Abstract
Proteolytic activation of membrane-bound transcription factors has emerged as an important mechanism for the regulation of gene expression. Two membrane-bound transcription factors regulated in this manner are the Saccharomyces cerevisiae proteins Mga2p and Spt23p, which direct transcription of the Δ9-fatty acid desaturase gene OLE1. We now show that a membrane-associated complex containing the highly conserved Npl4p, Ufd1p, and Cdc48p proteins mediates the proteasome-regulated cleavage of Mga2p and Spt23p. Mutations in NPL4, UFD1, and CDC48 cause a block in Mga2p and Spt23p processing, with concomitant loss of OLE1 expression. Taken together, our data indicate that the Npl4 complex may serve to target the proteasome to the ubiquitinated endoplasmic reticulum membrane-bound proteins Mga2p and Spt23p. Given the recent finding that NPL4 is allelic to the ERAD gene HRD4, we further propose that this NPL4 function extends to all endoplasmic reticulum-membrane–associated targets of the proteasome.
INTRODUCTION
Selective ubiquitin-proteasome–dependent degradation of regulatory proteins has emerged as a crucial component of many basic cellular pathways. Temporally regulated ubiquitin-mediated degradation of cyclins, cyclin-dependent kinase inhibitors, and other cell-cycle regulators plays a central role in eukaryotic cell cycle progression (reviewed by Tyers and Jorgensen, 2000). In addition, many oncoproteins, transcription factors, cell growth modulators, and signal transducers undergo regulated degradation in response to internal and external cues. Proteins are marked for degradation through covalent attachment of the small, highly conserved ubiquitin protein. Ubiquitination occurs through a well characterized enzymatic cascade involving classes of enzymes known as E1s (ubiquitin-activating enzymes), E2s (ubiquitin-conjugating enzymes), and E3s (ubiquitin protein ligases). Once a multi-ubiquitin chain is assembled, target proteins are quickly recognized and degraded by the 26S proteasome (reviewed by Hershko and Ciechanover, 1998; Voges et al., 1999).
Most identified targets of the ubiquitin-proteasome degradation pathway are soluble cytoplasmic and nucleoplasmic proteins. However, a major development in recent years has been the finding that endoplasmic reticulum (ER) membrane and luminal proteins are also targeted for destruction via the ubiquitin-proteasome pathway (reviewed by Bonifacino and Weissman, 1998). This ER-associated degradation pathway (termed ERAD) directs the degradation of misfolded ER proteins and unassembled/misassembled protein complexes, as well as ER-resident proteins whose levels are controlled by regulated proteolysis. ERAD involves three general steps: 1) recognition of target proteins, 2) retrotranslocation of target proteins across the ER membrane to the cytosol, and 3) polyubiquitination and degradation by the 26S proteasome. Genetic screens in yeast have identified a number of ER-resident factors required for ERAD function, including the proteins Der1p, Hrd1p/Der3p, and Hrd3p (Hampton et al., 1996; Knop et al., 1996; Bordallo et al., 1998). In addition, components of the Sec61p translocon (through which ERAD substrates are thought to retrotranslocate to the cytoplasm) and components of the cytosolic ubiquitination machinery are required for ERAD function (Hiller et al., 1996; Wiertz et al., 1996; Pilon et al., 1997; Plemper et al., 1997; Zhou and Schekman, 1999).
A second, ubiquitin-independent form of regulated membrane-associated proteolysis has also come to light in recent years. This process, termed regulated intramembrane proteolysis (Rip), results in the proteolytic activation of membrane-bound precursors of regulatory proteins (reviewed by Brown et al., 2000). The first and best understood example of Rip involves activation of the sterol regulatory element-binding proteins (SREBPs; Wang et al., 1994). Full-length SREBP transcription factors are anchored in the ER membrane by two transmembrane domains such that both the N and C termini are cytosolic. In the absence of sterols, two sequential proteolytic events liberate the cytosolic N-terminal transactivation domain, which then enters the nucleus and directs transcription of sterol and fatty acid synthesis genes (Sakai et al., 1996). Both cleavage events, the first within the ER luminal domain (site-1) and the second within the first membrane-spanning domain (site-2), are mediated by site-specific membrane-bound proteases (Rawson et al., 1997; Sakai et al., 1998). Interestingly, the site-2 protease is a member of a newly identified family of zinc-metalloproteases, which also mediate Rip in bacteria (Rudner et al., 1999). Three other regulatory proteins that undergo Rip activation include the Alzheimer's disease-linked ameloid precursor protein, the cell-signaling protein Notch, and the unfolded protein response-signaling kinase Ire1 (reviewed by Brown et al., 2000). In contrast to SREBP, cleavage of these Rip target proteins requires presenilin-1, a polytopic membrane protein that is postulated to be an aspartyl protease (Wolfe et al., 1999; De Strooper, 2000; Steiner et al., 2000).
The Saccharomyces cerevisiae partially redundant membrane-bound transcription factors Mga2p and Spt23p have recently been shown to undergo a unique form of proteolytic activation (Hoppe et al., 2000). Mga2p and Spt23p are required for transcription of the tightly regulated Δ9 fatty acid desaturase gene OLE1 (Zhang et al., 1999). Like SREBPs, these factors are initially made as inactive precursor proteins anchored to the ER/nuclear membrane, in this case via a C-terminal transmembrane domain. Proteolytic processing (fatty acid regulated in the case of Spt23p) then generates active, soluble transcription factors, which presumably enter the nucleus to direct OLE1 transcription. Unlike Rip targets, however, Mga2p and Spt23p processing has been shown to be dependent (either directly or indirectly) on the ubiquitin-proteasome degradation pathway. This unexpected finding challenges our current understanding of proteasome function and ERAD in particular. How the proteasome may direct the precise processing of these ER-membrane–bound factors (in a fatty acid-regulated manner), rather than the more traditional complete degradation of these proteins, remains unknown. The identification of upstream components of this regulated processing pathway will be crucial to addressing this question.
Mutations in the essential S. cerevisiae NPL4 gene result in membrane structural defects including nuclear envelope herniations/protrusions and ER proliferation, which likely lead to the nucleocytoplasmic trafficking defects that facilitated its identification (Bossie et al., 1992; DeHoratius and Silver, 1996). In this study, we present our finding that the Npl4p protein is part of an evolutionarily conserved complex required for the proteasome-dependent processing of Spt23p and Mga2p and subsequent activation of OLE1 transcription. These results, along with the recent finding that NPL4 corresponds to the ERAD gene HRD4 (Bays et al., 2001), suggest that Npl4p may be part of a general machinery responsible for delivery of the proteasome to ubiquitinated ER-associated target proteins.
MATERIALS AND METHODS
Yeast Strains and Manipulations
A summary of yeast strains used in this study is provided in Table 1. Media were prepared according to standard methods (Adams et al., 1997). Growth media were supplemented with unsaturated fatty acids (UFAs) as described by Stukey et al. (1989). Briefly, palmitoleic acid (16:1; Sigma, St. Louis, MO) and oleic acid (18:1; Sigma) were added to 0.25 mM each ([UFA]total = 0.5 mM); 1% Tergitol (Fluka) was included to aid in solubilization. Yeast transformations were performed with the use of the high-efficiency lithium acetate/single-stranded carrier DNA/polyethylene glycol method (Agatep et al., 1998).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FY23 | MATa ura3-52 leu2Δ1 trp1Δ63 | Winston et al., 1995 |
| MLY1640 | MATa leu2-3,-112, Δpep4::URA3 cdc48-3 | Latterich et al., 1995; Moir et al., 1982 |
| MLY2004 | MATα ura3-52 cdc48-2 | Moir et al., 1982 |
| PM373 | MATa ura3-52 leu2-3,-112 his4-519 ade1-100 ufd1-1 | Johnson et al., 1995 |
| PSY825 | MATa ura3-52 leu2Δ1 npl4-1 | DeHoratius and Silver, 1996 |
| PSY826 | MATa ura3-52 his3Δ200 npl4-2 | DeHoratius and Silver, 1996 |
| PSY1602 | MATa/α ura3-52/ura3-52 leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 TRP1/trp1Δ63 | This study |
| PSY2340 | MATa ura3-52 leu2Δ1 trp1Δ63 npl4-1 | This study |
| PSY2341 | MATa ura3-52 leu2Δ1 trp1Δ63 npl4-2 | This study |
| PSY2342 | MATa ura3-52 leu2Δ1 trp1Δ63 rpn4Δ::HIS3 | This study |
| PSY2343 | MATa ura3-52 leu2Δ1 his3Δ200 npl4Δ::NPL4-GFP::URA3 | This study |
| PSY2344 | MATa ura3-52 leu2Δ1 trp1Δ63 npl4Δ::NPL4-pA-6HIS::URA3 | This study |
| PSY2373 | MATα ura3-52 leu2Δ1 trp1Δ63 npl4-1 | This study |
| PSY2374 | MATa ura3-52 leu2Δ1 his3Δ200 npl4-1 | This study |
| PSY2377 | MATa ura3-52 leu2Δ1 npl4-1 SPT23::Tn3::LEU2 | This study |
| SZ66 | MATa his3Δ200 lys2-128g ura3 leu2Δ-hisG ole1Δ::LEU2 | Zhang et al., 1999 |
The npl4-1 and npl4-2 strains used in this study (PSY2340, PSY2373, PSY2374, and PSY2341) are FY23-backcrossed strains derived from PSY825 and PSY826, which were previously described (DeHoratius and Silver, 1996). To generate the rpn4Δ strain PSY2342, a polymerase chain reaction (PCR)-based method (Baudin et al., 1993) was used to replace the RPN4 gene with the HIS3 marker in the FY23-derived diploid strain PSY1602. After sporulation, a single rpn4Δ HIS+ spore was designated as PSY2342. To generate the Npl4p-green fluorescent protein (GFP)–expressing strain PSY2343, the PSY1602 diploid was transformed with pPS2009 linearized within the NPL4 sequence by NruI digestion. URA+ diploids were checked for proper expression of the Npl4p-GFP fusion protein by anti-GFP Western blotting (1:5000 dilution; Seedorf et al., 1999). A URA+ haploid strain was isolated by sporulation, reconfirmed for Npl4p-GFP expression, and designated PSY2343. The Npl4p-protein A (pA)–expressing strain PSY2344 was generated as for PSY2343, except that NruI-linearized pPS2015 was transformed directly into a wild-type haploid strain (FY23). Functionality of both Npl4p fusion proteins was confirmed by viability of the resulting haploid strains.
Plasmids
A summary of plasmids used in this study is provided in Table 2. To generate the NPL4-GFP integration plasmid (pPS2009), DNA encoding a C-terminal fragment of Npl4 was amplified by PCR from wild-type yeast genomic DNA. NotI and XhoI sites were incorporated at the 5′ and 3′ ends of this PCR fragment, respectively, to facilitate cloning and maintain the open reading frame. NotI/XhoI-digested PCR product was then cloned into NotI/XhoI-digested pPS967 (YIp NUF2-GFP URA3) backbone (Kahana et al., 1998). To generate the NPL4-pA-6His integration plasmid (pPS2015), the GFP fragment from pPS2009 was removed by XhoI/HindIII digestion and replaced by an XhoI/HindIII fragment containing four tandem repeats of the immunoglobulin (Ig) G-binding domain of protein A (2XpA) and a 6-His tag isolated from pPS1634, a derivative of pPS1656 (YCp NUF2-2XpA-6His URA3; Kahana et al., 1998). To generate a fusion of GFP to the N terminus of SPT23 under the control of the GAL1 promoter (pPS2348), the SPT23 open reading frame was PCR amplified as an SphI/XhoI fragment from pBDG769. The SphI/XhoI-digested PCR product was ligated into SphI/SalI-digested pCGF-1A (YEp pGAL1 GFP URA3; Kahana and Silver, 1996). A fusion of GFP to the N terminus of MGA2 under the control of the GAL1 promoter (pPS2351) was generated as for pPS2348 except that the MGA2 open reading frame was PCR amplified from pBDG965 as an HindIII/SalI fragment and cloned in-frame into HindIII/SalI-digested pCGF-1A.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pBDG769 | YEp SPT23 URA3 | Zhang et al., 1999 |
| pBDG965 | YEp MGA2 URA3 | Zhang et al., 1999 |
| pBDG982 | YEp OLE1 URA3 | Zhang et al., 1999 |
| pPS356 | YEp SON1(RPN4) URA3 | Nelson et al., 1993 |
| pPS703 | YEp URA3 | Christianson et al., 1992 |
| pPS806 | YEp NPL4 URA3 | DeHoratius and Silver, 1996 |
| pPS1914 | YEp HCS#1 URA3 | This study |
| pPS1915 | YEp HCS#9 URA3 | This study |
| pPS1916 | YEp HCS#40 URA3 | This study |
| pPS2009 | YIp NPL4-GFP URA3 | This study |
| pPS2015 | YIp NPL4-2XpA-6His URA3 | This study |
| pPS2019 | YEp MGA2(partial) URA3 | This study |
| pPS2348 | YEp pGAL1 GFP-SPT23 URA3 | This study |
| pPS2351 | YEp pGAL1 GFP-MGA2 URA3 | This study |
| YEp105 | YEp pCUP1 Myc-Ub TRP1 | Ellison and Hochstrasser, 1991 |
Microscopy
Live cells were viewed directly from midlog phase cultures grown in liquid synthetic dropout medium. Fluorescent signal was observed with the use of a Nikon fluorescence microscope equipped with a GFP and 4′,6-diamidino-2-phenylindole (DAPI) filter set (Chroma Technology, Brattleboro, VT) and a 100× DIC (Nomarski) objective. Images were captured by a Micromax digital camera (Princeton Instruments, Trenton, NJ) with Metamorph imaging software (Universal Imaging, Media, PA). To prepare fixed cells for microscopy, cell cultures were fixed in 3.7% formaldehyde for 5 min. Cells were washed twice in 0.1 M potassium phosphate buffer, pH 6.5, and permeabilized by 0.5% Triton X-100 in P solution (1.2 M sorbitol, 0.1 M potassium phosphate buffer, pH 6.5). After two washes with phosphate-buffered saline, DNA was stained with 5 ng/ml DAPI for 1 min at room temperature. Cells were washed two to three times with phosphate-buffered saline before viewing under the microscope. For Figure 2D, images were acquired and processed on a DeltaVision platform (Applied Precision, Inc., Issaquah, WA). Data were collected in 0.1-μm sections and subjected to five cycles of iterative deconvolution (Agard et al., 1989). A central plane from each cell is presented.
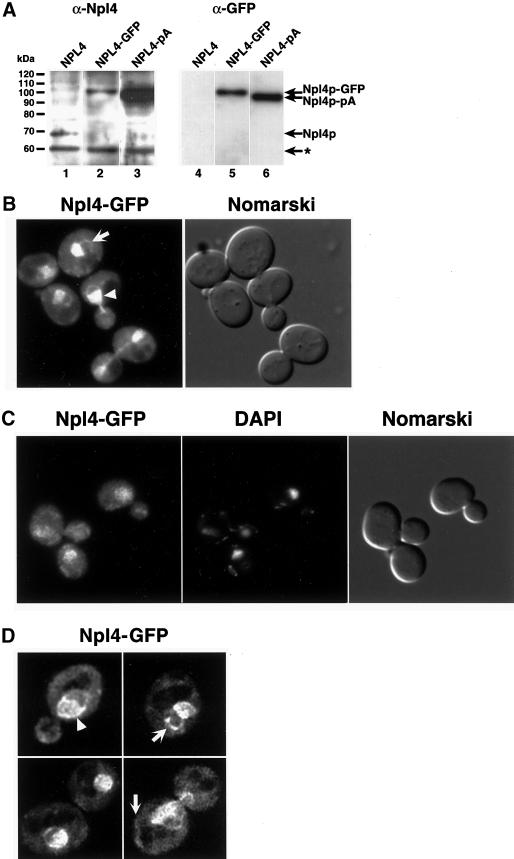
Figure 2.
Npl4p-GFP localizes to ER/nuclear membranes. (A) Anti-Npl4 (left) and anti-GFP (right) Western blots of whole cell extracts derived from strains expressing endogenous Npl4p (lanes 1 and 4), a genomically tagged Npl4p-GFP (lanes 2 and 5), and a genomically tagged Npl4p-protein A(pA) (lanes 3 and 6). The anti-Npl4 blot shows the precise replacement of endogenous Npl4p with the two different C-terminally tagged proteins (compare lanes 2 and 3 to lane 1). The robust reactivity of anti-Npl4 with Npl4-pA (lane 3) is a result of antibody interactions with Npl4p epitopes as well as the IgG-binding pA tag. The rabbit anti-GFP blot recognizes both Npl4p-GFP and Npl4p-pA (lanes 5 and 6, respectively). The identity of the different proteins is indicated by arrows. The asterisk indicates an unidentified cross-reacting protein. (B) Npl4p-GFP localization as observed by fluorescence microscopy. The live cells were viewed at midlog phase. Localization is mostly nuclear (white arrowhead) and cytoplasmic, with an apparent concentration at perinuclear membranes. The white arrow indicates an example of Npl4p-GFP localization to peripheral membranes. A Nomarski image of the cells is shown to the right. (C) Npl4p-GFP colocalization with DAPI-stained nuclear DNA. Cells were fixed in formaldehyde, permeabilized with Triton X-100, and DAPI stained before fluorescence microscopy. The majority of Npl4p-GFP localizes to a region overlapping with and surrounding the nucleus as observed by DAPI staining. A Nomarski image of the cells is shown to the right. (D) Npl4p-GFP expression as viewed by fluorescence microscopy on a DeltaVision platform. Sequential 0.1-μm fluorescence images were taken through a diploid yeast strain expressing Npl4p-GFP from both NPL4 loci and subjected to five rounds of deconvolution. The images presented represent a central plane from deconvolved data. The white arrowhead indicates an example of Npl4p-GFP concentration at perinuclear membranes. White arrows indicate examples of Npl4p-GFP localization to peripheral membranes.
Cell Fractionation, Microsome Purification, and Membrane Extraction
Microsome purification was performed essentially as described by Latterich and Schekman (1994). To determine the fractionation profile of Npl4p, equal protein from the crude lysate, high-speed supernatant, and microsomal fractions was analyzed by Western blot analysis with anti-Npl4 antibodies (DeHoratius and Silver, 1996). To determine the extraction profile of Npl4p from microsomal membranes, 6× 150 μg of purified wild-type microsomes were pelleted and resuspended in 1 ml of B88 buffer (20 mM HEPES, pH 6.8, 150 mM KOAc, 5 mM Mg(OAc)2, 250 mM sorbitol), 1 M KOAc in B88, 2 M KOAc in B88, 3 M urea in B88, 0.1 M Na2CO3, pH 11, in B88, or 1% Triton X-100 in B88. Samples were vortexed for 1 min at room temperature and separated into pellet and supernatant fractions by centrifugation. Pellets were resuspended in 150 μL of 1× protein sample buffer, and supernatant fractions were trichloroacetic acid precipitated and resuspended in 150 μL of 1× protein sample buffer. Each sample (20 μg) was separated by SDS-PAGE and Western blotted with anti-Npl4 (1:500 dilution), anti-Cdc48 (1:1000 dilution) (Latterich et al., 1995), anti-Sec17 (1:1000 dilution), or anti-Sec62 (1:10,000 dilution).
Npl4p-pA Purification and Identification of Copurifying Proteins
Npl4p-pA and copurifying proteins were isolated essentially as described by Siniossoglou et al. (1997). Briefly, yeast cells expressing either untagged or protein A-tagged Npl4 were grown in 1 l of YEPD (yeast extract, peptone, dextrose) at 25°C to OD600 = 1.5, spheroplasted, and frozen at −20°C overnight. After thawing on ice, spheroplasts were centrifuged, resuspended in 20 ml of lysis buffer (150 mM KCl, 20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1% Triton X-100, plus 2 mM phenylmethylsulfonyl fluoride and 2.5 μg/ml each pepstatin A, leupeptin, aprotinin, and chymostatin), and lysed on ice with 10 strokes by hand in a 40-ml Dounce homogenizer. The resulting homogenate was centrifuged at 13,000 × g and the supernatant was applied to a column containing 500 μL of prepared IgG-Sepharose beads. After washes, bound proteins were eluted with acetic acid, trichloroacetic acid precipitated, and resuspended in 30 μL of SDS protein sample buffer (1/5000 original lysate volume). Each sample (25 μL) was separated by SDS-PAGE, and proteins were visualized by Coomassie blue staining.
The bands of interest were excised and subjected to in-gel tryptic digestion as described by William et al. (1997). The resulting peptide mixtures were extracted and analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy at the Dana-Farber Cancer Institute Molecular Biology Core Facility (Boston, MA). The determined sizes of tryptic fragments were used to search for protein candidates in a composite, nonredundant protein sequence database (OWL) with the use of the Mascot search program (Perkins et al., 1999).
npl4-1 High-Copy Suppressor Screen
The npl4-1 strain PSY825 was transformed with a high-copy (2 μ URA3) yeast genomic library (gift of C. Connelly and P. Hieter), and suppressors were selected for growth at 33°C. Twenty isolates demonstrated plasmid-dependent rescue of growth and were further analyzed. PstI digestion indicated that three different inserts were represented in the 20 clones. HCS#1 (pPS1914) was found 14 times, and HCS#9 (pPS1915) and HCS#40 (pPS1914) were found three times each. Sequencing and subcloning revealed that both pPS1914 (HCS#1) and pPS1915 (HCS#9) contained a partial MGA2 open reading frame (lacking the coding sequence for the C-terminal 199 amino acids) responsible for npl4-1 high-copy suppression. The truncated MGA2 subclone (pPS2019) was generated by isolating and self-ligating the KpnI-digested pPS1914 backbone. The ability of pPS1916 (HCS#40) to rescue npl4-1 growth at 33°C was found to be due to the OLE1 gene.
npl4-1 Extragenic Supressor Screen
The npl4-1 strains PSY2373 and PSY2374 were transformed at midlog phase with an NotI-digested yeast genomic library that had been mutagenized in vivo in Escherichia coli with a Tn3-derived minitransposon carrying the yeast-selectable marker LEU2 (Kumar and Snyder, 2000), and suppressors were selected at 30°C. The resulting mutants (200) were tested for recessive/dominant behavior by crossing to the parent npl4-1 strain of the opposite mating type. In all cases, the resulting diploid was able to grow at 30°C, indicating that all isolated suppressors were dominant. One such mutant (PSY2377) showed a strict linkage of LEU+ and growth at 30°C upon sporulation and was further analyzed. Isolation and identification of genomic DNA flanking the site of transposon insertion in this mutant (Kumar and Snyder, 2000) showed that the transposon is integrated in-frame 2133 nucleotides into the coding sequence of the SPT23 gene on chromosome XI.
Northern Blot Analysis
To perform temperature shift analysis of WT, npl4-1, npl4-2, cdc48-3, ufd1-1 and rpn4Δ strains, cells were grown at 25°C to OD600 = 0.3 and then shifted to 37°C. After various lengths of time at 37°C, cells were harvested (a T = 0 time point was also taken before temperature shift). All samples were immediately frozen at −20°C such that all RNA preparations were performed in parallel. Total RNA was probed with an [α-32P]dCTP-labeled 1.2-kb internal SalI/PacI OLE1 fragment isolated from pBDG769 and a 300-bp ACT1 (actin) probe from pPS332. To determine relative OLE1 levels, OLE1 and ACT1 signals for each sample were quantitated by phosphorimager analysis. After subtracting background, an OLE1/ACT1 ratio was calculated for each sample and normalized relative to the OLE1/ACT1 ratio for wild-type cells grown at 25°C in the absence of UFAs.
Spt23p and Mga2p Ubiquitination
Immunoprecipitations of GFP-Mga2p and GFP-Spt23p were performed in wild-type and mutant strains coexpressing either pPS2348 (YEp pGAL1 GFP-SPT23 URA3) or pPS2351 (YEp pGAL1 GFP-MGA2 URA3) and YEp105 (YEp pCUP1 Myc-Ub TRP1; Ellison and Hochstrasser, 1991). Cotransformants were grown to OD600=0.4 before simultaneously inducing expression of the GFP-Spt23p/Mga2p fusion proteins and Myc-Ub by the addition of 2% galactose and 150 μM CuSO4, respectively. As a control, parallel cultures were incubated with 2% glucose in place of galactose to repress expression of the GFP-Spt23p/Mga2p fusion proteins. After 2 h of induction at 25°C, 3 OD · ml cells were collected and processed for immunoprecipitation as described by Franzusoff et al. (1991) with the use of 2.5 μg of polyclonal anti-GFP (Seedorf et al., 1999). Equal volumes of bound fractions were separated by SDS-PAGE and immunoblotted with either anti-GFP (1:5000 dilution; Seedorf et al., 1999) or monoclonal anti-Myc (9E10) antibodies (1:1000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA).
RESULTS
The Npl4 Protein Is Highly Conserved
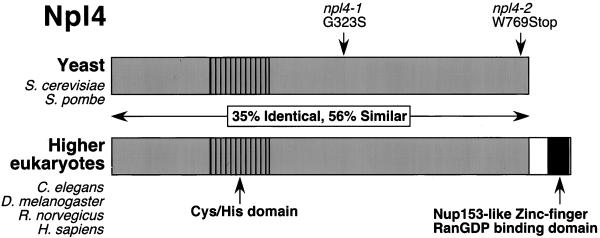
The essential S. cerevisiae Npl4p protein has been conserved throughout eukaryotic evolution. Figure 1 depicts a schematic of yeast (S. cerevisiae and Schizosaccharomyces pombe) and higher eukaryotic (Caenorhabditis elegans, Drosophila melanogaster, Rattus norvegicus, and Homo sapiens) Npl4 proteins. In total, identity and similarity among these proteins are ∼35 and 57%, respectively. The Npl4 proteins contain two notable features. First, several perfectly conserved cysteine and histidine residues in the N terminus may represent a novel Zn2+-binding domain that could mediate protein-DNA or protein-protein interactions. Second, the higher eukaryotic Npl4 proteins contain a C-terminal predicted Zn2+-finger Ran-GDP–binding domain similar to that found in the human nuclear pore protein Nup153 (Nakielny et al., 1999; Yaseen and Blobel, 1999).
Figure 1.
The Npl4 protein is evolutionarily conserved. The sequence homology between S. cerevisiae Npl4p protein (ScNpl4p) (accession number CAA85131), S. pombe predicted protein SPBC1711.10c (SpNpl4) (accession number CAB88240), C. elegans proteins F59E12.4 (CeNpl4[1]) (accession number AAB54254) and F59E12.5 (CeNpl4[2]) (accession number AAB54255), D. melanogaster predicted protein CG4673 (DmNpl4) (accession number AAF56480), R. norvegicus (rat) Npl4 (Meyer et al., 2000), and predicted H. sapiens (human) protein FLJ20657 (HsNpl4) (accession number NP 060391) is depicted schemati-cally. The D. melanogaster Npl4 protein contains a large amino acid insertion omitted from this alignment. Ten absolutely conserved cysteine and histidine residues in the N terminus may constitute a novel Zn2+-finger domain. A C-terminal extension of higher eukaryotic Npl4 homologues encodes a predicted Nup153-like zinc-finger RanGDP-binding domain. The location of residues affected by mutations in the S. cerevisiae npl4-1 and npl4-2 mutants are indicated. The npl4-1 allele substitutes serine for the conserved glycine at position 323, and the npl4-2 mutation results in the production of a truncated protein lacking the C-terminal 12 amino acids.
Two temperature-sensitive alleles of the S. cerevisiae NPL4 gene have been characterized in our laboratory. The first allele, npl4-1, was generated in the nuclear protein localization (NPL) screen (Bossie et al., 1992), and the second allele, npl4-2, was isolated from a library of temperature-sensitive yeast mutants by noncomplementation with npl4-1 (DeHoratius and Silver, 1996). Inspection of the sequence from the NPL4 promoter, open reading frame, and 3′-untranslated region from the wild-type and mutant strains revealed that npl4-1 harbors a G→A mutation that substitutes a serine for glycine at position 323. This glycine residue is absolutely conserved in all Npl4 homologues (Figure 1), and lies within a block of amino acids that show strong conservation. The npl4-2 allele contains a G→A mutation that places a premature stop codon 12 amino acids from the C terminus of wild-type Npl4p (Figure 1).
Npl4p Localizes to Perinuclear Membranes
Previous studies have suggested that Npl4p may be a component of the nuclear pore given its localization in fixed cells by indirect immunofluorescence to the nuclear rim (DeHoratius and Silver, 1996). To localize Npl4p in living yeast cells, we integrated DNA encoding a C-terminal GFP tag at the NPL4 genomic locus. Western blotting indicated that the resulting fusion protein was full-length and replaced endogenous untagged Npl4p (Figure 2A, lanes 2 and 5). Viability of the resulting strain confirms the functionality of Npl4p-GFP as it is the only form of Npl4p present in the cells. By fluorescence microscopy of living yeast cells, Npl4p-GFP appears to localize mainly to the nucleus (Figure 2B, arrowhead) and cytoplasm, possibly with a concentration at perinuclear membranes. In some cases, Npl4p-GFP signal can also be detected in what appears to correspond to cortical membranes (Figure 2B, arrow). To confirm that the membrane localization of Npl4p-GFP is perinuclear, we formaldehyde fixed and DAPI stained Npl4p-GFP-expressing yeast cells. As can be seen in Figure 2C, the majority of Npl4p-GFP signal can be seen in a region overlapping with or around the nuclear DNA. The diffuse cytoplasmic pool of Npl4p-GFP can also be seen in these cells. Finally, to analyze the Npl4p-GFP localization pattern in more detail, we viewed live cells on a DeltaVision platform. Images were captured in 0.1-μm sections through each cell and were subjected to deconvolution to minimize background and out-of-focus signal (Agard et al., 1989). After deconvolution, the presence of Npl4p-GFP at perinuclear (Figure 2D, arrowhead) and peripheral membranes (Figure 2D, arrows) was especially apparent. This perinuclear membrane signal of Npl4p-GFP is very similar to that expected for a protein that associates with ER and nuclear membranes (which are contiguous in yeast).
To biochemically confirm this Npl4p localization pattern, yeast extracts were separated into soluble and microsomal (ER/nuclear membrane) fractions (see MATERIALS AND METHODS). Immunoblotting against equal amounts of protein from each fraction with anti-Npl4 antibodies revealed that endogenous Npl4p is associated with microsomal membranes (Figure 3A, lane 2) and is present in soluble pools (Figure 3A, lane 3). This Npl4p fraction remained soluble even after a high-speed (100,000 × g) spin (Hitchcock, Krebber, Frietze, Lin, Latterich, and Silver, unpublished results). We estimate that ∼20% of cellular Npl4p is membrane associated based on the fact that similar levels of Npl4p are found in crude (Figure 3A, lane 1) and microsomal (Figure 3A, lane 2) fractions, whereas total microsomal protein isolated was 1/5 of the total cellular protein. Npl4p-GFP displayed the same fractionation profile (Hitchcock, Krebber, Frietze, Lin, Latterich, and Silver, unpublished results).
Figure 3.
Biochemical characterization of Npl4p localization and interactions. (A) Npl4p is found in both soluble and membrane-bound fractions. Equal protein from crude whole cell extract (lane 1), purified microsomes (lane 2), and ER/nuclear membrane-cleared extract (lane 3) derived from a wild-type yeast strain was separated by SDS-PAGE and Western blotted with anti-Npl4 antibodies. The band corresponding to Npl4p is indicated, and asterisks denote unidentified cross-reacting proteins. (B) The microsomal fraction of Npl4p is tightly membrane associated. Purified microsomes from a wild-type yeast strain were washed with either buffer alone (lanes 1 and 2), 1 M KOAc (lanes 3 and 4), 2 M KOAc (lanes 5 and 6), 3 M urea (lanes 7 and 8), 0.1 M Na2CO3, pH 11 (lanes 9 and 10), or 1% Triton X-100 (lanes 11 and 12). The samples were then separated into pellet (P; membrane associated; odd lanes) or soluble (S; even lanes) fractions and subjected to SDS-PAGE and Western blotting with anti-Npl4 and anti-Cdc48 antibodies. The extraction profiles of the peripheral ER membrane protein Sec17p and the ER integral membrane protein Sec62p were determined by Western blotting with anti-Sec17 and anti-Sec62 antibodies as controls. (C) Npl4p-protein A (pA) copurifies with Cdc48p and Ufd1p. Yeast whole cell extracts derived from either a strain expressing untagged Npl4p (lane 1) or Npl4p-pA (lane 2) were incubated with IgG-Sepharose beads. After extensive washing, proteins bound to the beads were eluted with acid and visualized by SDS-PAGE and Coomassie blue staining. Mass spectral analysis of excised protein bands revealed the identity of the Npl4p-pA copurifying p100 and p43 proteins as Cdc48p and Ufd1p, respectively.
Given that the Npl4p amino acid sequence does not contain any predicted transmembrane domains, we wanted to test the nature of the Npl4p interaction with microsomal membranes. Microsomal membranes derived from a wild-type yeast strain were washed with various buffers and separated into soluble (S) and pellet (P) fractions (Figure 3B). Immunoblotting with anti-Npl4 revealed that Npl4p could not be extracted into the soluble fraction by high salt (1 M and 2 M potassium acetate, lanes 4 and 6, respectively) and was only partially dissociated from membranes by 3 M urea (Figure 3B, lane 8) or 0.1 M sodium carbonate, pH 11 (Figure 3B, lane 10). Only in the presence of 1% Triton X-100 was Npl4p completely extracted (Figure 3B, lane 12). The Npl4p-interacting protein Cdc48p (Figure 3C) displayed a similar extraction profile as previously published (Latterich et al., 1995). As controls, these same fractions were also immunoblotted with antibodies against the peripheral ER membrane protein Sec17p and integral ER membrane protein Sec62p. As expected, Sec17p dissociated from the membranes upon the addition of 3 M urea (Figure 3B, lane 8) and 0.1 M sodium carbonate, pH 11 (Figure 3B, lane 10). In contrast, Sec63p was not dissociated from microsomal membranes under any conditions except when 1% Triton X-100 was added (Figure 3B, lane 12). These results suggest that a subset of cellular Npl4p protein behaves as a tightly associated peripheral ER/nuclear membrane protein.
Npl4p Complexes with Ufd1p and the AAA-ATPase Cdc48p
In an effort to identify other proteins with which Npl4p might interact, we integrated DNA encoding protein A (pA) at the NPL4 genomic locus, generating a NPL4-pA open reading frame. Expression of a full-length fusion protein that replaced endogenous Npl4p was confirmed by Western blotting (Figure 3A, lanes 3 and 6). The functionality of this construct was confirmed by viability of the resulting strain. Extracts were prepared from yeast cells expressing Npl4p-pA or untagged Npl4p as a control. After incubation with IgG-Sepharose and extensive washing, bound proteins were eluted, separated by SDS-PAGE, and visualized by Coomassie blue staining (Figure 3C). In the Npl4p-pA sample, three major eluted proteins were observed: p100, p90, and p43 (Figure 3C, lane 2). In the untagged sample, only background Coomassie blue staining was observed (Figure 3C, lane 1). The bands were excised from the gel and subjected to mass spectral analysis for identification. p90 was identified as the Npl4p-pA fusion protein, p100 was identified as Cdc48p, and p43 was identified as Ufd1p. Ufd1p is an essential protein of unknown function that was identified in a screen for yeast mutants that stabilize an artificial ubiquitin fusion protein (the ubiquitin fusion degradation [UFD] screen) (Johnson et al., 1995). The Cdc48p protein, along with its mammalian homologue p97, is a putative chaperone of the AAA-ATPase family that has been implicated in homotypic membrane fusion, ubiquitin-mediated protein turnover (including the UFD pathway), and cell cycle progression (Moir et al., 1982; Frohlich et al., 1991; Acharya et al., 1995; Latterich et al., 1995; Rabouille et al., 1995; Ghislain et al., 1996; Dai et al., 1998). The mammalian Npl4, Ufd1, and p97/Cdc48 proteins have also been recently demonstrated to interact (Meyer et al., 2000), indicating that this protein complex has been evolutionarily conserved.
Based on the interaction of Npl4p with Cdc48p, as well as the membrane localization of Npl4p, we tested whether npl4 mutants display defects in homotypic membrane fusion. Microsomal membranes were isolated from wild-type, npl4-1, and npl4-2 yeast strains and then tested for their ability to fuse in a previously described in vitro homotypic membrane fusion assay (Latterich and Schekman, 1994). Membranes derived from npl4 strains were able to fuse to levels comparable to wild-type membranes (Hitchcock, Krebber, Frietze, Lin, Latterich, and Silver, unpublished results), suggesting that Npl4p is not required for homotypic membrane fusion.
npl4 Mutants Are Suppressed by Overproduction of Components of the Fatty Acid Desaturation Pathway and the Proteasome-associated Gene RPN4
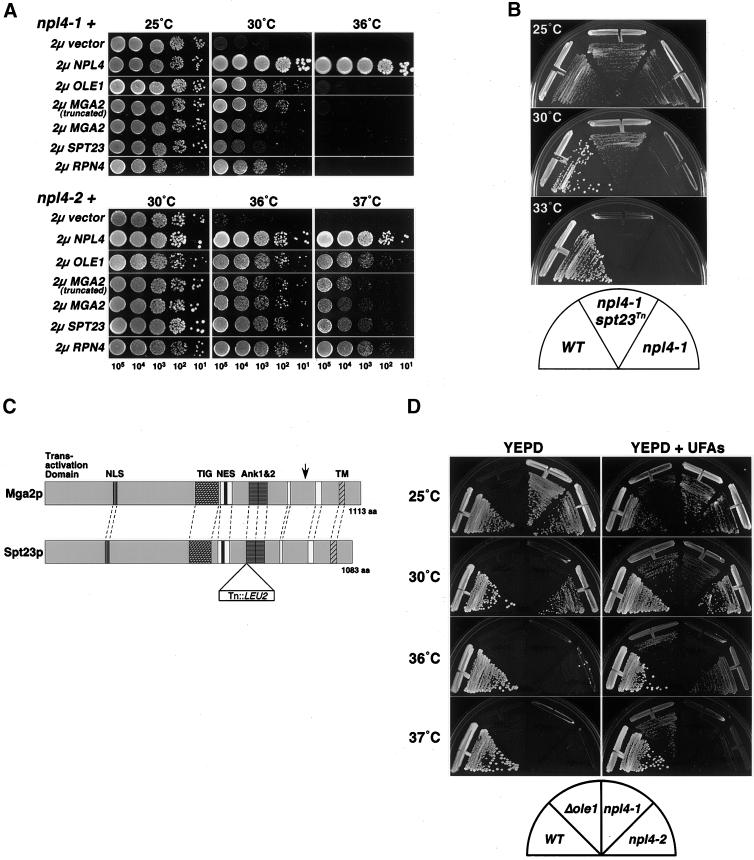
In an effort to uncover the primary defect of npl4 strains, a screen was performed to identify yeast genes that, when overexpressed, can rescue npl4-1 temperature sensitivity. The npl4-1 strain was transformed with a high-copy (2 μ URA3) yeast genomic library (gift of C. Connelly and P. Hieter) and screened for growth at the nonpermissive temperature (30°C). One gene isolated from this screen, OLE1, encodes the yeast Δ9-fatty acid desaturase, which is required for all de novo synthesis of UFAs (Stukey et al., 1990). As shown in Figure 4A (top), the temperature sensitivity of npl4-1 at 30°C is strongly rescued by 2 μ OLE1 as compared with the empty vector control. However, npl4-1 growth at the higher temperature (36°C) is not rescued, indicating that OLE1 is only able to complement npl4-1 temperature sensitivity at intermediate temperatures. Temperature sensitivity of npl4-2 at 36 and 37°C is also rescued by 2 μ OLE1 (Figure 4A, bottom), indicating that this effect is not allele specific.
Figure 4.
Yeast npl4 mutants are rescued by regulators of the UFA pathway and the proteasome. (A) Suppression of npl4-1 and npl4-2 temperature sensitivity by high-copy expression of OLE1, SPT23, MGA2, and RPN4. npl4-1 (PSY2340, top) and npl4-2 (PSY2341, bottom) strains transformed with either empty vector, or 2 μ vectors containing NPL4, OLE1, MGA2(aa1-914), MGA2, SPT23, or RPN4 were serially diluted onto YEPD media. The number of cells spotted in each dilution is indicated at the bottom. Plates were incubated at the indicated temperatures for 24–48 h. (B) Temperature sensitivity of a npl4-1 strain is extragenically suppressed by a transposon insertion allele of SPT23. Growth of wild-type (WT; FY23), npl4-1 (PSY2373), and npl4-1 SPT23:: Tn3:: LEU2 (PSY2377) strains on Leu− plates at the indicated temperatures. The FY23 and PSY2373 strains were transformed with an empty LEU2 vector to allow growth on Leu− dropout plates. (C) Schematic of the S. cerevisiae Mga2p and Spt23p proteins. These partially redundant factors are 38% identical and 54% similar. Sequence features include a putative basic nuclear localization signal (NLS), a putative leucine-rich nuclear export signal (NES), and two ankyrin repeats. The TIG domain is an Ig-like fold domain found in cell surface receptors and intracellular transcription factors including NF-κB. The C-terminal transmembrane domain (TM) is indicated. Blocks of strong amino acid conservation without assigned or predicted function are indicated by white boxes. The site of truncation in the isolated MGA2 high-copy suppressor clone (pPS2019) is indicated by an arrow in the Mga2p schematic. The site of transposon (Tn::LEU2) insertion in the npl4-1 extragenic suppressor allele of SPT23 (PSY2377) is indicated in the Spt23p schematic. (D) Temperature sensitivity of yeast npl4 mutants is partially rescued by supplementing growth media with UFAs. Wild-type (WT), ole1Δ, npl4-1, and npl4-2 strains were struck onto YEPD plates unsupplemented (left) or supplemented (right) with 0.5 mM UFAs (0.25 mM palmitoleic acid [16:1], 0.25 mM oleic acid [18:1], 1% Tergitol). Plates were incubated at the indicated temperatures for 48 h.
The other gene isolated from this npl4-1 high-copy suppressor screen, MGA2, encodes a transcription factor that has been shown to activate the transcription of OLE1 (Zhang et al., 1999). Mga2p and the redundant transcriptional activator Spt23p have recently been shown to be made as membrane-bound precursor proteins, which are then cleaved in a proteasome-dependent manner from the membrane (Hoppe et al., 2000). It is hypothesized that cleavage from the membrane is required for subsequent activation of OLE1 transcription by Mga2p and Spt23p. Interestingly, the two MGA2 clones isolated in our screen contained identically truncated forms of MGA2 lacking the coding sequence for the C-terminal 199 amino acids. The site of truncation, which eliminates the transmembrane domain (indicated by an arrow in the Mga2p schematic in Figure 4C) likely leads to expression of a soluble, truncated form of Mga2p. As with OLE1, the truncated MGA2 clone can rescue npl4-1 and npl4-2 temperature sensitivity at intermediate temperatures (Figure 4A, top and bottom). When tested directly, full-length MGA2 was able to suppress the temperature sensitivity of both npl4-1 and npl4-2 similarly to the truncated form of MGA2, although this suppression was slightly weaker for npl4-1 (Figure 4A top). In addition, the redundant gene SPT23 was tested for high-copy suppression of npl4 temperature sensitivity (Zhang et al., 1997; Zhang et al., 1999). In the case of npl4-1, SPT23 could only weakly suppress the growth defect at the intermediate temperature of 30°C (Figure 4A, top). In contrast, npl4-2 mutants were almost completely rescued by SPT23 (Figure 4A, bottom).
As additional evidence for the genetic interaction of NPL4 and SPT23, we have isolated a transposon-insertion allele of SPT23 that acts as a dominant extragenic suppressor of npl4-1 temperature sensitivity at 30°C but not higher temperatures (see Figure 4B and MATERIALS AND METHODS). Sequencing of transposon and flanking genomic DNA revealed that the transposon insertion results in the placement of an in-frame valine codon and stop codon 2130 nucleotides (corresponding to 710 amino acids) into the coding sequence of SPT23. The nature of this insertion would prevent expression of the predicted ankyrin repeats and transmembrane domain (see Spt23p schematic in Figure 4C). This finding indicates that single-copy expression of a truncated, soluble form of Spt23p can extragenically suppress npl4-1 temperature sensitivity at 30°C.
The results of these genetic screens suggest that the temperature sensitivity of npl4 mutants can be mitigated by increasing the levels of the OLE1 gene—either directly, by overexpressing OLE1 itself, or indirectly, by overexpressing or truncating (and presumably activating) transcription factors that direct OLE1 expression. Given that the primary function of Ole1p is to produce UFAs, we sought to determine whether npl4 mutants could be rescued by supplementing their growth media with UFAs. Indeed, growth of npl4-1 and npl4-2 strains at intermediate temperatures (30 and 36°C, respectively) is restored by supplemented palmitoleic (16:1) and oleic acid (18:1; Figure 4D). This effect was not due to the detergent used to solubilize the UFAs (Hitchcock, Krebber, Frietze, Lin, Latterich, and Silver, unpublished results). The ole1Δ strain was used as a control at all temperatures because its growth relies exclusively on the presence of supplemented UFAs.
Through the course of our studies, we have also discovered a genetic interaction between NPL4 and the proteasome regulator RPN4/SON1/UFD5. RPN4 was originally identified in our laboratory as an extragenic suppressor of C-terminal mutants in SEC63/NPL1 (Nelson et al., 1993) and was also isolated in the same genetic screen as UFD1 (Johnson et al., 1995). In addition, RPN4 has been shown to physically associate with the proteasome (Fujimuro et al., 1998) and to regulate proteasome gene transcription (Mannhaupt et al., 1999). When expressed in high copy, RPN4 strongly rescues the temperature sensitivity of both npl4-1 and npl4-2 mutant strains as compared with empty vector DNA (Figure 4A). In the case of npl4-1, RPN4 rescues growth at the intermediate temperature of 30°C (Figure 4A, top), whereas in the case of npl4-2, RPN4 is capable of rescuing growth at both 36 and 37°C (Figure 4A, bottom). Furthermore, npl4-1rpn4Δ double mutants display a synthetic slow-growth phenotype at 25°C compared with wild type and both single mutants alone (Hitchcock, Krebber, Frietze, Lin, Latterich, and Silver, unpublished results).
Npl4p Is Required for OLE1 Expression
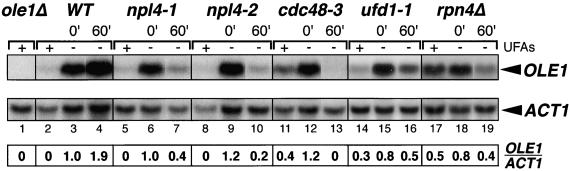
The genetic data presented above led us to the hypothesis that Npl4p may be required for the proteasome-dependent processing/activation of the Mga2p and Spt23p transcription factors and subsequent OLE1 expression (Hoppe et al., 2000). The isolation of truncated forms of MGA2 and SPT23 as suppressors of npl4 temperature sensitivity provided especially strong support for this hypothesis. As a first test, we asked whether OLE1 transcript levels are reduced in npl4 mutant cells compared with those of a control transcript (ACT1). Northern blot analysis was performed against total RNA isolated from wild-type, npl4-1, and npl4-2 strains before and after a shift to the nonpermissive temperature (37°C). As shown in Figure 5, OLE1 transcript levels are normal in both npl4-1 and npl4-2 cells at permissive temperature (Figure 5, compare lanes 3, 6, and 9). However, OLE1 levels are dramatically decreased in these mutants after a 60-min shift to 37°C (Figure 5, lanes 7 and 10). In contrast, OLE1 expression is stimulated in wild-type cells after a shift to 37°C (Figure 5, lane 4). The npl4-1 and npl4-2 strains were capable of repressing OLE1 expression to a similar extent as wild-type cells (Figure 5, lanes 2, 5, and 8). To quantitate the change in OLE1 expression, relative OLE1/ACT1 signal was calculated (see MATERIALS AND METHODS; Figure 5, bottom).
Figure 5.
NPL4 function is required for OLE1 transcription. Northern analysis was performed on total RNA isolated from ole1Δ (lane 1), wild-type (WT; lanes 2–4), npl4-1 (lanes 5–7), npl4-2 (lanes 8–10), cdc48-3 (lanes 11–13), ufd1-1 (lanes 14-16), and rpn4Δ cells (lanes 17–19) with OLE1- and ACT1-specific DNA probes. Cells were grown in YEPD media at 25°C with or without supplemented UFAs. For wild-type cells, the temperature-sensitive-strains npl4-1, npl4-2, and cdc48-3, and non–temperature sensitive strains ufd1-1 and rpn4Δ, samples were also collected after shifts to 37°C for 60 min. A normalized OLE1/ACT1 ratio was calculated (see MATERIALS AND METHODS) for each sample to allow for quantitation of the relative changes in OLE1 expression among samples.
Given the striking effect of NPL4 loss of function on OLE1 expression, we determined whether the NPL4 associated genes, CDC48, UFD1, and RPN4, are also required for OLE1 expression. As a test of the requirement for the Npl4p-interacting protein Cdc48p, Northern blot analysis was performed against total RNA isolated from the temperature-sensitive cdc48–3 strain. As with npl4-1 and npl4-2, OLE1 levels were normal at the permissive temperature of 25°C in cdc48-3 cells (Figure 5, compare lanes 3 and 12). However, OLE1 mRNA became undetectable after a 60-min shift to the nonpermissive temperature of 37°C (Figure 5, lane 13). To determine whether the Npl4p-interacting protein Ufd1p is also required for OLE1 expression, we monitored OLE1 mRNA levels in the ufd1-1 mutant strain. No significant defect in OLE1 expression was apparent in ufd1-1 cells at 25°C as compared with wild-type cells (Figure 5, compare lanes 3 and 15). Interestingly, the ufd1-1 strain had significantly lower levels of OLE1 mRNA after a 60-min shift to 37°C (Figure 5, compare lanes 4 and 16), despite the fact that this strain is not temperature sensitive. Finally, we tested the requirement for the RPN4 gene in OLE1 expression. OLE1 transcript levels were normal in the rpn4Δ strain at 25°C (Figure 5, compare lanes 3 and 18). Like the ufd1-1 strain, rpn4Δ cells are not temperature sensitive; however, after a shift to 37°C for 60 min, OLE1 levels were significantly decreased in the rpn4Δ strain as compared with wild type (Figure 5, compare lanes 4 and 19). The cdc48-3, ufd1-1, and rpn4Δ mutants were able to repress OLE1 mRNA expression in the presence of UFAs, although not quite as strongly as wild-type cells (Figure 5, lanes 2, 11, 14, and 17).
Npl4p Is Required for Efficient Processing of Mga2p and Spt23p
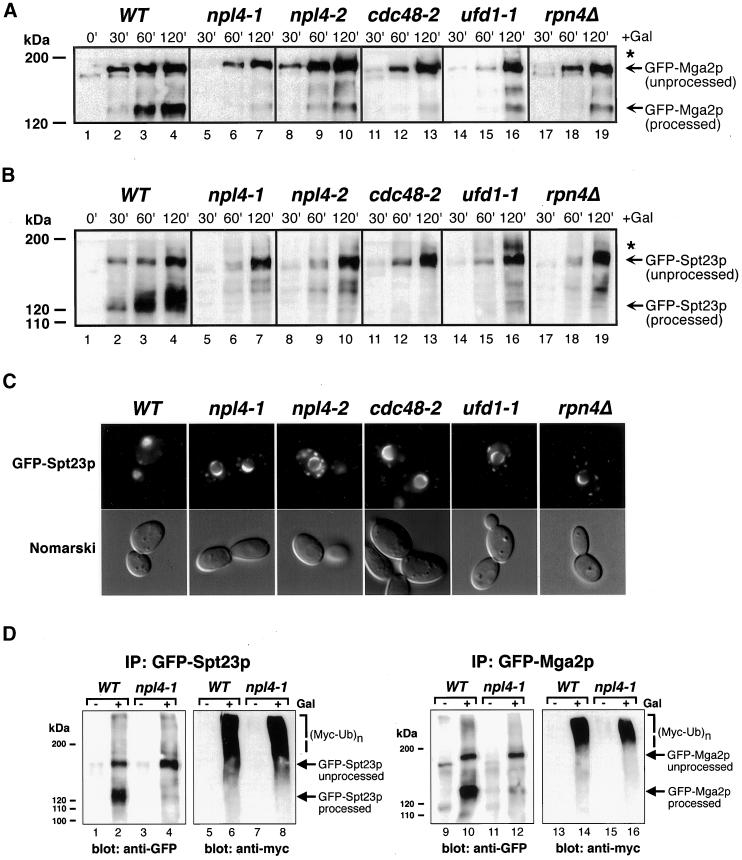
To more directly test our hypothesis that Npl4p function is upstream of Mga2p and Spt23p processing and activation, we expressed galactose-inducible N-terminally GFP-tagged Mga2p and Spt23p proteins in wild-type and npl4 mutant cells. At various time points into fusion protein induction, cells were collected and whole cell extracts were subjected to anti-GFP Western blot analysis. Within 60 min of induction in wild-type cells, both full-length and processed forms of GFP-Mga2p and GFP-Spt23p are detectable at significant levels (Figure 6, A and B, respectively, lanes 1–4). In contrast, very little processed GFP-Mga2p was detectable in npl4-1 and npl4-2 cells even after a 120-min induction (Figure 6A, lanes 7 and 10, respectively). Similarly, processed GFP-Spt23p was not detectable in npl4-1 and npl4-2 cells (Figure 6B, compare lanes 7 and 10 to lane 4). It should be noted that these experiments were performed at 25°C, indicating that efficient GFP-Mga2p and GFP-Spt23p processing is compromised in these strains even at the permissive temperature.
Figure 6.
NPL4 is required for efficient processing of ubiquitinated Mga2p and Spt23p fusion proteins. (A) Induction of GFP-Mga2p fusion protein in wild-type (WT; lanes 1–4), npl4-1 (lanes 5–7), npl4-2 (lanes 8–10), cdc48-2 (lanes 11–13), ufd1-1 (lanes 14-16), and rpn4Δ (lanes 17–19) cells at 25°C. Cells were collected for analysis by anti-GFP Western blot at the indicated time points into galactose induction. Asterisks indicate higher molecular weight GFP-Mga2p proteins. (B) Induction of GFP-Spt23p fusion protein in wild-type (WT; lanes 1–4), npl4-1 (lanes 5–7), npl4-2 (lanes 8–10), cdc48-2 (lanes 11–13), ufd1-1 (lanes 14-16), and rpn4Δ (lanes 17–19) cells at 25°C. Cells were collected for analysis by anti-GFP Western blot at the indicated time points into galactose induction. Asterisks indicate higher molecular weight GFP-Spt23p proteins. (C) Subcellular localization of GFP-Spt23p in wild-type (WT), npl4-1, npl4-2, cdc48-2, ufd1-1, and rpn4Δ cells after 120-min galactose inductions at 25°C. GFP-Spt23p signal is largely nucleoplasmic in wild-type cells, while remaining largely ER/nuclear envelope associated in the mutant strains. (D) GFP-Spt23p (left) and GFP-Mga2p (right) were immunoprecipitated (IP) from wild-type (WT) or npl4-1 yeast whole cell extracts expressing Myc-tagged ubiquitin. Expression of GFP-Spt23p and GFP-Mga2p was either induced for 2 h with galactose (lanes 2, 4, 6, 8, 10, 12, 14, and 16), or repressed with the addition of glucose (lanes 1, 3, 5, 7, 9, 11, 13, and 15) before immunoprecipitation. Samples run on the same gel were analyzed by anti-GFP (lanes 1–4 and 9–12) or anti-Myc (lanes 5–8 and 13–16) Western blotting.
We also tested whether the Npl4-associated genes CDC48, UFD1, and RPN4 are required for efficient GFP-Mga2p and GFP-Spt23p processing. A strain harboring the temperature-sensitive cdc48-2 mutation failed to accumulate processed GFP-Mga2p and GFP-Spt23p, similar to npl4 cells (Figure 6, A and B, lane 13). In addition, ufd1-1 and rpn4Δ cells were unable to accumulate processed forms of these fusion proteins (Figure 6, A and B, lanes 16 and 19, respectively). We noted from this Western analysis that the unprocessed form of GFP-Mga2p and GFP-Spt23p in these mutant strains was often accompanied by accumulation of higher molecular weight species (see asterisks in Figure 6, A and B).
We then observed these cells by fluorescence microscopy to determine the subcellular localization of GFP-Mga2p and GFP-Spt23p in npl4 and associated mutants. The results obtained with GFP-Spt23p–expressing cells are shown in Figure 6C. Strikingly, although GFP-Spt23p was mostly nucleoplasmic after 120 min of induction in wild-type cells, it was tightly associated with perinuclear envelopes in npl4-1, npl4-2, cdc48-2, ufd1-1, and rpn4Δ strains. GFP-Mga2p showed a similar localization pattern in these strains (Hitchcock, Krebber, Frietze, Lin, Latterich, and Silver, unpublished results). These results suggest that lack of processing of these fusion proteins in these mutant strains corresponds to an inability of this protein to dissociate from ER/nuclear envelope membranes.
Finally, based on our observation of higher molecular weight species of GFP-Mga2p and GFP-Spt23p in npl4 and associated mutant strains (Figure 6, A and B, asterisks), we tested whether these fusion proteins are ubiquitinated in the npl4-1 mutant. To this end, galactose-induced GFP-Mga2p and GFP-Spt23p proteins were immunoprecipitated from whole cell extracts derived from wild-type or npl4-1 yeast expressing a Myc epitope-tagged ubiquitin. Western analysis of bound fractions revealed significant levels of myc-immunoreactive proteins with sizes larger than the unprocessed GFP-Mga2p and GFP-Spt23p fusion proteins in both wild-type and npl4-1 cells (Figure 6D, lanes 6, 8, 14, and 16). Similar results were obtained with the npl4-2 mutant strain (Hitchcock, Krebber, Frietze, Lin, Latterich, and Silver, unpublished results). These data suggest that ubiquitination of GFP-Mga2p and GFP-Spt23p is not blocked in npl4 mutant cells.
DISCUSSION
We have presented evidence that the S. cerevisiae Npl4p protein is part of a highly conserved protein complex required for the proteasome-mediated processing and activation of the ER-membrane–bound transcription factors Mga2p and Spt23p. The striking conservation of the Npl4p protein and its biochemical interactions through evolution highlight the importance of Npl4p and the Npl4p-Ufd1p-Cdc48p/p97 complex to eukaryotic cell function. The yeast NPL4 gene was first identified in our laboratory in a genetic screen for nuclear transport mutants (Bossie et al., 1992; DeHoratius and Silver, 1996). Based on the nuclear transport defects and nuclear envelope herniations/protrusions of npl4 temperature-sensitive strains, as well as the observation that Npl4p localizes by immunofluorescence to the nuclear rim, it was concluded that Npl4p may be a component of the nuclear pore (DeHoratius and Silver, 1996). However, based on our current findings, it is likely that perturbation of membrane composition in npl4 cells leads to loss of ER/nuclear envelope integrity, which in turn causes the observed defects in nuclear transport.
Npl4p Localization and the Npl4p-Ufd1p-Cdc48p Complex
Our in vivo and biochemical localization studies have indicated that a subpopulation of Npl4p (∼20%) is tightly bound to ER/nuclear membranes, with the remainder constituting soluble cytoplasmic/nucleoplasmic pools. We have also shown that total cellular Npl4p protein exists in a tight complex with the Ufd1p and Cdc48p proteins. Although the mammalian homologues of these proteins have previously been demonstrated to interact (Meyer et al., 2000), our current study is the first to describe a function for this Npl4 complex. UFD1 was identified in a genetic screen for a pathway that directs proteasome-dependent turnover of an ubiquitin-fusion protein (UFD pathway; Johnson et al., 1995). Cdc48p is an AAA-ATPase first identified in a screen for cold-sensitive cell-division cycle mutants (Moir et al., 1982). Interestingly, we have observed that npl4 cells display a G2/M cell cycle arrest similar to that described for cdc48 mutants (Hitchcock, Krebber, Frietze, Lin, Latterich, and Silver, unpublished results). Cdc48p and its mammalian homologue p97 have also been implicated in additional cellular processes including homotypic membrane fusion and ubiquitin-mediated protein degradation (including the UFD pathway; Acharya et al., 1995; Latterich et al., 1995; Rabouille et al., 1995; Ghislain et al., 1996; Koegl et al., 1999). Cdc48p/p97 and other AAA-ATPases are thought to act as protein chaperones, either unfolding proteins or disassembling protein complexes; substrate specificity in turn is achieved through interactions of Cdc48p/p97 with different cofactor proteins (Hanson et al., 1997; Patel and Latterich, 1998; Golbik et al., 1999).
Npl4 and Processing of Mga2p and Spt23p
Maintenance of UFA levels in cellular membranes is crucial for proper membrane fluidity and dynamics, as well as membrane-bound organelle function. The molecular mechanisms by which eukaryotic cells monitor UFA levels and/or membrane fluidity remains unclear. However, the OLE1 gene, which encodes the Δ9-fatty acid desaturase enzyme responsible for all de novo synthesis of UFAs, appears to be the major target of such a signaling pathway (Stukey et al., 1990). Indeed, OLE1 activity is tightly regulated at both the transcriptional and posttranscriptional level in response to cellular fatty acid requirements (Bossie and Martin, 1989; Choi et al., 1996; Gonzalez and Martin, 1996).
The partially redundant SPT23 and MGA2 genes have been shown to be required for OLE1 expression, possibly as transcriptional activators (Zhang et al., 1997, 1999). A recent report by Hoppe et al. (2000) revealed a novel mechanism by which Mga2p and Spt23p activity can be regulated in response to UFA levels in the cell. Mga2p and Spt23p are made as nascent transmembrane proteins anchored in the ER/nuclear membrane; ubiquitin-proteasome–dependent proteolytic cleavage then allows the release of active, soluble transcriptional activators (Hoppe et al., 2000).
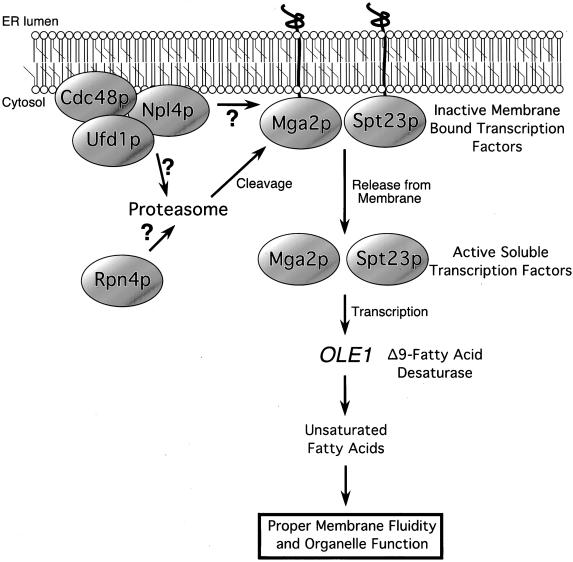
Our data support a model (depicted in Figure 7) that places NPL4 function upstream of this UFA regulatory pathway. In this model, the membrane-associated Npl4p-Ufd1p-Cdc48p complex functions in concert with the proteasome to proteolytically process the membrane-bound transcription factors Mga2p and Spt23p. The release of active Mga2p and Spt23p results in increased OLE1 transcription and subsequent UFA synthesis. When Npl4 activity is compromised, such as in npl4 temperature-sensitive mutants, Mga2p and Spt23p remain unprocessed and unable to activate OLE1 transcription; as a result, UFA levels decrease, perhaps causing global effects on cellular membrane dynamics and membrane-bound organelle function.
Figure 7.
A model for Npl4 function in the proteasome-dependent processing of the ER membrane-bound transcription factors Mga2p and Spt23p. Details are presented in the text.
In support of this model, the temperature sensitivity of npl4 mutants is rescued in part by overexpression of all downstream components of this pathway, namely, OLE1, MGA2, and SPT23, as well as UFAs themselves. Our isolation of truncated forms of MGA2 and SPT23 (lacking the C-terminal transmembrane domain) as npl4 suppressors is particularly interesting because these soluble proteins should bypass the requirement for Npl4p-dependent processing. NPL4 also displays genetic interactions with the proteasome-associated RPN4. RPN4 (also known as SON1/UFD5) has been proposed to copurify with the proteasome and to affect the transcription of proteasome subunits (Finley et al., 1998; Fujimuro et al., 1998; Glickman et al., 1998; Mannhaupt et al., 1999; Ng et al., 2000). In addition, the Npl4p-interacting proteins Cdc48p and Ufd1p have previously been implicated in proteasome-mediated degradation (Johnson et al., 1995; Ghislain et al., 1996; Dai et al., 1998).
As direct evidence for this model, we have demonstrated that OLE1 levels are drastically decreased in npl4 and cdc48 mutants after a short shift to the nonpermissive temperature. The non–temperature-sensitive strains ufd1-1 and rpn4Δ also display a defect in OLE1 expression at 37°C. Furthermore, we have shown that Mga2p and Spt23p fusion proteins are not efficiently processed in npl4, cdc48, ufd1, and rpn4Δ mutant cells even at permissive temperatures. We suggest that the normal levels of OLE1 transcript observed in these mutant strains at permissive temperature (conditions in which Mga2p and Spt23p fusion proteins are not efficiently processed) could be explained by sufficient residual processing of endogenous Mga2p and Spt23p proteins.
A model suggested by Hoppe et al. (2000) speculates that the Npl4p-Ufd1p-Cdc48p complex mediates events that occur downstream of the processing reaction, such as release from the proteasome or translocation of processed Mga2p and Spt23p into the nucleus (Hoppe et al., 2000). In contrast to this model, we have observed little to no processing of Mga2p and Spt23p proteins in these mutant backgrounds.
Npl4p-Ufd1p-Cdc48p Complex Function
The molecular mechanism by which the Npl4p complex mediates proteasome-dependent processing of the ER-associated proteins Mga2p and Spt23p remains unclear. We speculate that the Npl4p complex functions at a late step in the processing reaction, downstream of Mga2p and Spt23p ubiquitination. This speculation is based on our finding that Mga2p and Spt23p are still ubiquitinated in npl4 mutants. Furthermore, we suggest that the function of the Npl4p complex is not limited to the Mga2p and Spt23p activation pathway. If processing of Mga2p and Spt23p was the only essential function of NPL4, then npl4 mutants (like mga2Δspt23Δ mutants) should be completely rescued by the addition of exogenous UFAs. However, we have only observed partial rescue of npl4 mutants by supplemented UFAs, suggesting the presence of other essential targets of the Npl4p complex.
These speculations of a more general role for the Npl4p complex are confirmed by the recent finding by Bays et al. (unpublished results) that NPL4 is allelic to the HRD4 gene, which is required for degradation of several membrane-bound ERAD target proteins including hydroxymethyl glutaryl-CoA reductase. Importantly, Bays et al. (unpublished results) showed that ERAD substrates are still ubiquitinated in hrd4/npl4 mutants, consistent with our findings with Mga2p and Spt23p. Furthermore, degradation of a cytosolic proteasome target protein is unaffected in hrd4/npl4 mutants, suggesting that proteasome function is not compromised in these mutants (Bays et al., 2001). Therefore, it appears likely that the Npl4p/Hrd4p complex acts at a novel step in ER-associated proteasome-mediated processing/degradation, downstream of ubiquitination and upstream of proteasome function. One interesting possibility is that the Npl4p-Ufd1p-Cdc48p complex is responsible for recruitment of the proteasome to ubiquitinated ER target proteins, including Mga2p and Spt23p. Cdc48p chaperone activity may be important for this function. Indeed, the mammalian Cdc48p homologue p97 has been shown to bind ubiquitinated substrate proteins as well as to copurify with the proteasome (Dai et al., 1998). In this context the Npl4p-Ufd1p dimer could be thought of as a Cdc48p cofactor that targets Cdc48 chaperone activity to ubiquitinated ER membrane-bound substrates.
ACKNOWLEDGMENTS
We thank D. Finley, D. Garfinkel, E. Johnson, and T. Rapaport for plasmids, strains, and antibodies. We especially appreciate the critical comments concerning the manuscript by K. Auld, A. Brodsky, M. Damelin, J.K. Hood, P. Ko Ferrigno, and A.E. McBride and stimulating discussion with all members of the Silver and Latterich laboratories. We would also like to acknowledge P. Ko Ferrigno for expert advice on DeltaVision microscopy and J. Lee from the Dana-Farber Cancer Institute Molecular Biology Core Facility for mass spectral analysis of Npl4p-interacting proteins. This work was funded by National Institutes of Health (NIH) grants to P.A.S. and a Deutsche Forschungsgemeinschaft fellowship to H.K. A.L was supported by a NIH training grant to the University of California at San Diego. A.L.H. was supported by NIH training grants to Harvard Medical School and the Dana-Farber Cancer Institute.
REFERENCES
- Acharya U, Jacobs R, Peters JM, Watson N, Farquhar MG, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. doi: 10.1016/0092-8674(95)90269-4. [DOI] [PubMed] [Google Scholar]
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Agatep, R., Kirkpatrick, R.D., Parchaliuk, D.L., Woods, R.A., and Gietz, R.D. (1998). Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol: Technical Tips Online, http://tto.trends.com).
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays, N.W., Wilhovsky, S.K., Goradia, A., Hodgkiss-Harlow, K., and Hampton, R.Y. (2001). HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 12, in press. [DOI] [PMC free article] [PubMed]
- Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie MA, DeHoratius C, Barcelo G, Silver P. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie MA, Martin CE. Nutritional regulation of yeast delta-9 fatty acid desaturase activity. J Bacteriol. 1989;171:6409–6413. doi: 10.1128/jb.171.12.6409-6413.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Choi JY, Stukey J, Hwang SY, Martin CE. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1gene. J Biol Chem. 1996;271:3581–3589. doi: 10.1074/jbc.271.7.3581. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase Co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J Biol Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- De Strooper B. Alzheimer's disease: closing in on gamma-secretase. Nature. 2000;405:627. doi: 10.1038/35015193. , 629. [DOI] [PubMed] [Google Scholar]
- DeHoratius C, Silver PA. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4gene. Mol Biol Cell. 1996;7:1835–1855. doi: 10.1091/mbc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison MJ, Hochstrasser M. Epitope-tagged ubiquitin: a new probe for analyzing ubiquitin function. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- Finley D, Tanaka K, Mann C, Feldmann H, Hochstrasser M, Vierstra R, Johnston S, Hampton R, Haber J, McCusker J, Silver P, Frontali L, Thorsness P, Varshavsky A, Byers B, Madura K, Reed SI, Wolf D, Jentsch S, Sommer T, Baumeister W, Goldberg A, Fried V, Rubin DM, Toh-e A. Unified nomenclature for subunits of the Saccharomyces cerevisiaeproteasome regulatory particle. Trends Biochem Sci. 1998;23:244–245. doi: 10.1016/s0968-0004(98)01222-5. [DOI] [PubMed] [Google Scholar]
- Franzusoff A, Rothblatt J, Schekman R. Analysis of polypeptide transit through yeast secretory pathway. Methods Enzymol. 1991;194:662–674. doi: 10.1016/0076-6879(91)94048-h. [DOI] [PubMed] [Google Scholar]
- Frohlich KU, Fries HW, Rudiger M, Erdmann R, Botstein D, Mecke D. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J Cell Biol. 1991;114:443–453. doi: 10.1083/jcb.114.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro M, Tanaka K, Yokosawa H, Toh-e A. Son1p is a component of the 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 1998;423:149–154. doi: 10.1016/s0014-5793(98)00084-2. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin- mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiaeproteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbik R, Lupas AN, Koretke KK, Baumeister W, Peters J. The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol Chem. 1999;380:1049–1062. doi: 10.1515/BC.1999.131. [DOI] [PubMed] [Google Scholar]
- Gonzalez CI, Martin CE. Fatty acid-responsive control of mRNA stability: unsaturated fatty acid-induced degradation of the Saccharomyces OLE1transcript. J Biol Chem. 1996;271:25801–25809. doi: 10.1074/jbc.271.42.25801. [DOI] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRDgenes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Kahana JA, Schlenstedt G, Evanchuk DM, Geiser JR, Hoyt MA, Silver PA. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol Biol Cell. 1998;9:1741–1756. doi: 10.1091/mbc.9.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana JA, Silver PA. Use of the A. Victoria green fluorescent protein to study protein dynamics in vivo. In: Ausubel FM, Brent R, Kingston RE, Moore DE, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1996. pp. 9.7.22–29.27.27. [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Kumar A, Snyder M. Genome-wide transposon mutagenesis in yeast. In: Ausubel FM, Brent R, Kingston RE, Moore DE, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 2000. pp. 13.13.11–13.13.15. [DOI] [PubMed] [Google Scholar]
- Latterich M, Frohlich KU, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- Latterich M, Schekman R. The karyogamy gene KAR2and novel proteins are required for ER-membrane fusion. Cell. 1994;78:87–98. doi: 10.1016/0092-8674(94)90575-4. [DOI] [PubMed] [Google Scholar]
- Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian Ufd1 and Npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D, Stewart SE, Osmond BC, Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MK, Kurihara T, Silver PA. Extragenic suppressors of mutations in the cytoplasmic C terminus of SEC63 define five genes in Saccharomyces cerevisiae. Genetics. 1993;134:159–173. doi: 10.1093/genetics/134.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Spear ED, Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Latterich M. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Romisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, Goldstein JL, Brown MS. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Damelin M, Kahana J, Taura T, Silver PA. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Grandi P, Hurt EC. Affinity purification of protein A-tagged nuclear pore proteins from yeast. In: Celis J E, editor. Cell Biology: A Laboratory Handbook. 2nd ed. San Diego, CA: Academic Press; 1997. pp. 159–164. [Google Scholar]
- Steiner H, Kostka M, Romig H, Basset G, Pesold B, Hardy J, Capell A, Meyn L, Grim ML, Baumeister R, Fechteler K, Haass C. Glycine 384 is required for presenilin-1 function and is conserved in bacterial polytopic aspartyl proteases. Nat Cell Biol. 2000;2:848–851. doi: 10.1038/35041097. [DOI] [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. The OLE1 gene of Saccharomyces cerevisiaeencodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- William KR, LoPresti M, Stone K. Internal protein sequencing of SDS-PAGE separated proteins: optimization of an in-gel digest protocol. In: Marshak D, editor. Techniques VIII. San Diego, CA: Academic Press; 1997. pp. 79–90. [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiaestrains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- Yaseen NR, Blobel G. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc Natl Acad Sci USA. 1999;96:5516–5521. doi: 10.1073/pnas.96.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Burkett TJ, Yamashita I, Garfinkel DJ. Genetic redundancy between SPT23 and MGA2: regulators of Ty-induced mutations and Ty1 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4718–4729. doi: 10.1128/mcb.17.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Skalsky Y, Garfinkel DJ. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics. 1999;151:473–483. doi: 10.1093/genetics/151.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Schekman R. The engagement of Sec61p in the ER dislocation process. Mol Cell. 1999;4:925–934. doi: 10.1016/s1097-2765(00)80222-1. [DOI] [PubMed] [Google Scholar]