAPDH, or CaM protein levels, were increased in the hippocampus of the animals exposed to a novel context compared with naïve animals (Fig. 2 E and F). No significant differences of Ng levels were detected from striatal and cortical homogenates, suggesting the increase of Ng levels in hippocampus was specific (SI Appendix, Fig. S2 C–F).
To examine the temporal features of context-induced up-regulation of Ng levels in the hippocampus, we varied the exposure duration and the time of collection and tested the Ng levels in the hippocampus using Western blotting. A short exposure (2 min), which was not sufficient for contextual memory formation (SI Appendix, Fig. S1), did not induce changes in hippocampal Ng levels (Fig. 2 G–I), whereas a longer exposure (8 min), which was sufficient for contextual memory formation (SI Appendix, Fig. S1), led to increased Ng levels in the hippocampus (Fig. 2 G–I). This result showed that the temporal requirement of context exposure for increasing hippocampal Ng levels is consistent with the temporal requirement for contextual memory formation. Furthermore, the increase of hippocampal Ng levels after context exposure was transient, as the increase of Ng levels was detected immediately after the novel-context exposure (Fig. 2 J–L) and subsided 30–60 min after the initial exposure (Fig. 2 M–O), consistent with the temporal requirement of protein synthesis for contextual memory formation (Fig. 1).
This novel experience-induced up-regulation of Ng levels in the hippocampus requires protein synthesis. Intraperitoneal administration of anisomycin, 30 min before the novel-context exposure, blocked novel-context–induced up-regulation of hippocampal Ng protein levels (Fig. 2 P–R). Taken together, these results reveal rapid, experience-dependent up-regulation of translation of Ng, whose temporal expression pattern is consistent with the temporal profile of context exposure and protein synthesis required for contextual memory formation.
Neural Activity Induces Elevated New Protein Synthesis of Ng.
Our results thus far suggest that enhanced translation of the Nrgn mRNA leads to increased Ng protein levels. In support of this hypothesis, the increase of Ng protein levels and the increase of Nrgn mRNA in the polyribosome were linearly correlated (SI Appendix, Fig. S3). To directly test whether neural activity elevates Ng protein synthesis, we used dissociated primary neuronal culture, which enabled direct visualization of newly synthesized target proteins with cell biological approaches.
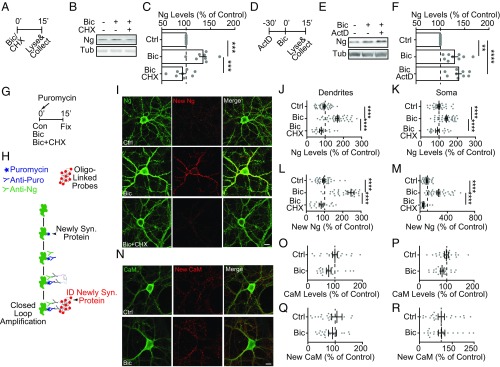
Treating the neuronal culture with bicuculline (Bic), a GABA receptor blocker, for 15 min to enhance excitatory neuronal activity, induced a significant increase of Ng levels in the total cell lysate compared with control, vehicle-treated sister cultures. The protein synthesis inhibitor cycloheximide (CHX), blocked the activity-dependent up-regulation of Ng protein levels, but the transcription inhibitor Actinomycin D (ActD) did not (Fig. 3 A–F), confirming the findings from the behavioral experiment that translation but not transcription is required for activity-dependent up-regulation of Ng (Fig. 2 and SI Appendix, Fig. S2).
Fig. 3.
Neural activity elevated de novo synthesis of Ng in cultured neurons. (A) The experimental schematics for the pharmacological treatments to the dissociated neuron cultures. (B) Representative Western blot images of Ng and Tub when stimulated with Bic (40 µM) or Bic in the presence of a protein synthesis inhibitor, CHX (100 µM). (C) Quantification of percentage of Ng in neuronal lysates (A) from treated neuron cultures (Bic, or Bic + CHX) normalized to vehicle-treated sister neuronal lysates (Ctrl), n = 8 of three independent cultures. One-way ANOVA, F(2, 21) = 15.54, P < 0.0001; followed by Tukey’s test, ***P < 0.001. (D) The experimental schematics for the pharmacological treatments to the dissociated neuron cultures. (E) Representative Western blot images of Ng and Tub when stimulated with Bic (40 µM) or Bic with pretreatment of a transcription inhibitor, ActD (5 µM). (F) Quantification of percentage of Ng in neuronal lysates from stimulated neuron cultures (Bic, Bic + ActD) normalized to vehicle-treated sister neuronal lysates (Ctrl), n = 9 of four independent cultures. One-way ANOVA, F(2, 24) = 14.76, P < 0.0001; followed by Tukey’s test, **P < 0.01, ****P < 0.0001. (G) The experimental schematics for the pharmacological treatments to the dissociated neuron cultures, purposed for PLA. (H) The schematics of the puromycin PLA. (I) Representative images of immunostaining of total (green) and puromycin+, newly synthesized (red) Ng from control (Ctrl), Bic, or Bic + CHX. (Magnification, 63×.) (Scale bar, 20 μm.) (J–M) Quantification of the immunostaining of total (J and K) and newly synthesized (L and M) of Ng in dendrites (J and L) and soma (K and M). Gray symbols, individual data points, mean ± SEM is represented, n = 18–35 neurons per condition, from two coverslips per culture, three independent cultures. One-way ANOVA: (J) F(2, 87) = 38.98, P < 0.0001; (K) F(2, 89) = 14.62, P < 0.0001; (L) F(2, 89) = 44.01, P < 0.0001; (M) F(2, 89) = 47.48, P < 0.0001; followed by Tukey’s test. ****P < 0.0001. (N) Representative images of immunostaining of total (green) and puromycin+, newly synthesized (red) CaM from control (Ctrl), Bic, or Bic + CHX. (Magnification, 63×.) (Scale bar, 20 μm.) (O–R) Quantification of the immunostaining of total (O and P) and newly synthesized (Q and R) of CaM in dendrites (O and Q) and soma (P and R). Gray symbols, individual data points, mean ± SEM is represented. Unpaired Student’s t test, no significance detected.
To directly test whether de novo protein synthesis of Ng was induced by elevated neural activity, we used puromycylation with the proximity-ligase assay (PLA) to visualize specific newly synthesized proteins (53) (Fig. 3H). First, we checked the total Ng levels using conventional immunofluorescent staining. Consistent with the Western blot results, Bic treatment induced a significant, protein synthesis-dependent increase of total Ng levels, both in the dendritic and somatic compartments, using Ng primary antibodies (Fig. 3 G and I–K). Then, using the PLA assay, we analyzed the levels of newly synthesized Ng proteins (Fig. 3 G and H). Bic treatment substantially increased the levels of newly synthesized Ng in a translation-dependent way, detected by the positive PLA signal of anti-puromycin antibodies and anti-Ng antibodies (Fig. 3 G–I, L, and M). As a negative control, the anti-Ng interaction partner, CaM, was not significantly affected in the same preparation both at the total level and at the newly synthesized level (Fig. 3 N–R). Taken together, these results indicate that increasing neuronal activity induces rapid de novo synthesis of Ng.
Activity-Dependent Increase of Ng Is Required for Contextual Memory Formation.
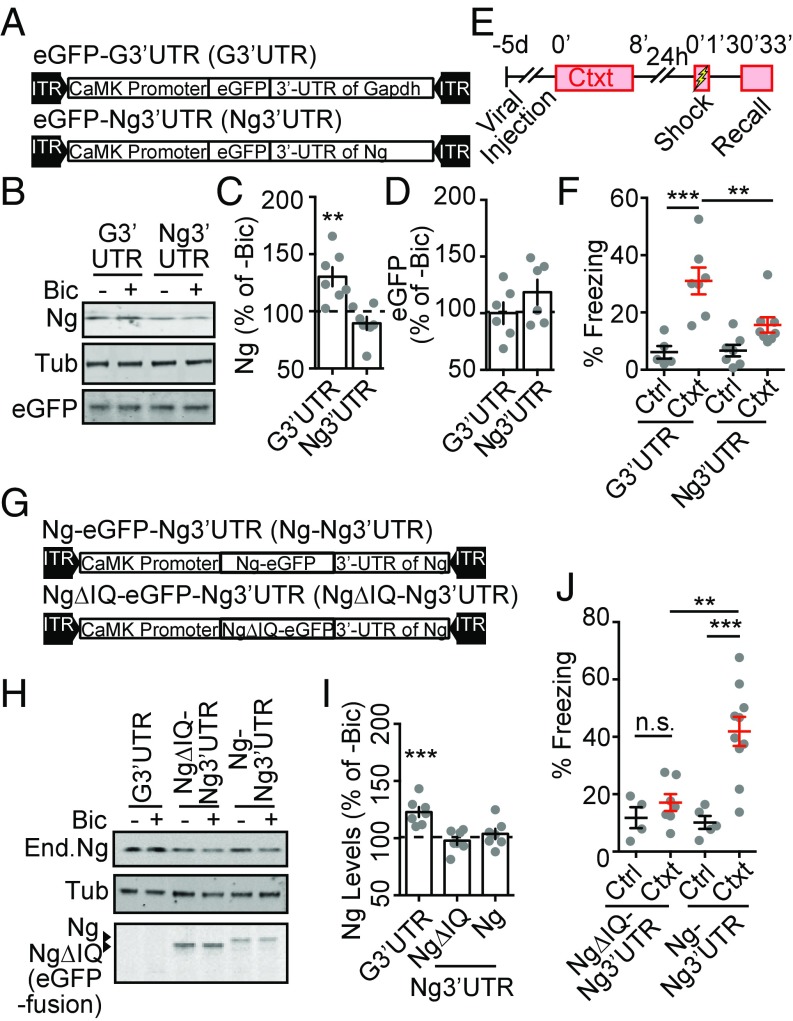
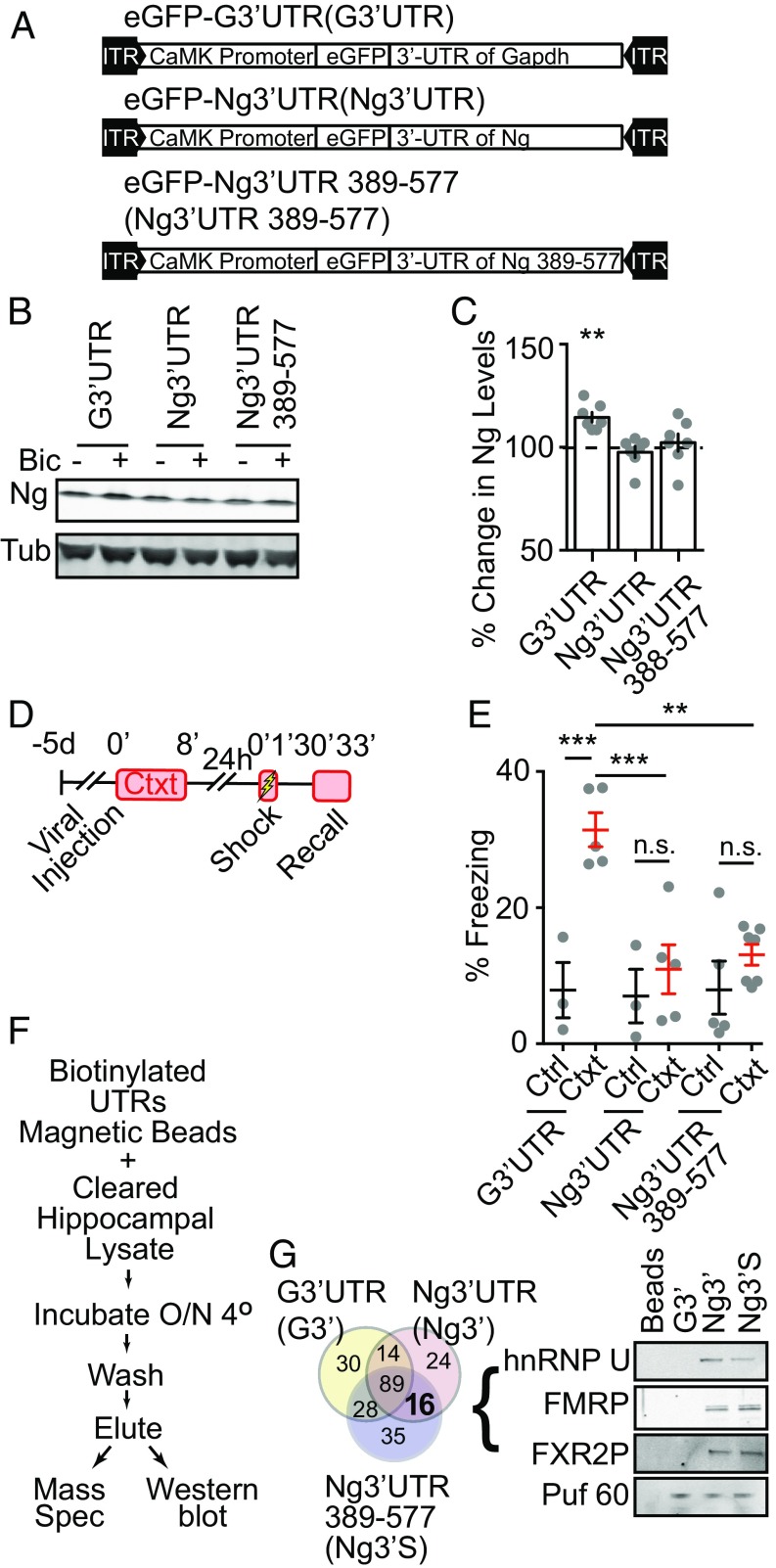
Novel-context exploration enhances neural activity in the hippocampus (54–56), which is thought to be critical for forming a stable hippocampal representation of the context. We then asked whether novel-context–induced increase of Ng contributes to contextual memory formation. To address this question, we developed a molecular reagent to perturb the activity-dependent translation of Ng. The 3′UTR of the Nrgn mRNA contains a potential dendritic targeting sequence that may also serve as an activity-dependent translational control element (40). We tested whether the activity-dependent translation of Ng is mediated by the 3′UTR of Nrgn. We constructed a recombinant adeno-associated viral vector (AAV) as a decoy that contained the 3′UTR of Nrgn following an eGFP ORF, under the control of a CaMKIIα promoter that drives the expression preferentially in excitatory neurons (57) (eGFP-Ng3′UTR) (Fig. 4A). A control AAV contained the 3′UTR of Gapdh with an eGFP ORF (eGFP-G3′UTR) (Fig. 4A). Using dissociated cultures, we tested the effect of these 3′UTR-expressing AAVs on activity-dependent regulation of Ng expression. The expression of both eGFP-Ng3′UTR and eGFP-G3′UTR did not affect the basal levels of Ng (Fig. 4 B and C). However, the expression of eGFP-Ng3′UTR blocked Bic-induced up-regulation of Ng protein levels, whereas the expression of eGFP-G3′UTR did not (Fig. 4 B and C). eGFP levels in eGFP-Ng3′UTR expressing neurons did not significantly increase when the neurons were stimulated with Bic (Fig. 4D), suggesting that the 3′UTR of Nrgn is necessary but not sufficient to mediate the activity-dependent translation of Ng. Thus, the exogenous 3′-UTR of the Nrgn mRNA likely functioned as a competitor for the regulatory factors that contribute to the activity-dependent translation of endogenous Ng.
Fig. 4.
The overexpression of 3′UTR of Nrgn inhibits activity-dependent up-regulation of Ng that is required for contextual memory formation. (A) Diagrams of expression cassettes in AAV vectors for expressing eGFP followed by the 3′UTR of Gapdh (eGFP-G3′UTR) and the 3′UTR of Nrgn (eGFP-Ng3′UTR) under the control of the CaMKII promoter. (B) Representative Western blot images of Ng and Tub from neuronal cultures infected with eGFP-G3′UTR or eGFP-Ng3′UTR, with (+) or without (−) Bic treatment. (C) Quantification of percentage of Ng from neuronal cultures infected with eGFP-G3′UTR or eGFP-Ng3′UTR, treated with Bic, normalized to vehicle-treated sister neuronal lysates, n = 7 from four independent cultures. One-way ANOVA, F(2, 15) = 11.24, P = 0.001, followed by Dunnett’s test, compared with control, **P < 0.01. (D) Quantification of percentage change of eGFP from neuronal cultures infected with eGFP-G3′UTR or eGFP-Ng3′UTR, treated with Bic, normalized to vehicle-treated sister neuronal lysates, n = 6 from three independent cultures. One-way ANOVA, F(2, 15) = 11.24, P = 0.0001, followed by Dunnett’s test, compared with control, P > 0.05. (E) The schematics for the contextual memory test after viral infusion. (F) Quantification of effects of the expression of eGFP-G3′UTR and eGFP-Ng3′UTR in DG on percentage of freezing during recall from control (Ctrl, black) and preexposed (Ctxt, red) mice; eGFP-G3′UTR (n = 5, Ctrl; n = 7, Ctxt); eGFP-Ng3′UTR (n = 7, Ctrl; n = 8, Ctxt). Two-way ANOVA, F(1, 23) = 6.102, P = 0.02, followed by Tukey’s test, **P < 0.01, ***P < 0.001. (G) Diagrams of expression cassettes in AAV vectors for expressing the 3′UTR of Nrgn (eGFP-Ng3′UTR) fused to Ng-eGFP (Ng-eGFP-Ng3′UTR) or NgΔIQ-eGFP (NgΔIQ-eGFP-Ng3′UTR) under the control of the CaMKII promoter. (H) Representative Western blot images of Ng and Tub from neuronal cultures infected with eGFP-G3′UTR, Ng-eGFP-Ng3′UTR or NgΔIQ-eGFP-Ng3′UTR, with (+) or without (−) Bic treatment. (I) Quantification of percentage of Ng from neuronal cultures infected with eGFP-G3′UTR, Ng-eGFP-Ng3′UTR, or NgΔIQ-eGFP-Ng3′UTR, treated with Bic, normalized to vehicle-treated sister neuronal lysates, n = 6 from three independent cultures. One-way ANOVA, F(3, 24) = 10.38, P = 0.0001, followed by Dunnett’s test, ***P < 0.001. (J) Quantification of effects of the expression of Ng-eGFP-Ng3′UTR and NgΔIQ-eGFP- Ng3′UTR, in the hippocampus DG region on percentage of freezing during recall from control (Ctrl) and preexposed (Ctxt) mice; NgΔIQ-eGFP-Ng3′UTR (n = 4, Ctrl; n = 7, Ctxt); Ng-eGFp-Ng3′UTR (n = 5, Ctrl; n = 10, Ctxt). Two-way ANOVA, F(1, 22) = 7.57, P = 0.01, followed by Tukey’s test, **P < 0.01, ***P < 0.001, n.s. nonsignificant.
Using these AAVs, we then tested whether activity-dependent up-regulation of endogenous Ng in the hippocampus is necessary for hippocampus-dependent memory formation. Given that the DG region in the hippocampus is important for parsing novel contextual information (58, 59), and that novel-context exposure elevates the Ng levels in the DG (Fig. 2G), we targeted eGFP-Ng3′UTR or eGFP-G3′UTR AAVs into the DG region bilaterally using stereotaxic injections (SI Appendix, Fig. S4A). Five days after AAV injection, the animals were subjected to behavioral tasks. Basal locomotor activity and anxiety were not different between animals injected with either eGFP-Ng3′UTR or eGFP-G3′UTR, tested in the open-field exploration and elevated plus maze (SI Appendix, Fig. S4 B–E). Using different cohorts of injected animals, we tested contextual memory formation (Fig. 4E). The expression of eGFP-G3′UTR in the DG did not affect contextual memory formation (Fig. 4F). However, the expression of eGFP-Ng3′UTR in the DG blocked contextual memory formation (Fig. 4F). These results are consistent with the hypothesis that activity-dependent translation of endogenous Ng is necessary for context memory formation.
As a decoy construct, the 3′UTR of the Nrgn mRNA may influence the activity-dependent translation of other genes. To test whether the learning deficits seen in eGFP-Ng3′UTR–expressing animals resulted from specific interference with Ng levels, we built a rescue construct, expressing Ng-eGFP ORF to achieve a higher level of Ng in the background of the exogenous Ng3′UTR (Ng-eGFP-Ng3′UTR), and a functionally null mutant (NgΔIQ-eGFP-Ng3′UTR) (21) was used as a control (Fig. 4G).
Using the dissociated culture, we confirmed that the expression of Ng-eGFP-Ng3′UTR and NgΔIQ-eGFP-Ng3′UTR blocked Bic-induced up-regulation of endogenous Ng levels (Fig. 4 H and I); and at the same time, the exogenous expression cassette allowed additional expression of Ng-eGFP or NgΔIQ-eGFP (Fig. 4H). We bilaterally injected Ng-eGFP-Ng3′UTR or NgΔIQ-eGFP-Ng3′UTR expressing AAVs into the DG region of the hippocampus (SI Appendix, Fig. S4F). Mobility and anxiety levels were indistinguishable between mice injected with either virus (SI Appendix, Fig. S4 G–J). Expression of NgΔIQ-eGFP in the Ng3′UTR expressing background did not rescue the behavioral deficit in contextual memory formation, whereas expression of Ng-eGFP in Ng3′UTR-expressing background rescued the memory deficit (Fig. 4J). Therefore, elevated Ng levels sufficiently rescued the contextual memory deficit caused by expressing Ng3′UTR, meaning that: (i) even if the translation of other transcripts can be influenced by the expression of Ng3′UTR, they are not critical for contextual memory formation; or (ii) their expression can be rescued by introducing exogenous Ng. Collectively, these results indicate that activity-dependent translation of Ng is necessary for contextual memory formation.
FMRP Interacts with the 3′UTR of the Nrgn mRNA.
To further investigate the molecular mechanism underlying activity-dependent up-regulation of Ng translation, we made deletions of the 3′UTR of Nrgn, and tested its effect on activity-dependent translation of Ng and contextual memory formation. We generated an AAV containing the nucleotides 389–577 of the Nrgn mRNA (eGFP-Ng3′UTR 389–577) (Fig. 5A), a small portion of the 5′-end of the 3′UTR of the Nrgn mRNA consisting of a cis-element for putative dendritic targeting and translational regulation (40). Expressing this small portion of the 3′UTR of the Nrgn mRNA was sufficient to block the Bic-induced increase of Ng in dissociated neuron culture, similar to the full-length 3′UTR of the Nrgn mRNA (Fig. 5 B and C). In addition, as was the case with eGFP-Ng3′UTR, expressing eGFP-Ng3′UTR 389–577 in the hippocampus prevented contextual fear-memory formation (Fig. 5 D and E). Thus, we narrowed down the critical element of the Ng3′UTR 389–577 for activity-dependent translation of Ng, which contributes to contextual memory formation. To identify the interaction partners that might contribute to activity-dependent translation of Ng, we generated in vitro-transcribed biotinylated RNA transcripts of the 3′UTR of the Gapdh mRNA (G3′UTR), the 3′UTR of the Nrgn mRNA (Ng3′UTR), and 389–577 of the 3′UTR of the Nrgn mRNA (Ng3′UTR 389–577), to affinity-purify potential interacting proteins from the hippocampal lysate (Fig. 5F). The affinity-purified components were subjected to MS for discovery-based analysis and Western blotting for further validation. From MS, we identified families of proteins that interact with each species of the 3′UTR transcripts (SI Appendix, Dataset S1). Analysis revealed 16 proteins that bind to both the Ng3′UTR and Ng3′UTR 389–577 transcripts. Notably, FMRP (gene name Fmr1) was identified as one of the interaction partners (Fig. 5G). One of the FMRP homologs, FXR2P, was also identified to interact with the Ng3′UTR 389–577 transcript. The interaction of FMRP and FXR2P with both the Ng3′UTR and Ng3′UTR 389–577 transcripts were validated in a second MS (SI Appendix, Dataset S1), and with Western blotting from independent samples. FMRP, FXR2P, and another identified interaction protein, hnRNP U, interacted directly or indirectly with the Ng3′UTR and Ng3′UTR 389–577 transcripts, but not with the G3′UTR transcript (Fig. 5G). A common RNA binding protein Puf 60 interacted with all three RNA transcripts (Fig. 5G). These results indicate that FMRP interacts with the 3′-UTR of Nrgn, revealing a potential role of FMRP in regulating activity-dependent translation of Ng.
Fig. 5.
FMRP interacts with the 3′-UTR of the Nrgn mRNA. (A) Diagrams of expression cassettes in AAV vectors for expressing the eGFP followed by 3′UTR of Gapdh (eGFP-G3′UTR), the 3′UTR of Nrgn (eGFP-Ng3′UTR) and the 5′-portion of the 3′UTR of Nrgn (eGFP-Ng3′UTR 389–577) under the control of the CaMKII promoter. (B) Representative Western blot images of Ng and Tub from neuronal cultures infected with eGFP-G3′UTR, eGFP-Ng3′UTR, and eGFP-Ng3′UTR 389–577 with (+) or without (−) Bic treatment. (C) Quantification of percentage of Ng from neuronal cultures infected with eGFP-G3′UTR, eGFP-Ng3′UTR, and eGFP-Ng3′UTR 389–577, treated with Bic, normalized to vehicle-treated sister neuronal lysates, n = 7 from four independent cultures. One-way ANOVA, F(3, 24) = 7.35, P = 0.001, followed by Dunnett’s test, **P < 0.01. (D) The schematics for the contextual memory test after viral infusion. (E) Quantification of effects of the expression of eGFP-G3′UTR, eGFP-Ng3′UTR, and eGFP-Ng3′UTR 389–577 in the hippocampus DG region on a percentage of freezing during recall from control (Ctrl) and preexposed (Ctxt) mice; eGFP-G3′UTR (n = 3, Ctrl; n = 5, Ctxt), eGFP-Ng3′UTR (n = 3, Ctrl; n = 5, Ctxt), and eGFP-Ng3′UTR 389–577 (n = 5, Ctrl; n = 7, Ctxt). Two-way ANOVA, F(2, 22) = 5.50, P = 0.01, followed by Tukey’s test, **P < 0.01, ***P < 0.001, n.s. nonsignificant. (F) The workflow for the RNA pull-down procedure from hippocampal lysates. (G) The Venn diagram (Left) shows different protein sets interacting with G3′UTR (light yellow), Ng3′UTR (light pink), and Ng3′UTR 389–577 (light purple), identified by MS; and Western blot images (Right) of indicated proteins from hippocampal lysate pulled down with proteins interacting with G3′UTR, Ng3′UTR, and Ng3′UTR 389–577 RNAs, compared with the beads-alone negative control.
FMRP Is Required for Novel-Context–Dependent Increase of Ng Translation and Activity-Dependent Transition of Nrgn mRNAs in the Polyribosome Pools.
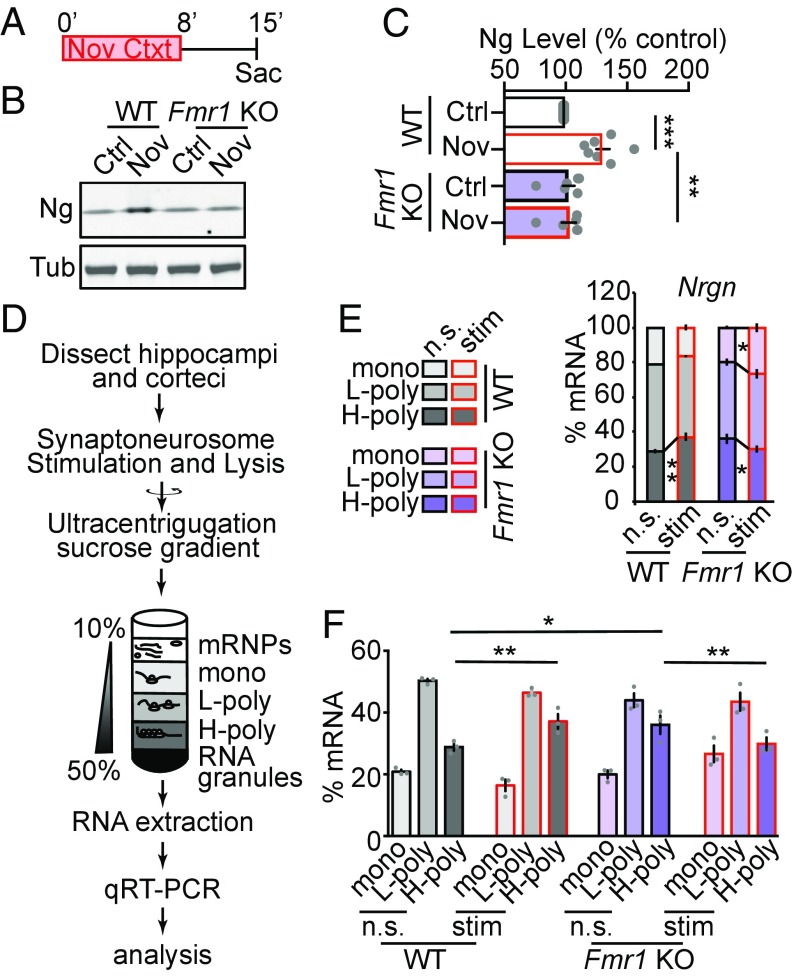
We first tested whether FMRP is critical for novel-context–dependent up-regulation of Ng. We exposed Fmr1 KO (Fmr1 -/y) and matched wild-type littermates to the novel context, and analyzed the Ng protein levels from hippocampal lysates (Fig. 6A). The basal levels of Ng protein in hippocampi were not significantly different between Fmr1 KO mice and wild-type littermate mice (Fig. 6 B and C). However, novel-context–induced up-regulation of Ng levels were absent in Fmr1 KO mice (Fig. 6 B and C). These results reveal that FMRP is required for experience-dependent up-regulation of Ng.
Fig. 6.
FMRP is required for novel-context–induced up-regulation of Ng and stimulation-dependent recruitment of Nrgn mRNA to polysomes in synaptoneurosomes. (A) The behavioral paradigm for protein analysis with the Fmr1 KO animals. (B) Representative Western blot images of Ng in hippocampal lysate from control (Ctrl) and novel-context–exposed (Nov) wild-type (WT) and Fmr1 knockout (Fmr1 KO) animals. Tub was used as a loading control. (C) Quantification of Ng (normalized to Tub) in hippocampal lysate from control (Ctrl) and novel-context–exposed (Nov) wild-type (WT) and Fmr1 knockout (Fmr1 KO) animals; WT (n = 7, Ctrl; n = 7, Nov); Fmr1 KO (n = 7, Ctrl; n = 7, Nov). Gray symbols represent individual data points from each animal. Mean ± SEM is represented in the bar graph (Ctrl, Black; Nov, Red). Two-way ANOVA, F(1, 24) = 11.45, P = 0.003, followed by Tukey’s test, **P < 0.01, ***P < 0.001. (D) The workflow for analyzing polyribosome association of mRNAs in stimulated synaptoneurosomes. (E and F) Quantification of the stacking percentage (E) and the separated percentage (F) of the Nrgn mRNA in monoribosome (mono), light polyribosome (L-poly), and heavy-polyribosome (H-poly) fractions from nonstimulated (n.s.) and NMDA and glutamate (50 μM and 10 μM, NMDA+Glu) stimulated (stim) synaptoneurosome preps from wild-type (WT) and Fmr1 knockout (Fmr1 KO) animals, n = 3 for each genotype. The data representation scheme is shown on the Left. Two-way ANOVA, F(5, 24) = 4.77, P = 0.004, followed by Fisher’s least-significant difference test. Mean ± SEM from three independent experiments is represented in the bar graphs, *P < 0.05, **P < 0.01.
To test the role of FMRP in regulating activity-dependent translation of Ng, we isolated synaptoneurosome fractions from wild-type and Fmr1 KO mice and induced NMDAR activity by applying NMDA and Glutamate (Glu) (45, 60). This procedure has been used successfully for analyzing specific mRNA species in the Fmr1 KO mouse model (61). The synaptoneurosomal extracts were then separated using a sucrose gradient to fractionate the polyribosome containing fractions. Total RNA was isolated from the collected fractions and Nrgn mRNA levels in each fraction were analyzed using qRT-PCR (Fig. 6D). Nrgn mRNAs were enriched in the H-polyribosome (H-poly) fraction after NMDA+Glu stimulation in synaptoneurosome from wild-type animals. This is an FMRP-signature effect seen for other candidate genes (61). This stimulation induced enrichment of the Nrgn mRNA in the H-poly fraction was absent in Fmr1 KO animals (Fig. 6E). The basal level of Nrgn mRNAs in the H-poly fraction was higher in the Fmr1 KO animals compared with the wild-type controls, which suggests that FMRP potentially prevents ribosome loading to Nrgn mRNAs at the basal level (Fig. 6F). Calm1 and Gap43 mRNAs, whose protein products are functionally related to Ng, did not show FMRP-signature effects upon NMDAR+Glu stimulation (SI Appendix, Fig. S5). Together, these results show that FMRP regulates activity-dependent synaptic translation of Ng, and lack of FMRP diminishes the activity-dependent effect.
FMRP Is Required for Context Memory Formation, via Regulating Novel-Context–Dependent Increase of Ng Translation.
Given that novel-context–dependent translation of Ng is required for context memory formation, and this process is disrupted in the Fmr1 KO mice, we asked whether memory formation is impaired in the Fmr1 KO mice (Fig. 7A). We found that the Fmr1 KO mice showed impaired contextual memory formation, compared with wild-type littermates (Fig. 7 A and B). Taken together (Figs. 6 and 7 A and B), these results suggest that FMRP mediates novel-context–induced translation of Ng, which is critical for context memory formation.
Fig. 7.
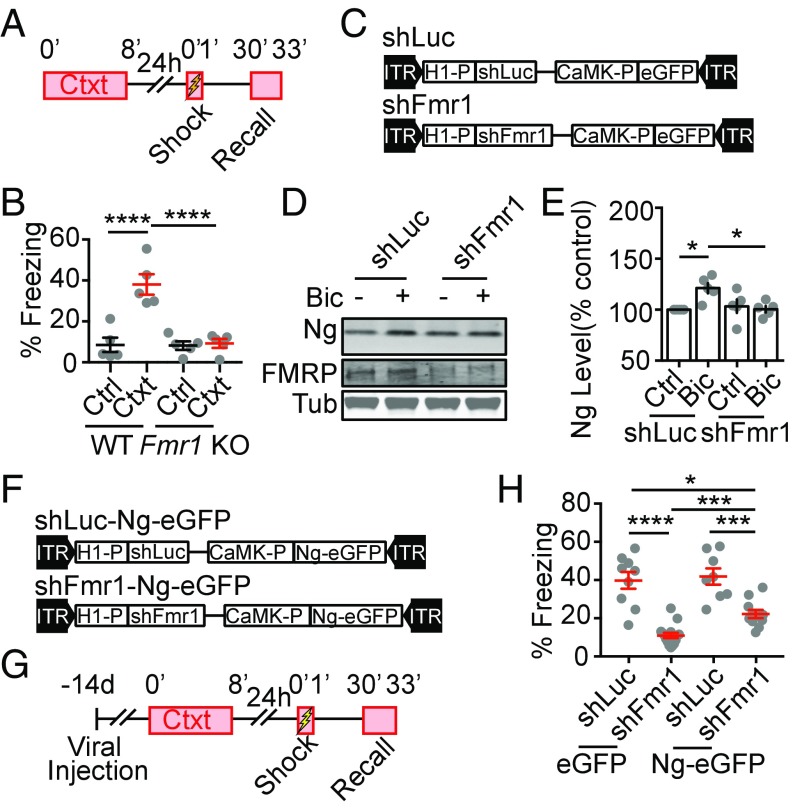
FMRP-mediated, novel-context–induced up-regulation of Ng is required for contextual memory formation. (A) The schematics for the contextual memory test. (B) Quantification of percentage of freezing during recall from control (Ctrl) and preexposed (Ctxt), wild-type (WT), and Fmr1 knockout (Fmr1 KO) mice; WT (n = 5, Ctrl; n = 5, Ctxt); Fmr1 KO (n = 5, Ctrl; n = 5, Ctxt). Two-way ANOVA, F(1, 16) = 17.86, P = 0.0006, followed by Tukey’s test, ****P < 0.0001. (C) Diagrams of cassettes in AAV vectors for expressing shRNAs targeted against Luciferase (shLuc) or Fmr1 (shFmr1) followed by an overexpression of eGFP under the control of the CaMK promoter. (D) Representative Western blot images of Ng, Tub, and FMRP from neuronal cultures infected with shLuc and shFmr1 with (+) or without (−) Bic treatment. (E) Quantification of percentage of Ng from neuronal cultures infected with shLuc and shFmr1, treated with Bic, normalized to vehicle-treated sister neuronal lysates, n = 5 from three independent cultures. Two-way ANOVA, F(1,16) = 7.436, P = 0.0149, followed by Tukey’s test, *P < 0.05. (F) Diagrams of cassettes in AAV vectors for expressing shRNAs targeted against Luciferase (shLuc) or Fmr1 (shFmr1) followed by an overexpression of Ng-eGFP under the control of the CaMK promoter. (G) The schematics for the contextual memory test. (H) Quantification of effects of the expression of shLuc (n = 9), shFmr1 (n = 15), shLuc-Ng-eGFP (n = 8), and shFmr1-Ng-eGFP (n = 11) in the hippocampal DG region on percentage of freezing during recall from preexposed mice. Two-way ANOVA, F(1, 39) = 2.523, P = 0.12, followed by Tukey’s test, ****P < 0.05.
Given that FMRP proteins are globally absent in the Fmr1 KO mice, it is unclear whether the memory deficit in the Fmr1 KO mice was due to specific disruption in the hippocampus. We next asked whether FMRP in the hippocampal DG region is specifically required for context memory formation via regulating Ng translation. We generated AAV-expressing shRNA that target the Fmr1 gene (Fig. 7C, shFmr1), using a published targeting sequence (62). A control AAV-expressing shRNA against luciferase (shLuc) was used as control (Fig. 7C). Expressing shFmr1 in cortical neurons effectively decreased FMRP levels (Fig. 7D and SI Appendix, Fig. S6A), and inhibited activity-dependent Ng up-regulation in the dissociated neuron culture (Fig. 7 D and E).
To test whether this hippocampal, FMRP-regulated Ng up-regulation is essential for context memory formation, we further developed AAVs that overexpress Ng-eGFP either with the background of shFmr1 or of shLuc as control (Fig. 7F). We bilaterally injected AAVs into the DG region of the hippocampus using stereotaxic injection, and tested contextual formation 14 d after the viral infusion (Fig. 7G). We found that expression of shFmr1 in the DG region of the hippocampus inhibited contextual memory formation, and this impairment of context memory can be partially rescued by overexpression of Ng-eGFP (Fig. 7H). shLuc and overexpression of Ng-eGFP with shLuc did not significantly affect context memory formation (Fig. 7H). Taken together, these results further support that FMRP-dependent, de novo protein synthesis of Ng in the hippocampus upon novel-context exposure is critical for context memory formation.
Discussion
Our present results confirm that novel-context–induced de novo protein synthesis is required for contextual memory formation. The rapid and transient time window flanks the contextual exposure and may thus be involved in memory encoding. Novel-context exposure recruits Nrgn mRNA into the actively translating mRNA pool and induces de novo Ng protein levels in the hippocampus. Using an AAV-mediated approach to expressing the 3′UTR of Nrgn to interfere with activity-dependent up-regulation of Ng and rescue, we showed that novel-context–dependent up-regulation of Ng is necessary for contextual memory formation. To understand the molecular mechanism, we used the biotinylated in vitro-transcribed RNA of the 3′UTR of Nrgn, and identified protein interaction partners. We found that FMRP interacts with the 3′UTR of Nrgn, among other proteins. Further analyses showed that FMRP is required for novel-context–dependent up-regulation of Ng in the hippocampus critical for contextual memory formation. Taken together, our data show that novel-context exposure induces rapid, activity-dependent translation of Ng in the hippocampus via an FMRP-dependent mechanism, necessary for durable contextual memory encoding.
Our study highlights: (i) our understanding of the immediate temporal requirement of activity-dependent translation in learning and memory; (ii) the functional contribution of activity-dependent translation of a target gene, Nrgn, in learning and memory; and (iii) the involvement of FMRP-dependent mechanism underlying the activity-dependent translation of Ng critical for contextual memory encoding.
Importantly, the experimental design that we adopted separates contextual memory formation from associative learning (42, 63) and allows the interrogation of contextual memory using the associative cue. This is different from a conventional contextual fear-conditioning paradigm, in which the context exposure and the fear association were presented concurrently. Using the conventional contextual fear-conditioning paradigm, Ryan et al. (9) showed that posttraining blockade of protein synthesis did not affect the implementation of the memory trace, but the memory recall using the natural cue was impaired. Our results reveal that protein synthesis inhibition after context exposure does not affect memory performance when the context exposure was separated from the associative cue (Fig. 1), suggesting that the protein synthesis after context exposure in the classic contextual fear-conditioning paradigm may be essential for strengthening the associative component of the task, but not necessary for the contextual memory encoding per se. The immediate de novo protein synthesis requirement for contextual memory formation (Fig. 1) suggests that de novo protein synthesis during the acquisition phase may play an important role in gating, facilitating, and durably encoding the memory.
Previous studies on FMRP have suggested that there is a pool of mRNAs containing hundreds of candidates that may undergo activity-dependent translation in response to behavioral stimulation (12, 15, 64, 65). However, little is known in terms of the extent, temporal dynamics, and the functional consequence of the activity-dependent translation of the candidate genes. Although some experiments have been done using reporter systems to monitor the activity-dependent translation of candidate genes (66, 67), studies of the endogenous translational targets under behavioral stimulation have been sparse. We detected a significant increase of Ng protein levels in total hippocampal lysate 15 min after the onset of the novel-context exposure, whereas other candidate genes, such as CaMKIIα and PSD-95, in the total protein levels were not increased in the hippocampus at the same time point (Fig. 2 J–L), suggesting activity-dependent translation of Ng in the hippocampus is an early responder to novel-context exposure. This also indicates that the regulation of the expression of the FMRP targets may not be homogeneous, and different genes or pools of genes can be regulated via different stimulation and in different temporal and spatial domains.
Our pull-down studies show that the 3′UTR of Nrgn interacts with protein complexes that contain FMRP and FXR2P, and previous studies have identified the Nrgn transcript as an FMRP-bound transcript using high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation (12), suggesting a direct interaction between FMRP and the Nrgn transcript. Although the discrete interaction site has not been previously described, we narrowed it down to Ng3′UTR 389–577, which can pull down FMRP and FXR2P and block memory formation. It is known that FMRP can repress mRNA translation, which can be released in an activity-dependent manner (68, 69), and absence of FMRP can lead to a general enhancement of the de novo protein synthesis rate and an absence of activity-dependent increase of protein synthesis (70). Given the presence of FXR2P in the complex, it is also possible that the interaction of FMRP-FXR2P with the 3′-UTR of the Nrgn transcript drives the activity-dependent translation of Ng, as seen in other studies (13).
De novo protein synthesis, in particular local protein synthesis upon synaptic activity, has been hypothesized to be important for synaptic plasticity and memory formation (71, 72). De novo protein synthesis is different from IEG expression in that no transcription is required (except, perhaps for Arc), and the targets include effector proteins that directly impact synaptic function. Absence of FMRP from animal models causes an array of learning and behavioral deficits (73), and the deficit can be restored by resetting certain signaling pathways, or prevent expression of elevated gene expression (14, 74). Our results highlight Nrgn as one important FMRP target for regulating cellular function essential for memory formation. On the other hand, given that the rescue is only partial, these results suggest that other factors influenced by decrease of FMRP also contribute to the full exertion of memory formation. It remains a significant area of research as to what other targets exist sharing similar temporal expression patterns and similar extent of activity-dependent translation to those of Ng, and what are their functional implications in memory formation. Furthermore, questions remain about the difference between Ng and other well-known translational targets in terms of the translation potency, temporal dynamics, location of action, and potentially cell-type specificity.
Notably, the expression of Ng is increased throughout hippocampal DG and CA1 regions upon novel-context exposure. The wide spread of expression suggests that this is most likely a gating mechanism responding to novelty, rather than an encoding mechanism for memory trace. De novo Ng translation coincides (if not precedes) with IEG transcription and expression (Fig. 2 and SI Appendix, Fig. S2). Given the fact that Ng regulates Ca2+/CaM-dependent signaling events, changes in Ng levels can have a profound impact on the downstream signaling events, including cyclic AMP-responsive element binding phosphorylation, voltage-gated Ca2+ channel inactivation, and CaMKII activation, which will be essential for memory encoding and stabilization relying on these events (75).
Taking these data together, we propose that activity-dependent local translation of Ng induced by novel experience via an FMRP-dependent mechanism enables durable memory encoding.
Materials and Methods
For commercially available resources, see SI Appendix, Table S2.
All animals were maintained in a vivarium with a light/dark cycle (7:00 AM–7:00 PM). Animal care and handling were performed according to NIH guidelines and with the approval of the Massachusetts Institute of Technology Institutional animal care and use committee and Division of Comparative Medicine.
AAV constructs were cloned in an AAV with AAV2 ITRs (76). AAV 2/9 serotype AAV vectors were produced as previously described (77–79).
For the context memory test with preexposure (42), C57BL/6Ncrl 7- to 9-wk-old male mice were habituated to the room. Animals were exposed to the context for the indicated time. Twenty-four hours later, animals were placed in the chamber, given an immediate shock, and removed from the chamber after a total of 1 min. Thirty minutes later, animals were reexposed to the chamber for 3 min to assay freezing. At least two cohorts of the animals in each experiment were blinded to the experimenter. Animal IDs were blinded for data analyses.
Mice were rapidly decapitated and submerged in liquid nitrogen for 4 s to rapidly cool brain tissue. Hippocampi were dissected on ice within 90 s and homogenized, and polyribosome enrichment was performed as described previously (Fig. 2B) (48, 49). qRT-PCR primer sequences and references are given in SI Appendix, Table S1. For Western blot, animals were killed by cervical dislocation in a separate room from the behavioral room. Brain regions were rapidly dissected on ice and flash-frozen in liquid nitrogen for further Western blot analysis.
In situ PLA was performed using the DuoLink II kit (Sigma) according to the instructions of the manufacturer. Coverslips were mounted with fluorescence mounting medium (Dako) to subject to confocal microscopy. C57BL/6Ncrl mice (8-wk-old) were anesthetized with isofluorane and transcardially perfused with 4% paraformaldehyde in PBS. After postfixation, brains were sectioned (45 μm) on a vibratome (Leica). Slices were processed using the standard procedure for immunohistochemistry. Images were taken on either a Zeiss 710 or Zeiss 810 confocal microscope with a 5× or a 63× objective, and processed in Imaris and Adobe Photoshop.
The RNA pull-down assay was based on (13). MS was performed independently two times and proteins found in both samples were marked in bold (SI Appendix, Dataset S1). Polyribosome profiling and RNA quantification from stimulated synaptoneurosomes procedures were described previously (45, 60, 80). Detailed materials and methods are in SI Appendix.
Statistical analysis was performed using Prism (Graphpad). Group differences were determined using either one-way or two-way ANOVA with the appropriate post hoc test. A one-sample t test was used for comparison of a group of data with a fixed value. Significance threshold was set at P = 0.05.
Supplementary Material
Acknowledgments
We thank Drs. Oliver M. Schluter, Samuel M. Cooke, Li-Huei Tsai, Dheeraj Roy, and members of the W.X. laboratory for their helpful comments, and Xioabai Ren and Yan Liu for their excellent technical support. This work was supported by the JPB foundation, Whitehall Foundation, and Stanley Center at the Broad Institute. K.Z. is supported by the Swiss National Foundation (Grant SNF171978).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.M.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716750115/-/DCSupplemental.
References
- 1.Alberini CM. The role of protein synthesis during the labile phases of memory: Revisiting the skepticism. Neurobiol Learn Mem. 2008;89:234–246. doi: 10.1016/j.nlm.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis HP, Squire LR. Protein synthesis and memory: A review. Psychol Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- 3.Flexner JB, Flexner LB, Stellar E. Memory in mice as affected by intracerebral puromycin. Science. 1963;141:57–59. doi: 10.1126/science.141.3575.57. [DOI] [PubMed] [Google Scholar]
- 4.Alberini CM, Ledoux JE. Memory reconsolidation. Curr Biol. 2013;23:R746–R750. doi: 10.1016/j.cub.2013.06.046. [DOI] [PubMed] [Google Scholar]
- 5.Bekinschtein P, et al. Persistence of long-term memory storage requires a late protein synthesis- and BDNF-dependent phase in the hippocampus. Neuron. 2007;53:261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Bourtchouladze R, et al. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- 7.Quevedo J, et al. Two time windows of anisomycin-induced amnesia for inhibitory avoidance training in rats: Protection from amnesia by pretraining but not pre-exposure to the task apparatus. Learn Mem. 1999;6:600–607. doi: 10.1101/lm.6.6.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flood JF, Bennett EL, Orme E, Rosenzweig MR. Relation of memory formation to controlled amounts of brain protein synthesis. Physiol Behav. 1975;15:97–102. doi: 10.1016/0031-9384(75)90285-1. [DOI] [PubMed] [Google Scholar]
- 9.Ryan TJ, Roy DS, Pignatelli M, Arons A, Tonegawa S. Memory. Engram cells retain memory under retrograde amnesia. Science. 2015;348:1007–1013. doi: 10.1126/science.aaa5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schafe GE, LeDoux JE. Memory consolidation of auditory Pavlovian fear conditioning requires protein synthesis and protein kinase A in the amygdala. J Neurosci. 2000;20:RC96. doi: 10.1523/JNEUROSCI.20-18-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barondes SH, Cohen HD. Memory impairment after subcutaneous injection of acetoxycycloheximide. Science. 1968;160:556–557. doi: 10.1126/science.160.3827.556. [DOI] [PubMed] [Google Scholar]
- 12.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández E, et al. FXR2P exerts a positive translational control and is required for the activity-dependent increase of PSD95 expression. J Neurosci. 2015;35:9402–9408. doi: 10.1523/JNEUROSCI.4800-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sidorov MS, Auerbach BD, Bear MF. Fragile X mental retardation protein and synaptic plasticity. Mol Brain. 2013;6:15. doi: 10.1186/1756-6606-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasciuto E, Bagni C. SnapShot: FMRP mRNA targets and diseases. Cell. 2014;158:1446–1446.e1. doi: 10.1016/j.cell.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 16.Wu L, et al. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 17.Jakkamsetti V, et al. Experience-induced Arc/Arg3.1 primes CA1 pyramidal neurons for metabotropic glutamate receptor-dependent long-term synaptic depression. Neuron. 2013;80:72–79. doi: 10.1016/j.neuron.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neuner-Jehle M, Denizot JP, Mallet J. Neurogranin is locally concentrated in rat cortical and hippocampal neurons. Brain Res. 1996;733:149–154. [PubMed] [Google Scholar]
- 19.Represa A, Deloulme JC, Sensenbrenner M, Ben-Ari Y, Baudier J. Neurogranin: Immunocytochemical localization of a brain-specific protein kinase C substrate. J Neurosci. 1990;10:3782–3792. doi: 10.1523/JNEUROSCI.10-12-03782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson JB, Sutcliffe JG, Fisher RS. Localization of the protein kinase C phosphorylation/calmodulin-binding substrate RC3 in dendritic spines of neostriatal neurons. Proc Natl Acad Sci USA. 1992;89:8581–8585. doi: 10.1073/pnas.89.18.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerendasy DD, Sutcliffe JG. RC3/neurogranin, a postsynaptic calpacitin for setting the response threshold to calcium influxes. Mol Neurobiol. 1997;15:131–163. doi: 10.1007/BF02740632. [DOI] [PubMed] [Google Scholar]
- 22.Zhabotinsky AM, Camp RN, Epstein IR, Lisman JE. Role of the neurogranin concentrated in spines in the induction of long-term potentiation. J Neurosci. 2006;26:7337–7347. doi: 10.1523/JNEUROSCI.0729-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang K-P, et al. Neurogranin/RC3 enhances long-term potentiation and learning by promoting calcium-mediated signaling. J Neurosci. 2004;24:10660–10669. doi: 10.1523/JNEUROSCI.2213-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pak JH, et al. Involvement of neurogranin in the modulation of calcium/calmodulin-dependent protein kinase II, synaptic plasticity, and spatial learning: A study with knockout mice. Proc Natl Acad Sci USA. 2000;97:11232–11237. doi: 10.1073/pnas.210184697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong L, Cherry T, Bies CE, Florence MA, Gerges NZ. Neurogranin enhances synaptic strength through its interaction with calmodulin. EMBO J. 2009;28:3027–3039. doi: 10.1038/emboj.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruano D, et al. Association of the gene encoding neurogranin with schizophrenia in males. J Psychiatr Res. 2008;42:125–133. doi: 10.1016/j.jpsychires.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Stefansson H, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coldren CD, et al. Chromosomal microarray mapping suggests a role for BSX and Neurogranin in neurocognitive and behavioral defects in the 11q terminal deletion disorder (Jacobsen syndrome) Neurogenetics. 2009;10:89–95. doi: 10.1007/s10048-008-0157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernal J, Rodriguez-Pena A, Iniguez MA, Ibarrola N, Munoz A. Influence of thyroid hormone on brain gene expression. Acta Med Austriaca. 1992;19:32–35. [PubMed] [Google Scholar]
- 30.Broadbelt K, Ramprasaud A, Jones LB. Evidence of altered neurogranin immunoreactivity in areas 9 and 32 of schizophrenic prefrontal cortex. Schizophr Res. 2006;87:6–14. doi: 10.1016/j.schres.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 31.Buaud B, et al. A high-fat diet induces lower expression of retinoid receptors and their target genes GAP-43/neuromodulin and RC3/neurogranin in the rat brain. Br J Nutr. 2010;103:1720–1729. doi: 10.1017/S0007114509993886. [DOI] [PubMed] [Google Scholar]
- 32.Enderlin V, et al. Aging decreases the abundance of retinoic acid (RAR) and triiodothyronine (TR) nuclear receptor mRNA in rat brain: Effect of the administration of retinoids. FEBS Lett. 1997;412:629–632. doi: 10.1016/s0014-5793(97)00845-4. [DOI] [PubMed] [Google Scholar]
- 33.Golini RS, et al. Daily patterns of clock and cognition-related factors are modified in the hippocampus of vitamin A-deficient rats. Hippocampus. 2012;22:1720–1732. doi: 10.1002/hipo.22007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang FL, Huang K-P, Wu J, Boucheron C. Environmental enrichment enhances neurogranin expression and hippocampal learning and memory but fails to rescue the impairments of neurogranin null mutant mice. J Neurosci. 2006;26:6230–6237. doi: 10.1523/JNEUROSCI.1182-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovalevich J, et al. Cocaine decreases expression of neurogranin via alterations in thyroid receptor/retinoid X receptor signaling. J Neurochem. 2012;121:302–313. doi: 10.1111/j.1471-4159.2012.07678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNamara RK, Huot RL, Lenox RH, Plotsky PM. Postnatal maternal separation elevates the expression of the postsynaptic protein kinase C substrate RC3, but not presynaptic GAP-43, in the developing rat hippocampus. Dev Neurosci. 2002;24:485–494. doi: 10.1159/000069359. [DOI] [PubMed] [Google Scholar]
- 37.Neuner-Jehle M, Rhyner TA, Borbély AA. Sleep deprivation differentially alters the mRNA and protein levels of neurogranin in rat brain. Brain Res. 1995;685:143–153. doi: 10.1016/0006-8993(95)00416-n. [DOI] [PubMed] [Google Scholar]
- 38.Ressler KJ, Paschall G, Zhou XL, Davis M. Regulation of synaptic plasticity genes during consolidation of fear conditioning. J Neurosci. 2002;22:7892–7902. doi: 10.1523/JNEUROSCI.22-18-07892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell. 2008;19:2311–2327. doi: 10.1091/mbc.E07-09-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat Neurosci. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- 41.Rudy JW, Barrientos RM, O’Reilly RC. Hippocampal formation supports conditioning to memory of a context. Behav Neurosci. 2002;116:530–538. doi: 10.1037//0735-7044.116.4.530. [DOI] [PubMed] [Google Scholar]
- 42.Frankland PW, et al. Consolidation of CS and US representations in associative fear conditioning. Hippocampus. 2004;14:557–569. doi: 10.1002/hipo.10208. [DOI] [PubMed] [Google Scholar]
- 43.Tolic L, et al. Determination of anisomycin in tissues and serum by LC-MS/MS: Application to pharmacokinetic and distribution studies in rats. RSC Adv. 2016;6:92479–92489. [Google Scholar]
- 44.Wanisch K, Wotjak CT. Time course and efficiency of protein synthesis inhibition following intracerebral and systemic anisomycin treatment. Neurobiol Learn Mem. 2008;90:485–494. doi: 10.1016/j.nlm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- 46.Guzowski JF, et al. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller S, et al. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- 48.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kubik S, Miyashita T, Guzowski JF. Using immediate-early genes to map hippocampal subregional functions. Learn Mem. 2007;14:758–770. doi: 10.1101/lm.698107. [DOI] [PubMed] [Google Scholar]
- 51.Tischmeyer W, Grimm R. Activation of immediate early genes and memory formation. Cell Mol Life Sci. 1999;55:564–574. doi: 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramamoorthi K, et al. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334:1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.tom Dieck S, et al. Direct visualization of newly synthesized target proteins in situ. Nat Methods. 2015;12:411–414. doi: 10.1038/nmeth.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitchigina V, Vankov A, Harley C, Sara SJ. Novelty-elicited, noradrenaline-dependent enhancement of excitability in the dentate gyrus. Eur J Neurosci. 1997;9:41–47. doi: 10.1111/j.1460-9568.1997.tb01351.x. [DOI] [PubMed] [Google Scholar]
- 55.Green EJ, McNaughton BL, Barnes CA. Exploration-dependent modulation of evoked responses in fascia dentata: Dissociation of motor, EEG, and sensory factors and evidence for a synaptic efficacy change. J Neurosci. 1990;10:1455–1471. doi: 10.1523/JNEUROSCI.10-05-01455.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frank LM, Stanley GB, Brown EN. Hippocampal plasticity across multiple days of exposure to novel environments. J Neurosci. 2004;24:7681–7689. doi: 10.1523/JNEUROSCI.1958-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dittgen T, et al. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci USA. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunsaker MR, Rosenberg JS, Kesner RP. The role of the dentate gyrus, CA3a,b, and CA3c for detecting spatial and environmental novelty. Hippocampus. 2008;18:1064–1073. doi: 10.1002/hipo.20464. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt B, Satvat E, Argraves M, Markus EJ, Marrone DF. Cognitive demands induce selective hippocampal reorganization: Arc expression in a place and response task. Hippocampus. 2012;22:2114–2126. doi: 10.1002/hipo.22031. [DOI] [PubMed] [Google Scholar]
- 60.Kuzniewska B, Chojnacka M, Milek J, Dziembowska M. Preparation of polysomal fractions from mouse brain synaptoneurosomes and analysis of polysomal-bound mRNAs. J Neurosci Methods. 2018;293:226–233. doi: 10.1016/j.jneumeth.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 61.Janusz A, et al. The fragile X mental retardation protein regulates matrix metalloproteinase 9 mRNA at synapses. J Neurosci. 2013;33:18234–18241. doi: 10.1523/JNEUROSCI.2207-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu H, et al. Emotion enhances learning via norepinephrine regulation of AMPA-receptor trafficking. Cell. 2007;131:160–173. doi: 10.1016/j.cell.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 64.Thomson SR, et al. Cell-type-specific translation profiling reveals a novel strategy for treating fragile X syndrome. Neuron. 2017;95:550–563.e5. doi: 10.1016/j.neuron.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernández E, Rajan N, Bagni C. The FMRP regulon: From targets to disease convergence. Front Neurosci. 2013;7:191. doi: 10.3389/fnins.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Butko MT, et al. Fluorescent and photo-oxidizing TimeSTAMP tags track protein fates in light and electron microscopy. Nat Neurosci. 2012;15:1742–1751. doi: 10.1038/nn.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ju W, et al. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- 68.Muddashetty RS, Kelić S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Napoli I, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 70.Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30:15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelleher RJ, 3rd, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 72.Jiang C, Schuman EM. Regulation and function of local protein synthesis in neuronal dendrites. Trends Biochem Sci. 2002;27:506–513. doi: 10.1016/s0968-0004(02)02190-4. [DOI] [PubMed] [Google Scholar]
- 73.Santos AR, Kanellopoulos AK, Bagni C. Learning and behavioral deficits associated with the absence of the fragile X mental retardation protein: What a fly and mouse model can teach us. Learn Mem. 2014;21:543–555. doi: 10.1101/lm.035956.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci. 2015;16:595–605. doi: 10.1038/nrn4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lisman J, Cooper K, Sehgal M, Silva AJ. Memory formation depends on both synapse-specific modifications of synaptic strength and cell-specific increases in excitability. Nat Neurosci. 2018;21:309–314. doi: 10.1038/s41593-018-0076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang X, et al. Progressive maturation of silent synapses governs the duration of a critical period. Proc Natl Acad Sci USA. 2015;112:E3131–E3140. doi: 10.1073/pnas.1506488112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han KS, Cooke SF, Xu W. Experience-dependent equilibration of AMPAR-mediated synaptic transmission during the critical period. Cell Rep. 2017;18:892–904. doi: 10.1016/j.celrep.2016.12.084. [DOI] [PubMed] [Google Scholar]
- 78.Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc. 2006;1:1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 79.Zolotukhin S, et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 80.Dziembowska M, et al. Activity-dependent local translation of matrix metalloproteinase-9. J Neurosci. 2012;32:14538–14547. doi: 10.1523/JNEUROSCI.6028-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.