Abstract
Background
Parathyroid hormone (PTH) is one of the principal regulators of calcium homeostasis. Although serum PTH level is mostly accounted by genetic factors, genetic background underlying PTH level is insufficiently known. Therefore, the aim of this study was to identify novel genetic variants associated with PTH levels.
Methods
We performed GWAS meta-analysis within two genetically isolated Croatian populations followed by replication analysis in a Croatian mainland population and we also combined results across all three analyzed populations. The analyses included 2596 individuals. A total of 7,411,206 variants, imputed using the 1000 Genomes reference panel, were analysed for the association. In addition, a sex-specific GWAS meta-analyses were performed.
Results
Polymorphisms with the lowest P-values were located on chromosome 4 approximately 84 kb of the 5′ of RASGEF1B gene. The most significant SNP was rs11099476 (P = 1.15 × 10−8). Sex-specific analysis identified genome-wide significant association of the variant rs77178854, located within DPP10 gene in females only (P = 2.21 × 10− 9). There were no genome-wide significant findings in the meta-analysis of males.
Conclusions
We identified two biologically plausible novel loci associated with PTH levels, providing us with further insights into the genetics of this complex trait.
Electronic supplementary material
The online version of this article (10.1186/s10020-018-0018-5) contains supplementary material, which is available to authorized users.
Keywords: Parathyroid hormone, Genome-wide association analysis, Meta-analysis
Background
Parathyroid hormone (PTH) plays a critical role in the regulation of bone mineral metabolism and calcium homeostasis (DeLuca, 1986). PTH regulates serum calcium levels by stimulating osteoclast activity within bone in order to release calcium. Circulating PTH enhances the reabsorption of calcium in distal nephrons and induces the synthesis of the vitamin D active metabolite 1,25-dihydroxyvitamin D (1,25(OH)2D3) within the kidney (Kumar & Thompson, 2011; Kumar et al., 1991; Khundmiri et al., 2016). The 1,25(OH)2D3 stimulates intestinal calcium absorption and moreover, has a synergistic effect with PTH in bone resorption by stimulating proliferation of osteoclasts (Kumar & Thompson, 2011; Kumar et al., 1991; Khundmiri et al., 2016).
Variations in PTH synthesis and secretion are regulated by serum levels of calcium and phosphate, as well as by 1,25(OH)2D3 (Kumar & Thompson, 2011; Gago et al., 2005). Decreases in serum levels of calcium and increases in serum levels of phosphate stimulate the secretion of PTH, while 1,25(OH)2D3 decreases PTH secretion (Silver & Levi, 2005). Regulation of PTH secretion in response to variations in serum calcium is mediated by the calcium-sensing receptors on the membrane of parathyroid cells (Kumar & Thompson, 2011; Brent et al., 1988). 1,25(OH)2D3 associates with the vitamin D receptor and thus represses the transcription of PTH. The secretion of PTH is also indirectly altered by 1,25(OH)2D3 and its regulation of calcium-sensing receptor expression (Kumar & Thompson, 2011). Serum phosphate regulates PTH mRNA and serum PTH levels independently of changes in either serum calcium or 1,25(OH)2D3 levels (Kilav et al., 1995).
The most common pathological condition of excessive secretion of parathyroid hormone is hyperparathyroidism. Primary hyperparathyroidism is due to hypersecretion of the parathyroid gland, while secondary hyperparathyroidism can result from conditions that lead to hypocalcemia, especially observed in patients with chronic kidney disease (Fraser, 2009). Hypoparathyroidism, parathyroid hormone deficiency, is an uncommon condition that occurs mostly due to surgical removal of the parathyroid gland (Abate & Clarke, 2017).
Both environmental and genetic factors influence serum PTH levels. It is estimated that 60% of the variation in PTH concentrations is genetically determined. (Hunter et al., 2001). However, the genetic background underlying PTH level is not yet well understood.
Only one high-density genome-wide association study (GWAS) of PTH concentration has been reported to date (Robinson-Cohen et al., 2017). Robinson-Cohen et al. identified five significantly associated loci, including the strongest associated SNP rs6127099 located upstream of CYP24A1, a gene that encodes the primary catabolic enzyme for 1.25 (OH)2D (Robinson-Cohen et al., 2017). The other significantly associated loci were intronic variant rs4074995 within RGS14 (regulator of G-protein signaling 14), rs219779 adjacent to CLDN14 (Claudin 14), rs4443100 located near RTDR1 (RSPH14, radial spoke head 14 homolog) and rs73186030 located near CASR (calcium-sensing receptor) gene (Robinson-Cohen et al., 2017). However, only three of these five loci (rs6127099, rs4074995 and rs219779) were replicated within an independent sample. Altogether, the five reported loci explained only 4.2% of the variance in circulating PTH, suggesting that additional genetic variants remain undiscovered.
The aim of our study is identification of novel loci associated with the parathyroid function, by performing a GWAS meta-analysis of plasma PTH levels within two genetically isolated Croatian populations (Korcula and Vis) following by replication analysis in the urban population of Split. To maximize the power of the study, we additionally performed meta-analysis for PTH plasma levels in all three Croatian populations. We also conducted gender-specific GWAS meta-analyses.
Methods
Study cohorts
This study was performed on samples from three Croatian populations: from the Dalmatian islands of Korcula and Vis and the mainland city of Split, within the large-scale project of “10,001 Dalmatians” (Rudan et al., 2009). A detailed description of the cohorts is provided in Table 1. The Korcula population is genetically isolated from Croatian Mainland, while Vis population is genetically isolated from Croatian Mainland and surrounding islands (Vitart et al., 2008). For all study populations, we excluded participants who underwent parathyroid surgery, as well as individuals who had PTH level < 5 pg/ml, which is near the minimum PTH assay detection limit (4.3 pg/ml). After these exclusions, the number of individuals available with PTH level and genotype data was 806 in Korcula, 831 in Vis and 959 in Split. In all three cohorts there were no participants who reported serious renal disease that could affect PTH concentration. The study was approved by the Research Ethics Committees in Croatia and Scotland and all participants provided informed consent. All analyses were in accordance with the relevant guidelines and regulations.
Table 1.
Characteristics of study participants
| Variables | Korcula | Vis | Split |
|---|---|---|---|
| N with PTH and GWAS data | 863 | 834 | 960 |
| N underwent parathyroid surgery | 1 | 0 | 1 |
| N with PTH level < 5 pg/ml | 56 | 3 | 3 |
| Sample size used in the analyses | 806 | 831 | 959 |
| Women, N (%) | 524 (65%) | 486 (58%) | 586 (61%) |
| Median age, (qL,qU) | 57 (47, 67) | 57 (45,69) | 52 (40, 61) |
| Median PTH, pg/ml (qL,qU) | 19.9 (13.7, 29.1) | 25.9 (18.4, 32.1) | 21.6 (17.2, 26.5) |
N: number of individuals; qL: lower quartile, qu: upper quartile
Genotyping and imputation
Additional file 1: Table S1 shows cohort-summary information on genotyping, imputation and quality control procedures. The final numbers of single nucleotide polymorphisms (SNPs) included in analyses were 9,182,797 for the Korcula sample, 8,865,173 for the Vis sample and 8,777,560 for the Split sample. The number of overlapping SNPs present in all three cohorts was 7,411,206.
Measurement of PTH
Plasma PTH levels were determined by radio-immunoassay method (RIA) in the Laboratory of Biochemistry, Department of Nuclear Medicine, University Hospital Split. RIA ran on the Scintillation counter liquid samples, Capintec, and 125I served as a marker. The concentrations of PTH in the plasma were determined using commercial kits (DIAsource hPTH -120 min-IRMA Kit, DIAsource ImmunoAssays S.A, Belgium). The reference range of plasma PTH levels is 12.26–35.50 pg/ml.
Statistical analyses
We performed genome-wide association analysis within each data set and then conducted a meta-analysis of two genetically isolated cohorts (Korcula and Vis) followed by replication analysis in the cohort of the mainland city of Split. To maximize the study power, we also performed a further meta-analysis of all three cohorts.
Genome-wide association analyses
Association analysis for the Split sample was carried out using a combination of R-package GenABEL and SNPTEST software, while for the Korcula and Vis samples analyses were conducted using R-packages GenABEL and VariABEL (Aulchenko et al., 2007; Marchini et al., 2007; Struchalin et al., 2012).
PTH levels were adjusted for age and sex using linear regression analysis and the calculated residuals were inverse-Gaussian transformed to achieve a normal distribution. GWAS was performed on transformed residuals using linear mixed model which accounts for population structure and relatedness. Association statistics for each SNP, including effect size estimates (β-estimates), standard errors and p-values were calculated under an additive genetic model.
Prior to performing the meta-analysis we calculated genomic inflation factors (lambdas) in individual data sets. No adjustments were necessary (λKorcula = 1.026, λVis = 1.001, λSplit = 0.99).
Meta-analysis
Meta-analysis was carried out using the R-package MetABEL (R: A Language and Environment for Statistical Computing, 2018). Meta-analysis was conducted using the inverse-variance fixed-effects method on overlapping SNPs based on the β-estimates and standard errors from each study. Meta-analyses showed no significant evidence for inflated statistics (both λKorcula − Vis and λKorcula − Vis − Split were 1.01), thus no genomic correction was applied. To visualize results of the meta-analysis, Manhattan and quantile-quantile (QQ) plots were created using R-package qqman (Turner, 2014). Regional association plots for loci of interest (±400 kb) were produced using Locus Zoom based on hg19 genome build and 1000 genomes EUR population as the linkage disequilibrium (LD) population (Pruim et al., 2010). Forest plots for the most associated SNP were created using R-package MetABEL. To confirm the genotyping quality for the most associated SNPs in the regions, cluster plots were visually inspected using the Illumina GenomeStudio software package. If the SNP of interest was not directly genotyped, but imputed, then cluster plots were examined for directly genotyped SNPs in high LD with the SNP of interest (r2 > 0.8), located on the same chromosome and less than 400 kb apart. A genome-wide significance of association was defined as p − value ≤ 5 × 10−8. Power calculations were performed using Quanto version 1.2.4 for quantitative traits (WJ MJ, 2006).
Sex-specific analyses
In order to identify sex-specific effects we performed GWAS analyzing males and females separately in each cohort. We used the same procedures as in the primary analyses with the exception of the gender covariate. Association results were meta-analyzed using the inverse-variance fixed-effects method. The total sample sizes were 1596 in women and 1000 in men.
Results
Meta-analyses
In each population, a separate genome-wide association study of PTH levels was conducted. We meta-analyzed two genetically isolated cohorts, Korcula and Vis (Additional file 1: Figure S1), and then replicated results in the Split population. A total of 1637 individuals were included in the meta-analysis and 959 in the replication analysis (Table 1). The most associated SNP was rs4616742 (reference allele C, β = 0.18, SE = 0.04, P = 4.42 × 10−7). The SNP is located near protein coding gene RASGEF1B (RASGEF Domain Family Member 1B). All top SNPs, located on the chromosome 4 near RASGEF1B gene with P < 10−6 from meta-analysis of ‘genetically isolated populations’ reached P < 0.05 in the Split population.
To maximize the study power, we performed a meta-analysis of all three cohorts. In total, 2596 individuals were included in the meta-analysis (Table 1). The results are shown in Fig. 1a. As seen from the quantile-quantile plot there was no early deviation from expected P values (Fig. 1b). Four SNPs, representing one locus, reached genome-wide significance. As in the meta-analysis of two genetically isolated cohorts, SNPs with the lowest P-values were located on chromosome 4 near RASGEF1B. The most associated SNP was rs11099476 (P = 1.15 × 10−8), which explained 1.14% of the variance in PTH. We found the T allele of the rs11099476 to be associated with higher PTH level (β = 0.16, SE = 0.03). Effect sizes were in the same direction across all three cohorts (Fig. 1c). The regional association plot for rs11099476 is given in Fig. 2a. The identified SNP, rs11099476, is in high LD with the top SNP from meta-analysis of ‘genetically isolated populations’, rs4616742 (r2 = 0.9). These results indicated that associated locus is becoming more significant as the sample size increases and confirmed the consistency of our top finding.
Fig. 1.
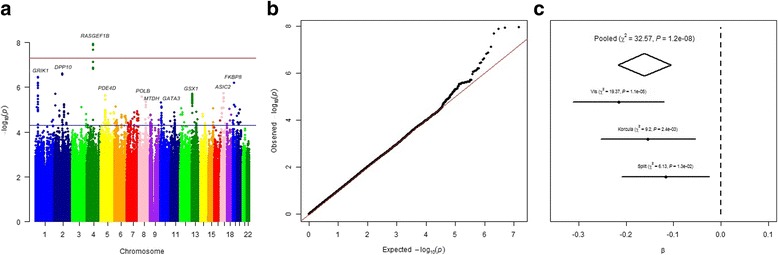
a Manhattan plot of SNPs for PTH levels in the meta-analysis of three cohorts. The y axis shows the −log10 P values of 7,411,206 SNPs, and the x axis shows their chromosomal positions. The blue line indicates the threshold for suggestive hits (P = 5 × 10−5), and the red line represents the threshold for genome-wide significance (P = 5 × 10−8). b Quantile-quantile plot in the meta-analysis of three cohorts. c Forest plot of rs11099476 effect estimates in individual populations and the combined meta-analysis
Fig. 2.
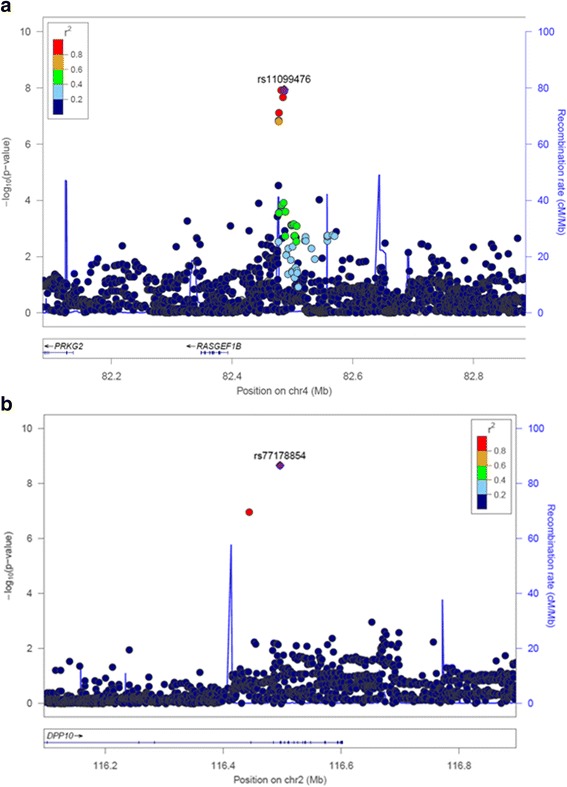
a Regional association plot for the chromosome 4 locus rs11099476 in the meta-analysis of three cohorts. b Regional association plot for the chromosome 2 locus rs77178854 in the sex-stratified meta-analysis of three cohorts among females. SNPs are plotted by position against association with PTH (−log10 P values). The purple diamond highlights the most significant, whereas the colors of other variant represent LD with most significant SNP
Analysis also revealed several suggestive loci (P < 5 × 10−6), including rs77178854 in the DPP10 gene (P = 2.46 × 10−7), rs481121 near the GRIK3 gene (P = 3.58 × 10−7), rs76615278 in the FKBP8 gene (P = 6.34 × 10−7), rs1875872 near the ASIC2 gene (P = 1.94 × 10−6), rs9512841 near the GSX1 gene (P = 2.01 × 10−6), rs191686630 in the PDE4D gene, rs3136797 in the POLB gene (P = 2.68 × 10−6), rs499177 near the MTDH gene and rs58726672 near the GATA3 gene (P = 4.77 × 10−6) (Table 2).
Table 2.
Associations of top single nucleotide polymorphisms(P < 5 × 10−6) with PTH concentrations
| SNP | Chr | Position | Nearest Gene | Effect Allele | Other Allele | EAF Korcula | EAF Vis | EAF Split | GnomAD EAF | β | SE | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11099476 | 4 | 82,486,056 | RASGEF1B | T | A | 0.57 | 0.55 | 0.54 | 0.59 | 0.16 | 0.03 | 1.15×10−8 |
| rs77178854 | 2 | 116,496,539 | DPP10 | C | G | 0.97 | 0.98 | 0.99 | 0.98 | 0.58 | 0.11 | 2.46×10−7 |
| rs481121 | 1 | 37,203,485 | GRIK1 | A | G | 0.56 | 0.56 | 0.58 | 0.49 | 0.14 | 0.03 | 3.58×10−7 |
| rs76615278 | 19 | 18,654,588 | FKBP8 | G | A | 0.84 | 0.83 | 0.82 | * | 0.20 | 0.04 | 6.34×10−7 |
| rs1875872 | 17 | 31,795,716 | ASIC2 | A | G | 0.62 | 0.65 | 0.65 | 0.65 | 0.14 | 0.03 | 1.94×10−6 |
| rs9512841 | 13 | 28,309,646 | GSX1 | G | A | 0.51 | 0.52 | 0.53 | 0.58 | 0.13 | 0.03 | 2.01×10− 6 |
| rs191686630 | 5 | 58,477,398 | PDE4D | A | T | 0.11 | 0.16 | 0.21 | * | 0.19 | 0.04 | 2.36×10−6 |
| rs3136797 | 8 | 42,226,805 | POLB | C | G | 0.98 | 0.99 | 0.98 | 0.99 | 0.57 | 0.12 | 2.68×10−6 |
| rs499177 | 8 | 98,472,201 | MTDH | T | C | 0.46 | 0.57 | 0.45 | 0.44 | 0.13 | 0.03 | 4.66×10−6 |
| rs58726672 | 10 | 8,407,822 | GATA3 | C | T | 0.98 | 0.98 | 0.98 | 0.98 | 0.57 | 0.13 | 4.77×10−6 |
Top SNPs were defined as the SNP with lowest P value within a 500 kb window
Chr: chromosome; EAF: effect allele frequency; GnomAD EAF: effect allele frequency from Genome Aggregation Database; β: effect size; SE: standard error
*variants without frequency information in Genome Aggregation Database
Sex-specific analyses
We searched for gender-specific loci by performing sex-specific GWAS meta-analysis, analyzing females and males separately in each cohort. The results for females are shown in Fig. 3. The top hit detected in the meta-analysis of all three cohorts, rs77178854, located within DPP10 gene, reached genome-wide significance (reference allele C, β = 0.82, SE = 0.14, P = 2.21 × 10−9) in females (Table 3). Effect sizes were in the same direction across all three cohorts (Fig. 3c). Regional association plot of the identified SNP is shown in Fig. 2b. No single locus reaching genome wide significance was identified in males (Additional file 1: Table S2).
Fig. 3.
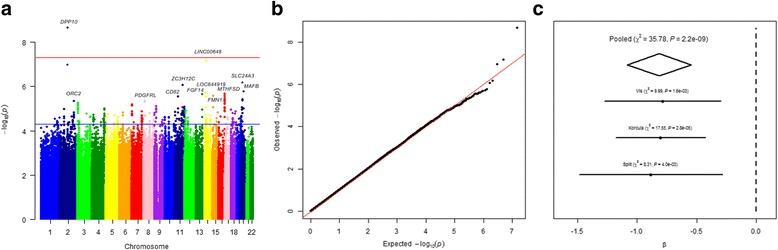
a Manhattan plot of SNPs for PTH levels in the sex-stratified meta-analysis of three cohorts among females. The y axis shows the −log10 P values of 7,411,206 SNPs, and the x axis shows their chromosomal positions. The blue line indicates threshold for suggestive hits (P = 5 × 10−5), and red line represents the threshold for genome-wide significance (P = 5 × 10−8). b Quantile-quantile plot in the sex-stratified meta-analysis of three cohorts among females. c Forest plot of rs77178854 effect estimates in individual populations and the combined meta-analysis among females
Table 3.
Associations of top single nucleotide polymorphisms(P < 5 × 10−6) with PTH concentrations among females
| SNP | Chr | Position | Nearest Gene | Effect Allele | Other Allele | EAF Korcula | EAF Vis | EAF Split |
GnomAD EAF | β | SE | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs77178854 | 2 | 116,496,539 | DPP10 | C | G | 0.98 | 0.97 | 0.99 | 0.98 | 0.82 | 0.14 | 2.21×10−9 |
| rs1890709 | 14 | 49,101,833 | LINC00648 | A | G | 0.38 | 0.30 | 0.33 | 0.31 | 0.20 | 0.04 | 7.12×10−8 |
| rs16981087 | 20 | 19,739,954 | SLC24A3 | G | C | 0.80 | 0.78 | 0.77 | 0.81 | 0.22 | 0.04 | 6.99×10−7 |
| rs661171 | 11 | 110,016,519 | ZC3H12C | G | T | 0.74 | 0.70 | 0.71 | 0.70 | 0.20 | 0.04 | 8.94×10−7 |
| rs74629672 | 20 | 39,105,870 | MAFB | T | A | 0.95 | 0.94 | 0.94 | 0.96 | 0.43 | 0.09 | 1.68×10−6 |
| rs1349573 | 14 | 41,403,160 | LOC644919 | G | A | 0.05 | 0.05 | 0.06 | 0.03 | 0.45 | 0.10 | 1.94×10−6 |
| rs3866634 | 16 | 86,567,929 | MTHFSD | G | A | 0.93 | 0.92 | 0.93 | 0.91 | 0.32 | 0.07 | 2.14×10−6 |
| rs7997888 | 13 | 102,759,325 | FGF14 | A | G | 0.04 | 0.04 | 0.04 | 0.15 | 0.49 | 0.10 | 2.19×10−6 |
| rs5024438 | 15 | 33,077,401 | FMN1 | G | A | 0.72 | 0.70 | 0.79 | * | 0.23 | 0.05 | 2.76×10−6 |
| rs77796218 | 11 | 44,580,581 | CD82 | C | T | 0.97 | 0.96 | 0.97 | 0.98 | 0.49 | 0.10 | 2.84×10−6 |
| rs13406545 | 2 | 201,792,123 | ORC2 | T | A | 0.15 | 0.19 | 0.18 | 0.23 | 0.21 | 0.05 | 4.54×10−6 |
| rs2588129 | 8 | 17,462,468 | PDGFRL | A | G | 0.04 | 0.02 | 0.02 | 0.11 | 0.54 | 0.12 | 4.57×10−6 |
Top SNPs were defined as the SNP with lowest P value within a 500 kb window
Chr: chromosome; EAF: effect allele frequency; GnomAD EAF: effect allele frequency from Genome Aggregation Database; β: effect size; SE: standard error
*variants without frequency information in Genome Aggregation Database
Discussion
In this GWAS meta-analysis of three Croatian populations we identified a novel genome-wide significant locus associated with plasma PTH level near gene RASGEF1B on chromosome 4. We also identified a sex-specific significant association in females in the DPP10 gene.
The significance of the identified polymorphism rs11099476 was most influenced by the Vis population, which is isolated from the Croatian Mainland and surrounding islands, then by the Korcula population which is isolated from the Croatian Mainland and the least contributed by the mainland city of Split population (Fig. 2c). However, although the locus significance was most affected by the isolated populations, significance has been amplified in the meta-analysis of all three cohorts compared to meta-analysis of ‘genetically isolated populations’.
The identified common variant rs11099476 accounts for 1.14% of population variance in plasma PTH. RASGEF1B is the guanine nucleotide exchange factor with specificity for Rap2A, a member of Rap subfamily of Ras-like G proteins (Yaman et al., 2009). Rap2 subfamily contains Rap2A and Rap2B, which share about 90% sequence homology (Paganini et al., 2006). Rap2A protein binds GDP to GTP and exhibits a low intrinsic GTPase activity in the presence of Mg2+ (Lerosey et al., 1991), while Rap2B increases intracellular calcium level and phosphorylation level of extracellular signal-related kinase (ERK) 1/2 (Di et al., 2015). Variations near RASGEF1B gene have been associated with height (He et al., 2015; Allen et al., 2010). Height is positively correlated with calcium absorption efficiency which is important determinant of calcium balance (Abrams et al., 2005). Some evidence of association was also found for variations in this gene and bone density, hip, and cystatin C in serum (Kiel et al., 2007; Kottgen et al., 2010). PTH is a significant negative predictor of bone mineral density at the hip (Sneve et al., 2008). Cystatin C in serum is a biomarker of kidney function, and chronic kidney disease (Kottgen et al., 2010). Disturbed kidney function can influence PTH stimulated calcium reabsorption and synthesis of 1,25(OH)2D3 (Kumar & Thompson, 2011; Kumar et al., 1991; Khundmiri et al., 2016). However, to understand the mechanism underlying the observed association further functional studies of RASGEF1B will be needed.
Although no signals other than rs11099476 reached genome-wide significance, several candidate loci showed suggestive evidence of association. Particularly interesting is the variant near GATA3 gene since mutations in this gene are the cause of hypoparathyroidism with sensorineural deafness and renal dysplasia (Van Esch et al., 2000). Of note, in a previous large GWA meta-analysis, variant near GATA3 gene was found to be associated with serum calcium (O'Seaghdha et al., 2013).
Given the reported differences in PTH level between males and females, we performed sex-specific analyses (Wei et al., 2015; Serdar et al., 2017). Our study supports the sex-specificity underlying PTH level. Sex-stratified analysis in women identified a novel locus associated with PTH. The identified locus is the intron variant rs77178854, located within DPP10 gene. DPP10 encodes a membrane protein that is a member of the serine proteases family, which binds specific voltage-gated potassium channels and alters their expression and biophysical properties. It is highly expressed in brain, pancreas, spinal cord and adrenal gland (Allen et al., 2003), and may serve as a prognostic marker in colorectal cancer (Park et al., 2013). It is interesting to note that Aigner et al. showed that high serum PTH concentrations were associated with distal colorectal cancer in women but not in men (Aigner et al., 2015). The existence of DPP10 in endocrine cells indicate that the protein might also have an additional role in the regulation of hormone secretion (Bezerra et al., 2015), which also supports our finding. Further studies of DPP10 will be needed to clarify this result.
The only previously published high-density GWAS for PTH levels did not identify RASGEF1B or DPP10 at a genome-wide significant level, despite having a sample size of over 29,155 participants (22, 653 in discovery stage and 6502 in replication analysis) (Robinson-Cohen et al., 2017). The possible explanation could be an increased relative effect of these loci in our populations due to the reduced genetic and environmental heterogeneity found in two out of three cohorts (i.e., Korcula and Vis) (Rudan et al., 2008) compared to the urban populations used in the analysis of Robinson-Cohen et al. (Robinson-Cohen et al., 2017). Previously reported CYP24A1, RGS14 and CLDN14 variants associated with PTH level (Robinson-Cohen et al., 2017) had the same directions of effect in our study as originally reported but did not show significant associations, probably due to limited sample size of our study or specificity of isolated populations (Additional file 1: Table S3).
The greatest strengths of our study include a comprehensive set of genetic variants examined and ethnically homogeneous sample. We had sufficient data to confidently detect an association for the identified RASGEF1B locus, since our meta-analysis had 92% power to detect associated SNP with an effect size of 0.19 and minor allele frequency of 0.45 at the genome-wide level of significance. Furthermore, meta-analysis performed in females only had 86% power to detect DPP10 locus with an effect size of 0.82 and minor allele frequency of 0.02 at the genome-wide level of significance. Meta-analysis performed in males only had 84% power to detect SNPs with an effect size of 0.35 and minor allele frequency of 0.3 at the genome-wide level of significance. The main limitation of our study is the modest sample size used in the analysis, reducing statistical power for detecting additional associations with smaller effect sizes or minor allele frequencies. Nevertheless, we have identified novel, previously unsuspected and biological plausible associations with PTH variation. Further replication analysis would be required to confirm our findings and to discover additional genetic variants underlying PTH levels in order to explain more of the variability in PTH variations.
Conclusions
In summary, in a GWA meta-analysis of PTH levels we identified a novel significant locus rs11099476 located near a guanine nucleotide exchange factor RASGEF1B. The finding appears to be consistent based on analyses of meta-GWAS across all three analyzed cohorts and meta-GWAS across two isolated populations followed by replication analysis in the mainland cohort. Our work also includes the first gender-specific GWAS performed to date and revealed significant association for an intron variant rs77178854 located within the DPP10 gene in women, indicating the possibility that sex-specificity is underlying PTH level. To conclude, findings from this study improve the current knowledge of the genetic factors regulating PTH levels and their validation in independent populations would be beneficial.
Additional file
Supporting Information. (XLSX 54 kb)
Acknowledgments
We would like to thank all participants of this study and acknowledge invaluable support of the local teams in Zagreb and Split, especially that of the Institute for Anthropological Research, Zagreb, Croatia.
Funding
The Croatian Science Foundation funded this work under the project “Identification of new genetic loci implicated in regulation of thyroid and parathyroid function” (grant no. 1498).
The “10 001 Dalmatians” project was funded by grants from the Medical Research Council (UK), European Commission Framework 6 project EUROSPAN (Contract No.LSHG-CT-2006-018947), the Republic of Croatia Ministry of Science, Education and Sports research grant (216–1080315-0302), the Croatian Science Foundation (grant 8875), CEKOM (Ministry of Economy, Entrepreneurship and Crafts) and the Research Centre of Excellence in Personalized Medicine (Ministry of Science and Education).
Availability of data and materials
The datasets generated during the current study are available from the corresponding author on reasonable request.
Abbreviations
- GWAS
genome-wide association study
- PTH
Parathyroid hormone
- RIA
radio-immunoassay method
- SNP
single nucleotide polymorphism
Authors’ contributions
TZ conceived the study idea; OP, CH, TZ, VBP, IK and MB formed the biobank “10 001 Dalmatians”; VT, DB and AP performed the measurements of parathyroid hormones and verified the relevance of the results; TB performed imputation of the data; AM, MP, IG researched the data; AM performed the statistical analysis and drafted the manuscript; TZ, CH, MB, MP, IG, VBP and OP contributed to the discussion, review and editing of the manuscript; all authors approved the final version of the manuscript.
Ethics approval and consent to participate
The study was approved by the Research Ethics Committees in Croatia and Scotland and all participants provided informed consent.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s10020-018-0018-5) contains supplementary material, which is available to authorized users.
Contributor Information
Antonela Matana, Email: antonela.boljat@mefst.hr.
Dubravka Brdar, Email: d.brdar.dr@gmail.com.
Vesela Torlak, Email: veselakbsplit@yahoo.com.
Thibaud Boutin, Email: tboutin@exseed.ed.ac.uk.
Marijana Popović, Email: mpopovic@mefst.hr.
Ivana Gunjača, Email: igunjaca@mefst.hr.
Ivana Kolčić, Email: ivana.kolcic@mefst.hr.
Vesna Boraska Perica, Email: vboraska@mefst.hr.
Ante Punda, Email: ante.punda@mefst.hr.
Ozren Polašek, Email: ozren.polasek@mefst.hr.
Maja Barbalić, Email: mbarbali@mefst.hr.
Caroline Hayward, Email: caroline.hayward@igmm.ed.ac.uk.
Tatijana Zemunik, Phone: 38521557888, Email: tzemunik@mefst.hr.
References
- Abate EG, Clarke BL. Review of hypoparathyroidism. Front Endocrinol. 2017;7 [DOI] [PMC free article] [PubMed]
- Abrams SA, Griffin IJ, Hawthorne KM, Liang LL. Height and height Z-score are related to calcium absorption in five- to fifteen-year-old girls. Journal of Clinical Endocrinology & Metabolism. 2005;90:5077–5081. doi: 10.1210/jc.2005-0537. [DOI] [PubMed] [Google Scholar]
- Aigner E, Stadlmayr A, Huber-Schonauer U, Zwerina J, Husar-Memmer E, Niederseer D, Eder SK, Stickel F, Pirich C, Schett G, et al. Parathyroid hormone is related to dysplasia and a higher rate of distal colorectal adenoma in women but not men. Hormones & Cancer. 2015;6:153–160. doi: 10.1007/s12672-015-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen HL, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, Bhattacharyya S, Tinsley J, Zhang YM, Holt R, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet. 2003;35:258–263. doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, Van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Bezerra GA, Dobrovetsky E, Seitova A, Fedosyuk S, Dhe-Paganon S, Gruber K. Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels. Sci Rep. 2015;5 [DOI] [PMC free article] [PubMed]
- Brent GA, Leboff MS, Seely EW, Conlin PR, Brown EM. Relationship between the concentration and rate of change of calcium and serum intact parathyroid-hormone levels in normal humans. Journal of Clinical Endocrinology & Metabolism. 1988;67:944–950. doi: 10.1210/jcem-67-5-944. [DOI] [PubMed] [Google Scholar]
- DeLuca HF. The metabolism and functions of vitamin D. Adv Exp Med Biol. 1986;196:361–375. doi: 10.1007/978-1-4684-5101-6_24. [DOI] [PubMed] [Google Scholar]
- Di JH, Huang H, Qu DB, Tang JJ, Cao WJ, Lu Z, Cheng Q, Yang J, Bai J, Zhang YP, Zheng JN. Rap2B promotes proliferation, migration, and invasion of human breast cancer through calcium-related ERK1/2 signaling pathway. Sci Rep. 2015;5 [DOI] [PMC free article] [PubMed]
- Fraser WD. Hyperparathyroidism. Lancet. 2009;374:145–158. doi: 10.1016/S0140-6736(09)60507-9. [DOI] [PubMed] [Google Scholar]
- Gago EV, Cadarso-Suarez C, Perez-Fernandez R, Burgos RR, Mugica JD, Iglesias CS. Association between vitamin D receptor Fokl polymorphism and serum parathyroid hormone level in patients with chronic renal failure. J Endocrinol Investig. 2005;28:117–121. doi: 10.1007/BF03345353. [DOI] [PubMed] [Google Scholar]
- He MA, Xu M, Zhang B, Liang J, Chen P, Lee JY, Johnson TA, Li HX, Yang XB, Dai JC, et al. Meta-analysis of genome-wide association studies of adult height in east Asians identifies 17 novel loci. Hum Mol Genet. 2015;24:1791–1800. doi: 10.1093/hmg/ddu583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter D, De Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, Spector TD. Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J Bone Miner Res. 2001;16:371–378. doi: 10.1359/jbmr.2001.16.2.371. [DOI] [PubMed] [Google Scholar]
- Khundmiri SJ, Murray RD, Lederer E. PTH and vitamin D. Comprehensive Physiology. 2016;6:561–601. doi: 10.1002/cphy.c140071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, Karasik D. Genome-wide association with bone mass and geometry in the Framingham heart study. Bmc Medical Genetics. 2007;8 [DOI] [PMC free article] [PubMed]
- Kilav R, Silver J, Navehmany T. Parathyroid-hormone gene-expression in Hypophosphatemic rats. J Clin Investig. 1995;96:327–333. doi: 10.1172/JCI118038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao XY, Yang Q, Smith AV, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–U334. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Cahan DH, Madias NE, Harrington JT, Kurtin P, Dawsonhughes BF. Vitamin-D and calcium-transport. Kidney Int. 1991;40:1177–1189. doi: 10.1038/ki.1991.332. [DOI] [PubMed] [Google Scholar]
- Kumar R, Thompson JR. The regulation of parathyroid hormone secretion and synthesis. J Am Soc Nephrol. 2011;22:216–224. doi: 10.1681/ASN.2010020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerosey I, Chardin P, Degunzburg J, Tavitian A. The product of the Rap2 gene, member of the Ras superfamily - biochemical-characterization and site-directed mutagenesis. J Biol Chem. 1991;266:4315–4321. [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- O'Seaghdha CM, Wu HS, Yang Q, Kapur K, Guessous I, Zuber AM, Kottgen A, Stoudmann C, Teumer A, Kutalik Z, et al. Meta-analysis of genome-wide association studies identifies six new loci for serum calcium concentrations. PLoS Genet. 2013;9 [DOI] [PMC free article] [PubMed]
- Paganini S, Guidetti GF, Catricala S, Trionfini P, Panelli S, Balduini C, Torti M. Identification and biochemical characterization of Rap2C, a new member of the rap family of small GTP-binding proteins. Biochimie. 2006;88:285–295. doi: 10.1016/j.biochi.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Park HS, Yeo HY, Chang HJ, Kim KH, Park JW, Kim BC, Baek JY, Kim SY, Kim DY. Dipeptidyl peptidase 10, a novel prognostic marker in colorectal Cancer. Yonsei Med J. 2013;54:1362–1369. doi: 10.3349/ymj.2013.54.6.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R: A Language and Environment for Statistical Computing 2018 [http://www.R-project.org/].
- Robinson-Cohen C, Lutsey PL, Kleber ME, Nielson CM, Mitchell BD, Bis JC, Eny KM, Portas L, Eriksson J, Lorentzon M, et al. Genetic variants associated with circulating parathyroid hormone. J Am Soc Nephrol. 2017;28:1553–1565. doi: 10.1681/ASN.2016010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudan I, Carothers AD, Polasek O, Hayward C, Vitart V, Biloglav Z, Kolcic I, Zgaga L, Ivankovic D, Vorko-Jovic A, et al. Quantifying the increase in average human heterozygosity due to urbanisation. Eur J Hum Genet. 2008;16:1097–1102. doi: 10.1038/ejhg.2008.48. [DOI] [PubMed] [Google Scholar]
- Rudan I, Marusic A, Jankovic S, Rotim K, Boban M, Lauc G, Grkovic I, Dogas Z, Zemunik T, Vatavuk Z, et al. "10 001 Dalmatians:" Croatia launches its National Biobank. Croatian Medical Journal. 2009;50:4–6. doi: 10.3325/cmj.2009.50.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serdar MA, Can BB, Kilercik M, Durer ZA, Aksungar FB, Serteser M, Coskun A, Ozpinar A, Unsal I. Analysis of changes in parathyroid hormone and 25 (oh) vitamin D levels with respect to age, gender and season: a data mining study. Journal of Medical Biochemistry. 2017;36:73–83. doi: 10.1515/jomb-2017-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J, Levi R. Regulation of PTH synthesis and secretion relevant to the management of secondary hyperparathyroidism in chronic kidney disease. Kidney Int. 2005;67:S8–S12. doi: 10.1111/j.1523-1755.2005.09501.x. [DOI] [PubMed] [Google Scholar]
- Sneve M, Emaus N, Joakimsen RM, Jorde R. The association between serum parathyroid hormone and bone mineral density, and the impact of smoking: the Tromso study. Eur J Endocrinol. 2008;158:401–409. doi: 10.1530/EJE-07-0610. [DOI] [PubMed] [Google Scholar]
- Struchalin MV, Amin N, Eilers PHC, van Duijn CM, Aulchenko YS. An R package "VariABEL" for genome-wide searching of potentially interacting loci by testing genotypic variance heterogeneity. BMC Genet. 2012;13 [DOI] [PMC free article] [PubMed]
- Turner SD: qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv DOI: 101101/005165 2014.
- Van Esch H, Groenen P, Nesbit MA, Schuffenhauer S, Lichtner P, Vanderlinden G, Harding B, Beetz R, Bilous RW, Holdaway I, et al. GATA3 haplo-insufficiency causes human HDR syndrome. Nature. 2000;406:419–422. doi: 10.1038/35019088. [DOI] [PubMed] [Google Scholar]
- Vitart V, Rudan I, Hayward C, Gray NK, Floyd J, Palmer CN, Knott SA, Kolcic I, Polasek O, Graessler J, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40:437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- Wei QS, Chen ZQ, Tan X, Su HR, Chen XX, He W, Deng WM. Relation of age, sex and bone mineral density to serum 25-Hydroxyvitamin D levels in Chinese women and men. Orthop Surg. 2015;7:343–349. doi: 10.1111/os.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ MJ: QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies.: University of Southern California; 2006.
- Yaman E, Gasper R, Koerner C, Wittinghofer A, Tazebay UH. RasGEF1A and RasGEF1B are guanine nucleotide exchange factors that discriminate between rap GTP-binding proteins and mediate Rap2-specific nucleotide exchange. FEBS J. 2009;276:4607–4616. doi: 10.1111/j.1742-4658.2009.07166.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information. (XLSX 54 kb)
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.


