Abstract
Characterizing the stability of the gut microbiome is important to exploit it as a therapeutic target and diagnostic biomarker. We metagenomically and metatranscriptomically sequenced the faecal microbiomes of 308 participants in the Health Professionals Follow-Up Study. Participants provided four stool samples—one pair collected 24–72 h apart and a second pair ~6 months later. Within-person taxonomic and functional variation was consistently lower than between-person variation over time. In contrast, metatranscriptomic profiles were comparably variable within and between subjects due to higher within-subject longitudinal variation. Metagenomic instability accounted for ~74% of corresponding metatranscriptomic instability. The rest was probably attributable to sources such as regulation. Among the pathways that were differentially regulated, most were consistently over- or under-transcribed at each time point. Together, these results suggest that a single measurement of the faecal microbiome can provide long-term information regarding organismal composition and functional potential, but repeated or short-term measures may be necessary for dynamic features identified by metatranscriptomics.
Understanding the temporal dynamics of the healthy adult gut microbiome is integral in leveraging these microbial communities to promote human health. Large-scale changes in microbial composition have been associated with host health overall1–3, but inferring causality and developing personalized therapies will require large-scale prospective cohort studies. Furthermore, to exploit the faecal microbiome as a predictive biomarker or eventually as a diagnostic tool in clinical settings, it is critical to be able to discriminate between normal versus pathological variation over time4.
Previous efforts have provided excellent characterization of the ecological stability of the adult faecal microbiome4–13. All measures of stability in microbial communities must be in the context of relative differences since, despite daily variability in species’ relative abundances, microbial communities in the gut microbiome have been observed to be generally consistent over time, even on the scale of years or decades4,5,14,15. This relative stability appears to be due to individually persistent strains within individual hosts5,16. Moreover, specific inter-individual differences in community structures appear to be preserved over the long-term17, allowing an individual’s faecal microbiome to be uniquely distinguished from that of others to serve as a faecal microbial fingerprint13. Nonetheless, despite the relative stability of the community profile over the long-term, recent studies have shown that host lifestyle or exposures such as a sudden change in diet, the initiation of antibiotics or the acquisition of pathogenic species can lead to profound disruptions in the microbiome9,18,19. When such pressures are lifted, the host’s faecal microbiome generally recovers to a composition comparable to its original state9.
An understanding of microbiome stability as it affects taxonomic and functional features is vital for applying it diagnostically or prognostically in long-term public health studies. Different molecular features, including strain membership, species abundances, functional profiles or metatranscription, may all prove to be informative regarding host health conditions and they are likely to differ dramatically in their relative stability within and between subjects over time20. The Human Microbiome Project (HMP), for example, found that in the absence of perturbations from disease or overt xenobiotics, metagenomic functional profiles were more comparable between individuals, while strains were stable within subjects17. Even fewer studies have focused on the metatranscriptome, an indicator of different aspects of microbial functional activity21,22. Finally, the dynamics of response in any of these features to known dietary and xenobiotic perturbations are themselves not yet fully known23,24. Thus, the magnitude of changes in microbial composition, functional potential and gene expression that occur over various time intervals and their utility for molecular epidemiology are unclear.
To address these knowledge gaps, we deeply characterized the faecal microbiome among 308 individuals enroled in the Men’s Lifestyle Validation Study (MLVS), nested within the ongoing population-based Health Professionals Follow-up Study (HPFS)—a prospective cohort of 51,529 men followed since 1986. This cohort provided an unparalleled opportunity to apply insights from community ecology and epidemiology to characterize the stability of the faecal microbiome structure and function over time. Specifically, we hypothesized that inter-individual differences in faecal microbiome communities and metagenomes would persist over short- (24–72 h) and intermediate-term (6 months) intervals, but that metatranscriptomes would demonstrate greater variability over time. The work described here characterizing faecal microbial stability is accompanied by further research on molecular function in the faecal metagenome and metatranscriptome population25. These studies will provide a foundation for leveraging the faecal microbiome as a biomarker for population-based cohort studies and provide insight into how metagenomics and metatranscriptomics relate to each other over time.
Results
The faecal metagenome is more stable than the metatranscriptome
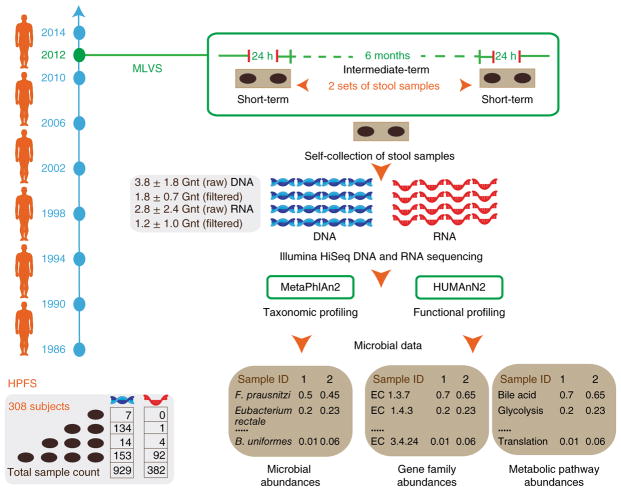
We first quantified the longitudinal stability of metagenomic taxonomic profiles, functional profiles and metatranscriptomes in MLVS subjects. Our approach is detailed in Fig. 1. Participants provided up to four stool samples—a set of stool samples collected 24–72 h apart followed by collection of a second set approximately 6 months later. DNA was extracted from 929 samples collected from all 308 men. RNA was extracted and reverse-transcribed to complementary DNA from 378 samples from participants who provided a stool at both sampling time points and who did not report the use of antibiotics within the past year. All samples were then sequenced using the Illumina HiSeq platform (see Methods). Raw sequence data were filtered to remove low-quality and human host reads and features with low overall relative abundance. Metagenomic and metatranscriptomic reads were profiled for functional and taxonomic composition using HUMAnN2 (ref. 26) and MetaPhlAn2 (ref. 27), respectively.
Fig. 1. Experimental design.
308 participants from the MLVS, nested within the HPFS cohort, were recruited to assess the stability of microbiome communities, metagenomes and metatranscriptomes. Participants provided up to four stool samples using a previously validated self-sampling method21. One set of stool samples was collected 24–72 h apart followed by a second set approximately 6 months later. Metagenomic and metatranscriptomic reads were generated to provide taxonomic, functional metagenomic and transcriptional features for stability assessment.
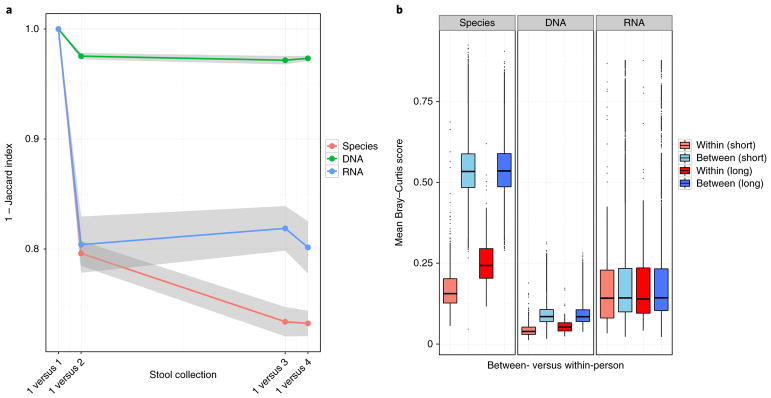
We compared the within-person stability of the faecal metatranscriptome (functional elements from RNA) with the stability of taxonomic composition (species from DNA) and the metagenome (functional elements from DNA) over short (24–72 h) and intermediate (6 month) time intervals. This was assessed by calculating two β-diversity metrics—the Jaccard index (for membership) and Bray–Curtis (BC) dissimilarity (for abundance)—between an individual’s samples collected over the study period. In accordance with our earlier pilot study21, the metagenomic functional potential (DNA) was more stable than the metatranscriptome (RNA), which was in turn more stable than taxonomic profile abundances (species) (Fig. 2a).
Fig. 2. Inter-individual differences in organismal composition and functional potential appear to be preserved, unlike the more variable metatranscriptomes.
a, 1 – Jaccard Index (fraction of shared features) between all possible pairwise combinations of the first faecal sample with the other three samples collected from each individual (n = 308). 95% confidence intervals are shown in grey. b, Bray–Curtis β-diversity scores within and between subjects for short- (24–72 h; n = 308 individuals) and intermediate-term intervals (6 months; n = 160 individuals). Here, species represents taxonomic profile abundances, DNA represents metagenomic functional profiles, and RNA represents metatranscriptomes. Boxplot whiskers include observations within 1.5 interquartile range of the upper and lower quartiles.
In addition, we tested whether the stability of microbiome features was consistent over short- and intermediate-term intervals. Species, DNA and RNA features that remained stable within a subject over a 24–72 h period tended to be similarly stable over 6 months (Spearman’s r =0.99, two-tailed P <2.2×10−16), suggesting the presence of a subset of features that can be consistently measured over time.
Between-person exceeds within-person metagenomic variation
As in previous studies of the faecal microbiome17, individuals’ microbial community structure and metagenomic functional potential were more self-similar (red boxes) over time than between subjects (blue boxes) (Fig. 2b). This remained consistent over short- (24–72 h) and intermediate-term (6 month) periods (within-person variation versus between-person variation for all four Mann–Whitney U tests: P <2.2×10−16). In contrast, we found that within- and between-subject variation over short- and intermediate-term periods was comparable for metatranscriptomes (short term Mann–Whitney U test: P = 0.25; intermediate term Mann–Whitney U test: P =0.50).
To additionally quantify the proportion of variation that can be attributed to between-subject variation for the purposes of prospective population-based cohort studies, we estimated intra-class correlation coefficients (ICCs). The majority of species and genes (86.8 and 92.8%, respectively) retained ICCs greater than 0.40 over the longest sampling interval in our study. In contrast, only a small fraction of transcripts (0.79%) had an ICC greater than 0.40 (Supplementary Table 1).
Taken together, these data suggest that specific inter-individual differences in faecal microbiome organismal composition and functional potential appear to be preserved over intervals up to at least six months and thus may be more reliably associated with long-term health outcomes using a limited number of measurements. In contrast, the more dynamic features of the metatranscriptome may be more informative regarding short-term events.
Feature abundance and prevalence are associated with stability
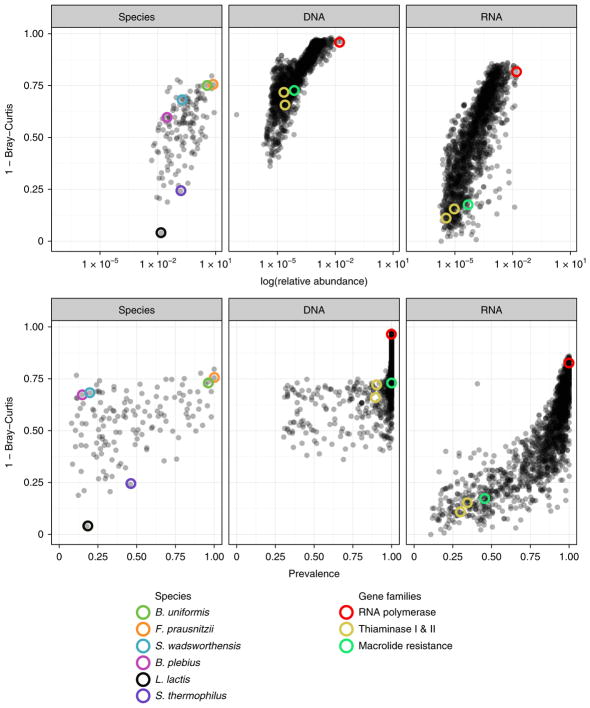
Feature consistency over short- and intermediate-term periods led us to consider if there may be properties inherent to features that contribute to stability across all hosts. Across a range of body sites, overall taxonomic stability of the human microbiome is correlated with feature abundance and prevalence13. We extended these results by exploring the correlations between these two ecological properties and the stability of individual organisms in the faecal microbiome, as well as the stability of the genomic and transcriptomic levels of individual enzymes. As in previous analyses where dissimilarity metrics have been used for individual features5, using the complement of within-person BC dissimilarity score (1 – BC) over the longest interval in our study (time point 1 versus time point 4; ~6 months), we similarly found that baseline feature prevalence and abundance were also strongly and positively correlated with not only organismal composition, but also genomic and transcriptional stability (Fig. 3). Many species that were highly abundant and prevalent in this population were highly stable, including the well-characterized Bacteroides uniformis (dark green circle) and Faecalibacterium prausnitzii (orange circle)28. In contrast, species that were of low prevalence and abundance were less stable, including those that were probably carried from the oral cavity including Lactococcus lactis (black circle) and Streptococcus thermophilus (dark purple circle)21,29. Similarly, enzymes such as RNA polymerase (red circle) (EC (Enzyme Commission) 2.7.7.6), which are essential for prokaryotic function, were universally prevalent, highly abundant and stably expressed at the genomic and transcriptional level.
Fig. 3. Stability of individual species, genes and transcripts over 6 months is correlated with average baseline relative abundance and prevalence.
Each point represents an individual feature—species (n = 139), genes (n = 1,952) or transcripts (n = 1,803). Coloured circles highlight species and gene families discussed in the main text, according to the figure legend.
Notably, however, there were select features for which the stability levels did not correspond with their prevalence or abundance. For example, Sutterella wadsworthensis (blue circle), was in the bottom quintile for prevalence and the bottom half for abundance, but it was in the top quintile for stability. This may be due to the role of this species’ lone production of the adenosine 3′-phosphate-5′-phosphosulfate-independent sulfuryl transferase (EC 2.8.2.22)—an important microbial enzyme involved in the detoxification of phenolic compounds30,31. Similarly, Bacteroides plebius (light purple circle), found to occupy a unique role in starch utilization, was found in the bottom quintile for prevalence but in the top quintile for stability32. Additionally, as observed in the HMP, there were genes that were universally prevalent at the DNA level but of relatively low genomic abundance17. Many of these enzymes had some of the most variable gene expression. For example, the pair of enzymes thiaminase I and II (EC 2.5.1.2 and EC 3.5.99.2), which are involved in the microbial degradation of thiamin, possessed DNA sequences detected in 100% of participants, but were within the bottom 5% of stable transcripts33. Similarly, the enzyme ErmC (EC 2.1.1.184), which encodes one of the macrolide resistance genes was universally prevalent at the genomic level but was unstable at the RNA level and of intermediate stability at the genomic level34.
Host factors that may influence faecal microbiome stability
In addition to examining determinants of stability for specific features, we assessed whether there were lifestyle factors or behaviours at the time of stool collection that could explain global instability of the faecal microbiome for each individual. There have been inconsistent data on the effects of antibiotics and bowel preparation on the faecal microbiome composition and limited data on temporal change in the metagenome19,24,35–38. In addition, ecological theory suggests that species-rich communities may have a greater compensatory capacity for disturbances in the ecosystem39,40.
In an exploratory analysis, we examined the association of putative host or environmental factors that may influence the stability of microbial community structure or pathways over a longer period. To do this, we calculated BC dissimilarity values for each individual over the longest sampling interval (around six months) using species, genes and transcripts as our response variables. As shown in Table 1, there were few factors that consistently explained instability. However, common predictive factors across feature types included mean Shannon index values (α-diversity) and exposure to a bowel laxative preparation within the two months before collection. Antibiotic use during the six-month period between sample collection seemed to affect genomic instability to a slightly greater extent than species instability. However, the overall low correspondence between antibiotic usage and dissimilarity suggests that faecal ecological variability may either be attributed to additional exogenous factors not measured here or to endogenous and perhaps stochastic host–microbial interactions.
Table 1.
Association of putative factors that may influence the stability of human faecal microbiome communities and pathways over a six-month period
| Species (n = 160) | DNA (n = 160) | RNA (n = 93) | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Median dissimilarity over 6 monthsa | Parameter estimate | P valueb | Parameter estimate | P valueb | Parameter estimate | P valueb |
| Mean Shannon index (α-diversity) | −0.096 | 0.006 | −0.037 | <0.0001 | −0.076 | 0.13 |
| Acid-lowering medication usec | 0.020 | 0.33 | 0.007 | 0.07 | 0.008 | 0.78 |
| Bowel preparationd | 0.087 | 0.01 | 0.015 | 0.04 | 0.13 | 0.03 |
| Body mass index (kg m−2) | 0.002 | 0.51 | 0.0007 | 0.15 | 0.0035 | 0.33 |
| Age | −0.0004 | 0.84 | 0.0002 | 0.69 | 0.005 | 0.08 |
| Any antibiotic usee | 0.020 | 0.33 | 0.0004 | 0.92 | NA | NA |
| New antibiotic usee | 0.033 | 0.34 | 0.022 | 0.002 | NA | NA |
| Bristol category changef | 0.0004 | 0.56 | −0.00001 | 0.52 | 0.0002 | 0.12 |
In an exploratory analysis, linear models were used to determine which of the measured factors predicted individuals’ dissimilarity in faecal microbiome communities and faecal microbiome pathways between the first and last sample. All models were adjusted by the mean total number of reads that passed filtration per identification. n refers to the number of participants in each analysis. Statistically significant values appear in bold.
As estimated by the BC dissimilarity score between the first and fourth stool sample.
All models were adjusted for median read depth (pass-filter reads).
Use of proton-pump inhibitors or H2 receptor antagonists more than once per week in the past two months.
Any reported bowel preparation (two months before collection).
The reference level was no antibiotic use. 'Any antibiotic use' was defined as any use in the past 12 months before the second stool collection. 'New antibiotic use' was defined as new use of antibiotics before the second collection. We did not assess transcriptomics in individuals who reported antibiotic use.
Bristol scores were assigned to three categories: 1–2 = hard; 3–4 = normal; 5–7 = soft or liquid.
Relating metagenomic and metatranscriptomic stability over time
Next, we considered the degree to which transcriptomic stability is related to metagenomic stability, since the central dogma suggests that DNA copy number changes are likely to influence transcript levels. Recent evidence from a pilot study involving a subset of these participants (n = 8) found that many gene and transcript relative abundances were well correlated and that a substantial fraction (41%) of microbial transcripts were basally regulated21. Again using the complement of the within-person BC dissimilarity score over six months as our metric of stability, we found that genomic instability appears to explain a large proportion of transcriptomic instability (Spearman’s r = 0.86; r2 = 0.74 = 74%; P <2.2×10−16). To further investigate whether this trend is driven by a small number of highly expressed genes, we divided our dataset into genes that were prevalently expressed (defined as transcripts detected in >90% of samples; n = 1,049) and genes that were variably detected (n =754). When comparing r2 values for these two groups, we did not observe an appreciable difference (0.56 versus 0.48). However, we found that among only the transcripts in the bottom 10% of prevalence (n = 176), a similar strength in association was not seen (r2 =0.01), suggesting that measured expression of these genes is influenced by other factors.
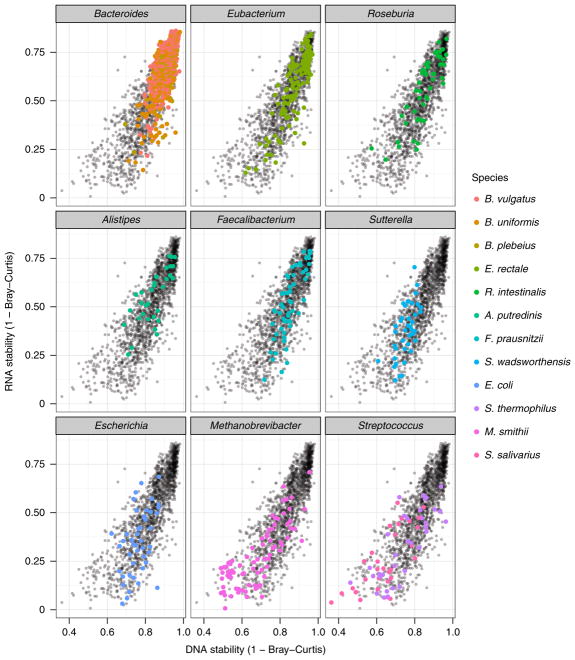
In addition, we investigated whether dominant expression of stable versus unstable genes varied according to species. For each transcript, a dominant contributing species was identified based on the maximal average contributing organism for each transcript from HUMAnN2 (see Methods). We then ranked species according to the total number of genes that they dominantly expressed (Fig. 4). As shown, Bacteroides species are dominant contributors to the greatest number of genes expressed, as well as dominant contributors to the expression of the most stable transcripts. In contrast, Streptococcus species are dominant contributors to the expression of some of the most unstable transcripts. Interestingly, these are not typically viable gut residents21, suggesting that some species play more variable roles because they are rare, transitory, more prone to influence by environmental factors or possibly under-sampled.
Fig. 4. Relating metagenomic and metatranscriptomic stability over time.
Genomic instability appears to explain a large proportion of transcriptomic instability over intermediate-term (6 month) periods (r2= 0.74); the metatranscriptome should typically be no more stable than the metagenome. In addition, we investigated whether dominant expression of stable genes versus expression of unstable genes varied according to species. For each transcript (n = 1,803), a dominant contributing species was identified based on the maximal average contribution to each transcript. We then ranked the species according to the total number of genes that they dominantly expressed and (for presentation purposes) selected those contributing to 30 or more and overlaid them on the scatterplot in colour (grouped by genus). Each point represents an individual feature.
Stability of gene expression over time
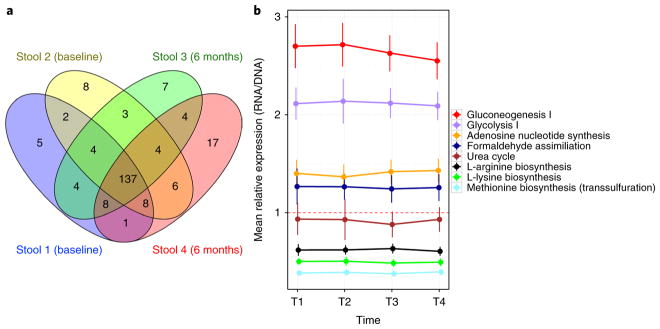
The previous section highlighted that much of the transcriptional variability over time for each feature was associated with changes in the metagenome. Thus, next we queried what proportion of the remainder of transcriptional variation could be explained by differences in the transcriptional expression of features over time. In an approach similar to that performed in the pilot study21, we identified significantly differentially transcribed pathways, defined as having a mean log RNA/DNA abundance ratio >2 for all four time points and then explored where they overlapped. Of the 218 pathways that were significantly differentially expressed, 79% were consistently over- or under-expressed across three or four time points (Fig. 5a).
Fig. 5. Exploring the stability of gene expression.
a, For a large proportion of transcriptional pathways over- or under-expressed over time in the stool, this differential expression may be consistent. The Venn diagram shows consistency of differentially transcribed pathways (mean RNA/DNA ratios >2 across at least one time point; n = 218) across 4 stool samples. b, A subset (n = 8) of stably transcribed pathways is highlighted, according to those with the lowest coefficient of variance of mean RNA/DNA expression ratios across the four time points. Many of the stable pathways appear to be involved in cellular housekeeping, such as carbon metabolism (over-expressed; above dashed red line; >1) and amino acid synthesis (under-expressed; below dashed red line; <1). Error bars indicate 95% confidence intervals.
To better understand which pathways were most stably expressed, for each time point, we computed the mean RNA/DNA ratio (see 'RNA/DNA normalization' in the Methods of the accompanying manuscript by48 for each pathway across the 85 participants who gave 4 samples for transcriptomic analysis. We then calculated the coefficient of variation for these means and ranked the pathways by their coefficient of variation (Fig. 5b). The top eight resulting most stable pathways in terms of transcriptional regulation appear to be involved in cellular housekeeping, such as carbon metabolism (stably over-expressed) and amino acid synthesis (stably under-expressed).
Discussion
Here, we quantify faecal microbiome stability for prospective population-based cohort studies, with a particular focus on the relationship between metatranscriptomic features and the metagenome (both organismal composition and functional potential). See also ref. 48 for a discussion of the molecular and functional activity. As expected from single organisms and human studies, metatranscriptomic variation exceeded that of the metagenome. In addition, unlike the metagenome, metatranscriptomic variation within individuals over time was higher, to the point that it was comparable to variation between individuals. Taken in combination with our accompanying report describing a relatively stable 'core' metatranscriptome48, these findings, combined with previous reports21,23,24, suggest a model in which gut microbial community function mirrors that of differentiated human cell types: a subset of transcripts are prevalently active in the gut from varied microbes and conditions, while the remainder respond dynamically to intrinsic or external factors, such as diet.
In addition, we found the striking result that approximately 74% of the instability in the faecal metatranscriptome can be explained by instability in the faecal metagenome. These findings are similar to those from our pilot study, which showed that at a single time point, more than half of the variation in microbial community gene expression can be explained by metagenomic composition. This is best illustrated by our finding that some of the most highly stable (as well as highly abundant and prevalent) species, such as B. uniformis, were responsible for the dominant expression of a large number of stable genes. In contrast, Streptococcus salivarius—an unstable species that is probably transiting from the oral microbiome21—dominantly expresses many of the most variable genes. Similar findings were found in relation to the regulation of gene expression. Of the genes we identified as significantly differentially transcribed, nearly 80% were consistently over- or under-expressed. Many of these stably expressed pathways appeared to be essential for cellular function or involved in glycolysis, the synthesis of amino acids or the production of short-chain fatty acids. Taken together, it appears that much of the metatranscriptomic variation in stools over time might reflect changes in gene family copy number, driven by factors such as the proliferation of microbes, introduction of new species or host-induced shifts in community structures. In contrast, in the face of relative metagenomic stability, substantial deviations from a common genomic profile may be associated with shifts in disease state41,42.
Of course, we observed exceptions to these patterns, which may additionally explain some of the instability in the function of the faecal microbiome. Some species, such as S. wadsworthensis and Methanobrevibacter smithii, despite their stable presence, appear to be dominant contributors to the expression of unstable genes. Especially in the case of functionally unique taxa such as the archaea, this suggests specialized roles associated with targeted, highly variable activity over time. In addition, we found gene families with levels of RNA stability that were discordant with DNA stability, as well as several pathways that were differentially transcribed (up or down) only at a single time point. Given the large relative time scales of even our shortest gap (1–3 days) compared with transcriptional regulation (minutes), these changes are difficult to interpret without further data, but they may indicate pathways particularly susceptible to modulation by xenobiotics, diet or other targeted perturbations23,24. These outlying genes with high metagenomic stability but variable metatranscriptomic levels, or the stable microbes leading to the expression of unstable genes, are likely to play a critical role in our understanding of the impact of host behaviour on the faecal microbiome48.
Finally, for the purposes of prospective, longitudinally followed cohort studies, our study is consistent with previous work and indicates that many long-term features of the faecal microbiome are well-represented in a single self-collected stool sample4,5,17,21. These include strain composition, general taxonomic abundances and core metagenomic functional profiles. Furthermore, our results suggest that, as expected, metatranscriptomes vary over time and are appropriate markers for short-term exposures, and not necessarily long-term exposure–disease relationships. They do, however, remain somewhat personalized, and subsets of microbial transcripts are individually more stable. It remains to be determined which of these microbiome features, if any, represent the best biomarkers for diagnosis or prognosis of health conditions in diverse human populations.
Moving forward, the large-scale, population-based collection of stools is critical to exploring the factors that promote a stable core functional microbiome, yet yield unique species and metagenomic profiles. Prospective cohort studies characterizing the microbiome in relation to lifestyle data, such as dietary and medication information, have already begun to explore inter-individual variation and will be essential in mechanistically understanding the interactions between the faecal microbiome and the host in the context of health and disease17,43–45. Continuing to improve our understanding of the stability of the faecal microbiome function is critical not only in determining the ecological dynamics of the human microbiome25, but also for the promotion of human health.
Methods
Study population
The HPFS is an ongoing prospective cohort study that began in 1986 among 51,529 US male podiatrists, dentists, osteopathic physicians, veterinarians, pharmacists and optometrists aged 40 to 75 years at enrolment. In this study, participants returned questionnaires every 2–4 years with greater than 90% follow-up to provide information about lifestyle and dietary factors, medication use, and diagnoses of colorectal cancer and other diseases. The MLVS was established among 700 men aged 52–81 years (median 69 years) nested within HPFS who had completed the 2010 food frequency questionnaire and previously provided a blood sample. Men with coronary artery disease, stroke or transient ischaemic attack, cancer (except squamous or basal cell skin cancer) or major neurological disease (amyotrophic lateral sclerosis, Alzheimer’s, Parkinson’s, epilepsy or multiple sclerosis) were excluded. The 308 individuals sampled in this study were recruited into the MLVS starting in July 2012 and ending in July 2013. Participant recruitment and protocols were approved by the Harvard T. H. Chan School of Public Health Institutional Review Board #HSPH 22067-102. All participants provided informed consent for the study.
Sample size rationale
A tiered study design with metagenomic sequencing and a smaller batch of metatranscriptomic sequencing was used to determine both the composition and metabolic potential of the faecal microbiota. From the HMP findings of ~300 taxa per sample, our sample was estimated to yield relative abundances with standard deviation quartiles of (0.016, 0.027, 0.058) among rare taxa (present in < 50% of samples) and (0.081, 0.10, 0.15) among common taxa (arcsine square root-transformed relative abundance units).
Sample collection
Participants provided up to four stool samples—a set of samples from two consecutive bowel movements 24–72 h apart followed by collection of a second set of two such samples approximately 6 months later. Our collection protocol had previously been validated (see Supplementary Discussion)21. Briefly, participants deposited each bowel movement into a plastic commode collection bowl and then, using a specially designed spoon attached to a collection tube cap, faeces were scooped into a tube containing RNAlater. At each stool collection, participants documented the date and time of defecation as well as the Bristol score46. In addition, concurrent with each set of samples, participants completed a brief questionnaire collecting information regarding their recent use of antibiotics, gastric-acid-reducing medications, laxatives and probiotics, as well as other relevant exposures. At each time point, participants stored the specimen in the RNAlater fixative at ambient temperature until the specimen collected from the second bowel movement was produced. Each set of stool specimens was then placed in a special mailing kit and returned at ambient temperature by overnight mail. Upon receipt, the collection tubes were immediately placed into −80 °C freezers.
Sample handling and nucleic acid extraction
DNA and RNA extraction, processing and sequencing were as published21. Briefly, 100 mg stool aliquots were centrifuged at maximum speed to remove the excess of RNAlater. To the pellet, 110 μl of Tris-EDTA buffer with Proteinase K (Qiagen) and lysozyme (Sigma–Aldrich) (15 mg ml−1) were added with incubation on a laboratory shaker for 10 min. The mechanical lysis was performed by the addition of a 1.2 ml RLT buffer with 2-mercaptoethanol (Qiagen) and 1 ml of 0.1 mm glass beads (BioSpec Products), followed by bead beating for 3 min. The debris were removed by centrifugation and the supernatant was used in Qiagen AllPrep spin columns according to the manufacturer’s protocol (Qiagen). The only exception was in the preparation of the RNA: 60 μl DNase solution was added to buffer RW1 and incubated at room temperature for 15 min, followed by another wash with RW1 buffer. A NanoDrop 1000 (Thermo Fisher Scientific) was used to determine the DNA and RNA concentrations, quality and purity. SUPErase In was added to the aliquot before freezing. RNA was extracted and reverse-transcribed to complementary DNA only from stool samples spanning 6 months from participants who did not report the use of antibiotics within the past year (Fig. 1). RNA sequencing (RNA-Seq) libraries were depleted for ribosomal RNA using Ribo-Zero (Epicentre) as previously described21.
DNA was extracted from all 929 samples. RNA was extracted from 378 samples derived from a subset of 96 individuals who provided all four stool samples spanning the 6-month study interval and who did not report the use of antibiotics within the past year.
Library prep, sequencing and taxonomic/functional profiling
We used the Nextera XT DNA Library Preparation Kit for whole-genome sequencing. For RNA-Seq, we used RNAtag-Seq—a method to create an RNA-Seq library containing large numbers of RNA samples that are bar-coded and pooled before library construction47. This approach does not require poly(A) capture/enrichment or random priming.
Using Illumina HiSeq paired-end (2 × 101 nucleotides) shotgun sequencing, 929 metagenomes and 378 metatranscriptomes were obtained. Six RNA samples were not sequenced due to platform constraints. After sequencing and before biocomputing quality control, the sequencing depth (mean ±s.d.) for DNA was 3.8 ± 1.6 giganucleotides (Gnt). After quality control, it was 1.8 ± 0.7 Gnt. For RNA, the mean sequencing depth was 2.8 ± 2.4 Gnt before quality control and 1.2 ± 1.0 Gnt after. The quality control step included the removal of human sequences, quality trimming and depletion of duplicate reads using HMP protocols (HMP) with KneadData (http://huttenhower.sph.harvard.edu/kneaddata).
We then performed taxonomic profiling with MetaPhlAn2 (ref. 27) and functional profiling of genes and transcripts using HUMAnN2 (ref. 26). Outlier identification was performed and eight samples were removed because their ordination scores in either dimension (NMDS1 or NMDS2) were outside the range of the median ± 3 interquartile ranges. Notably, four of these samples were provided by a participant who reported a history of colectomy (Supplementary Fig. 1). Six RNA samples were removed because they did not have matching DNA-sequenced samples. We subsequently filtered all taxonomic features with a relative abundance less than 10−4 (0.01%) in greater than 10% of all samples. Similarly, for DNA and RNA, we filtered all gene families with a relative abundance less than 10−5 (0.001%) in greater than 10% of all samples.
Statistical analysis
Variability in the relative abundance values for community composition, metagenomes and metatranscriptomes within and between samples was determined by calculating the BC dissimilarity metric for each individual over time17. Short-term intervals, as previously described, were defined as consecutive bowel movements (24–72 h apart), whereas the intermediate interval was six months—the longest sampling interval in the study. Similarly, the Jaccard index was calculated within samples across time, where comparisons were drawn between the first sample and the remaining three5. With these metrics, higher values signify more variable features, whereas lower values indicate more stable features.
To estimate the reliability or reproducibility in measuring relative abundance values of features over time, we calculated ICCs. Relative abundance values were first arcsine square root-transformed to variance-stabilize data and better approximate normality and then, using linear mixed-effects models with restricted maximum likelihood estimation, we divided the between-person variance by the sum of the within- and between-person variances48. In a previous study, in simulations on proportion data, this approach performed similarly to generalized linear mixed model-based estimates of ICCs using multiplicative models fitted by penalized quasi-likelihood estimation and from additive models fitted by Markov chain Monte Carlo sampling48.
We explored whether several putative factors (bowel preparation use, antibiotic use, acid-lowering medication, and so on) were associated with within-person community, metagenome and metatranscriptome stability across the longest interval in our study using linear models. The Shannon index was calculated to be the mean of the within-sample diversity at time point 1 and time point 4. Acid-lowering medication use was determined by the use of proton-pump inhibitors or H2 receptor antagonists more than once a week for two months. Bowel preparation was determined as any use of bowel preparation in the two months before stool collection. Antibiotic use was defined as the oral or intravenous administration of antibiotics in the 12 months before stool collection. Those who never used antibiotics were set as the reference level. 'Any antibiotics' users were defined as participants using antibiotics at any point during the 12 months before the second stool collection and 'new antibiotics' users were defined as non-users of antibiotics before collection time point 1 who had initiated the use of antibiotics after the first collection and before the second. The Bristol category change was determined by categorization of participant-reported Bristol stool scores into hard (1–2), normal (3–4) and soft or liquid (5–7) categories and then calculating the difference between the categories over time46.
We examined associations between the baseline mean relative abundance and prevalence of each feature versus feature stability, and how metagenomic variability correlated with metatranscriptomic variability for each feature over time. As part of this, we investigated whether dominant expression of stable genes versus the expression of unstable genes varied according to species. For each transcript, a dominant contributing species was identified based on the maximal average contribution to each transcript. We then ranked the species according to the total number of genes that they dominantly expressed and (for presentation purposes) selected those contributing to 30 or more and overlaid them on a scatterplot in colour (grouped by genus). Examples of this process are presented in Figs. 3 and 4.
To test the stability of gene expression over time, we identified 740 pathways found in the 340 samples for which we had DNA and RNA data across all four time points. In total, 218 differentially transcribed pathways were defined as having a mean log10(RNA/DNA) significantly different from zero at each later time point using a linear model. We subjected these nominal P values to false discovery rate correction following the Benjamini–Hochberg method with α = 0.05. We then explored the consistency of the differential regulation of each pathway by exploring the overlap at each time point.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
Sequence data have been deposited in the Sequence Read Archive under BioProject ID: PRJNA354235. Data from the Health Professionals Follow-up Study, including metadata not included in the current manuscript but collected as a part of the MLVS, can be obtained through written application. As per standard controlled access procedure, applications to use HPFS resources will be reviewed by our External Collaborators Committee for scientific aims, evaluation of the fit of the data for the proposed methodology and verification that the proposed use meets the guidelines of the Ethics and Governance Framework and the consent that was provided by the participants. Investigators wishing to use HFPS or MLVS cohort data are asked to submit a brief (two pages) description of the proposed project ('letter of intent') to E.B.R. (HPFS Director; erimm@hsph.harvard.edu).
Supplementary Material
Acknowledgments
We thank the participants who graciously participated in this research, K. Stewart and G. Gupta at the Massachusetts General Hospital (MGH) who assisted with recruitment for the study, and S. Sawyer (Brigham and Women’s Hospital), M. Atar (MGH), C. Dulong (MGH and the Harvard T. H. Chan School of Public Health) and T. Poon (Broad Institut) for their assistance with project logistics, sample handling, nucleic acid extractions and sequencing. This work was supported by National Institutes of Health grants U54DE023798, UM1 CA167552, U01CA152904, R01 HL35464, R01CA202704 and K24DK098311, as well as by the Starr Cancer Consortium. A.T.C. was in part supported by the Stuart and Suzanne Steele MGH Research Scholars Program. J.I. was in part supported by the Nebraska Tobacco Settlement Biomedical Research Development Fund. R.S.M. was supported by a Howard Hughes Medical Institute Medical Research Fellowship and an AGA–Eli and Edythe Broad Student Research Fellowship.
Footnotes
Author contributions
J.I., A.T.C. and C.H. designed and managed the study. R.S.M., D.A.D., K.L.I., G.T.B., C.D., E.B.R. and J.I. collected the samples and generated the data. R.S.M., G.S.A.-A., D.A.D., J.L.-P., A.S., P.L., A.D.J., H.K., G.T.B., M.S., L.H.N. and H.M. analysed the data. R.S.M., G.S.A.-A., D.A.D., K.L.I., J.I., C.H. and A.T.C. prepared and wrote the manuscript.
Competing interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41564-017-0096-0.
References
- 1.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 3.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv7. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;41:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flores GE, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffery IB, Lynch DB, O’Toole PW. Composition and temporal stability of the gut microbiota in older persons. ISME J. 2016;10:170–182. doi: 10.1038/ismej.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David LA, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajilić-Stojanović M, Heilig HGHJ, Tims S, Zoetendal EG, De Vos WM. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol. 2013;15:1146–1159. doi: 10.1111/1462-2920.12023. [DOI] [PubMed] [Google Scholar]
- 11.Zoetendal EG, Akkermans ADL, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzosa EA, et al. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA. 2015;112:E2930–E2938. doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubos RJ, Schaedler RW. Reversible changes in the susceptibility of mice to bacterial infections. J Exp Med. 1956;104:53–65. doi: 10.1084/jem.104.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaedler RW, Dubos RJ. Reversible changes in the susceptibility of mice to bacterial infections. J Exp Med. 1956;104:67–84. doi: 10.1084/jem.104.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schloissnig S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;93:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium THMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;86:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 21.Franzosa EA, et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci USA. 2014;111:E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNulty NP, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abu-Ali GS, et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat Microbiol. 2018 doi: 10.1038/s41564-017-0084-4. https://doi.org/10.1038/s41561-017-0084-4. [DOI] [PMC free article] [PubMed]
- 26.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong DT, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841–852. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahony J, McDonnell B, Casey E, van Sinderen D. Phage–host interactions of cheese-making lactic acid bacteria. Annu Rev Food Sci Technol. 2016;7:267–285. doi: 10.1146/annurev-food-041715-033322. [DOI] [PubMed] [Google Scholar]
- 30.Kim DH, Konishi L, Kobashi K. Purification, characterization and reaction mechanism of novel arylsulfotransferase obtained from an anaerobic bacterium of human intestine. Biochim Biophys Acta. 1986;872:33–41. doi: 10.1016/0167-4838(86)90144-5. [DOI] [PubMed] [Google Scholar]
- 31.Malojćić G, et al. A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2008;105:19217–19222. doi: 10.1073/pnas.0806997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hehemann JH, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins AH, Schyns G, Potot S, Sun G, Begley TP. A new thiamin salvage pathway. Nat Chem Biol. 2007;3:492–497. doi: 10.1038/nchembio.2007.13. [DOI] [PubMed] [Google Scholar]
- 34.Bussiere DE, et al. Crystal structure of ErmC’, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry. 1998;37:7103–7112. doi: 10.1021/bi973113c. [DOI] [PubMed] [Google Scholar]
- 35.Jalanka J, et al. Effects of bowel cleansing on the intestinal microbiota. Gut. 2015;64:1562–1568. doi: 10.1136/gutjnl-2014-307240. [DOI] [PubMed] [Google Scholar]
- 36.O’Brien CL, Allison GE, Grimpen F, Pavli P. Impact of colonoscopy bowel preparation on intestinal microbiota. PLoS ONE. 2013;8:e62815. doi: 10.1371/journal.pone.0062815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014;124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakobsson HE, et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCann KS. The diversity–stability debate. Nature. 2000;405:228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- 40.Schindler DE, et al. Population diversity and the portfolio effect in an exploited species. Nature. 2010;465:609–612. doi: 10.1038/nature09060. [DOI] [PubMed] [Google Scholar]
- 41.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhernakova A, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falony G, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 45.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. 1997;32:920–924. doi: 10.3109/00365529709011203. [DOI] [PubMed] [Google Scholar]
- 47.Shishkin AA, et al. Simultaneous generation of many RNA-Seq libraries in a single reaction. Nat Methods. 2015;12:323–325. doi: 10.1038/nmeth.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakagawa S, Schielzeth H. Repeatability for Gaussian and non-Gaussian data: a practical guide for biologists. Biol Rev. 2010;85:935–956. doi: 10.1111/j.1469-185X.2010.00141.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in the Sequence Read Archive under BioProject ID: PRJNA354235. Data from the Health Professionals Follow-up Study, including metadata not included in the current manuscript but collected as a part of the MLVS, can be obtained through written application. As per standard controlled access procedure, applications to use HPFS resources will be reviewed by our External Collaborators Committee for scientific aims, evaluation of the fit of the data for the proposed methodology and verification that the proposed use meets the guidelines of the Ethics and Governance Framework and the consent that was provided by the participants. Investigators wishing to use HFPS or MLVS cohort data are asked to submit a brief (two pages) description of the proposed project ('letter of intent') to E.B.R. (HPFS Director; erimm@hsph.harvard.edu).