FIG. 2.
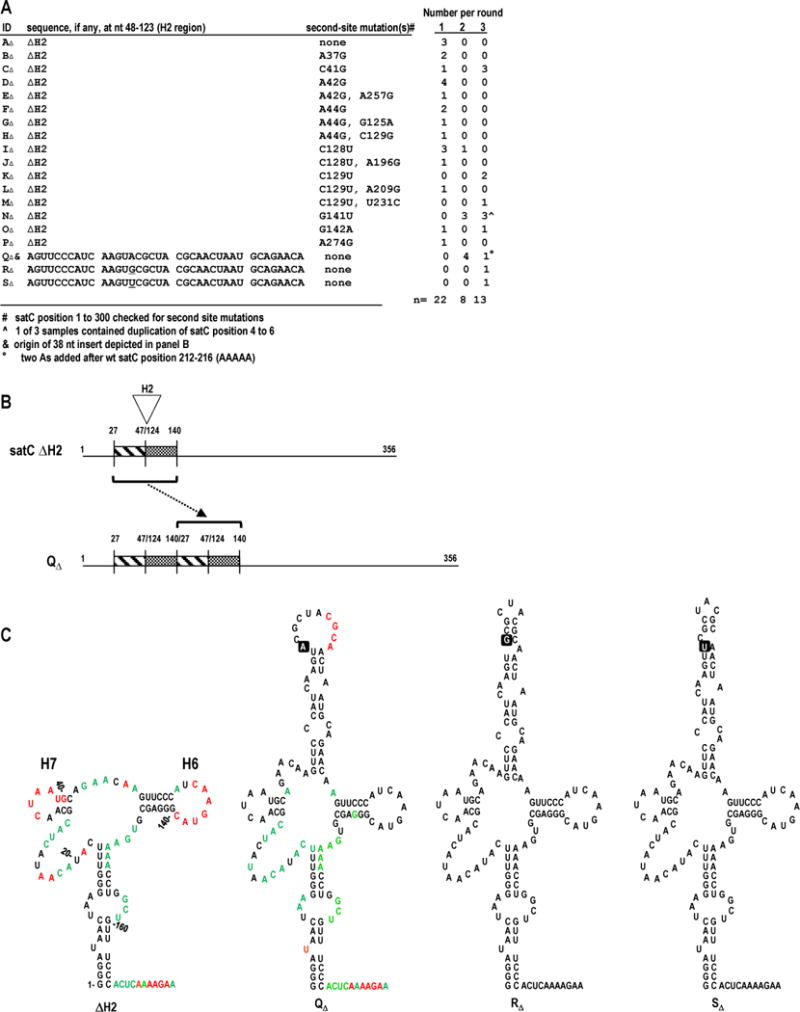
Progeny generated during in planta passaging of satC with H2 deleted (ΔH2) recover 38 nt in the H2 region. (A) satCΔH2 transcripts were mixed with TCV genomic RNA and passaged through 30 plants (Round 1). RNA was extracted, pooled, and passaged through 6 plants two sequential times (Rounds 2 and 3). satC progeny were cloned following each round. Samples AΔ through PΔ retained the deletion. Second-site mutations in samples BΔ through PΔ are noted. (B) Model depicting duplication leading to QΔ. Duplication (bracket) of nt 27-47 and 124-140 (H2-flanking sequences) from satCΔH2 resulted in a 38 nt H2 region. (C) Secondary structure predictions for the 5′ region of samples satCΔH2, QΔ, RΔ, and SΔ. SHAPE results are shown on the structures of satCΔH2 and QΔ (see Fig. 1B for nucleotide coloring). The single-nucleotide difference among QΔ, RΔ, and SΔ is boxed. Numbering is according to wt satC.
