Abstract
In well over half of the individuals with rare disease who undergo clinical or research next-generation sequencing, the responsible gene cannot be determined. Some reasons for this relatively low yield include unappreciated phenotypic heterogeneity; locus heterogeneity; somatic and germline mosaicism; variants of uncertain functional significance; technically inaccessible areas of the genome, incorrect mode of inheritance investigated; and, inadequate communication between clinicians and basic scientists with knowledge of particular genes, proteins or biological systems. To facilitate such communication and improve the search for patients or model organisms with similar phenotypes and variants in specific candidate genes we have developed the Matchmaker Exchange (MME). MME was created to establish a federated network connecting databases of genomic and phenotypic data using a common application programming interface (API). To date, seven databases can exchange data using the API (GeneMatcher, PhenomeCentral, DECIPHER, MyGene2, matchbox, Australian Genomics Health Alliance Patient Archive, and Monarch Initiative), the later included for model organism matching. This protocol guides usage of the MME for rare disease gene discovery.
Keywords: Matchmaker Exchange, candidate genes, GeneMatcher, PhenomeCentral, DECIPHER, MyGene2, matchbox, Australian Genomics Health Alliance Patient Archive, Monarch Initiative
INTRODUCTION
In the last decade improvements in DNA sequencing technology have led to the use of next-generation sequencing (NGS) methods in many areas of science, including discovery research and clinical testing. Moreover, the use of exome sequencing (ES) has allowed the discovery of more than a thousand disease genes associated with rare Mendelian disorders (Chong et al., 2015; Boycott et al, 2017). Despite these advances, in more than half of the individuals with a rare Mendelian phenotype who undergo clinical or research ES, the responsible gene cannot be identified (Yang et al., 2014; Chong et al., 2015; Retterer et al., 2015). Some of these cases harbor variants in one or more novel candidate genes and the necessary evidence for causally implicating the disease gene often comes down to identifying other affected individuals with similar phenotype and functionally impactful variants in the same candidate gene. Many databases have addressed this need by developing platforms that use genomic and phenotypic matching algorithms to identify cases with similar phenotypes and variants in the same gene (Washington et al., 2009; Gonzalez et al., 2012, Swaminathan et al., 2012, Gonzalez et al., 2013, Robinson et al., 2014; Zemojtel et al., 2014; Buske et al., 2015a; Lancaster et al., 2015; Sobreira et al., 2015a). MME was created to establish a federated network connecting these databases of genomic and phenotypic data using a common application programming interface (API) and allowing data exchange among them. Here we provide two protocols including 1. Using an existing matchmaker exchange database to search for a match, and 2. Connecting a database to the MME. In the commentary section we describe the consent requirements for MME and how patients can use the MME.
BASIC PROTOCOL 1
USING AN EXISTING MATCHMAKER DATABASE TO SEARCH FOR A MATCH
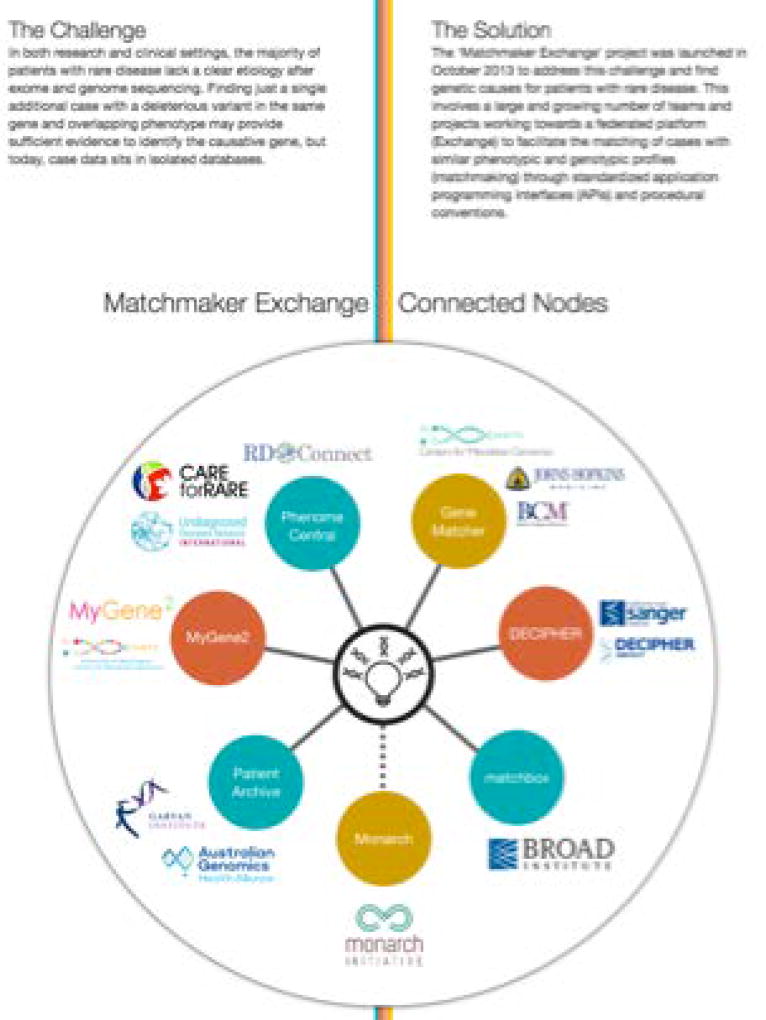
The main MME Web site is accessible via the Internet at (http://www.matchmakerexchange.org/) (Figure 1). Its home page contains information on how to get started with matchmaking, available resources needed to support matchmaking, who the participants are and how to contact us (Figure 1). It also contains an overview of MME (Figure 2). In the HOW TO GET STARTED tab in the home page users can identify themselves as an individual with a case to match, an individual with a dataset to deposit or an individual with a database to connect to the MME.
Figure 1.
Matchmaker Exchange Homepage.
Figure 2.
Matchmaker exchange overview.
To date, seven databases can Exchange data using the API: GeneMatcher (https://genematcher.org/), PhenomeCentral (https://phenomecentral.org/), DECIPHER (https://decipher.sanger.ac.uk/), MyGene2 (https://www.mygene2.org/), matchbox (https://seqr.broadinstitute.org/), Australian Genomics Health Alliance Patient Archive (https://mme.australiangenomics.org.au/#/home), and Monarch Initiative (https://monarchinitiative.org/) (Figure 2). To initiate a search the users should first identify themselves as clinicians, laboratories or patients (Figure 3). In the HOW TO GET STARTED tab in the home page users identified as clinicians or laboratories can find a page with detailed description of each database connected to MME including: 1 - The database location and types of data stored by each database (Figure 4); 2 - Parameters used for matching and score output (Figure 5); 3 - MME connections (Figure 6); 4 - Types of users (Figure 7); and 5 - MME matching and notification protocols. After the review of this information the user can choose what database to use as their primary point of entry into the MME, which is where they will deposit their data and initiate queries of the other databases. The queries allow a gene symbol with or without a specific variant and/or variant attributes (e.g. de novo, loss of function, etc.) a disease name or phenotypic features, to be sent as a query to get a match notification about any similar cases. Matching algorithms are defined by the matchmaker services and have evolved over time.
Figure 3.
How to get started.
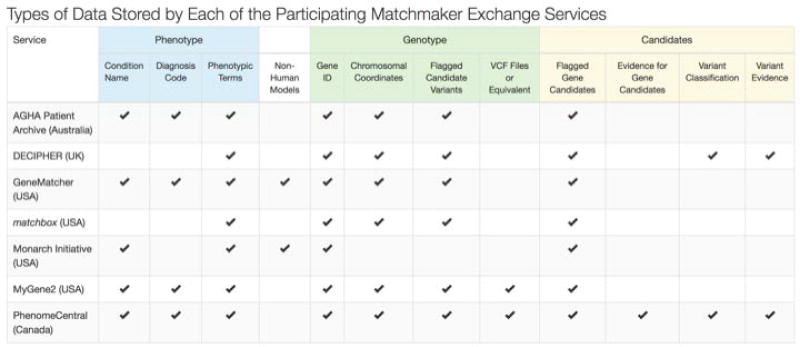
Figure 4.
Database location and types of data stored by each database.
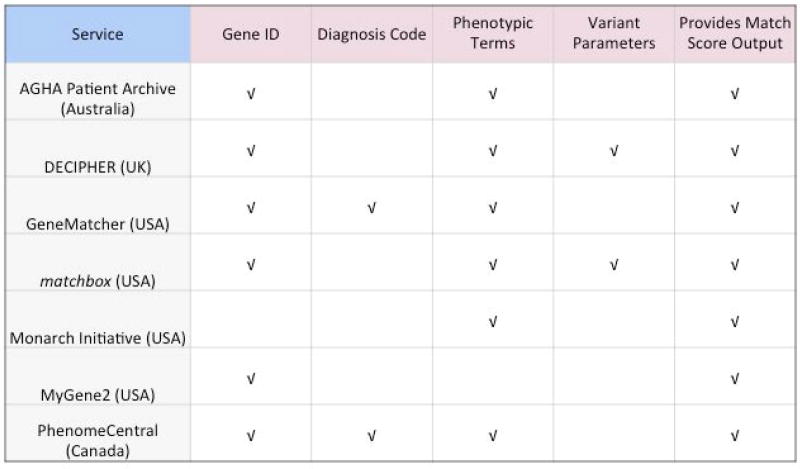
Figure 5.
Parameters used for matching and score output.
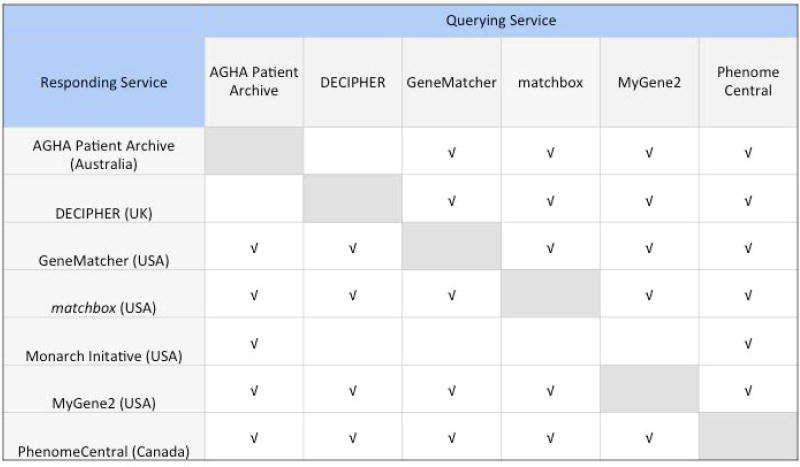
Figure 6.
Matchmaker exchange connections.
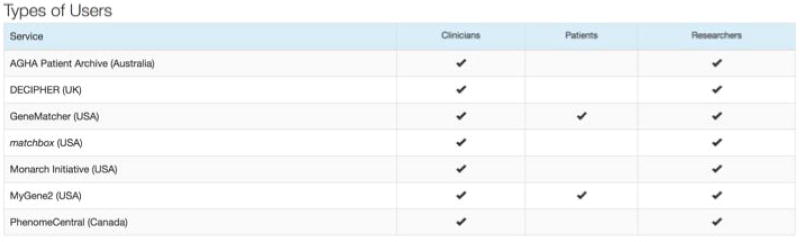
Figure 7.
Types of users allowed in each Matchmaker exchange database.
Necessary Resources
The minimal resources required for matchmaking is an internet connected device and a gene candidate symbol, a set of Human Phenotype Ontology (HPO) terms to describe the patient’s phenotype, or both.
From GeneMatcher to other MME databases
Create a GeneMatcher account at https://genematcher.org/.
Create a submission at “Create a new submission” in the home page (Figure 8).
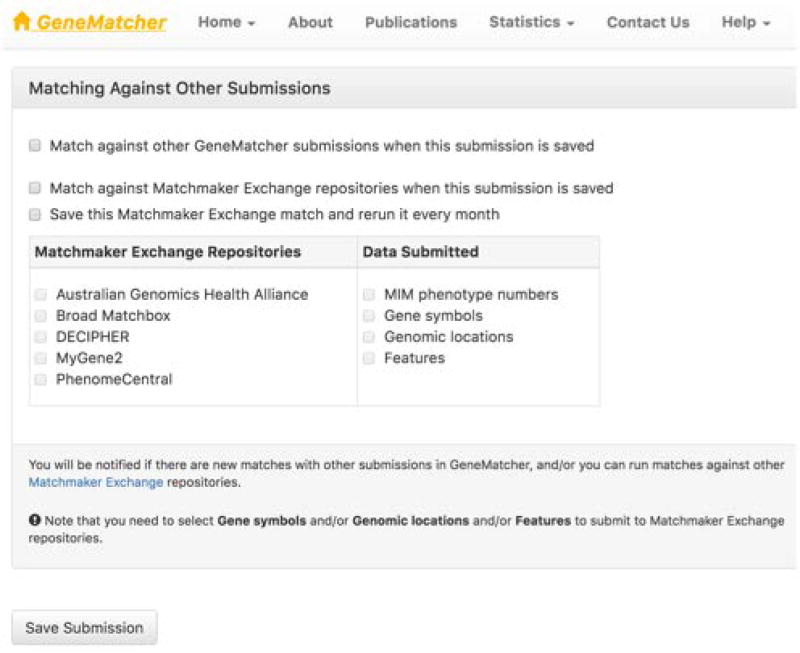
- Evaluate the options and check the relevant boxes before fitting the “Save Submission” button (Figure 9):
-
Fill out the Submission Identification information, including your name, institution, and email address, as well as a unique identifier to tag this entry.
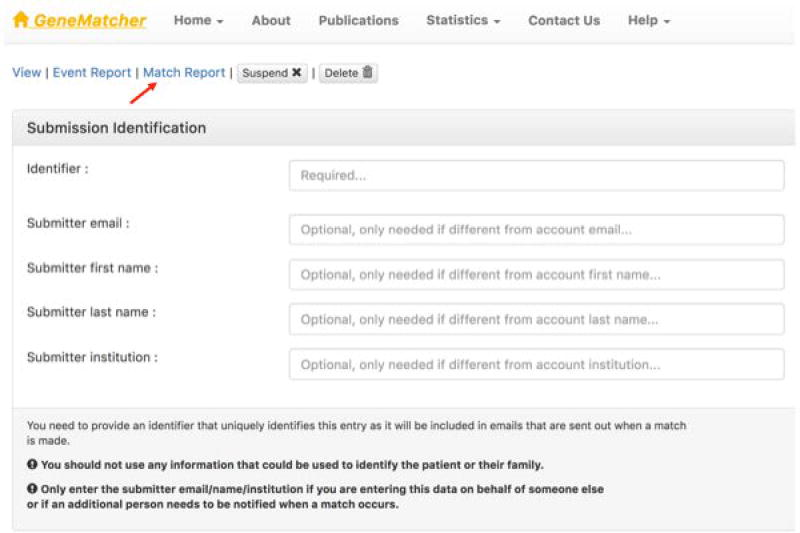
Users will be notified of all matches via email and matches are also displayed at the Match Report tab (Figure 10).
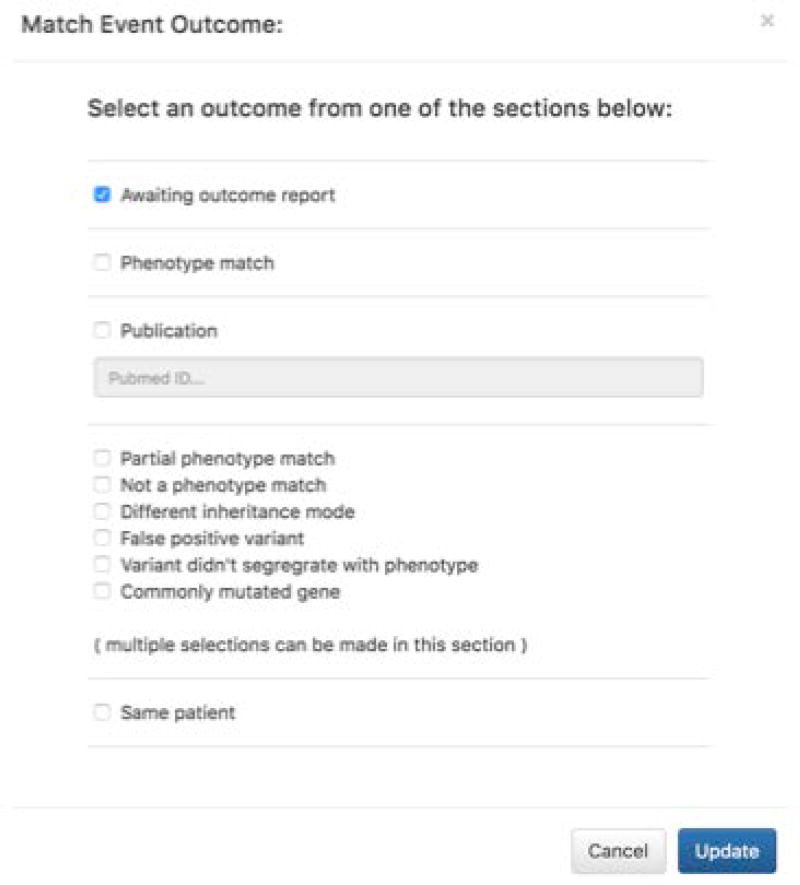
In the Match Report page, record the outcome of the match follow up as shown in Figure 11.
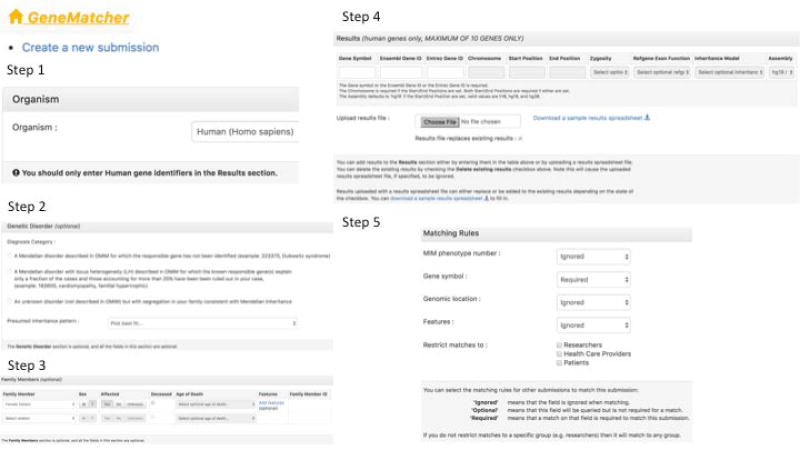
Figure 8.
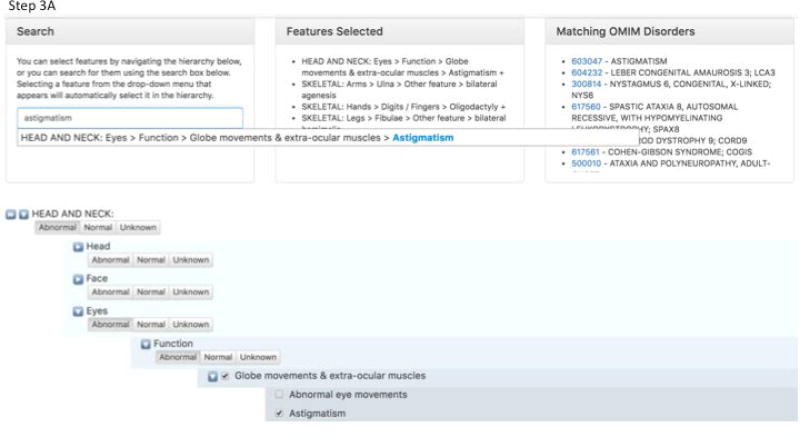
How to create a general submission in GeneMatcher. Step 1 - Identify the organism being investigated. Step 2 - Classify the phenotype being investigated. Step 3 - Give pedigree information and add the features of the affected family members. Step 4 - Add gene name information, and variant information. Step 5 - Choose what data to match on.
Step 3A - By clicking at “Add features” the user is taken to a page were features can be added.
Figure 9.
How to send data from GeneMatcher to other Matchmaker exchange databases.
Figure 10.
In GeneMatcher, matches are displayed at the Match Report tab.
Figure 11.
Recording the outcome of a match follow up in GeneMatcher.
From PhenomeCentral (PC) to other MME databases
Create a PhenomeCentral account at https://phenomecentral.org/
Create a patient at “Create… New Patient” in the homepage.
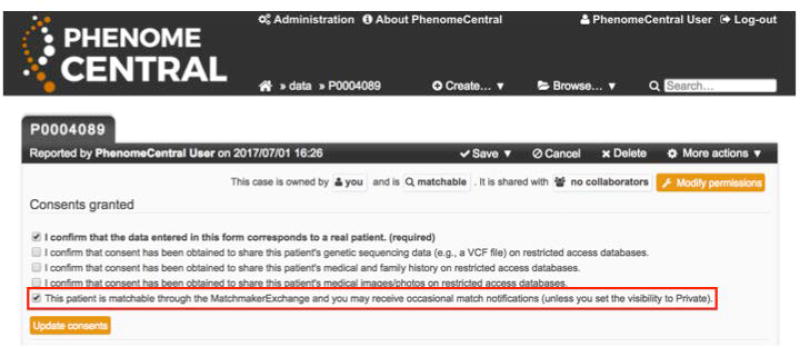
Grant consent to sharing the patient over the MME by checking the corresponding box and saving the patient record (Figure 12); this allows the patient to be matched against by other databases.
- Queries are initiated in several ways:
-
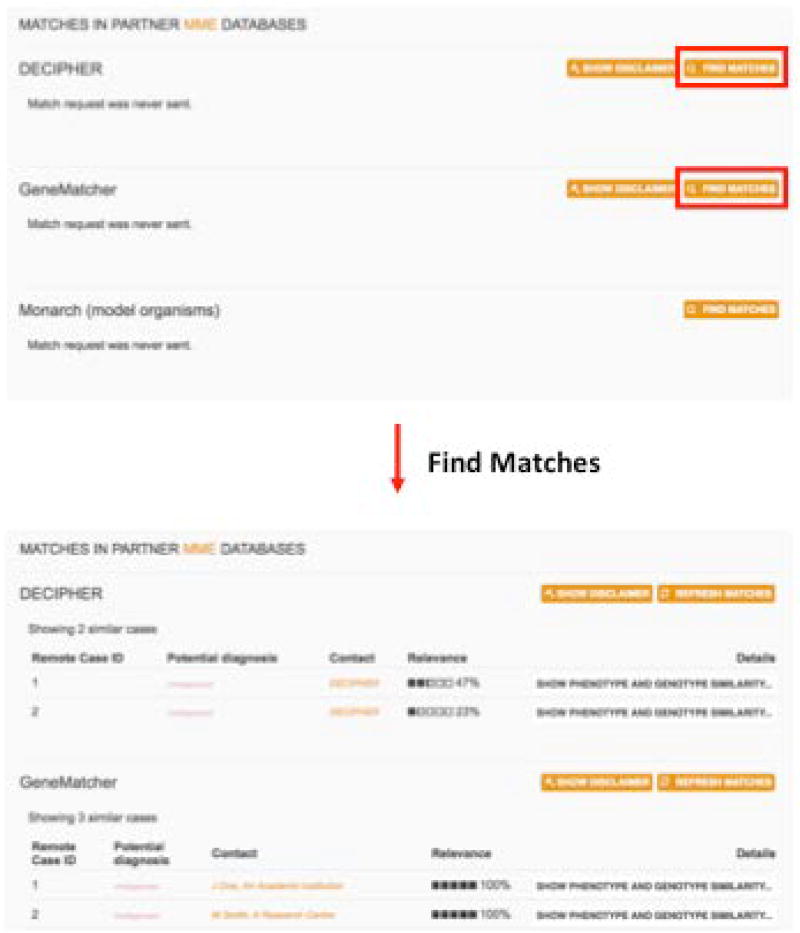
4aUsers can initiate queries and view matches by going to a patient record, scrolling to the “Matches in partner MME databases” section, and clicking “Find matches” next to the desired database (or “Refresh matches” to send a new request) (Figure 13).
-
4bQueries are periodically re-sent for all patients to all connected databases, and a genetic counsellor reviews the quality of the matches and decides whether to send a notification email to the PhenomeCentral user.
-
4a
Figure 12.
PhenomeCentral users can enable matching over the Matchmaker exchange by checking the corresponding consent box when creating or editing a patient record.
Figure 13.
PhenomeCentral users can find and view matches in other Matchmaker exchange databases by viewing a patient record, scrolling to the “Matches in partner Matchmaker exchange databases” section, and clicking “Find matches” next to the desired database (or “Refresh matches” to send a new request). Matches can then be reviewed and expanded to see a phenotype and genotype comparison. If the match is promising, the Contact link can be clicked to send an email to the other researcher or view a webpage with more information.
From DECIPHER to other MME databases
-
Create a DECIPHER account at https://decipher.sanger.ac.uk.
Membership of DECIPHER is open to academic centers of clinical genetics and research institutions that intend to deposit and share consented patient data. Users can either join an established project or apply for a new project; criteria for joining DECIPHER can be viewed here: https://decipher.sanger.ac.uk/join#info.
Create a submission by creating a patient record and adding genotype and phenotype information. To patient match via MME, the patient record must be marked as consented for open sharing (requires patient consent) and must include at least one open-access sequence variant.
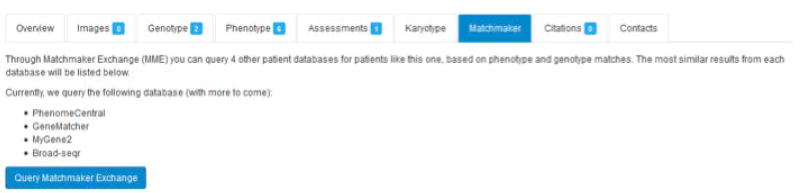
In the patient record, click on the “Matchmaker” tab and then click on the “Query Matchmaker Exchange” button (Figure 14).
By clicking on “Query Matchmaker Exchange”, connected databases will be queried for patient records that share similarity with the linked anonymous consented patient record, and the results will be displayed on the Matchmaker tab in the patient record.
Figure 14.
Matchmaker exchange tab in a patient record in the DECIPHER database.
From MyGene2 to other MME databases
Create a clinician or researcher MyGene2 account at https://www.mygene2.org
Create a submission at “Create new case.”
If the case created is not classified as a known gene for a known phenotype, the data will be automatically submitted to other MME databases.
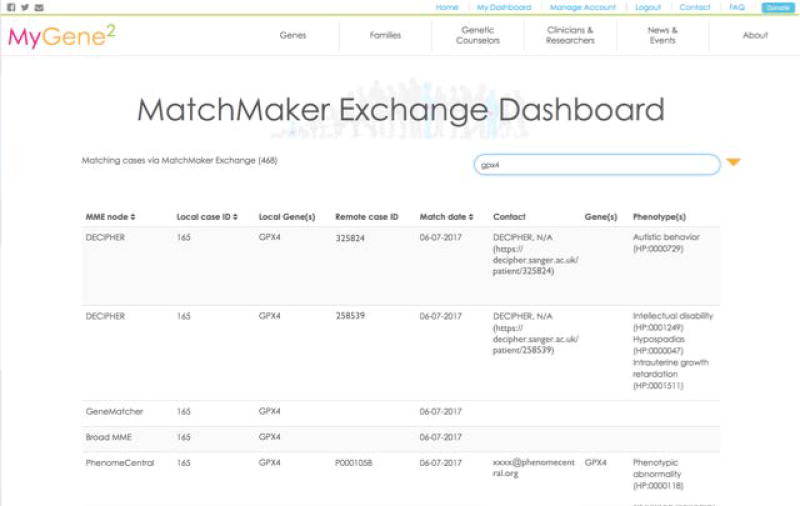
The users will be notified of all matches via email and matches are also summarized via the Matchmaker Exchange Dashboard (Figure 15).
Figure 15.
Matchmaker Exchange Dashboard in the MyGene2 database.
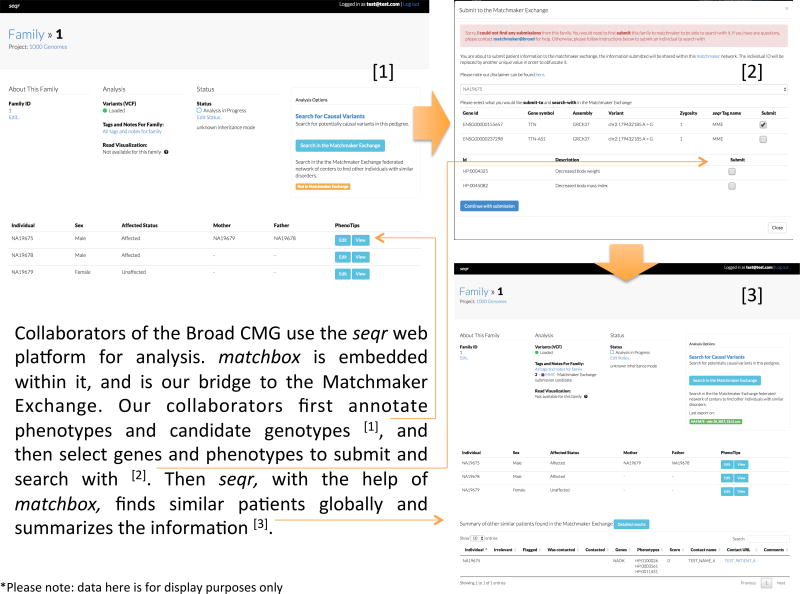
From matchbox to other MME databases
The use of matchbox at the Broad Institute is currently only supported for users storing their rare disease genomic data in seqr (https://seqr.broadinstitute.org/) at the Broad Institute (Figure 16). To collaborate with the Broad Center for Mendelian Genomics and use seqr and matchbox software tools, contact cmgbroadinstitute.org.
Figure 16.
Collaborators of the Broad CMG use the seqr web platform for analysis. matchbox is embedded within it, and is our bridge to the Matchmaker Exchange. Our collaborators first annotate phenotypes and candidate genotypes [1], and then select genes and phenotypes to submit and search with [2]. Then seqr, with the help of matchbox, finds similar patients globally and summarizes the information [3].
From Australian Genomics Health Alliance Patient Archive to other MME databases
Create an account with the AGHA Patient Archive at https://mme.australiangenomics.org.au/
Create a new patient at the dashboard.
Add the phenotype and genotype profile of the proband.
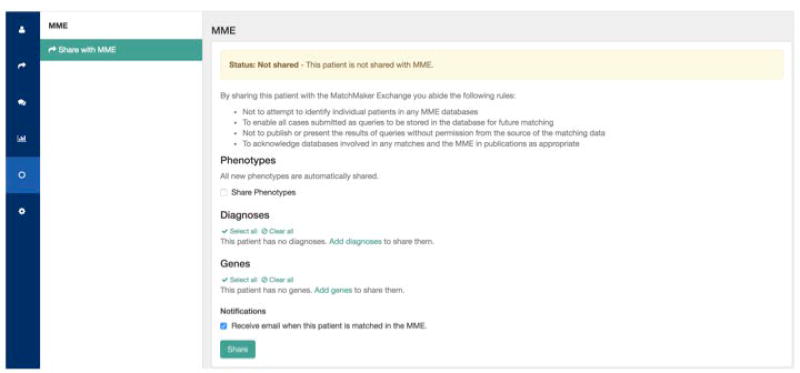
Share the patient with the MME; users have the choice of sharing phenotypes, genes and diagnoses (Figure 17), as well as to opt in or out for being notified on external matches.
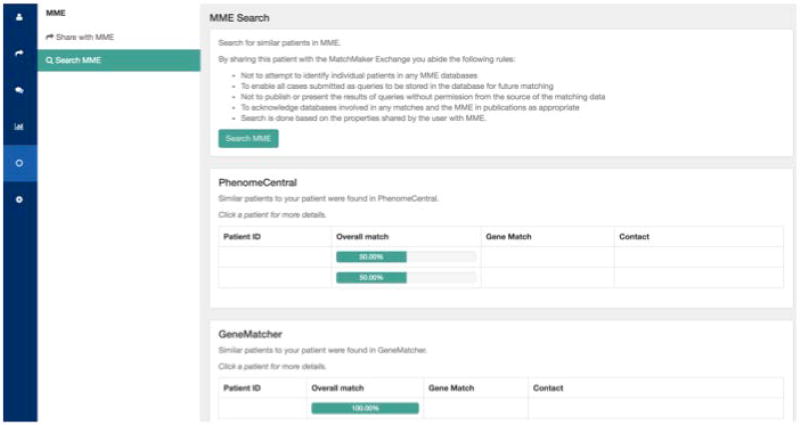
Once the patient has been shared, the user can query the other MME databases; all databases will be queried at once.
Results will be displayed on demand (Figure 18).
Subject to the notification setting chosen when sharing the patient, the users will be notified by email when external matches occur.
Figure 17.
How to send data from Australian Genomics Health Alliance Patient Archive to other Matchmaker exchange databases.
Figure 18.
Display of Matchmaker exchange matches in the Australian Genomics Health Alliance Patient Archive.
From Monarch Initiative to other MME databases
The Monarch Initiative does not send queries to the other MME databases. It receives queries and returns genotype and HPO phenotype based-matches with known human diseases (e.g. ClinVar, Orphanet, etc) and animal models to assist in evidence gathering.
BASIC PROTOCOL 2
CONNECTING A DATABASE TO MATCHMAKER EXCHANGE
If the user has a database to connect to the MME he/she will find the requirements and the contact email (apimatchmakerexchange.org) by clicking at “I have a database to connect” (Figure 3).
This protocol will walk you through the main steps to connect an existing genomic and/or phenotypic database to MME allowing the exchange of data with the other MME databases.
Necessary Resources
The only resource required is an internet-connected computer.
Application programming interface (API)
The databases of the MME communicate with each other using a common API that exchanges data in JSON. This API is documented and maintained collaboratively at the MME GitHub repository (https://github.com/ga4gh/mme-apis) by the technical working group.
For a new database to join the MME, there are two main technical steps that must be undertaken:
The database must first implement the MME API. At a high level, connecting a database to the MME involves exposing an API endpoint “/match” that allows another MME database to query your database for similar patients. The queried database replies with a list of other similar patients that it hosts. The algorithm that decides similarity is defined by the database being queried. A database can either develop the necessary software to implement the API themselves or use one of the existing open-source implementations that simplify the process of connecting a database to the MME. There are currently two portable, open-source solutions available: the MME reference server (https://github.com/MatchmakerExchange/reference-server) and the matchbox system (https://github.com/macarthur-lab/matchbox). Other open-source implementations of the MME API can be found here: https://github.com/ga4gh/mme-apis/wiki/Implementations.
Once the API is implemented, the database must exchange some basic information with the other databases it wants to connect to. MME is a peer-to-peer federated network, where each database connects to two or more other databases within the network. Because of the sensitivity of the information being shared, most MME databases require requests from other databases to be authenticated with a pre-shared key (PSK). These keys are usually shared via encrypted email messages. This process of connecting to other databases can be time consuming, but it assures each database full control over who it shares data with.
COMMENTARY
Background Information
The main evidence to identify novel disease genes associated with rare Mendelian phenotypes is the identification of other affected individuals with similar phenotype and variants in the candidate genes. Historically, clinicians and researches have been identifying these additional cases by personal communication, presentation of abstracts in scientific meetings, publication of case reports, etc. More recently, the rapid development of informatics tools has made this process faster and more efficient. Projects such GeneMatcher, PhenomeCentral, DECIPHER, MyGene2, matchbox, Australian Genomics Health Alliance Patient Archive, Monarch Initiative and others have allowed the discovery of many novel disease genes by connecting clinicians, researchers and patients from all around the world.
MME was created in October 2013 to facilitate the connection among these databases allowing the users to store their data in one database and query the others without the need to create multiple accounts and store the same data in more than one database.
Value of federation
A federated model was chosen to support the MME. The federated architecture provides several benefits to other, more centralized approaches such as:
Data locality — each database hosts, maintains ownership and is responsible for the safekeeping of their own data. It also enables them to comply with any national or regional restrictions on data storage and sharing;
Data privacy — data security and privacy management are not entirely in the hands of one, centralized database. Each database has full control over which other databases it wishes to connect to, and what information is exchanged with each of these other databases;
Network robustness — because there is no central server, there is no single point of failure for the MME; if one database goes offline, all connections between other databases will be unaffected;
Algorithm development and data maintenance — each database has full autonomy to develop and evolve their matching algorithms independently. A federated system also makes it easier to support updates to patient phenotype and updated genomic interpretations over time.
The federated approach allows each MME database to maintain its autonomy and primary purpose, while contributing valuable data to the MME and the genomics community. Users no longer need to submit the same datasets into multiple databases to find matches, and they will have more options for databases in which to store data, including databases in their own jurisdiction if certain regulations prohibit data from leaving a region. Also, users may decide to put some cases into one database and other cases into another database depending on the focus (clinical and/or data types) of each database (Philippakis et al., 2015).
Consent requirements
The MME consent policy (Informed Consent Policy) can be found at the web page under OUR RESOURCE LIBRARY (Figure 3). This approach was developed in collaboration with the Global Alliance for Genomics and Health (GA4GH) Regulatory and Ethics Working Group (REWG) members and their Consent Task Team and is detailed in Dyke et al 2017.
The approach has two levels depending on the type of data being deposited in the system as well as two data types (clinical vs research). This allows patient consent needs to be defined based on the probability of occurrence and seriousness of potential harm of re-identification in the matchmaking process as well as the expectations of the patient/study subject. For level 1, when only a candidate gene +/− non-sensitive structured phenotype data are provided, written patient consent is not required if matching is submitted in support of clinical care (e.g. a diagnostic clinical exome report with a reported novel candidate gene) given that matching efforts are consistent with clinical care but may be required for research subjects. For level 2, when variant-level data and/or sensitive phenotypic data is provided, written consent is typically required because the likelihood of re-identification and/or the potential for harm is higher.
Patient use of MME
Although all matchmaker services support the entry of patient data by clinicians and researchers, two MME databases support direct use of their databases by patients and their families. These include GeneMatcher and MyGene2, both of which currently support patient-led matching within their databases (though now not yet across the MME platform) to help identify other individuals with similar profiles. Patients or family members should be aware that a health care provider (including the clinical laboratory that performed the test) may have already created a submission with the same novel disease gene candidate which could result in a match based on the same patient within a given database.
As of today, the other MME databases do not yet accept queries directly from patients. Patients or family members wishing to query all MME databases should contact their healthcare provider who ordered their testing, to create a submission to allow query across the entire MME platform.
Critical Parameters
Optimizing for success in matchmaking
To enable the most successful matches and facilitate efficient follow-up communication after match notifications have been sent by email, users are encouraged to submit detailed phenotypic and suspected inheritance information at the initial point of entry of their candidate gene, as well as restrict genes to highly likely candidates. For example, usually only a single or small number of candidate genes are submitted per patient/research subject such as those candidate genes in which a variant arose de novo or has biallelic variants absent from (or extremely rare in) general population databases or has some functional evidence supporting the gene’s potential role in disease.
By providing phenotypic data during submission, matches will be ranked with higher scores based on matching algorithms that incorporate phenotype. And finally, a data depositor may be more likely to respond to a match notification email if they can immediately see a phenotypic and inheritance match provided in the notification email.
Troubleshooting
If troubleshooting on how to connect a database to MME, the user should contact us at api@matchmakerexchange.org.
If troubleshooting on how to find a match in the MME databases, the user should contact the database where the data are primarily stored.
Anticipated Results
After the submission of a query to one or more of the MME databases the users will receive an email informing them of the presence or absence of a match in the queried database(s).
If there is currently no match in any of the databases, the user will be notified and the submission will only be stored in the database from which the query originated, not the external databases queried. In the future, if the users would like to repeat the query they would need to send the submission again. In several databases, the users have the option to automatically resend the data from their submissions to the MME on a periodic basis.
If there is an instant match, all data depositors involved in the match will be notified according to notification procedures maintained by each database and described in detail here: http://www.matchmakerexchange.org/assets/files/MME_Matching_Protocols_and_Notification_Method_2017-04-03.pdf
The email about the match typically includes the information that matched and the email contact of the users involved. Sometimes a score is included to define the strength of a match or the relevant rank if multiple cases match. The users contact each other to exchange additional detailed information about the gene, variant, mode of inheritance, phenotype, etc.
Time Considerations
When a MME match occurs, the submitters will automatically and immediately (in few seconds/minutes) receive an email notification.
Acknowledgments
The authors acknowledge the contributions of the entire MME as well as GA4GH, IRDiRC and the Centers for Mendelian Genomics and Care4Rare Canada in advancing this collaborative initiative.
Contract grant sponsors: NIH (grants 1U54HG006542, UM1HG008900, 5R24OD011883, U54HG006493), Genome Canada (CAN-SHARE)
Footnotes
- Matchmaker Exchange - http://www.matchmakerexchange.org/
- GeneMatcher - https://genematcher.org/
- PhenomeCentral - https://phenomecentral.org/
- DECIPHER - https://decipher.sanger.ac.uk/
- MyGene2 - https://www.mygene2.org/
- Matchbox - https://seqr.broadinstitute.org/
- Australian Genomics Health Alliance Patient Archive - https://mme.australiangenomics.org.au/#/home
- Monarch Initiative - https://monarchinitiative.org/
Contributor Information
Nara L M Sobreira, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University, 733 North Broadway Street Suite 569 Baltimore, Maryland 21205, Phone: 433-287-1104, nsobrei2@jhmi.edu.
Harindra Arachchi, The Broad Institute of Harvard and MIT, Cambridge, MA, 02139 USA, harindra@broadinstitute.org.
Orion Buske, Centre for Computational Medicine, Hospital for Sick Children, Toronto, ON, M5G 0A4, Canada, Phone: +1(416)813-8899, buske@cs.toronto.edu.
Jessica X. Chong, Department of Pediatrics, University of Washington, Seattle, WA 98195, USA, Phone: 206-221-4075, Fax: 206-221-3795, jxchong@uw.edu
Ben Hutton, Wellcome Trust Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridgeshire, CB10 1SA, UK, Phone: 01223 834244, ben.hutton@sanger.ac.uk.
Julia Foreman, Wellcome Trust Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SA, UK, Phone: 01223 492392, jf11@sanger.ac.uk.
François Schiettecatte, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University, fschiettecatte@gmail.com.
Tudor Groza, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, The Kinghorn Cancer Centre, Level 6, 370 Victoria Street, Darlinghurst, NSW 2010, Australia; St Vincent's Clinical School, Faculty of Medicine, University of New South Wales, Australia, Phone: + 61 (0)2 9355 5717, t.groza@garvan.org.au.
Julius O.B. Jacobsen, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London, EC1M 6BQ, UK, j.jacobsen@qmul.ac.uk
Melissa Haendel, Director of the Ontology Development Group, Associate Professor, OHSU Library, Department of Medical Informatics & Clinical Epidemiology, Phone: 503-407-5970, haendel@ohsu.edu.
Kym M Boycott, Children’s Hospital of Eastern Ontario Research Institute, University of Ottawa, 401 Smyth Rd, Ottawa, Ontario, K0A 2P0, Canada, Phone:1-613-737-7600 (4139), kboycott@cheo.on.ca.
Ada Hamosh, Dr. Frank V. Sutland Professor, McKusick-Nathans Institute of Genetic Medicine (IGM), Clinical Director, IGM. Scientific Director, OMIM. Johns Hopkins University. Blalock 1007 600 N. Wolfe St Baltimore, MD 21287-4922, Phone: 410-614-3313, Fax: 410-614-9246, ahamosh@jhmi.edu.
Heidi L. Rehm, The Broad Institute of Harvard and MIT, Cambridge, MA, 02139 USA, Phone: 617-768-8291, hrehm@broadinstitute.org
LITERATURE CITED
- Boycott KM, Rath A, Chong JX, Hartley T, Alkuraya FS, Baynam G, Brookes AJ, Brudno M, Carracedo A, den Dunnen JT, Dyke SOM, Estivill X, Goldblatt J, Gonthier C, Groft SC, Gut I, Hamosh A, Hieter P, Höhn S, Hurles ME, Kaufmann P, Knoppers BM, Krischer JP, Macek M, Jr, Matthijs G, Olry A, Parker S, Paschall J, Philippakis AA, Rehm HL, Robinson PN, Sham PC, Stefanov R, Taruscio D, Unni D, Vanstone MR, Zhang F, Brunner H, Bamshad MJ, Lochmüller H. International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. Am J Hum Genet. 2017 May 4;100(5):695–705. doi: 10.1016/j.ajhg.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske OJ, Girdea M, Dumitriu S, Gallinger B, Hartley T, Trang H, Misyura A, Friedman T, Beaulieu C, Bone WP, Links AE, Washington NL, et al. PhenomeCentral: a Portal for Phenotypic and Genotypic Matchmaking of Patients with Rare Genetic Diseases. Hum Mutat. 2015 Oct;36(10):931–40. doi: 10.1002/humu.22851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, et al. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MA, Van Booven D, Hulme W, Ulloa RH, Lebrigio RF, Osterloh J, Logan M, Freeman M, Zuchner S. Whole Genome Sequencing and a New Bioinformatics Platform Allow for Rapid Gene Identification in D. melanogaster EMS Screens. Biology. 2012;1(3):766–77. doi: 10.3390/biology1030766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MA, Lebrigio RFA, Van Booven D, Ulloa RH, Powell E, Speziani F, Tekin M, Schule R, Zuchner S. GEnomes Management Application (GEM.app): A new software tool for large-scale collaborative genome analysis. Hum Mutat. 2013;34(6):842–846. doi: 10.1002/humu.22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster O, Beck T, Atlan D, Swertz M, Dagleish R, Brookes AJ. Cafe Variome: general-purpose software for making genotype-phenotype data discoverable in restricted or open access contexts. Hum Mutat. 2015 Oct;36(10):957–64. doi: 10.1002/humu.22841. [DOI] [PubMed] [Google Scholar]
- Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, Brunner HG, Buske OJ, Carey K, Doll C, Dumitriu S, Dyke SO, den Dunnen JT, Firth HV, Gibbs RA, Girdea M, Gonzalez M, Haendel MA, Hamosh A, Holm IA, Huang L, Hurles ME, Hutton B, Krier JB, Misyura A, Mungall CJ, Paschall J, Paten B, Robinson PN, Schiettecatte F, Sobreira NL, Swaminathan GJ, Taschner PE, Terry SF, Washington NL, Züchner S, Boycott KM, Rehm HL. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat. 2015 Oct;36(10):915–21. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, et al. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2015 Jul;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- Robinson PN, Köhler S, Oellrich A, Sanger Mouse Genetics Project. Wang K, Mungall CJ, Lewis SE, Washington N, Bauer S, Seelow D, Krawitz P, Gilissen C, et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014;24(2):340–8. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New Tools for Mendelian Disease Gene Identification: PhenoDB Variant Analysis Module; and GeneMatcher, a Web-Based Tool for Linking Investigators with an Interest in the Same Gene. Hum Mutat. 2015a;36(4):425–31. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan GJ, Bragin E, Chatzimichali EA, Corpas M, Bevan AP, Wright CF, Carter NP, Hurles ME, Firth HV. DECIPHER: web-based, community resource for clinical interpretation of rare variants in developmental disorders. Hum Mol Genet. 2012;21(R1):R37–R44. doi: 10.1093/hmg/dds362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7(11):e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Köhler S, Mackenroth L, Jäger M, Hecht J, Krawitz P, Graul-Neumann L, Doelken S, Ehmke N, Spielmann M, Oien NC, Schweiger MR, et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci Transl Med. 2014;6(252):252ra123. doi: 10.1126/scitranslmed.3009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]