Abstract
Background
Stratified human keratinocytes (SHKs) are an essential part of mucosal innate immune response that modulates adaptive immunity to microbes encountered in the environment. The importance of these SHKs in mucosal integrity and development has been well characterized, however their regulatory immunologic role at different mucosal sites, has not. In this study we compared the immune gene expression of SHKs from five different anatomical sites before and after HPV16 transfection using microarray analyses.
Methods
Individual pools of human keratinocytes from foreskin, cervix, vagina, gingiva, and tonsils (HFKs, HCKs, HVKs, HGKs and HTLKs) were prepared. Organotypic (raft) cultures were established for both normal and HPV16 immortalized HFKs, HCKs, HVKs, HGKs and HTLKs lines which stably maintained episomal HPV16 DNA. Microarray analysis was carried out using the HumanHT-12 V4 gene chip (Illumina). Immune gene expression profiles were obtained by global gene chip (GeneSifter) and Ingenuity pathway analysis (IPA) for each individual site, with or without HPV16 transfection.
Results
We examined site specific innate immune response gene expression in SHKs from all five different anatomical sites before and after HPV16 transfection. We observed marked differences in SHK immune gene repertoires within and between mucosal tracts before HPV 16 infection. In addition, we observed additional changes in SHKs immune gene repertoire patterns when these SHKs were productively transfected with HPV16. Some immune response genes were similarly expressed by SHKs from different sites. However, there was also variable expression of non-immune response genes, such as keratin genes, by the different SHKs.
Conclusions
Our results suggest that keratinocytes from different anatomical sites are likely hard wired in their innate immune responses, and that these immune responses are unique depending on the anatomical site from which the SHKs were derived. These observations may help explain why select HPV types predominate at different mucosal sites, cause persistent infection at these sites, and on occasion, lead to HPV induced malignant and benign tumor development.
Electronic supplementary material
The online version of this article (10.1186/s10020-018-0022-9) contains supplementary material, which is available to authorized users.
Keywords: Immune responses, Immune pathways, HPV, Keratinocytes, Microarrays
Background
Stratified human keratinocytes (SHKs) are important immunologic components of both healthy and diseased mucosal surfaces in addition to their established role as physical epithelial barriers to infection. Accumulating evidence shows that SHKs from various mucosal tracts are important in mucosal development, inflammation, and HPV-induced cancer development (Wu et al. 2011; Saenz et al. 2008; Swamy et al. 2010; Nestle et al. 2009; Strid et al. 2009). Responses of SHKs can cause immune dysregulation (Swamy et al. 2010; Nestle et al. 2009; Strid et al. 2009; Albanesi et al. 2005; Tonel and Conrad 2009), however they also support the maintenance of the mucosal microbiome, via defensin expression, and they preserve mucosal homeostasis (Chung and Dale 2004; Frohm et al. 1997). Taken together, the importance of SHKs in both innate and adaptive immunity at mucosal sites is compelling (Swamy et al. 2010). Unresolved is the mechanism(s) that render these cells resistant or permissive to select viruses within a given viral family, such as human papillomaviruses (HPVs).
Towards understanding how oral cavity-derived keratinocytes influence adaptive immunity, Wu et al. (Wu et al. 2011) performed a global gene expression analysis that showed the significant effect of murine oral keratinocytes on adaptive immunity (Wu et al. 2011).
We previously reported that human oral tissues are permissive to HPV16 infection and that HPV replication can spread via the oral cavity (Israr et al. 2016). Here we compared the immune gene expression of SHKs using microarray analyses of pooled human keratinocytes from tonsil, foreskin, uterine cervix, vagina, and gingiva, before and after HPV16 transfection. We also compared immune gene network expression by SHKs taken from each of these anatomical sites to determine how they respond to HPV16 transfection. We observed marked differences in SHK immune gene expression within a given mucosal tract and by those derived from different mucosal tracts. In addition, we observed additional changes in SHK immune gene expression patterns when these SHKs were productively transfected with HPV16.
Methods
SHKs cultures, generation of HPV16 positive cell lines
Individual pools of primary SHKs from foreskin, cervix, vagina, gingiva, and tonsils (HFKs, HCKs, HVKs, HGKs and HTLKs) were grown as previously reported (McLaughlin-Drubin et al. 2004; McLaughlin-Drubin and Meyers 2005). Organotypic (raft) cultures were established as described (Meyers et al. 1997; McLaughlin-Drubin et al. 2005). Each pool of SHKs was prepared from 3 to 5 individual healthy donors. Both normal and HPV16 immortalized HFKs, HCKs, HVKs, HGKs and HTLKs lines which stably maintained episomal HPV16 DNA, and were seeded onto rat tail type 1 collagen matrices with J2 3 T3 feeder cells (Israr et al. 2016).
HPV16 infection of these SHKs could be more closely analogous to a natural infection than transfection. However, HPV16 infection of keratinocytes results in low efficiency and high variability between cell lines. Second, cells often lose more virus during further propagation (i.e.) in raft cultures, while HPV16 transfection provides high uptake efficiency, high cell line consistency, and rare heterogeneity. In addition, during transfection cells stably maintain the HPV genome as an episome, which is a critical step for a productive HPV16 life cycle and persistent HPV infection. Furthermore, integration of the HPV16 genome into keratinocyte DNA suppresses virus expression, and prevents formation of small, circular HPV16 genomes that can be packaged and transmitted to a new host keratinocyte (McBride and Warburton 2017).
RNA isolation and microarrays analysis
Triplicate cultures, of each SHK pool were harvested, and total RNA isolated using an RNeasy mini kit (Qiagen). Microarray analysis was carried out using the HumanHT-12 V4 gene chip (Illumina) containing 47231 probes targeting > 25,000 human genes. Expression data (baseline + HPV infected for each site) was log transformed and normalized to the median. Initial comparisons were performed using GeneSifter software and a volcano plot constructed (p = 0.05). Genes with ≥ 2 fold expression changes were considered significant in these analyses.
Comparison of gene expression by SHKs from different mucosae
A list of “immunologic genes” was obtained by querying http://ctdbase.org/ using key words “immu*, interleukin, cytokine, and defensin”. Duplicates were removed, resulting in 2576 unique genes. A search for these genes in GeneSifter was done, sorted by expression using Kruskal-Wallis, (p cutoff = 0.02) resulting in 81 differentially expressed genes. A principle component analysis (PCA) was performed (dots = genes; lines with arrows = tissue types; axis 3 principle components) and the tissue types were separated using PC1, PC2, and PC3, to show differential gene expression patterns for these immunologic genes.
We then performed a pairwise analysis (normalized to all medians, p-value cutoff< 0.05) comparing individual SHKs from a single mucosal tract before and after HPV16 transfection. Among all differentially expressed genes (DEGs), a subset of genes known to be involved in immune responses/associated with immune regulation was selected for analysis. Expression data was analyzed using Ingenuity pathway analysis software: (IPA®, QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis). Networks were generated using the Core analysis tool (Kramer et al. 2014).
To better visualize the immunological genes uniquely, and/or commonly expressed by SHKs from each of the different anatomical sites, with or without HPV transfection, we applied these data sets to software at http://bioinformatics.psb.ugent.be/webtools/Venn/ to generate a 5 intersection Venn/Euler diagram (Fig. 2e) showing which DEGs were in expressed at each intersection.
Fig. 2.
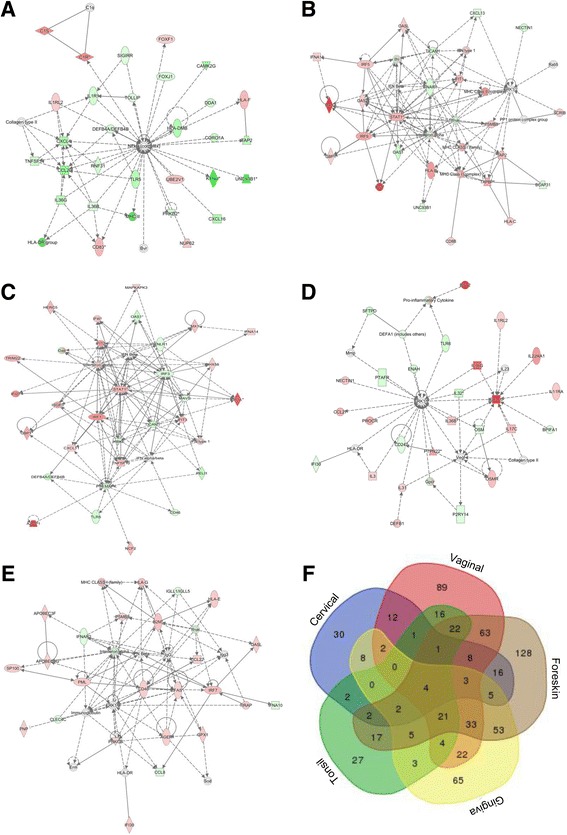
Immune gene network expression by HPV16+ stratified human keratinocytes from different anatomical sites vary significantly. Pairwise analysis, normalized to all medians (p-value cutoff < 0.05) was performed individually to comparing SHKs expression at a single mucosal site before and after HPV16 transfection. Differentially expressed genes were selected from a larger list of genes (immune responses/regulation) for the analysis. Data were analyzed using Ingenuity pathway analysis software (IPA®, QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis). Networks were generated using the Core analysis tool (a-e) (Kramer et al. 2014), and a Venn/Euler diagram was generated (f) to visualize common vs. unique gene expression by applying the data sets of SHKs (different sites, resting vs HPV16+ cells) to software at http://bioinformatics.psb.ugent.be/webtools/Venn/. The number of genes that were uniquely or commonly expressed by cells from each anatomical site are shown within each intersection (f)
Results and discussion
Keratins and immune gene mRNA expression by SHKs with or without HPV16 transfection
First we examined site specific keratins expression in all five different anatomical sites with or without HPV16 transfection that is consistent with published reports (Chu and Weiss 2002). As anticipated, all of the SHKs expressed some of keratin genes, see Additional file 1: Figure S1. However, there was variability in the repertoire of keratin genes that were expressed by different SHKs, suggesting that the cultures of pooled primary keratinocytes grown in rafts maintains the individual gene expression profiles characteristic of the site(s) from which the cells were obtained (Chu and Weiss 2002). As expected, the majority of immune genes having significant levels of expression were expressed at similar levels by all SHK cultures studied, see Fig. 2f. Significantly all 3 GAPDH probe sets, both HPRT-1 probe sets and all 3 B-actin probe sets were detected in all samples at equal levels, as anticipated, since all expression data was normalized prior to analysis. In addition to several keratins present in all tissue types (heat map), IFNAR1 and IFNGR1 and IFNGR2 were detected at equivalent levels in all samples tested, as well as IL-10 and the IL-10RB. Also, both the IL-4R and the IL-13R were expressed by all keratinocytes studied, and the expression of these genes was unaffected by introducing HPV16.
Of note, expression of immune response genes varied by site of SHK origin, see heat map and principal component analysis (Fig. 1a, b). Cluster analysis grouped gingival, vaginal and tonsil cells together, with the latter two being more closely related. Foreskin and cervical cells grouped together, but clearly showed many differences. Within the upper digestive mucosal tract, gingival and tonsil cells also showed differences, for example mRNA expression for individual members of multiple classes of immune genes (CCL-20, CXCL2 and CXCL6, interleukins IL-1, IL-8, and IL1F9 (IL-36γ)), and cell surface receptors/adhesion molecules (IL1RII, IL-13Rα, CD99). Several defensins were expressed by SHKs from these tissues, but not by cervical or foreskin SHKs.
Fig. 1.
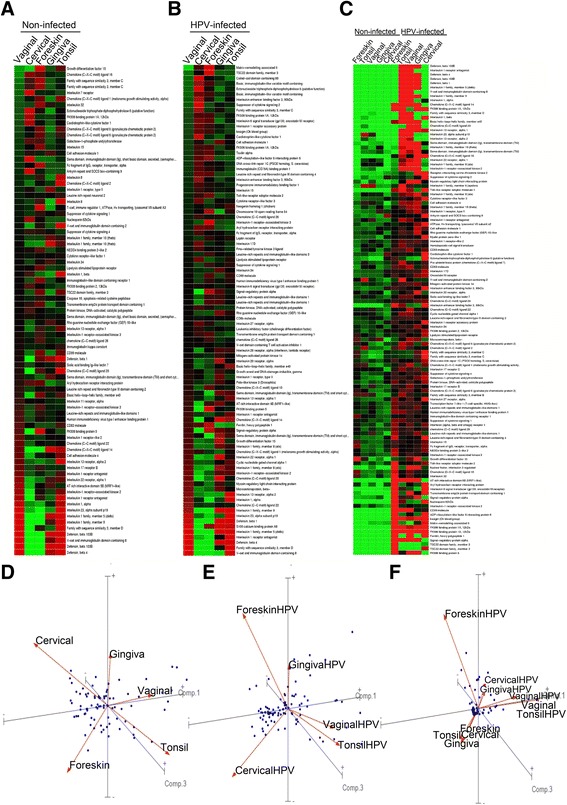
Immune gene mRNA repertoires of stratified human keratinocytes from different anatomical sites vary significantly. Results are for SHKs grown in organotypic cultures from a pool of 3–5 individuals and represent 3 independent samples of each of the mucosal sites shown (baseline and HPV infected). Data were log transformed, and normalized to the median. Eighty one “immunologic genes” are shown, and a principle component analysis (PCA) performed (dots = genes; lines with arrows = tissue types; axis 3 PCA). Tissue types were separated (PC1, PC2, and PC3), to demonstrate differential genetic expression patterns for these genes. Heat maps and the PCA for SHKs from each site are shown for resting (a, d), HPV16 transfected (b, e), and HPV16− or HPV16+ keratinocytes from all sites separately grouped together (c, f)
We examined of all of the chemokines and interleukins present on the gene microarray, and found a limited number of immunologic signaling molecules being expressed in our keratinocyte cultures, largely irrespective of the derivation of the SHKs we studied. A single CCL chemokine, specifically CCL-20 was detected in all samples, together with its receptor CCR6. 2 CXC chemokines, CXCL14 and CXCL16 were also expressed by keratinocytes as well as the CXC receptors CXCR1 and CXCR7. IL8 was also expressed by all keratinocytes studied, while IL-23a was absent from both cervical and foreskin cultures. IL1-A and IL1-B were detected in all keratinocytes studied, albeit at varying levels. However, the antagonist of both 1A and 1B, IL1RN, the IL-20 receptor, and the IL17D receptor were highly expressed by all keratinocytes studied. IL-18 was equally expressed by all keratinocytes studied. In contrast, the IL-1 family member IL1F9, now called IL-36γ, and its antagonist IL1F5 were differentially expressed by different keratinocytes obtained from different anatomical sites.
HPV16 transfected cells also showed marked differences between cell types (Fig. 1b). Under these conditions, (cut off significance = 0.02), 92 different genes were identified. Similar to uninfected cells, vaginal and tonsil cells clustered together. However, gingival cell immune gene expression was quite different, while foreskin and cervical cells clustered together, although foreskin and cervical SHKs were quite different from each other. Among the most highly expressed genes by gingiva, vagina, and tonsil cells was IL-36γ. We previously reported that this cytokine was highly expressed by laryngeal papilloma cells (HPV6/11 infected) (DeVoti et al. 2008; DeVoti et al. 2014). Interestingly, IL36γ was not elevated in HPV16+ foreskin and cervical cells. Thus, SHKs from different mucosae show differential immune genes expression before and after HPV transfection.
Comparing SHKs from different sites as a group before or after HPV transfection (Fig. 1c) showed that SHKs before HPV16 transfection behaved more similarly to each other than to their HPV16+ counterparts which also behaved more similarly to each other (Fig. 1c). Two exceptions were noted, HPV16+ SHKs from foreskin did not express a similar repertoire of immune response genes than HPV16+ cells from other anatomical sites expressed, and HPV16− SHKs vaginal cells expressed similar immune gene profiles as HPV+ cells from other mucosae (Fig. 1c).
Immune gene mRNA networks expressed by SHKs from different sites
We compared mRNA expression of an expanded list of immune genes and regulators by Ingenuity software, at each anatomical site with or without HPV16 transfection (Fig. 2a-e). There were significant differences in gene pathways used by SHKs in response to HPV16 transfection at each site. Differentially expressed genes in these comparisons are show in Additional file 2: Table S1. Fig. 2f (Venn/Euler diagram) shows that there were more differences in genes expressed by SHKs from each site, compared to genes expressed in common by SHKs from all sites.
In summary, immune gene expression by SHKs in health and disease has previously been studied predominantly in the oral cavity (gingiva, tonsil), the skin, and the uterine cervix (Wu et al. 2011; Chung and Dale 2004; Frohm et al. 1997; Nees et al. 2001; Adami et al. 2014). In some of these studies, mucosal biopsies from different epithelial tissues contained cells other than keratinocytes, and they showed novel cytokine expression at different anatomical sites (Frohm et al. 1997). However, there is only a single report of a genome wide analysis of oral mucosa-derived SHKs in mice that demonstrated the essential role of these cells in regulating adaptive immunity (Wu et al. 2011).
Several important points can be drawn from the results presented in this communication. The repertoire of immune response genes expressed by resting SHKs from different mucosal tract show significant differences when compared to each other (Fig. 1). These expression patterns can distinguish one mucosa from another. Even within the same mucosal tract there are marked differences in immune gene expression. These responses are likely to be “hard wired” as SHKs from different sites do not regress to express a common immune gene repertoire in organotypic culture (Fig. 1). HPV16− SHKs from different anatomical sites as a group cluster together and look more similar to each other than do their HPV16+ counterparts, and vice versa. However, there were a few notable exceptions (Fig. 1c). We speculate that the mucosal immune micromilieu at different anatomical sites may influence which immune genes the keratinocytes express as it is likely the stem cells that ultimately differentiate into keratinocytes are the same for all keratinocytes at each site. We have not performed microarrays of naturally infected SHKs from these different anatomical sites, however, our in vitro transfected cervical cell lines showed similar gene expression profiles as previously published in human HPV16 cervical cancers (Nees et al. 2001; Santin et al. 2005; Perez-Plasencia et al. 2007).
Additionally, the immune gene repertoires of HPV16+ SHKs from different sites also differ from each other based on the origin of the SHKs (Fig. 1b). Thus, immune gene expression by these cells appears to also be based on mucosal SHK origin (Fig. 1b). Finally, immune gene networks associated with HPV16+ SHKs from the different anatomical sites, compared to their HPV16 non-transfected counterparts, was not uniform and significantly varied based on the anatomical origin of origin (Fig. 2a-f). The significant variation in immune gene repertoires and networks (Fig. 2f) may have bearing on why individual members of the large HPV family of DNA viruses predominate as the cause of persistent infection and disease within and between different mucosal tracts (Cubie 2013).
Conclusions
In this study, we identify the differences and similarities in mRNA gene expression made by stratified keratinocytes obtained from five different anatomical sites, grown in organotypic cultures before and after HPV16 transduction. Our results show that keratinocytes within and outside of a given mucosal tract show different immune gene repertoires that are site specific, and that introducing HPV16 into these cells also alters their immune gene expression in a site specific manner. Thus, keratinocytes from different anatomical sites are likely hard wired in their innate immune responses, before and after the introduction of HPV16, and that these immune responses are unique depending on the anatomical site from which they were derived.
The potential medical significance of understanding and manipulating differential immunologic gene expression by keratinocytes from different mucosal anatomical sites is intriguing. It is known that at some mucosal sites, HPV 16 predominates as a potential pathogen and causes persistent infection leading to malignancy, while others, like HPV6 and 11 rarely if ever cause disease at that mucosal site. For example, tonsil and base of tongue, HPV-induced disease is caused by HPV16, but not HPV6 or 11. In contrast, HPV 6 and 11 commonly cause persist infection and “benign” respiratory papilloma development in the larynx and upper airway, while HPV16 rarely does at this anatomical site. It is interesting to speculate that differential immune gene expression by keratinocytes from a given mucosal site may convey resistance vs. susceptibility to different HPVs dependent on the repertoire of innate immune responses genes they express at a given mucosal site. This may be so given that the keratinocyte HPV receptor appears to be the same for all HPVs (Schafer et al. 2015; Raff et al. 2013). Thus, an explanation of why certain keratinocytes from a given mucosal site are resistant to a some HPVs, but susceptible to others, and vice versa for keratinocytes at different mucosal sites, may be related to the kind of innate immune signaling that a given keratinocyte expresses at a given mucosal site, before and after HPV infection. If this were the case, understanding how keratinocytes confer resistance or susceptibility to a given HPV at a given mucosal site could potentially open a novel preventative and/or therapeutic modality to block a specific HPV from causing persistent infection, and ultimately benign or malignant tumor development at a specificceptible anatomical site. These observations may help explain why select HPV types predominate at different mucosal sites, and can cause persistent infection, that on occasion, can lead to HPV induced malignancy.
Additional files
Figure S1. Heat map showing the relative gene expression level of keratins among five different stratified human keratinocytes with or without HPV16 transfection. To generate a keratins heat map. All keratin genes, KRT-1 to KRT-86 on the illumina human HT-12v4 expression array were aligned and assigned colors corresponding to the relative expression levels, comparing triplicate cultures of (Strid et al. 2009) types of normal human keratinocytes (NHK) with triplicate cultures from each type transfected with HPV16. KRT-12, − 18, − 20, − 22, KRT-25 to KRT-77, KRT-79, and KRT-82 to KRT-86 were excluded because all 10 triplicates had average expression levels that were below the 95th percentile of all genes on the chip. (XLSX 13 kb)
Table S1. Genes that are differentially expressed by stratified human keratinocytes from different anatomical sites after HPV16 transfection. (DOCX 16 kb)
Acknowledgements
We thank Robert Brucklacher, Genome Sciences and Bioinformatics Core, Penn State College of Medicine, Hershey for technical assistance in microarrays.
Funding
Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research (NIDCR) of the National Institutes of Health under Award Numbers DE017227 to Vincent Bonagura and DE018305 to Craig Meyers.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DEGs
Differentially expressed genes
- HCKs
Human cervical keratinocytes
- HFKs
Human foreskin keratinocytes
- HGKs
Human gingival keratinocytes
- HPVs
Human papillomaviruses
- HTLKs
Human tonsil keratinocytes
- HVKs
Human vaginal keratinocytes
- IPA
Ingenuity pathway analysis
- PCA
Principle component analysis
- SHKs
Stratified human keratinocytes
Authors’ contributions
MI performed the cell and organotypic cultures and microarrays in Dr. Meyers laboratory before moving to Dr. Bonagura laboratory, DR performed the heat map and principal component data analysis, LF-N performed the Ingenuity analysis. CM conceptualized and supported Dr. Israr’s efforts to generate keratinocyte organotypic cultures, and the HPV16 transfection and microarray experiments. VRB conceptualized and supported the experiments that analyzed keratinocyte immune gene expression in the keratinocyte microarray experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This research falls under Northwell Health IRB protocol 13-526B approval date July 7, 2017, expiration date July 6, 2018. Tissue discarded research not involving greater than minimal risk.
Competing interests
The authors declare they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Mohd Israr, David Rosenthal and Lidia Frejo-Navarro are shared co-first authors.
Craig Meyers and Vincent R. Bonagura are shared senior authors
Electronic supplementary material
The online version of this article (10.1186/s10020-018-0022-9) contains supplementary material, which is available to authorized users.
Contributor Information
Mohd Israr, Email: Misrar@northwell.edu.
David Rosenthal, Email: DRosenthal@northwell.edu.
Lidia Frejo-Navarro, Email: lidia.frejo@genyo.es.
James DeVoti, Email: JDevoti@northwell.edu.
Craig Meyers, Email: cmeyers@pennstatehealth.psu.edu.
Vincent R. Bonagura, Email: VBonagura@northwell.edu
References
- Adami GR, et al. Gene expression based evidence of innate immune response activation in the epithelium with oral lichen planus. Arch Oral Biol. 2014;59:354–361. doi: 10.1016/j.archoralbio.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanesi C, Scarponi C, Giustizieri ML, Girolomoni G. Keratinocytes in inflammatory skin diseases. Curr Drug Targets Inflamm Allergy. 2005;4:329–334. doi: 10.2174/1568010054022033. [DOI] [PubMed] [Google Scholar]
- Chu PG, Weiss LM. Keratin expression in human tissues and neoplasms. Histopathology. 2002;40:403–439. doi: 10.1046/j.1365-2559.2002.01387.x. [DOI] [PubMed] [Google Scholar]
- Chung WO, Dale BA. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect Immun. 2004;72:352–358. doi: 10.1128/IAI.72.1.352-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubie HA. Diseases associated with human papillomavirus infection. Virology. 2013;445:21–34. doi: 10.1016/j.virol.2013.06.007. [DOI] [PubMed] [Google Scholar]
- DeVoti JA, et al. Immune dysregulation and tumor-associated gene changes in recurrent respiratory papillomatosis: a paired microarray analysis. Mol Med. 2008;14:608–617. doi: 10.2119/2008-00060.DeVoti. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVoti JA, et al. Decreased Langerhans cell responses to IL-36gamma: altered innate immunity in patients with recurrent respiratory papillomatosis. Mol Med. 2014;20:372–380. doi: 10.2119/molmed.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohm M, et al. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J Biol Chem. 1997;272:15258–15263. doi: 10.1074/jbc.272.24.15258. [DOI] [PubMed] [Google Scholar]
- Israr M, Biryukov J, Ryndock EJ, Alam S, Meyers C. Comparison of human papillomavirus type 16 replication in tonsil and foreskin epithelia. Virology. 2016;499:82–90. doi: 10.1016/j.virol.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J, Tugendreich S., Jr Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AA, Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017;13(4):e1006211. doi: 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Bromberg-White JL, Meyers C. The role of the human papillomavirus type 18 E7 oncoprotein during the complete viral life cycle. Virology. 2005;338:61–68. doi: 10.1016/j.virol.2005.04.036. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Christensen ND, Meyers C. Propagation, infection, and neutralization of authentic HPV16 virus. Virology. 2004;322:213–219. doi: 10.1016/j.virol.2004.02.011. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Meyers C. Propagation of infectious, high-risk HPV in organotypic “raft” culture. Methods Mol Med. 2005;119:171–186. doi: 10.1385/1-59259-982-6:171. [DOI] [PubMed] [Google Scholar]
- Meyers C, Mayer TJ, Ozbun MA. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol. 1997;71:7381–7386. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nees M, et al. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J Virol. 2001;75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat Rev Immunol. 2009;9:679–691. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Plasencia C, et al. Genome wide expression analysis in HPV16 cervical cancer: identification of altered metabolic pathways. Infect Agent Cancer. 2007;2:16. doi: 10.1186/1750-9378-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff AB, et al. The evolving field of human papillomavirus receptor research: a review of binding and entry. J Virol. 2013;87:6062–6072. doi: 10.1128/JVI.00330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin AD, et al. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology. 2005;331:269–291. doi: 10.1016/j.virol.2004.09.045. [DOI] [PubMed] [Google Scholar]
- Schafer G, Blumenthal MJ, Katz AA. Interaction of human tumor viruses with host cell surface receptors and cell entry. Viruses. 2015;7:2592–2617. doi: 10.3390/v7052592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strid J, Tigelaar RE, Hayday AC. Skin immune surveillance by T cells--a new order? Semin Immunol. 2009;21:110–120. doi: 10.1016/j.smim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Swamy M, Jamora C, Havran W, Hayday A. Epithelial decision makers: in search of the ‘epimmunome’. Nat Immunol. 2010;11:656–665. doi: 10.1038/ni.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonel G, Conrad C. Interplay between keratinocytes and immune cells--recent insights into psoriasis pathogenesis. Int J Biochem Cell Biol. 2009;41:963–968. doi: 10.1016/j.biocel.2008.10.022. [DOI] [PubMed] [Google Scholar]
- Wu T, et al. Genome-wide analysis reveals the active roles of keratinocytes in oral mucosal adaptive immune response. Exp Biol Med (Maywood) 2011;236:832–843. doi: 10.1258/ebm.2011.010307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Heat map showing the relative gene expression level of keratins among five different stratified human keratinocytes with or without HPV16 transfection. To generate a keratins heat map. All keratin genes, KRT-1 to KRT-86 on the illumina human HT-12v4 expression array were aligned and assigned colors corresponding to the relative expression levels, comparing triplicate cultures of (Strid et al. 2009) types of normal human keratinocytes (NHK) with triplicate cultures from each type transfected with HPV16. KRT-12, − 18, − 20, − 22, KRT-25 to KRT-77, KRT-79, and KRT-82 to KRT-86 were excluded because all 10 triplicates had average expression levels that were below the 95th percentile of all genes on the chip. (XLSX 13 kb)
Table S1. Genes that are differentially expressed by stratified human keratinocytes from different anatomical sites after HPV16 transfection. (DOCX 16 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


