Abstract
Trisomy of chromosome 21 (TS21) is the most common autosomal aneuploidy compatible with postnatal survival with a prevalence of 1 in 700 newborns. Its phenotype is highly complex with constant features, such as mental retardation, dysmorphic traits and hypotonia, and variable features including heart defects, susceptibility to Alzheimer’s disease (AD), type 2 diabetes, obesity and immune disorders. Overexpression of genes on chromosome-21 (Hsa21) is responsible for the pathogenesis of Down syndrome (DS) phenotypic features either in a direct or in an indirect manner since many Hsa21 genes can affect the expression of other genes mapping to different chromosomes. Many of these genes are involved in mitochondrial function and energy conversion, and play a central role in the mitochondrial dysfunction and chronic oxidative stress, consistently observed in DS subjects.
Recent studies highlight the deep interconnections between mitochondrial dysfunction and DS phenotype. In this short review we first provide a basic overview of mitochondrial phenotype in DS cells and tissues. We then discuss how specific Hsa21 genes may be involved in determining the disruption of mitochondrial DS phenotype and biogenesis. Finally we briefly focus on drugs that affect mitochondrial function and mitochondrial network suggesting possible therapeutic approaches to improve and/or prevent some aspects of the DS phenotype.
Keywords: Down syndrome/trisomy of chromosome 21, Mitochondrial dysfunction, Mitochondrial dynamics, Chromosome 21 genes, Down syndrome therapy
Background
Down syndrome (DS) is a genetic disorder caused by trisomy of chromosome 21 (TS21) in which the specific phenotypic manifestations may result from the balance among genetic, environmental and stochastic events (Rachidi and Lopes 2007; Reeves et al. 2001). Several studies have demonstrated that TS21 negatively affects mitochondrial function (Valenti et al. 2011; Brooksbank and Balazs 1984; Arbuzova et al. 2002). The downregulation of nuclear encoded mitochondrial genes (NEMGs) has been demonstrated in TS21 fetal heart samples (Conti et al. 2007) and in fetal brains (Mao et al. 2005). The mitochondrial energy production apparatus appears to be less efficient in DS fetal fibroblasts than in controls (Valenti et al. 2011; Valenti et al. 2010; Piccoli et al. 2013). In human primary lines of DS fetal fibroblasts, TS21 demonstrated to perturb the expression of genes involved in mitochondrial pathways, to decrease oxygen consumption and ATP content, to increase mtCa2+ load and ROS production (Piccoli et al. 2013; Izzo et al. 2017b). A contemporary dysregulation of ATP translocators (ANT1, ANT2 and ANT3) (Piccoli et al. 2013), ATP synthase and adenilate kinase (Valenti et al. 2010) was observed in the same cells. Studies of mitochondrial morphology in trisomic fetal fibroblasts demonstrated that these organelles were significantly damaged if compared with those of euploid cells. Furthermore a comparison between tissues from DS fetuses with and without heart defects revealed that TS21 fibroblasts derived from fetuses with cardiopathy presented a more severe mitochondrial dysfunction (Piccoli et al. 2013), thus suggesting that mitochondrial dysfunction contributes to generating a more severe phenotype. This concept might be extended to other phenotypic traits.
Many other studies performed on different DS models, tissues and animals agree with such a mitochondrial scenario in DS. Protein levels of mitochondrial complexes I, III and V were decreased in the brain of DS subjects (Kim et al. 2001) and, moreover, several mitochondrial DNA mutations were found in DS brain tissue (Coskun and Busciglio 2012). Decreased mitochondrial redox activity and membrane potential have been observed in human DS astrocytes and neuronal cultures (Arbuzova et al. 2002; Busciglio et al. 2002; Helguera et al. 2013) as well as in the brain of the Ts1Cje mouse model (Shukkur et al. 2006).
A substantial alteration in mitochondrial morphology was observed in primary cultures of TS21 human fetal fibroblasts. Mostly, they exhibited reduced or damaged cristae, which were broken, shorter, concentric or highly swollen (Piccoli et al. 2013; Izzo et al. 2017b). Furthermore the mitochondrial network in DS human fibroblasts from Down syndrome fetuses (DS-HFFs) was highly fragmented when compared with euploid cells (N-HFFs) (Fig. 1) with an increased number of shorter mitochondria and a smaller average mitochondrial volume (Izzo et al. 2017b). An increased fragmentation of mitochondrial network was observed also in primary cultures of TS21 astrocytes and neurons (Helguera et al. 2013). It is known that the perturbation of mitochondrial dynamics, as well as of ultrastructure and volume, are mechanistically linked to mitochondrial function (Izzo et al. 2017b).
Fig. 1.
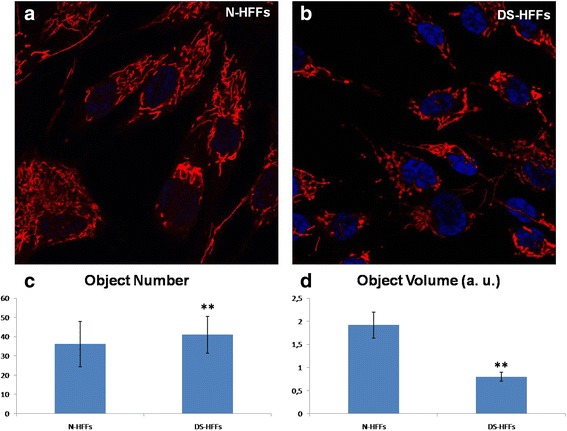
Confocal microscopy of the MitoTracker Red-related fluorescence in N-HFFs versus DS-HFFs. Representative images show that the mitochondrial network is less fragmented in (a) N-HFFs than in (b) DS-HFFs. c The number of mitochondria, measured using the Fiji software (http://www.fiji.sc), is significantly higher in trisomic cells compared with non-trisomic cells. (D) The average mitochondrial volume is significantly lower in DS cells compared with non-trisomic cells. The bars show mean values ± SEM of two non-trisomic and two trisomic cell cultures. Fifteen randomly selected cells for each sample/experimental condition were analyzed. **p ≤ 0.05
Cells were obtained from the ‘Telethon Bank of Foetal Biological Samples’ at the University of Naples. All experimental protocols were approved by the local Institutional Ethics Committee. Abbreviations: N-HFFs: Euploid human fetal fibroblasts; DS-HFFs: Trisomic human fetal fibroblasts; a.u.: arbitrary units.
The observed alterations of the mitochondrial network correlate with the decreased expression of two genes involved in the mitochondrial fusion process, namely OPA1 and MFN2 (Izzo et al. 2017b). A decrease in OPA1 expression is also consistent with the alterations of the proper structure of cristae as OPA1, in addition to its role in mitochondrial fusion, is involved in the maintenance and remodeling of cristae morphology (Frezza et al. 2006; Zick et al. 2009).
The common denominator of most of the altered mitochondrial mechanisms is the peroxisome proliferator activated receptor gamma, coactivator 1 alpha (PGC-1α or PPARGC1A) - a key modulator of mitochondrial function (Scarpulla et al. 2012) – which is significantly downregulated in DS samples (Piccoli et al. 2013) (Conti et al. 2007). Its role in regulating mitochondrial biogenesis and respiratory function is mediated through interaction with transcriptional partners, such as NRF1, ERRa, PPARs and YY1 that modulate the expression of most NEMGs (Scarpulla et al. 2012). Also these PGC-1α transcriptional partners as well as most NEMGs have been found downregulated in DS fetal heart tissue (Conti et al. 2007) and fibroblasts (Piccoli et al. 2013).
In this review, we describe a possible link between the overexpression of Hsa21 genes and mitochondrial dysfunction in DS. We also briefly speculate about possible strategies to restore mitochondrial function and therefore to exert protective effects against DS-associated pathologies.
Hsa21 genes candidate for inducing mitochondrial alterations in DS
A comprehensive meta-analysis from 45 DS gene expression studies (Vilardell et al. 2011) identified 77 Hsa21 genes mostly upregulated across all the studies. These genes are the ones most likely involved in the DS phenotype. Seven of the genes included in this list, namely NRIP1, SUMO3, DYRK1A, RCAN1, SOD1, APP and CBS are directly or indirectly involved in mitochondrial function (Table 1); therefore their dysregulation might account for mitochondrial alterations observed in DS, as discussed below.
Table 1.
List of genes mapping to Hsa21 functionally associated to the mitochondrial abnormalities in DS
| Hsa21 Genes involved in Mitochondrial function | Effects on mitochondrial phenotype |
|---|---|
| NRIP1/RIP140 | Decreases respiratory efficiency and alters morphology of mitochondria. |
| SUMO3 | Modulates NRIP1 repressive activity and attenuates the transcriptional activity of PGC-1α. |
| DYRK1A | Controls PGC-1α via the calcineurin/NFAT pathway. |
| DSCR1/RCAN1 | Controls PGC-1α via the calcineurin/NFAT pathway and is associated with calcium overloading. |
| SOD1 | Is associated with oxidative stress. |
| APP | Induces mitochondrial oxidative stress and mitochondrial dysfunction. |
| CBS | Influences the mitochondrial redox activity negatively regulating methylation processes. |
| ETS2 | Promotes the activation of a mitochondrial death pathway. |
| ITSN1 | Regulates the mitochondrial apoptotic pathway. |
| PREP1 | Inhibits the OXPHOS negatively regulating PGC-1α and mitochondrial fusion genes OPA1 and MFN2. |
| hsa-mir-155 | Affects mitochondrial biogenesis by targeting TFAM. |
| hsa-let-7c | May affect mitochondrial function by targeting ANT1. |
OXPHOS Oxidative phosphorylation
We have recently demonstrated that the over-expression of a transcriptional corepressor gene mapping to Hsa21, the nuclear receptor interacting protein 1 (NRIP1/RIP140), is responsible for decreased respiratory efficiency and altered morphology of mitochondria in DS (Izzo et al. 2014). Many literature reports corroborate the results of this study (Powelka et al. 2006; Fritah et al. 2010). NRIP1 was found overexpressed in the hearts and fibroblasts from DS fetuses as well as in other DS tissues (Conti et al. 2007; Piccoli et al. 2013; Vilardell et al. 2011) and NRIP1 protein was found upregulated in the DS hippocampus (Gardiner 2006). NRIP1 is known to bind and repress several members of the nuclear receptor superfamily, thus regulating gene expression of NEMGs in adipose tissue, heart, muscle and liver (Fritah et al. 2010; Nautiyal et al. 2013). By these actions the gene significantly affects oxidative metabolism and mitochondrial biogenesis (Powelka et al. 2006). NRIP1 activity on mitochondrial pathways is mainly exerted through the repressive control on the transcriptional coactivator PGC-1α and its targets (Scarpulla 2011) including the transcription factors NRF1, ERRα, PPARs, which are repressed by NRIP1 and induced by PGC-1α in a dose dependent manner (Chen et al. 2012). The PPAR family of nuclear receptor genes consists of three isoforms, PPAR-α, PPAR-β/δ, and PPAR-γ expressed in different tissues, which induce gene expression through specific interaction with transcription factors. PPAR-α and PPAR-δ are primarily regulators of lipid oxidation, whereas PPAR-γ promotes lipid synthesis and storage. PPAR-α cooperates with PGC-1α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes (Vega et al. 2000). In addition to their effects on lipid metabolism, PPAR-γ and PPAR-δ promote mitochondrial biogenesis in a cell type-specific manner (Hock and Kralli 2009). Till date several PPAR agonists have been tested for their neuroprotective effects against mitochondrial abnormalities in neurodegenerative diseases (Lee et al. 2009; Agarwal et al. 2017; Skerrett et al. 2015). Notably, in all these studies PPAR-γ agonists induced the expression of endogenous PGC-1α, suggesting that PPAR-γ affects mitochondrial biogenesis indirectly by enhancing the transcription of PGC-1α. Because PGC-1α coactivates PPAR-γ, this element also enables PGC-1α to enhance its own expression in an autoregulatory fashion (Hock and Kralli 2009).
More than one hundred NEMGs have been found upregulated after PGC-1α induction in human osteoblast-like cells (Schreiber et al. 2004). At least 1/3 of these genes are NRIP1 targets. By contrast, PGC-1α null mice (Leone et al. 2005; Mitra et al. 2012), as well as knock-in NRIP1 mice (Seth et al. 2007), show decreased expression of the same mitochondrial genes in multiple tissues. It appears therefore that NRIP1 and PGC-1α have mutually antagonizing roles in the regulation of metabolism in the tissues where they are co-expressed. Accordingly, we have observed that NRIP1 is significantly upregulated whereas PGC-1α is significantly downregulated in DS fetal hearts and fibroblasts at both mRNA and protein level (Conti et al. 2007; Piccoli et al. 2013). Furthermore, after NRIP1 silencing the expression levels of PGC-1α and NEMGs and consequently the mitochondrial function are restored (Izzo et al. 2014).
Another Hsa21 gene, SUMO3 affects mitochondrial function by modulating the NRIP1 repressive activity, as two conserved lysines, Lys756 and Lys1154, located in distinct repression domains of NRIP1, are subject to reversible SUMOylation. The SUMO acceptor lysines have a significant effect on the nuclear distribution of NRIP1 contributing to its corepressor activity (Rytinki and Palvimo 2008). SUMOylation also attenuates the transcriptional activity of PGC-1α, by promoting its transfer outside the nucleus (Rytinki and Palvimo 2009). Even though SUMOylating effects on NRIP1 and PGC-1α have been demonstrated for SUMO-1 and SUMO-2, we presume an identical effect by SUMO-3 that share with SUMO-2 a nearly identical structure (Huq and Wei 2005; Mukhopadhyay and Riezman 2007). SUMO-3 overexpression in DS might therefore be responsible for a concurrent improvement of NRIP1 function and decrease of PGC-1α activity.
The Hsa21 genes, DYRK1A and DSCR1/RCAN1, have been proposed as possible candidates for mitochondrial abnormalities as they control PGC-1α via the calcineurin/NFAT pathway (Arron et al. 2006). It is possible that NFAT proteins may exert a positive effect by interacting functionally with MEF2C or MEF2D on the PGC-1α promoter (Handschin et al. 2003). Accordingly, in DS fetal hearts and fibroblasts, we have observed that whereas NFATc3 and NFATc4 were downregulated, DYRK1A and RCAN1 were upregulated if compared with euploid samples (Piccoli et al. 2013).
DYRK1A has also been proposed as relevant candidate gene in various DS phenotypic traits being involved in multiple cellular pathways including neuronal differentiation, nuclear factor activation and basic cellular metabolism (Park et al. 2009).
RCAN1, also known as calcipressin, has been found chronically overexpressed in the brain of both DS patients and sporadic AD patients (Sun et al. 2011; Wu and Song 2013). Interestingly, RCAN1 overexpression has been linked to oxidative stress and mitochondrial dysfunction (Chang and Min 2005; Ermak et al. 2012) and is strictly related to calcium overloading (Sun et al. 2014), as it affects mitochondrial permeability transition pore (mPTP) function. RCAN1-induced mPTP opening leads to a series of consequences, including Ca2+ retention incapability, massive swelling of mitochondria and rupture of the outer membrane (Sun et al. 2014). In agreement with these data, deregulation of Ca2+ homeostasis and Ca2+ mediated signaling has been described in cells derived from trisomic patients or in murine models of DS (Caviedes et al. 2006; Yamato et al. 2009). Mitochondrial Ca2+ concentration was significantly higher in fetal fibroblasts from DS fetuses than in euploid fetal fibroblasts (Piccoli et al. 2013). Trisomic fibroblasts showed also swelled mitochondria with damaged membranes (Izzo et al. 2017b).
The overexpression of the brain-specific RCAN1.1S isoform in mice promotes early age dependent memory and synaptic plasticity deficits, tau pathology, and dysregulation of dynamin-related protein 1 (DRP1) activity associated with altered mitochondrial dynamics and oxidative stress (Wong et al. 2015). Duan et al. found that RCAN1 induces mitochondrial autophagy and improves cell survival in cardiomyocytes (Duan et al. 2015).
A number of studies have provided evidence that the DS phenotype is associated with oxidative stress. The generation of oxidative stress in DS may be attributed to over-expression of some Hsa21 genes even though molecular signals, such as citrate pathway, may play an important role as key metabolic regulators affecting lipid metabolism and ROS production in DS, with a mechanism not yet completely understood (Convertini et al. 2016).
The redox imbalance in DS has been long attributed to overexpression of Cu, Zn superoxide dismutase SOD1, which has been investigated in many in vitro, ex vivo and animal studies (Brooksbank and Balazs 1984; Epstein et al. 1987). Levels of SOD1 in cells from DS patients are approximately 50% greater than normal (Groner et al. 1994). SOD1, the major SOD in mammalian cells, catalyses the dismutation of superoxide radicals to H2O2 and O2 and is an important antioxidant defence system (Fridovich 1995).
Another gene mapping to chromosome 21 and consistently overexpressed in DS, the amyloid β precursor protein APP, has been proposed to contribute to oxidative stress. Overexpression of APP induces mitochondrial oxidative stress and activates the intrinsic apoptotic cascade (Bartley et al. 2012). In addition, amyloid-β fragments, particularly Aβ42, exert direct toxic effects within cells, including Ca2+ dysregulation, mitochondrial dysfunction, and induction of oxidative stress (Chen and Yan 2007; Chen and Yan 2010; Demuro et al. 2010). Recent studies have demonstrated that APP protein progressively accumulates within mitochondrial matrix leading to increased free radicals and impaired mitochondrial metabolism (Manczak et al. 2010). In addition APP have been shown to translocate to the mitochondria when overexpressed in a human cortical neuronal cell line (Anandatheerthavarada et al. 2003).
The overexpression of genes coding for specific enzymes translates directly into biochemical aberrations that affect multiple metabolic pathways including mitochondrial pathways. An example is given by the alterations that overexpression of the enzyme cystathionine β-synthase (CBS) induces on the methionine/homocysteine pathway in children with DS (Pogribna et al. 2001; Infantino et al. 2011). CBS overexpression indirectly deprives the methionine synthase reaction of one of its precursors, the homocysteine. The significant decrease in plasma methionine levels observed in children with DS may affect the synthesis of S-adenosylmethionine (SAM), the primary methyl donor for cellular methylation reactions. This event is destined to impact mitochondrial redox activity since methylation is a necessary event in mitochondria and relies on the availability and uptake of the methyl donor SAM (Infantino et al. 2011).
Three more Hsa21 genes not included in the list from a meta-analysis by Vilardell (Vilardell et al. 2011) were found to be involved in apoptotic events, namely ETS-2, ITSN1 and PKNOX1/PREP1.
Studies in ETS-2 transgenic mice show that ETS-2 overexpression induces apoptosis of thymus, spleen, and brain cells (Wolvetang et al. 2003). Furthermore ETS-2 promotes the activation of a mitochondrial death pathway in DS neurons (Helguera et al. 2005). Overexpression of ETS-2 induces cytochrome c cytoplasmic translocation and apoptotic features in normal human cortical neurons (Helguera et al. 2005).
The Intersectin-1 s (ITSN1) gene regulates the mitochondrial apoptotic pathway in endothelial cells (Predescu et al. 2007).
Finally, PKNOX1/PREP1, another Hsa21 gene encoding for a homeodomain transcription factor, regulates multiple aspects of embryonic development regulating the homeobox protein Pbx activity (Ferretti et al. 2006). DS human fibroblasts, which express more PREP1, are more sensitive to genotoxic stress. These data suggest that PREP1 may be involved in the apoptotic phenotype of DS tissues (Micali et al. 2009). Recently it has been demonstrated that PREP1 regulates mitochondrial oxidative phosphorylation components via direct and indirect mechanisms (Kanzleiter et al. 2014).
Indirectly PREP1 controls the stability of p160 Myb-Binding Protein, a powerful negative regulator of PGC-1α activity (Fan et al. 2004). In the muscle of Prep1 ablated mice, Pgc-1α expression was increased with consequent increase in mitochondrial capacity (Kanzleiter et al. 2014). Furthermore, it has been shown that PREP1 ablation in muscle leads to an increase in abundance of mitochondrial OXPHOS proteins and an increased citrate synthase activity together with an improved maximal oxidative capacity. ChiP-seq identified Prep1 binding sites in the promoter regions of genes encoding 16 mitochondrial proteins that were also upregulated in skeletal muscle in response to Prep1 ablation. This suggests that PREP1 is a direct negative trascriptional regulator of mitochondrial proteins in addition to its indirect effects via p160-PGC-1α (Kanzleiter et al. 2014). Furthermore, Opa1 and Mfn2, two genes involved in mitochondrial fusion process, were significantly increased in the Prep1 ablated mice (Kanzleiter et al. 2014). According to these results, PREP1 negatively regulates mitochondrial proteins thus affecting both mitochondrial function and dynamics.
Hsa21 miRNAs involved in mitochondrial phenotype
Hsa21 encodes several classes of non-coding RNAs, the most enriched being long non-coding RNAs, while miRNAs are the less represented (Letourneau and Antonarakis 2012). Recent bioinformatic annotations of miRNA database have indicated that Hsa21 harbors 14 miRNAs (Xu et al. 2013), two of them possibly involved in mitochondrial function.
It was recently reported that the Hsa21 miR-155-5p affects mitochondrial biogenesis by targeting the mitochondrial transcription factor A (TFAM) a gene that was found downregulated in trisomic hearts (Quinones-Lombrana and Blanco 2015). TFAM is a nuclear encoded protein that controls the transcription and maintenance of mtDNA and therefore mitochondrial biogenesis.
Another Hsa21 miRNA potential candidate for mitochondrial anomalies is let-7c. By bioinformatics analysis it appears to have several targets among genes that were found downregulated in trisomic fetal hearts and involved in mitochondrial function. Among these targets, we have identified and validated SLC25A4/ANT1, a gene that plays a major role in mitochondrial function as it codes for the main translocator of ADP/ATP across the mitochondrial membrane (Izzo et al., 2017a).
These results support the hypothesis that both miR-155 and let-7c dysregulation might have a potential impact on mitochondrial phenotype.
Conclusions and perspectives
It is now becoming evident that counteracting mitochondrial dysfunction in DS is possible by targeting the NRIP1- PGC-1α axis.
It is puzzling that, although drugs capable of modulating the activity of PGC-1α and/or the downstream PPAR proteins have been available for a long time, few therapeutic approaches have been so far undertaken to correct the overall mitochondrial dysfunction in DS patients. It is known that PGC-1α activity is mainly controlled by PPARs, AMP-activated kinases (AMPKs) and the NAD-dependent deacetylase SIRT1 (Canto and Auwerx 2009). Direct phosphorylation by AMPK promotes PGC-1α dependent induction at the PGC-1α promoter level (Jager et al. 2007), while SIRT1 stimulates PGC-1α activity through deacetylation, thereby inducing mitochondrial biogenesis (Rodgers et al. 2005). Pharmacological activators for these proteins, such as metformin, via AMPK induction, as well as resveratrol, via SIRT1 induction, have been tested in mouse models for neurodegenerative diseases in which mitochondrial alterations play a central role such as AD, Parkinson’s disease, Huntington’s diseases, (Jager et al. 2007; Dong et al. 2007; Lagouge et al. 2006). Recent studies indicated that metformin treatment exerts multiple positive effects on mitochondrial activity in a human cell model of DS (Izzo et al. 2017b), while Resveratrol and Epigallocatechin-3-gallate (EGCG) reverse the severe impairment of mitochondrial bioenergetics and biogenesis in hippocampal progenitor cells from the DS mouse model Ts65Dn (Valenti et al. 2016).
Other pharmacological activators of PGC-1α pathway, including thiazolidinediones, pioglitazones, and bezafibrates, which selectively stimulate PPARs (Landreth et al. 2008), should be tested to restore mitochondrial function in DS.
This will open new therapeutic perspectives to improve the neurological phenotypes in DS and to prevent associated pathologies, such as AD, diabetes, and hypertrophic cardiopathy.
Acknowledgements
We thank Mario Senesi for technical support.
Funding
This work was supported by POR Campania FSE 2007–2013, Campania FSE 2014–2020, Project CREME from Campania Region to L.N.
Abbreviations
- AD
Alzheimer’s disease
- ANT
Adenine nucleotide translocator
- DS
Down Syndrome
- DS-HFFs
Human fibroblasts from Down syndrome fetuses
- Hsa21
Chromosome 21
- mPTP
mitochondrial permeability transition pore
- mtDNA
mitochondrial DNA
- mΔΨ
mitochondrial membrane potential
- NEMGs
Nuclear-encoded mitochondrial genes
- N-HFFs
Human non trisomic fetal fibroblasts
- OCR
Oxygen consumption rate
- OXPHOS
Oxidative phosphorylation
- ROS
Intracellular reactive oxygen species
- TMRM
Mitotropic probe tetramethylrhodamine methyl ester
- TS21
Hsa21 trisomy
Authors’ contributions
AI and NM drafted the manuscript. MN, SP & GC contributed to study correlation between Hsa21 genes and mitochondrial morphology. RG, FB & RC contributed to study correlation between Hsa21 genes/miRNAs and molecular mechanisms of mitochondrial dysfunction. MB & VS contributed to figure preparation and manuscipt and reference formatting. AC is the corresponding author. She contributed to draft the manuscript. LN supervised and critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Cells were obtained from the ‘Telethon Bank of Foetal Biological Samples’ at the University of Naples. All experimental protocols were approved by the local Institutional Ethics Committee.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Agarwal S, Yadav A, Chaturvedi RK. Peroxisome proliferator-activated receptors (PPARs) as therapeutic target in neurodegenerative disorders. Biochem Biophys Res Commun. 2017;483:1166–1177. doi: 10.1016/j.bbrc.2016.08.043. [DOI] [PubMed] [Google Scholar]
- Anandatheerthavarada HK, Biswas G, Robin MA, Avadhani NG. Mitochondrial targeting and a novel transmembrane arrest of Alzheimer's amyloid precursor protein impairs mitochondrial function in neuronal cells. J Cell Biol. 2003;161:41–54. doi: 10.1083/jcb.200207030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuzova S, Hutchin T, Cuckle H. Mitochondrial dysfunction and Down's syndrome. BioEssays. 2002;24:681–684. doi: 10.1002/bies.10138. [DOI] [PubMed] [Google Scholar]
- Arron JR, Winslow MM, Polleri A, Chang CP, Wu H, Gao X, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- Bartley MG, Marquardt K, Kirchhof D, Wilkins HM, Patterson D, Linseman DA. Overexpression of amyloid-beta protein precursor induces mitochondrial oxidative stress and activates the intrinsic apoptotic cascade. J Alzheimers Dis. 2012;28:855–868. doi: 10.3233/JAD-2011-111172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooksbank BW, Balazs R. Superoxide dismutase, glutathione peroxidase and lipoperoxidation in Down's syndrome fetal brain. Brain Res. 1984;318:37–44. doi: 10.1016/0165-3806(84)90060-9. [DOI] [PubMed] [Google Scholar]
- Busciglio J, Pelsman A, Wong C, Pigino G, Yuan M, Mori H, et al. Altered metabolism of the amyloid beta precursor protein is associated with mitochondrial dysfunction in Down's syndrome. Neuron. 2002;33:677–688. doi: 10.1016/S0896-6273(02)00604-9. [DOI] [PubMed] [Google Scholar]
- Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviedes P, Caviedes R, Rapoport SI. Altered calcium currents in cultured sensory neurons of normal and trisomy 16 mouse fetuses, an animal model for human trisomy 21 (down syndrome) Biol Res. 2006;39:471–481. doi: 10.4067/S0716-97602006000300009. [DOI] [PubMed] [Google Scholar]
- Chang KT, Min KT. Drosophila Melanogaster homolog of down syndrome critical region 1 is critical for mitochondrial function. Nat Neurosci. 2005;8:1577–1585. doi: 10.1038/nn1564. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang Y, Chen J, Chen X, Cao W, Chen S, et al. Roles of transcriptional corepressor RIP140 and coactivator PGC-1alpha in energy state of chronically infarcted rat hearts and mitochondrial function of cardiomyocytes. Mol Cell Endocrinol. 2012;362:11–18. doi: 10.1016/j.mce.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Chen JX, Yan SD. Pathogenic role of mitochondrial [correction of mitochondral] amyloid-beta peptide. Expert Rev Neurother. 2007;7:1517–1525. doi: 10.1586/14737175.7.11.1517. [DOI] [PubMed] [Google Scholar]
- Chen JX, Yan SS. Role of mitochondrial amyloid-beta in Alzheimer's disease. J Alzheimers Dis. 2010;20(Suppl 2):S569–S578. doi: 10.3233/JAD-2010-100357. [DOI] [PubMed] [Google Scholar]
- Conti A, Fabbrini F, D'Agostino P, Negri R, Greco D, Genesio R, et al. Altered expression of mitochondrial and extracellular matrix genes in the heart of human fetuses with chromosome 21 trisomy. BMC Genomics. 2007;8:268. doi: 10.1186/1471-2164-8-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convertini P, Menga A, Andria G, Scala I, Santarsiero A, Castiglione Morelli MA, et al. The contribution of the citrate pathway to oxidative stress in down syndrome. Immunology. 2016;149:423–431. doi: 10.1111/imm.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun PE, Busciglio J. Oxidative stress and mitochondrial dysfunction in Down's syndrome: relevance to aging and dementia. Curr Gerontol Geriatrics Res. 2012;2012:383170. doi: 10.1155/2012/383170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuro A, Parker I, Stutzmann GE. Calcium signaling and amyloid toxicity in Alzheimer disease. J Biol Chem. 2010;285:12463–12468. doi: 10.1074/jbc.R109.080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Gao D, Zhang X. Mitochondria biogenesis induced by resveratrol against brain ischemic stroke. Med Hypotheses. 2007;69:700–701. doi: 10.1016/j.mehy.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Duan H, Li Y, Yan L, Yang H, Wu J, Qian P, et al. Rcan1-1L overexpression induces mitochondrial autophagy and improves cell survival in angiotensin II-exposed cardiomyocytes. Exp Cell Res. 2015;335:99–106. doi: 10.1016/j.yexcr.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Epstein CJ, Avraham KB, Lovett M, Smith S, Elroy-Stein O, Rotman G, et al. Transgenic mice with increased cu/Zn-superoxide dismutase activity: animal model of dosage effects in down syndrome. Proc Natl Acad Sci U S A. 1987;84:8044–8048. doi: 10.1073/pnas.84.22.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermak G, Sojitra S, Yin F, Cadenas E, Cuervo AM, Davies KJ. Chronic expression of RCAN1-1L protein induces mitochondrial autophagy and metabolic shift from oxidative phosphorylation to glycolysis in neuronal cells. J Biol Chem. 2012;287:14088–14098. doi: 10.1074/jbc.M111.305342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, et al. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Villaescusa JC, Di Rosa P, Fernandez-Diaz LC, Longobardi E, Mazzieri R, et al. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol Cell Biol. 2006;26:5650–5662. doi: 10.1128/MCB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- Fritah A, Christian M, Parker MG. The metabolic coregulator RIP140: an update. Am J Physiol Endocrinol Metab. 2010;299:E335–E340. doi: 10.1152/ajpendo.00243.2010. [DOI] [PubMed] [Google Scholar]
- Gardiner K. Transcriptional dysregulation in down syndrome: predictions for altered protein complex stoichiometries and post-translational modifications, and consequences for learning/behavior genes ELK, CREB, and the estrogen and glucocorticoid receptors. Behav Genet. 2006;36:439–453. doi: 10.1007/s10519-006-9051-1. [DOI] [PubMed] [Google Scholar]
- Groner Y, Elroy-Stein O, Avraham KB, Schickler M, Knobler H, Minc-Golomb D, et al. Cell damage by excess CuZnSOD and Down's syndrome. Biomed Pharmacother. 1994;48:231–240. doi: 10.1016/0753-3322(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helguera P, Pelsman A, Pigino G, Wolvetang E, Head E, Busciglio J. Ets-2 promotes the activation of a mitochondrial death pathway in Down's syndrome neurons. J Neurosci. 2005;25:2295–2303. doi: 10.1523/JNEUROSCI.5107-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helguera P, Seiglie J, Rodriguez J, Hanna M, Helguera G, Busciglio J. Adaptive downregulation of mitochondrial function in down syndrome. Cell Metab. 2013;17:132–140. doi: 10.1016/j.cmet.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- Huq MD, Wei LN. Post-translational modification of nuclear co-repressor receptor-interacting protein 140 by acetylation. Mol Cell Proteomics. 2005;4:975–983. doi: 10.1074/mcp.M500015-MCP200. [DOI] [PubMed] [Google Scholar]
- Infantino V, Castegna A, Iacobazzi F, Spera I, Scala I, Andria G, et al. Impairment of methyl cycle affects mitochondrial methyl availability and glutathione level in Down's syndrome. Mol Genet Metab. 2011;102:378–382. doi: 10.1016/j.ymgme.2010.11.166. [DOI] [PubMed] [Google Scholar]
- Izzo A, Manco R, Bonfiglio F, Cali G, De Cristofaro T, Patergnani S, et al. NRIP1/RIP140 siRNA-mediated attenuation counteracts mitochondrial dysfunction in down syndrome. Hum Mol Genet. 2014;23:4406–4419. doi: 10.1093/hmg/ddu157. [DOI] [PubMed] [Google Scholar]
- Izzo A, Manco R, de Cristofaro T, Bonfiglio F, Cicatiello R, Mollo N, et al. Overexpression of chromosome 21 miRNAs may affect mitochondrial function in the hearts of down syndrome fetuses. Int J Genomics. 2017;2017:8737649. doi: 10.1155/2017/8737649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo A, Nitti M, Mollo N, Paladino S, Procaccini C, Faicchia D, et al. Metformin restores the mitochondrial network and reverses mitochondrial dysfunction in down syndrome cells. Hum Mol Genet. 2017b; [DOI] [PubMed]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzleiter T, Rath M, Penkov D, Puchkov D, Schulz N, Blasi F, et al. Pknox1/Prep1 regulates mitochondrial oxidative phosphorylation components in skeletal muscle. Mol Cell Biol. 2014;34:290–298. doi: 10.1128/MCB.01232-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Vlkolinsky R, Cairns N, Fountoulakis M, Lubec G. The reduction of NADH ubiquinone oxidoreductase 24- and 75-kDa subunits in brains of patients with down syndrome and Alzheimer's disease. Life Sci. 2001;68:2741–2750. doi: 10.1016/S0024-3205(01)01074-8. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Landreth G, Jiang Q, Mandrekar S, Heneka M. PPARgamma agonists as therapeutics for the treatment of Alzheimer's disease. Neurotherapeutics. 2008;5:481–489. doi: 10.1016/j.nurt.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Kang HJ, Park HH, Hong SC, Kim JK, Cho JH. Effect of peroxisome proliferator-activated receptor gamma agonists on myofibroblast differentiation and collagen production in nasal polyp-derived fibroblasts. Ann Otol Rhinol Laryngol. 2009;118:721–727. doi: 10.1177/000348940911801007. [DOI] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau A, Antonarakis SE. Genomic determinants in the phenotypic variability of down syndrome. Prog Brain Res. 2012;197:15–28. doi: 10.1016/B978-0-444-54299-1.00002-9. [DOI] [PubMed] [Google Scholar]
- Manczak M, Mao P, Calkins MJ, Cornea A, Reddy AP, Murphy MP, et al. Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer's disease neurons. J Alzheimers Dis. 2010;20(Suppl 2):S609–S631. doi: 10.3233/JAD-2010-100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Wang X, Spitznagel EL, Jr, Frelin LP, Ting JC, Ding H, et al. Primary and secondary transcriptional effects in the developing human down syndrome brain and heart. Genome Biol. 2005;6:R107. doi: 10.1186/gb-2005-6-13-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali N, Ferrai C, Fernandez-Diaz LC, Blasi F, Crippa MP. Prep1 directly regulates the intrinsic apoptotic pathway by controlling Bcl-XL levels. Mol Cell Biol. 2009;29:1143–1151. doi: 10.1128/MCB.01273-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Nogee DP, Zechner JF, Yea K, Gierasch CM, Kovacs A, et al. The transcriptional coactivators, PGC-1alpha and beta, cooperate to maintain cardiac mitochondrial function during the early stages of insulin resistance. J Mol Cell Cardiol. 2012;52:701–710. doi: 10.1016/j.yjmcc.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Nautiyal J, Christian M, Parker MG. Distinct functions for RIP140 in development, inflammation, and metabolism. Trends Endocrinol Metab. 2013;24:451–459. doi: 10.1016/j.tem.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Park J, Song WJ, Function CKC. Regulation of Dyrk1A: towards understanding down syndrome. Cell Mol Life Sci. 2009;66:3235–3240. doi: 10.1007/s00018-009-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli C, Izzo A, Scrima R, Bonfiglio F, Manco R, Negri R, et al. Chronic pro-oxidative state and mitochondrial dysfunctions are more pronounced in fibroblasts from down syndrome foeti with congenital heart defects. Hum Mol Genet. 2013;22:1218–1232. doi: 10.1093/hmg/dds529. [DOI] [PubMed] [Google Scholar]
- Pogribna M, Melnyk S, Pogribny I, Chango A, Yi P, James SJ. Homocysteine metabolism in children with down syndrome: in vitro modulation. Am J Hum Genet. 2001;69:88–95. doi: 10.1086/321262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powelka AM, Seth A, Virbasius JV, Kiskinis E, Nicoloro SM, Guilherme A, et al. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J Clin Invest. 2006;116:125–136. doi: 10.1172/JCI26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Predescu SA, Predescu DN, Knezevic I, Klein IK, Malik AB. Intersectin-1s regulates the mitochondrial apoptotic pathway in endothelial cells. J Biol Chem. 2007;282:17166–17178. doi: 10.1074/jbc.M608996200. [DOI] [PubMed] [Google Scholar]
- Quinones-Lombrana A, Blanco JG. Chromosome 21-derived hsa-miR-155-5p regulates mitochondrial biogenesis by targeting mitochondrial transcription factor a (TFAM) Biochim Biophys Acta. 2015;1852:1420–1427. doi: 10.1016/j.bbadis.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi M, Lopes C. Mental retardation in down syndrome: from gene dosage imbalance to molecular and cellular mechanisms. Neurosci Res. 2007;59:349–369. doi: 10.1016/j.neures.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Reeves RH, Baxter LL, Richtsmeier JT. Too much of a good thing: mechanisms of gene action in down syndrome. Trends Gene. 2001;17:83–88. doi: 10.1016/S0168-9525(00)02172-7. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rytinki MM, Palvimo JJ. SUMOylation modulates the transcription repressor function of RIP140. J Biol Chem. 2008;283:11586–11595. doi: 10.1074/jbc.M709359200. [DOI] [PubMed] [Google Scholar]
- Rytinki MM, Palvimo JJ. SUMOylation attenuates the function of PGC-1alpha. J Biol Chem. 2009;284:26184–26193. doi: 10.1074/jbc.M109.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Steel JH, Nichol D, Pocock V, Kumaran MK, Fritah A, et al. The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab. 2007;6:236–245. doi: 10.1016/j.cmet.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukkur EA, Shimohata A, Akagi T, Yu W, Yamaguchi M, Murayama M, et al. Mitochondrial dysfunction and tau hyperphosphorylation in Ts1Cje, a mouse model for down syndrome. Hum Mol Genet. 2006;15:2752–2762. doi: 10.1093/hmg/ddl211. [DOI] [PubMed] [Google Scholar]
- Skerrett R, Pellegrino MP, Casali BT, Taraboanta L, Landreth GE. Combined liver X receptor/peroxisome proliferator-activated receptor gamma agonist treatment reduces amyloid beta levels and improves behavior in amyloid precursor protein/Presenilin 1 mice. J Biol Chem. 2015;290:21591–21602. doi: 10.1074/jbc.M115.652008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wu Y, Chen B, Zhang Z, Zhou W, Tong Y, et al. Regulator of calcineurin 1 (RCAN1) facilitates neuronal apoptosis through caspase-3 activation. J Biol Chem. 2011;286:9049–9062. doi: 10.1074/jbc.M110.177519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wu Y, Herculano B, Song W. RCAN1 overexpression exacerbates calcium overloading-induced neuronal apoptosis. PLoS One. 2014;9:e95471. doi: 10.1371/journal.pone.0095471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti D, de Bari L, de Rasmo D, Signorile A, Henrion-Caude A, Contestabile A, et al. The polyphenols resveratrol and epigallocatechin-3-gallate restore the severe impairment of mitochondria in hippocampal progenitor cells from a down syndrome mouse model. Biochim Biophys Acta. 2016;1862:1093–1104. doi: 10.1016/j.bbadis.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Valenti D, Manente GA, Moro L, Marra E, Vacca RA. Deficit of complex I activity in human skin fibroblasts with chromosome 21 trisomy and overproduction of reactive oxygen species by mitochondria: involvement of the cAMP/PKA signalling pathway. Biochem J. 2011;435:679–688. doi: 10.1042/BJ20101908. [DOI] [PubMed] [Google Scholar]
- Valenti D, Tullo A, Caratozzolo MF, Merafina RS, Scartezzini P, Marra E, et al. Impairment of F1F0-ATPase, adenine nucleotide translocator and adenylate kinase causes mitochondrial energy deficit in human skin fibroblasts with chromosome 21 trisomy. Biochem J. 2010;431:299–310. doi: 10.1042/BJ20100581. [DOI] [PubMed] [Google Scholar]
- Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/MCB.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell M, Rasche A, Thormann A, Maschke-Dutz E, Perez-Jurado LA, Lehrach H, et al. Meta-analysis of heterogeneous down syndrome data reveals consistent genome-wide dosage effects related to neurological processes. BMC Genomics. 2011;12:229. doi: 10.1186/1471-2164-12-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolvetang EJ, Wilson TJ, Sanij E, Busciglio J, Hatzistavrou T, Seth A, et al. ETS2 overexpression in transgenic models and in down syndrome predisposes to apoptosis via the p53 pathway. Hum Mol Genet. 2003;12:247–255. doi: 10.1093/hmg/ddg015. [DOI] [PubMed] [Google Scholar]
- Wong H, Levenga J, Cain P, Rothermel B, Klann E, Hoeffer C. RCAN1 overexpression promotes age-dependent mitochondrial dysregulation related to neurodegeneration in Alzheimer's disease. Acta Neuropathol. 2015;130:829–843. doi: 10.1007/s00401-015-1499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Song W. Regulation of RCAN1 translation and its role in oxidative stress-induced apoptosis. FASEB J. 2013;27:208–221. doi: 10.1096/fj.12-213124. [DOI] [PubMed] [Google Scholar]
- Xu Y, Li W, Liu X, Ma H, Tu Z, Dai Y. Analysis of microRNA expression profile by small RNA sequencing in down syndrome fetuses. Int J Mol Med. 2013;32:1115–1125. doi: 10.3892/ijmm.2013.1499. [DOI] [PubMed] [Google Scholar]
- Yamato F, Takaya J, Yasuhara A, Teraguchi M, Ikemoto Y, Kaneko K. Elevated intracellular calcium in neutrophils in patients with down syndrome. Pediatrics Int. 2009;51:474–477. doi: 10.1111/j.1442-200X.2008.02761.x. [DOI] [PubMed] [Google Scholar]
- Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]


