Abstract
Melilotus albus and Melilotus officinalis are widely used in forage production and herbal medicine due to the biological activity of their coumarins, which have many biological and pharmacological activities, including anti-HIV and anti-tumor effects. To comprehensively evaluate M. albus and M. officinalis coumarin content (Cou), morphological variation, and molecular phylogeny, we examined the Cou, five morphological traits and the molecular characterization based on the trnL-F spacer and internal transcribed spacer (ITS) regions of 93 accessions. Significant (p < 0.05) variation was observed in the Cou and all five morphological traits in both species. Analysis of population differentiation (Pst) of the phenotypic traits showed that powdery mildew resistance (PMR) had the greatest Pst, meaning that this trait demonstrated the largest genetic differentiation among the accessions. The Pst values of dry matter yield (DMY) and Cou were relatively high. Biplot analysis identified accessions with higher DMY and higher and lower Cou. Analysis of molecular sequence variation identified seven haplotypes of the trnL-F spacer and 13 haplotypes of the ITS region. Based on haplotype and sequence analyses, the genetic variation of M. officinalis was higher than that of M. albus. Additionally, ITS sequence analysis showed that the variation among accessions was larger than that among species across three geographical areas: Asia, Europe, and North America. Similarly, variation among accessions for both the trnL-F and ITS sequences were larger than the differences between the geographical areas. Our results indicate that there has been considerable gene flow between the two Melilotus species. Our characterization of Cou and the morphological and genetic variations of these two Melilotus species may provide useful insights into germplasm improvement to enhance DMY and Cou.
Keywords: chloroplast sequence, coumarin, Melilotus, morphological trait, molecular phylogenetics, nuclear ITS
1. Introduction
Coumarins, a class of structurally unique and important natural compounds, exist in many plant species in Nature [1,2,3] and present potential medicinal value [4]. Many medicines containing coumarin-based compounds such as warfarin and phenprocoumon are often not specifically linked to coumarin in medical papers [5], and instead, are usually described as having anticoagulant properties or interacting with drugs with anticoagulant properties [4]. The coumarin-derivative prehispanolone from Chinese motherwort (Leonurus cardiaca) reduces fibrinogen and blood viscosity and inhibits platelet aggregation [6]. These functions make coumarins important medicinal compounds. Coumarin is a plant secondary metabolite in Melilotus [7], and its content varies significantly among different Melilotus species, with mean contents ranging from 0.06 to 0.753% of dry matter [8]. Melilotus belongs to the family Leguminosae, in the subfamily Papilionoideae, of the clover tribe (Tribus Trifolieae Bronn) [9]. Melilotus, commonly referred to as wild alfalfa, consists of 19 species that originated in Eurasia [10]. In China, Melilotus is mainly distributed in Liaoning, Shanxi, Gansu and Inner Mongolia [11]. This plant demonstrates good drought, cold and salt tolerance and is used as pharmaceutical material, fodder, green manure, and a soil conservation crop [12]. The most widely-cultivated species, M. albus and M. officinalis (Figure S1), are primarily used as fodder but also as medicinal plants [13]. The morphology of the two species is very and classification is largely based on flower color: M. albus has white flowers, and M. officinalis has yellow flowers [14]. Melilotus contains various chemical components such as coumarin [15], which possesses high medicinal value, but reliable and accurate methods for differentiating M. albus and M. officinalis during the vegetative stage are not available. M. albus is generally cross-fertile with a low degree of self-fertility in both the annual and biennial types.
Genetic variation in germplasm is the foundation of successful plant breeding, and wild germplasm is a valuable source of new alleles associated with desirable characteristics, such as higher yield, higher or lower coumarin content (Cou), and resistance to pests and diseases. It is essential that plant genetic resources are properly conserved and characterized. There are many examples of the successful use of native or exotic germplasm in plant breeding. To facilitate the use of germplasm resources, basic information on germplasm must be obtained. DNA sequences represent sources of information for biological systematics [16]. DNA sequences are often used to infer phylogenetic trees, which permit visualization of the diversity and relationships in different populations [17,18]. The chloroplast trnL-F DNA spacer has higher variability than many other regions in the plant genome [19], and the nuclear ribosomal internal transcribed spacer (ITS) region has alternating gene and spacer regions and tandem repeat structures [20,21,22,23]. A combined cpDNA and ITS approach has been successfully used to analyze systematics in various plants [24,25,26,27]. Sequences of matK, trnL-F and ITS were used to evaluate the relationships in Orchidaceae [28,29,30]. Phylogenetic analysis of the Lamiaceae based on sequences of the trnL intron, trnL-F intergenic spacer, and rps16 intron revealed that Lamioideae was divided into nine tribes [31]. A study combining molecular and morphological data found that Eremospatha, Laccosperma, and Oncocalamus (Palmae) constitute a monophyletic group [32,33]. These three genera share certain morphological characteristics (such as rachis and inflorescences) [34] and possess distinctive traits (including reproductive systems) [32]. Our previous analysis using a chloroplast gene showed that 18 species of the genus Melilotus formed a monophyletic clade [35]. Analysis of the genetic diversity and population structure using SSR markers in accessions of the genus Melilotus revealed substantial variation among the 18 species and among or within populations [36,37]. Despite the importance of M. albus and M. officinalis as crops in Eurasia, Africa, and North America, there is a lack of information regarding Cou, genetic variation, and systematics of these plant species. In the present work, a total of 93 accessions, including 42 accessions of M. albus and 51 accessions of M. officinalis, were collected. To evaluate potential genetic variation and to screen for high dry matter yield (DMY) and higher/lower Cou for future Melilotus breeding, we conducted coumarin analysis, morphological evaluation based on five traits and molecular characterization based on the trnL-F spacer and ITS region.
2. Results
2.1. Coumarin Content and Morphological Variation
Analysis of variance indicated significant (p < 0.05) genotypic differences among the accessions and within each species for all measured traits (PH, plant height; DMY; LSR leaf:stem ratio; Cou; PMR, powdery mildew resistance; and GH, growth habit) (Table 1). There was a wide range of variation for each of the traits. There were some differences between M. officinalis accessions and M. albus accessions. The population differentiation (Pst) parameters for all traits varied within a relatively narrow range, from 0.3405 to 0.5251 in M. officinalis and 0.3690 to 0.5434 in M. albus. The Pst parameter of PMR showed the largest variation (0.5251 in M. officinalis and 0.5434 in M. albus) among all accessions, and the DMY and Cou values were relatively high (0.4127 and 0.4022 in M. officinalis and 0.4642 and 0.4094 in M. albus, respectively) compared with the GH value (Table 1).
Table 1.
Ranges, means, and population differentiation (Pst) for traits measured in M. officinalis and M. albus accessions †.
| Trait | M. Officinalis | M. Albus | ||||||
|---|---|---|---|---|---|---|---|---|
| Range | Mean | Significance | Pst | Range | Mean | Significance | Pst | |
| PH | 20–100 | 62 | * | 0.4254 | 11–161 | 65 | * | 0.4103 |
| DMY | 4–41 | 17 | * | 0.4127 | 0.5–43 | 16 | * | 0.4642 |
| LSR | 0.7–1.8 | 1.17 | * | 0.4238 | 0.3–3.4 | 1.00 | * | 0.4138 |
| Cou | 0.3–1.5 | 0.83 | * | 0.4022 | 0.2–1.3 | 0.73 | * | 0.4094 |
| PMR | 1–4 | 2.75 | * | 0.5251 | 1–4 | 2.16 | * | 0.5434 |
| GH | 1–2 | 1.96 | * | 0.3405 | 1–2 | 1.56 | * | 0.3690 |
* Significantly different at p < 0.05; † PH, plant height (cm); DMY, dry matter yield (g/plant); LSR, leaf:stem ratio; Cou, coumarin content (%); PMR, powdery mildew resistance (0, 1 to 4 scale); GH, growth habit (1, annual; 2, biennial); Pst, population differentiation.
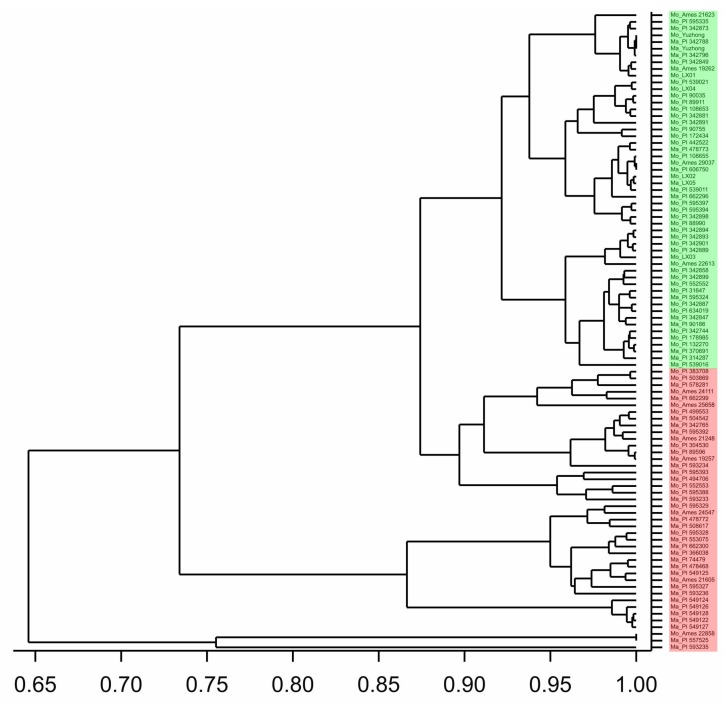
Cluster analysis of M. officinalis and M. albus accessions based on six traits was performed using UPGMA, and a dendrogram was inferred (Figure 1). All accessions were divided into two major clusters (green part and red part). The grouping was generated according to species type; most M. officinalis accessions grouped together, as did M. albus accessions (Figure 1).
Figure 1.
Dendrogram of M. officinalis and M. albus accessions generated based on six traits using UPGMA as implemented in GenStat software. Two parts were clustered: the green part was largely M. officinalis accessions, and the red part was largely M. albus accessions. Mo: M. officinalis, Ma: M. albus.
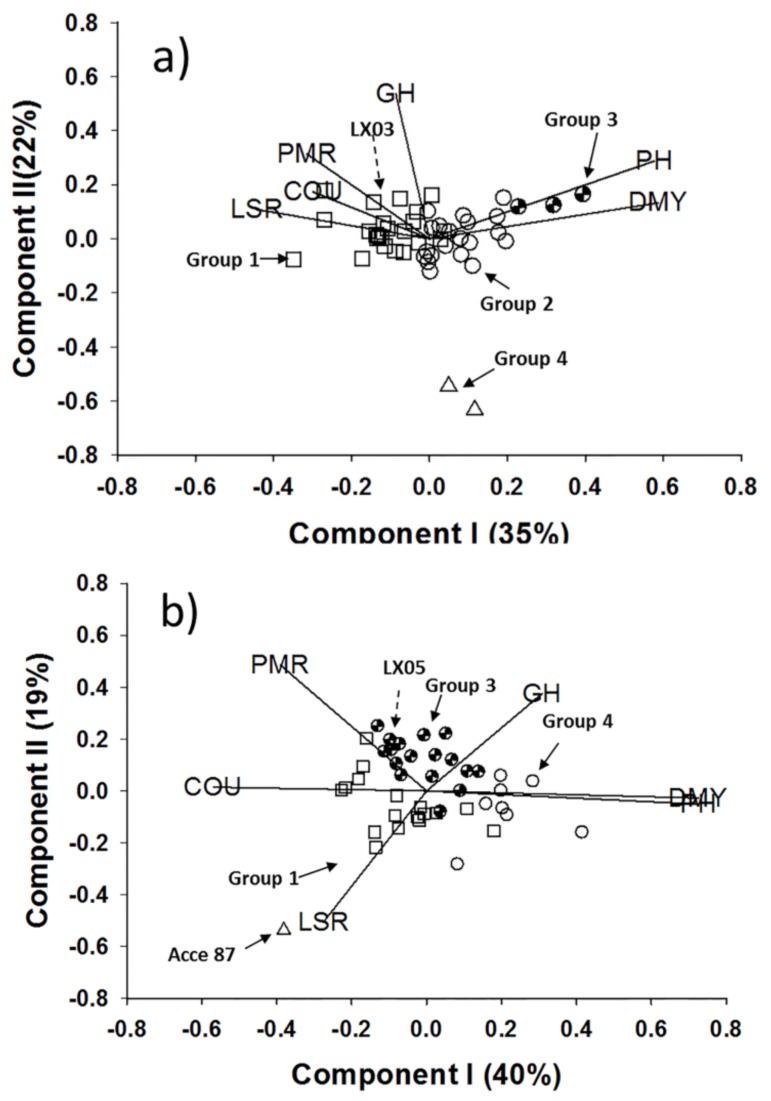
Biplots (Figure 2a,b) provided a graphical summary of the multivariate data based on the Cou and five morphological traits measured for all germplasm accessions and checks (commercial cultivars) of M. officinalis and M. albus. The directional vectors in each biplot, which were generated from the cluster analysis, show the associations among traits and provide a basis to compare accession groups and trait expression. In both M. officinalis and M. albus, there was a negative association between DMY and Cou. There were a number of germplasm accessions in both species that had higher DMY values than those of the commercial lines LX03 and LX05; this was particularly evident for the accessions in Group 3 for M. officinalis and Group 4 for M. albus (Figure 2a,b).
Figure 2.
Biplots generated using the accession-by-trait matrix approach for the six traits. PH, plant height; DMY, dry matter yield; LSR, leaf:stem ratio; Cou, coumarin content; and PMR, powdery mildew resistance, measured for (a) M. officinalis accessions and (b) M. albus accessions. In each biplot, the variation along components I and II is expressed as a percentage. The different symbols indicate the accession groups generated from the cluster analysis. The two local commercial lines LX03 and LX05 are indicated. The vectors represent the six traits.
The within-group accession means for the six traits measured for M. officinalis indicated that Group 1 contained plants with higher Cou and higher LSR values relative to the other groups, whereas Group 3 contained taller plants with higher DMY values and lower Cou (Table 2). For M. albus, accession Group 4 contained taller plants with higher DMY values and lower Cou, whereas Acce 87 contained plants with higher Cou and higher LSR values (Table 2).
Table 2.
Within-group accession means for each of the six traits based on the four clusters generated from the cluster analysis of M. officinalis and M. albus germplasm entries evaluated in Yuzhong, China.
| Species | Groups | No. in Group | PH | DMY | LSR | COU | PMR | GH |
|---|---|---|---|---|---|---|---|---|
| M. officinalis | Group 1 | 23 | 55 | 12 | 1.255 | 0.908 | 3.39 | 2 |
| Group 2 | 20 | 43 | 20 | 1.104 | 0.79 | 2.15 | 2 | |
| Group 3 | 3 | 92 | 35 | 1.04 | 0.526 | 2.67 | 2 | |
| Group 4 | 2 | 48 | 17 | 0.98 | 0.673 | 1.5 | 1 | |
| M. albus | Group 1 | 17 | 58 | 13 | 0.974 | 0.769 | 2.29 | 1 |
| Acce 87 | 1 | 11 | 0.52 | 3.421 | 1.158 | 1 | 1 | |
| Group 3 | 17 | 63 | 14 | 0.828 | 0.771 | 2.53 | 2 | |
| Group 4 | 8 | 94 | 29 | 1.115 | 0.511 | 1.25 | 1.875 |
PH, plant height (cm); DMY, dry matter yield (g/plant); LSR, leaf:stem ratio; Cou, coumarin content (%); PMR, powdery mildew resistance (0, 1 to 4 scale); GH, growth habit (1, annual; 2, biennial); Acce 87 is the code of accession PI 557525.
2.2. Alignments and Sequence Variation
Among the 93 M. albus and M. officinalis accessions, the average lengths of the trnL-F spacer and nrITS regions were 459 and 714 bases, respectively; however, the trnL-F spacer contains indels. For the aligned trnL-F and nrITS sequences, the conserved sites consisted of 452 and 700 bases, respectively, while there were 9 and 14 variable sites, with six and eight parsimony-informative characteristics, respectively (Table S1).
Sequencing of the trnL-F spacer in samples of both species from among the 93 accessions identified seven different sequences (Table S2a), with a total of nine variable sites in the trnL-F spacer. Three sites were indels, representing five types, and the other six sites were nucleotide substitutions. Total alignment of the ITS regions identified 13 different sequences (Table S2b). The ITS region included 14 variable sites, all of which were substitutions. All trnL-F and nrITS haplotype sequences were deposited in the GenBankdatabase under accession numbers KF758417-KF758423 and KF758424-KF758436, respectively.
2.3. Phylogenetic Analyses
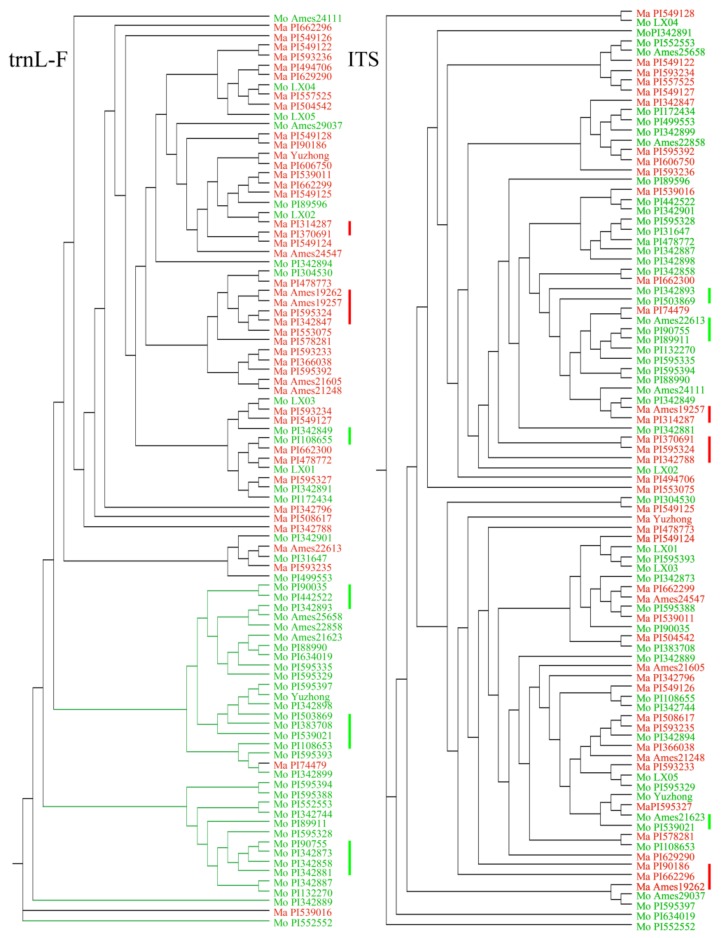
Bayesian trees based on the ITS and trnL-F sequences are shown in Figure 3. In the ITS tree, the two species were not very well separated. In the trnL-F tree based on the 459-base alignment, the accessions tended to separate into two species groups. The combined tree (created in MEGA 6.0) showed greater similarity to the trnL-F tree than to the ITS tree (Figure S2). Additionally, the combination of morphological data (Figure 2 and Table 2) and Bayesian trees suggested that some M. officinalis accessions (derived from M. officinalis Group 1, in Table 2) with high Cou clustered together, while some M. albus accessions (derived from M. albus Group 3, in Table 2) with relatively high Cou clustered together (Figure 3). A total of seven clusters, marked by green bars, belonged to accessions with high Cou, and five clusters, marked by red bars, belonged to accessions with relatively high Cou (Figure 3). The trnL-F tree shared six clusters, and 65% of M. officinalis accessions clustered together. Therefore, compared with ITS, trnL-F was a better genetic maker to distinguish M. officinalis from M. albus.
Figure 3.
Bayesian tree constructed with MrBayes based on the trnL-F and ITS datasets. M. albus accessions are indicated with red, M. officinalis accessions are indicated with green; the red bars indicate accessions from M. albus Group 3 in Table 2, and the green bars indicate accessions from M. officinalis Group 1 in Table 2.
2.4. Phylogenetic Relationships of Haplotypes
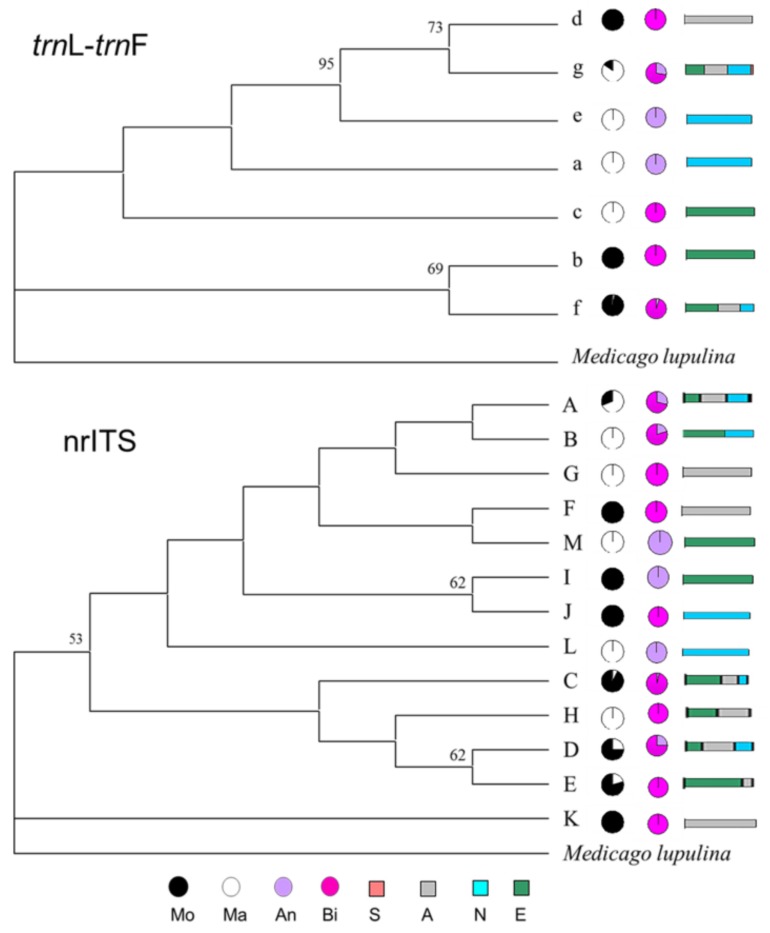
The haplotype phylogenetic trees of the trnL-F spacer and ITS regions of Melilotus are shown in Figure 4, with Medicago lupulina as the out group. In the trnL-F tree, haplotypes b and f formed a clade: The accessions in haplotype b were all biennial M. officinalis that were collected in Europe, and almost all accessions in haplotype f were biennial M. officinalis from Asia, Europe, and North America (Figure S3).
Figure 4.
Phylogenetic relationships of haplotypes resolved in M. albus and M. officinalis. The single maximum parsimonious tree is presented. Bootstrap values (>50%) are denoted above (maximum parsimony). Pie and column charts indicate the frequency of accessions within each haplotype, and unique alleles are indicated by different colors. The first pie chart column shows species distribution, the second column shows growth habit (GH), and the third column shows geographical distribution. trnL-F, phylogenetic tree based on cpDNA trnL-F haplotypes; nrITS, phylogenetic tree based on nrITS haplotypes (Mo, M. officinalis; Ma, M. albus; An, Annual; Bi, Biennial; S, South America; N, North America; A, Asia; E, Europe).
Haplotypes a and e were annual M. albus that originated from North America, and haplotype c was a biennial M. albus from Europe. Haplotypes d and g formed a subclade: haplotype d was a biennial M. officinalis derived from Asian stock, and haplotype g comprised annual and biennial M. officinalis and M. albus from Asia, Europe, and North America. In the ITS tree, haplotype K formed a clade consisting of biennial M. officinalis from Asia. Haplotypes B, G, M, L, and H were M. albus, and haplotypes F, I, and J were M. officinalis. Haplotypes G, F, J, H, and E were biennial, and haplotypes M, I and L were annual.
2.5. Genetic Variation within and between Species
The genetic variation of 42 accessions of M. albus and 51 accessions of M. officinalis were analyzed (Table 3). The aligned trnL-F sequence dataset was 459/439 bases in length and contained seven haplotypes: Four M. albus and three M. officinalis. The ITS region was 714 bases in length and included 13 haplotypes: seven M. albus, nine M. officinalis, and three shared by the two species. The haplotype variation and nucleotide variation of the two sequences were higher in M. officinalis than in M. albus.
Table 3.
Summary of genetic variation for the 93 accessions of Melilotus.
| Sequence | Dataset | Length | N | h | Hd ± SD | π ± SD |
|---|---|---|---|---|---|---|
| trnL-F | M. albus | 459 | 44 | 4 | 0.171 ± 0.075 | 0.0017 ± 0.0015 |
| M. officinalis | 439 | 49 | 3 | 0.357 ± 0.064 | 0.0050 ± 0.0016 | |
| All | 459 | 93 | 7 | 0.592 ± 0.021 | 0.0069 ± 0.0013 | |
| ITS | M. albus | 714 | 44 | 7 | 0.490 ± 0.091 | 0.0010 ± 0.0009 |
| M. officinalis | 714 | 49 | 9 | 0.703 ± 0.645 | 0.0017 ± 0.0014 | |
| All | 714 | 93 | 13 | 0.701 ± 0.036 | 0.0017 ± 0.0014 |
N, number of sequences; h, number of haplotypes; Hd, haplotype diversity; π, nucleotide diversity; SD, standard deviation.
2.6. Molecular Variation within and between Areas
The accessions were divided into four groups according to their origin: Asia, Europe, North America, and South America (Figure S3). AMOVA conducted with the Melilotus accessions based on the trnL-F spacer showed that most of the variation occurred among species within each geographical area. In contrast, for the nrITS region, most of the variation was present among accessions with low variation among species. AMOVA conducted on three areas (Asia, Europe and North America) showed that the variation was significant among accessions with trnL-F and ITS sequences, and only a small amount of variation was present among regions (Figure S3, Table 4).
Table 4.
Analyses of molecular variance (AMOVAs) for Melilotus accessions based on trnL-F and nrITS sequences.
| Region | Sequence | Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation | Fst |
|---|---|---|---|---|---|---|---|
| A | trnL-F | Among species | 1 | 3.199 | 0.206 | 55.820 | 0.558 |
| Among accessions | 31 | 4.737 | 0.163 | 44.180 | |||
| Total | 32 | 7.935 | 0.370 | ||||
| ITS | Among species | 1 | 0.549 | 0.016 | 4.710 | 0.047 | |
| Among accessions | 31 | 9.355 | 0.323 | 95.290 | |||
| Total | 32 | 9.903 | 0.339 | ||||
| E | trnL-F | Among species | 1 | 5.417 | 0.347 | 75.020 | 0.750 |
| Among accessions | 34 | 3.583 | 0.116 | 24.980 | |||
| Total | 35 | 9.000 | 0.463 | ||||
| ITS | Among species | 1 | 2.419 | 0.134 | 30.320 | 0.303 | |
| Among accessions | 34 | 10.152 | 0.308 | 69.680 | |||
| Total | 35 | 12.571 | 0.442 | ||||
| N | trnL-F | Among species | 1 | 3.841 | 0.400 | 78.650 | 0.786 |
| Among accessions | 26 | 2.714 | 0.109 | 21.350 | |||
| Total | 27 | 6.556 | 0.509 | ||||
| ITS | Among species | 1 | 0.882 | 0.065 | 18.850 | 0.188 | |
| Among accessions | 26 | 6.733 | 0.281 | 81.150 | |||
| Total | 27 | 7.615 | 0.346 | ||||
| A & E & N | trnL-F | Among regions | 3 | 1.463 | 0.012 | 3.850 | 0.038 |
| Among accessions | 92 | 23.295 | 0.268 | 96.150 | |||
| Total | 93 | 95.000 | 24.758 | ||||
| ITS | Among regions | 3 | 1.855 | 0.014 | 3.840 | 0.038 | |
| Among accessions | 92 | 29.808 | 0.339 | 96.160 | |||
| Total | 93 | 31.663 | 0.352 |
A, Asian; E, Europe; N, North America; d.f., degrees of freedom; Fst, Fixation index.
3. Discussion
Coumarins are interesting due to their biological functions, which include antioxidant [38], anti-inflammatory [39], antibacterial [40], and termiticidal properties. These compounds have medicinal value due to their therapeutic properties, including edema reduction and possible anticancer activity [41]. With its variable Cou (ranging from 0 (M. segetalis accessions) to 0.943% (M. indicus accessions)) [8], Melilotus represents a cheap, abundant medicinal plant resource yielding high levels of coumarin. However, the genetic variation within M. albus and M. officinalis has not been fully investigated prior to this study. Knowledge of the distribution and amount of genetic variation and the systematic relationships among species is important for plant breeding systems utilizing germplasm resources. The initial objective in breeding Melilotus is to identify agronomically adapted low/high-coumarin germplasm to be used as initial breeding material. Historically, a number of Melilotus varieties were released throughout China, Canada, and the United States (largely before the 1980s). However, distinct varieties are not readily available today due to widespread hybridization among the different varieties, and no new varieties have been released in recent years.
This study indicated significant (p < 0.05) differences among accessions in terms of Cou and the morphological traits of PH, DMY, LSR, PMR, and GH. There was also genetic variation between the two species for these traits. Interspecific variation in the two Melilotus species (93 accessions with 87 strains) was studied using morphological characteristics. The results showed that M. albus and M. officinalis had greater variation among accessions. Fst was affected by genetic drift, migration rate, and selection [42]. PMR demonstrated the largest genetic differentiation capacity among the accessions, while DMY and Cou values were relatively high. The potentially higher genetic differentiation capacity of DMY and Cou provides a genetic basis for breeding cultivars with high DMY and high/low Cou. Cluster analysis of the six traits indicated that all M. albus accessions or all M. officinalis accessions did not cluster into a single group (Figure 1). Thus, hybridization has occurred between M. albus and M. officinalis.
M. albus and M. officinalis, which have low to high coumarin levels, have been identified in different accessions and environments [14,43]. A comprehensive investigation of coumarin concentrations in 149 accessions belonging to 15 Melilotus species showed that the mean Cou ranged from 0.06 to 0.753% of dry matter [8]. Our study identified a negative correlation between the DMY and Cou in two species. There were a number of germplasm accessions with higher DMY values and lower Cou than the commercial lines LX03 and LX05 or accessions with lower DMY values and higher Cou than these commercial lines. Furthermore, 40 half sib (HS) families obtained from cross-pollinating Group 3 (Table 2) individuals of M. officinalis were assessed for their performance across two contrasting locations to identify materials with superior agronomic performance [44]. Any plant improvement program that aims to develop new cultivars of Melilotus for agriculture should aim for high DMY and high/low Cou (high Cou for medicine and low Cou for forage). This information is important for breeding programs targeting high/low Cou in Melilotus.
Sequence variation studies showed that the trnL-F spacer had three indels containing a total of 26 bases representing variable sites. Rarely, indels were detected in M. officinalis; this phenomenon may be due to hybridization with M. albus. Additionally, the Bayesian tree for the trnL-F spacer showed that M. officinalis accessions and M. albus accessions tended to separate into two species groups. Therefore, the trnL-F spacer could be used as a barcode to identify M. albus and M. officinalis because different indels in the two species effectively clustered them into two groups [44].
The higher sequence variability in the ITS region compared with cpDNA, a phenomenon that has also been observed in many other taxa [45,46], may lead to incongruence in the phylogenetic tree. The genetic variation in M. officinalis was higher than that in M. albus in both sequences, as revealed by analyzing haplotypes and nucleotide variation (Table 3). The amplification of nine microsatellite DNA loci from Melilotus also showed that both the allelic variation and expected heterozygosity were slightly lower for M. albus than for M. officinalis, with heterozygote deficits at several loci [47]. M. officinalis originated in Europe, and M. albus originated in Western Asia [48], suggesting that the different geographical and evolutionary histories of these species may also cause differences in their genetic variation. Genetic variation analyses by region showed that genetic variation in the ITS sequence among accessions was higher than that among species in all three regions, but genetic variation in the trnL-F sequence among accessions was lower. Analysis of the three regions indicated that the genetic variation among accessions was higher than that among species for both markers. The difference in variation between the two sequences in all three regions was mainly attributable to the indels in the trnL-F sequence. Germplasm exchange and introduction throughout the world are very frequent, which may also influence the genetic variation in plant species in different regions. Haplotype phylogenetic analyses showed that the haplotypes of the two species were mixed together in the trnL-F and ITS trees. The results indicated that M. albus and M. officinalis have a close relationship. Karyotype analyses [49] and interspecific phylogenic relationships [35] also showed that M. albus and M. officinalis are closely related within the Melilotus genus. In addition, incomplete lineage sorting [50] and hybridization between, and within, species [51] may cause phylogenetic incongruence. The positions of M. albus and M. officinalis in the phylogenetic tree indicate that they have shared haplotypes, although they have obvious morphological differences. Therefore, there is a certain amount of gene flow between the two species. It might be argued that interspecific hybridization, a widespread phenomenon that has markedly contributed to variation and speciation in the plant kingdom [52,53,54,55], contributed to the genetic introgression of M. albus and M. officinalis. In this report, some accessions sharing similar phenotype trait data and Cou clustered together by trnL-F and ITS sequences. Therefore, the combination of Cou, morphological variation and molecular phylogenetics identified accessions of M. albus and M. officinalis based on first-year establishment data for medicinal purposes and low Cou for forage.
4. Materials and Methods
4.1. Germplasm
A total of 93 accessions were evaluated in this study, comprising 42 accessions of M. albus and 51 accessions of M. officinalis (Table S3). All seeds of accessions were obtained from the National Plant Germplasm System (NPGS, USA). In addition to the germplasm accessions, two cultivars (LX03, M. officinalis and LX05, M. albus) obtained from the local seed industry (Tongwei Longzhong Forage Seed Company, Tongwei county, Gansu province, China) were included in our study. The collection contained 36 wild, 15 cultivated, and 12 bred strains, as well as 30 accessions with uncertain pedigree according to the germplasm bank passport information (Table S3).
4.2. Field Experiment
4.2.1. Preparation of Plant Material
Sixty seeds from each accession were sown in plastic pots (13 cm × 12 cm) containing a potting mix of sandy soil and peat (1:1 by volume). After germination, the seedlings were planted into pots with the soil mixture in a glass house. Twenty seedlings of each accession, uniform in size, were transplanted into the field after eight weeks. The field trial was established at Yuzhong (35°57′ N, 104°09′ E) in Gansu Province, China, in June 2012. The long-term annual rainfall at Yuzhong is 382 mm, with an evaporation of 1343 mm and an average temperature of 6.7 °C throughout the year.
4.2.2. Field Trial
The field trial had a randomized complete block design with two replicates. Each accession was represented by 10 plants per replicate grown at a plant spacing of 30 cm × 30 cm. Prior to planting, the experimental area was prepared as a seedbed. Post planting, all trial management was conducted according to local procedures, and weeding occurred regularly. No irrigation was applied.
4.2.3. Agronomic Traits
As most Melilotus species are biennial, we did not consider first-year establishment data to be important for our breeding program; all measurements were carried out during the second year. Data were collected for the following traits: PH, DMY, LSR, Cou, and PMR. PH (cm) was measured with ten plants per accession per replicate at the early flowering stage, and the aboveground plant parts were harvested. The plant material was air-dried to a constant weight at room temperature and then separated by hand into leaf and stem fractions. The mass composition of whole plants (DMY) was calculated from the weights of the leaves and stems, and the LSR was calculated. The Cou (% of DMY) was measured following the UV method of Tang [56], in which 1.0 g of well-mixed plant material was extracted for 40 min in 50 mL of 50% ethanol using an ultrasonic bath. After ultrasound-assisted extraction, all of the filtrate was combined, and its volume was reduced using a rotating vacuum evaporator. Then, 10 mL of methanol was added, and the Cou was quantified using a UV-VIS spectrophotometer via light absorption at 310 nm, which is the maximum absorption of coumarin residues. The PMR of the plants was scored in the field following the method of Xie [57]. The plants were scored under field conditions during early autumn, which is a period of high powdery mildew infection, using a scale of 0 for no visible symptoms to 4 for a highly susceptible reaction.
4.2.4. Analysis of Variance and Pattern Analysis
To evaluate accession differentiation at the phenotype level, the variance component among accessions divided by the sum of variance components among and within accessions was computed for each trait (Pst). A key objective in the breeding program for both M. officinalis and M. albus is to improve the DMY and the Cou. Therefore, a combination of cluster analysis and principal component analysis (PCA) [58,59] was conducted to summarize the accession-by-trait data matrix for the traits PH, DMY, LSR, Cou, PMR, and GH. Cluster analysis was conducted by performing UPGMA clustering in GenStat 18th Edition (https://www.vsni.co.uk/webstore/old-webstore/software/ genstat/). Biplots were generated from the PCA to enable the assessment of genotypic variation among germplasm accessions on a multivariate scale. Only traits for which there was significant (p < 0.05) genotypic variation among the accessions were included in the pattern analysis.
4.3. Molecular Phylogeny
4.3.1. DNA Extraction, Amplification, and Sequencing
Young leaves were collected from twenty plants of each accession, frozen in liquid nitrogen, and stored at −80 °C until they were used for DNA isolation via the SDS extraction method [60]. Preliminary universal primer scanning was conducted on 10 individuals sampled from 10 different accessions. ITS1, 5.8s, and ITS2 were amplified as a single molecule using the flanking primers EC-1 (5′-GAGGAAGGAGAAGTCGTAAC-3′) and EC-2 (5′-GTTCGCTCGCCGTTACTAAG-3′) [61]. The trnL-F spacer was amplified using the primer pair trnL (CGAAATCGGTAGACGCTACG) and trnF (ATTTGAACTGGTGACACGAG) according to Taberlet et al. [62]. PCR amplification [63] was carried out in a 30 μL volume containing 12 μL of deionized water, 15 μL of Takara Taq DNA polymerase master mix, 1 μL of each primer (5 pmol/mL), and 1 μL of template DNA (40 ng/mL). The cycling conditions for amplification consisted of a single cycle at 94 °C for 3 min for initial denaturation; 36 cycles at 94 °C for 30 s, 52 °C for 45 s, and 72 °C for 1 min; and a final extension at 72 °C for 10 min. The PCR products were checked for fragment length and separation in 1% agarose gels and then sequenced using a Big Dye kit with the appropriate primers in an ABI 3730 DNA sequencer at Shanghai Shenggong Biotechnological, Ltd. (Shanghai, China).
All raw sequences were checked by Chromas software (Shanghai Shenggong Biotechnological, Ltd. Shanghai, China), and for sequences that were inaccurate, the corresponding PCR samples were re-prepared and sequenced again. If sequence quality was acceptable, the sequences were aligned by using DNAMAN to ensure their identity as Melilotus sequences.
4.3.2. Phylogenetic Analyses
The sequences were aligned in ClustalW (within MEGA 6.0) [64] and manually adjusted using MEGA 6.0 [65]. Maximum parsimony analyses involved a heuristic search strategy with 1000 replicates of random sequence addition in combination with tree bisection and reconnection (TBR) branch swapping in MEGA 6.0. A phylogenetic tree was constructed with ITS and trnL-F combined sequences in MEGA 6.0. Separate ITS and trnL-F phylogenetic trees were inferred by Bayesian inference via MrBayes-3.2.6 [66] (two runs of 10,000,000 generations with four chains for ITS and three chains for trnL-F) under a GTR model with base frequencies, gamma shape parameter, and proportion of invariants estimated from the data (referred to as GTR + I + G). All character states were treated as unordered and equally weighted. Informative insertions and deletions (indels) were coded as binary characters (0, 1) according to Graham et al. (2000). A strict consensus tree was constructed from the most parsimonious trees. Haplotype diversity (Hd) and nucleotide diversity (π) analyses were conducted using DnaSP v5 [67]. Fu’s Fs test [68] for all sequences was used to test each haplotype group to determine if all samples followed neutral evolution. Inter-accession differentiation between, and within, the two species was evaluated by AMOVA [69] using Arlequin software, version 3 [70]. The sequences of the outgroup M. lupulina were downloaded from GenBank (KX167243.1 and KU600399.1). The multiple sequence alignments and phylogenetic trees were submitted to TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S22217).
Acknowledgments
This work was financially supported by the Program for Changjiang Scholars and Innovative Research Team in University (IRT_17R50), the National Natural Science Foundation of China (31572453), the Open Project Program of the State Key Laboratory of Grassland Agro-ecosystems (SKLGAE201702), and the 111 project (B12002). We thank Carol Baskin from Kentucky University for kindly revising the manuscript; Shugao Fan, Tingna Liu, Xiaohui Huang, Yu Chang, Zhiqian Li and many other colleagues for their assistance with germplasm evaluation and data collection; Zhaofeng Wang and Qisheng Feng for useful advice; and the anonymous reviewers for their helpful comments and detailed suggestions for our revisions.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/23/4/810/s1. Figures S1–S3; Tables S1–S3.
Author Contributions
J.Z., Y.W. and Z.J. conceived and designed the experiments; H.D., K.L. and Z.Y. performed the experiments; F.W. and Z.D. analyzed the data; Z.J. and A.S. contributed reagents/materials/analysis tools; F.W. and J.Z. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Hawryl M.A., Soczewinski E., Dzido T.H. Separation of coumarins from Archangelica officinalis in high-performance liquid chromatography and thin-layer chromatography systems. J. Chromatogr. A. 2000;886:75–81. doi: 10.1016/S0021-9673(00)00321-6. [DOI] [PubMed] [Google Scholar]
- 2.De Vincenzi M., Mancini E., Dessi M. Monographs on botanical flavouring substances used in foods. Part VI. Fitoterapia. 1997;68:49–61. [Google Scholar]
- 3.Celeghini R., Vilegas J.H., Lanças F.M. Extraction and quantitative HPLC analysis of coumarin in hydroalcoholic extracts of Mikania glomerata Spreng: (“guaco”) leaves. J. Brazil Chem. Soc. 2001;12:706–709. doi: 10.1590/S0103-50532001000600003. [DOI] [Google Scholar]
- 4.Yarnell E., Abascal K. Plant coumarins: Myths and realities. Altern. Complement. Ther. 2009;15:24–30. doi: 10.1089/act.2009.15104. [DOI] [Google Scholar]
- 5.Christensen T.D., Maegaard M., Sørensen H.T., Hjortdal V.E., Hasenkam J.M. Self-versus conventional management of oral anticoagulant therapy: Effects on INR variability and coumarin dose in a randomized controlled trial. Am. J. Cardiovasc. Drug. 2007;7:191–197. doi: 10.2165/00129784-200707030-00005. [DOI] [PubMed] [Google Scholar]
- 6.Lee C.M., Jiang L.M., Shang H.S., Hon P.M., He Y., Wong H.N. Prehispanolone, a novel platelet activating factor receptor antagonist from Leonurus heterophyllus. Br. J. Pharmacol. 1991;103:1719–1724. doi: 10.1111/j.1476-5381.1991.tb09853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans P., Kearney G. Melilotus albus (Medik) is productive and regenerates well on saline soils of neutral to alkaline reaction in the high rainfall zone of south-western Victoria. Aust. J. Exp. Agric. 2003;43:349–355. doi: 10.1071/EA02079. [DOI] [Google Scholar]
- 8.Nair R.M., Whittall A., Hughes S.J., Craig A.D., Revell D.K., Miller S.M., Powell T., Auricht G.C. Variation in coumarin content of Melilotus species grown in South Australia. N. Z. J. Agric. Res. 2010;53:201–213. doi: 10.1080/00288233.2010.495743. [DOI] [Google Scholar]
- 9.Bowman G., Shirley C., Cramer C. Sustainable Agriculture Network Handbook Series Book. Volume 3. Sustainable Agriculture Network; Beltsville, MD, USA: 1998. Managing cover crops profitably; pp. 22–23. [Google Scholar]
- 10.Aboel-Atta A.M.I. Isozymes, RAPD and ISSR variation in Melilotus indica (L.) All. and M. siculus (Turra) BG Jacks.(Leguminosae) Acad. J. Plant Sci. 2009;2:113–118. [Google Scholar]
- 11.Yang Y.F. The pioneer pasture Melilotus that have drought and barren resistance. Beijing Agric. 2003;8:25–26. [Google Scholar]
- 12.Hirsch A.M., Lum M.R., Krupp R.S.N., Yang W., Karlowski W.M. Melilotus Alba Desr., White Sweetclover, a Mellifluous Model Legume. Horizon Scientific Press; Poole, UK: 2000. pp. 627–642. [Google Scholar]
- 13.Pleşca-Manea L., Pârvu A.E., Pârvu M., Taămaş M., Buia R., Puia M. Effects of Melilotus officinalis on acute inflammation. Phytother. Res. 2002;16:316–319. doi: 10.1002/ptr.875. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson G.A. An agronomic and taxonomic review of the genus Melilotus Mill. Can. J. Plant Sci. 1969;49:1–20. doi: 10.4141/cjps69-001. [DOI] [Google Scholar]
- 15.Cong J.M., Chen F.Q., Sun C.L. Study on Comprehensive Development of Metlilotus suaverolens L. J. Anhui Agric. Sci. 2012;5:2962–2963. [Google Scholar]
- 16.Griffiths B.S., Donn S., Neilson R., Daniell T.J. Molecular sequencing and morphological analysis of a nematode community. Appl. Soil Ecol. 2006;32:325–337. doi: 10.1016/j.apsoil.2005.07.006. [DOI] [Google Scholar]
- 17.Subbotin S.A., Halford P.D., Warry A., Perry R.N. Variations in ribosomal DNA sequences and phylogeny of Globodera parasitising solanaceous plants. Nematology. 2000;2:591–604. doi: 10.1163/156854100509484. [DOI] [Google Scholar]
- 18.Gozel U., Lamberti F., Duncan L., Agostinelli A., Rosso L., Nguyen K., Adams B.J. Molecular and morphological consilience in the characterisation and delimitation of five nematode species from Florida belonging to the Xiphinema americanum-group. Nematology. 2006;8:521–532. doi: 10.1163/156854106778613958. [DOI] [Google Scholar]
- 19.Tian B., Liu R., Wang L., Qiu Q., Chen K., Liu J. Phylogeographic analyses suggest that a deciduous species (Ostryopsis davidiana Decne., Betulaceae) survived in northern China during the Last Glacial Maximum. J. Biogeogr. 2009;36:2148–2155. doi: 10.1111/j.1365-2699.2009.02157.x. [DOI] [Google Scholar]
- 20.Hughes C.E., Bailey C.D., Harris S.A. Divergent and reticulate species relationships in Leucaena (Fabaceae) inferred from multiple data sources: Insights into polyploid origins and nrDNA polymorphism. Am. J. Bot. 2002;89:1057–1073. doi: 10.3732/ajb.89.7.1057. [DOI] [PubMed] [Google Scholar]
- 21.Arnheim N., Krystal M., Schmickel R., Wilson G., Ryder O., Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc. Natl. Acad. Sci. USA. 1980;77:7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmer E., Martin S., Beverley S., Kan Y., Wilson A.C. Rapid duplication and loss of genes coding for the alpha chains of hemoglobin. Proc. Natl. Acad. Sci. USA. 1980;77:2158–2162. doi: 10.1073/pnas.77.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hillis D.M., Dixon M.T. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Abbott R.J., Zheng W., Chen P., Wang Y., Liu J. History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae) Mol. Ecol. 2009;18:709–721. doi: 10.1111/j.1365-294X.2008.04055.x. [DOI] [PubMed] [Google Scholar]
- 25.Oskoueiyan R., Kazempour Osaloo S., Maassoumi A.A., Nejadsattari T., Mozaffarian V. Phylogenetic status of Vavilovia formosa (Fabaceae-Fabeae) based on nrDNA ITS and cpDNA sequences. Biochem. Syst. Ecol. 2010;38:313–319. doi: 10.1016/j.bse.2010.01.011. [DOI] [Google Scholar]
- 26.Zhang W., Kan S.L., Zhao H., Li Z.Y., Wang X.Q. Molecular Phylogeny of Tribe Theeae (Theaceae ss.) and Its Implications for Generic Delimitation. PLoS ONE. 2014;9:e98133. doi: 10.1371/journal.pone.0098133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardoso D., Paganucci de Queiroz L., Cavalcante de Lima H., Suganuma E., van den Berg C., Lavin M. A molecular phylogeny of the vataireoid legumes underscores floral evolvability that is general to many early-branching papilionoid lineages. Am. J. Bot. 2013;100:403–421. doi: 10.3732/ajb.1200276. [DOI] [PubMed] [Google Scholar]
- 28.Kores P.J., Molvray M., Weston P.H., Hopper S.D., Brown A.P., Cameron K.M., Chase M.W. A phylogenetic analysis of Diurideae (Orchidaceae) based on plastid DNA sequence data. Am. J. Bot. 2001;88:1903–1914. doi: 10.2307/3558366. [DOI] [PubMed] [Google Scholar]
- 29.Berg C.V.D., Higgins W.E., Dressler R.L., Whitten W.M., Soto Arenas M.A., Culham A., Chase M.W.A. Phylogenetic analysis of Laeliinae (Orchidaceae) based on sequence data from internal transcribed spacers (ITS) of nuclear ribosomal DNA. Lindleyana. 2000;15:96–114. [Google Scholar]
- 30.Whitten W.M., Williams N.H., Chase M.W. Subtribal and generic relationships of Maxillarieae (Orchidaceae) with emphasis on Stanhopeinae: Combined molecular evidence. Am. J. Bot. 2000;87:1842–1856. doi: 10.2307/2656837. [DOI] [PubMed] [Google Scholar]
- 31.Scheen A.C., Bendiksby M., Ryding O., Mathiesen C., Albert V.A., Lindqvist C. Molecular phylogenetics, character evolution, and suprageneric classification of Lamioideae (Lamiaceae) Ann. Mo. Bot. Gard. 2016;97:191–217. doi: 10.3417/2007174. [DOI] [Google Scholar]
- 32.Faye A., Pintaud J.C., Baker W., Sonké B., Couvreur T. A plastid phylogeny of the African rattans (Ancistrophyllinae, Arecaceae) Syst. Bot. 2014;39:1099–1107. doi: 10.1600/036364414X682634. [DOI] [Google Scholar]
- 33.Sunderland T.C.H. A taxonomic revision of the rattans of Africa (Arecaceae: Calamoideae) Phytotaxa. 2012;51:1–76. doi: 10.11646/phytotaxa.51.1.1. [DOI] [Google Scholar]
- 34.Baker W.J., Dransfield J., Hedderson T.A. Phylogeny, character evolution, and a new classification of the calamoid palms. Syst. Bot. 2000;25:297–322. doi: 10.2307/2666644. [DOI] [Google Scholar]
- 35.Di H.Y., Duan Z., Luo K., Zhang D.Y., Wu F., Zhang J.Y., Liu W.X., Wang Y.R. Interspecific phylogenic relationships within genus Melilotus based on nuclear and chloroplast DNA. PLoS ONE. 2015;10:e0132596. doi: 10.1371/journal.pone.0132596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu F., Zhang D.Y., Ma J.X., Luo K., Di H.Y., Liu Z.P., Zhang J.Y., Wang Y.R. Analysis of genetic diversity and population structure in accessions of the genus Melilotus. Ind. Crop. Prod. 2016;85:84–92. doi: 10.1016/j.indcrop.2016.02.055. [DOI] [Google Scholar]
- 37.Yan Z.Z., Wu F., Luo K., Zhao Y.F., Yan Q., Zhang Y.F., Wang Y.R., Zhang J.Y. Cross-species transferability of EST-SSR markers developed from the transcriptome ofMelilotusand their application to population genetics research. Sci. Rep. 2017;7:17959. doi: 10.1038/s41598-017-18049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bajerová P., Adam M., Bajer T., Ventura K. Comparison of various techniques for the extraction and determination of antioxidants in plants. J. Sep. Sci. 2014;37:835–844. doi: 10.1002/jssc.201301139. [DOI] [PubMed] [Google Scholar]
- 39.Witaicenis A., Seito L.N., da Silveira Chagas A., de Almeida L.D., Luchini A.C., Rodrigues-Orsi P., Cestari S.H., Di Stasi L.C. Antioxidant and intestinal anti-inflammatory effects of plant-derived coumarin derivatives. Phytomedicine. 2014;21:240–246. doi: 10.1016/j.phymed.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Céspedes C.L., Avila J.G., Martínez A., Serrato B., Calderón-Mugica J.C., Salgado-Garciglia R. Antifungal and antibacterial activities of Mexican tarragon (Tagetes lucida) J. Agric. Food Chem. 2006;54:3521–3527. doi: 10.1021/jf053071w. [DOI] [PubMed] [Google Scholar]
- 41.Musa M.A., Cooperwood J.S., Khan M.O.F. A review of coumarin derivatives in pharmacotherapy of breast cancer. Curr. Med. Chem. 2008;15:2664–2679. doi: 10.2174/092986708786242877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Podolsky R.H., Holtsford T.P. Population-Structure of Morphological Traits in Clarkia-Dudleyana. 1. Comparison of F-St between Allozymes and Morphological Traits. Genetics. 1995;140:733–744. doi: 10.1093/genetics/140.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo K., Jahufer M.Z.Z., Wu F., Di H.Y., Zhang D.Y., Meng X.C., Zhang J.Y., Wang Y.R. Genotypic variation in a breeding population of yellow sweet clover (Melilotus officinalis) Front. Plant Sci. 2016;7:972. doi: 10.3389/fpls.2016.00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu F., Ma J.X., Meng Y.Q., Zhang D.Y., Muvunyi B.P., Luo K., Di H.Y., Guo W.L., Wang Y.R., Feng B.C., et al. Potential DNA barcodes for Melilotus species based on five single loci and their combinations. PLoS ONE. 2017;12:e0182693. doi: 10.1371/journal.pone.0182693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fior S., Karis P.O., Casazza G., Minuto L., Sala F. Molecular phylogeny of the Caryophyllaceae (Caryophyllales) inferred from chloroplast matK and nuclear rDNA ITS sequences. Am. J. Bot. 2006;93:399–411. doi: 10.3732/ajb.93.3.399. [DOI] [PubMed] [Google Scholar]
- 46.Mitsui Y., Chen S.T., Zhou Z.K., Peng C.I., Deng Y.F., Setoguchi H. Phylogeny and biogeography of the genus Ainsliaea (Asteraceae) in the Sino-Japanese region based on nuclear rDNA and plastid DNA sequence data. Ann. Bot. 2008;101:111–124. doi: 10.1093/aob/mcm267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winton L.M., Krohn A.L., Conn J.S. Microsatellite markers for the invasive plant species white sweetclover (Melilotus alba) and yellow sweetclover (Melilotus officinalis) Mol. Ecol. Notes. 2007;7:1296–1298. doi: 10.1111/j.1471-8286.2007.01860.x. [DOI] [Google Scholar]
- 48.Zhang B.L., Wen C.R. Melilotus. Agriculture Press; Liaoning, China: 1978. [Google Scholar]
- 49.Clarke A.E. The Number and Morphology of Chromosomes in the Genus Melilotus. University of California Press; Berkeley, CA, USA: 1934. [Google Scholar]
- 50.Maddison W.P., Knowles L.L. Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 2006;55:21–30. doi: 10.1080/10635150500354928. [DOI] [PubMed] [Google Scholar]
- 51.Käss E., Wink M. Phylogenetic relationships in the Papilionoideae (family Leguminosae) based on nucleotide sequences of cpDNA (rbcL) and ncDNA (ITS 1 and 2) Mol. Phylogent. Evol. 1997;8:65–88. doi: 10.1006/mpev.1997.0410. [DOI] [PubMed] [Google Scholar]
- 52.Rieseberg L.H. Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 1997;28:359–389. doi: 10.1146/annurev.ecolsys.28.1.359. [DOI] [Google Scholar]
- 53.Rieseberg L.H., Carney S.E. Plant hybridization. New Phytol. 1998;140:599–624. doi: 10.1046/j.1469-8137.1998.00315.x. [DOI] [PubMed] [Google Scholar]
- 54.Arnold M.L., Bulger M.R., Burke J.M., Hempel A.L., Williams J.H. Natural hybridization: How low can you go and still be important? Ecology. 1999;80:371–381. doi: 10.1890/0012-9658(1999)080[0371:NHHLCY]2.0.CO;2. [DOI] [Google Scholar]
- 55.Barton N.H. The role of hybridization in evolution. Mol. Ecol. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- 56.Tang C.N. Master’s Thesis. Northwest University; Xian, China: 2010. Study on Extraction, Separation and Purification Technology of Coumarin from Melilotus. [Google Scholar]
- 57.Xie C., Sun Q., Ni Z., Yang T., Nevo E., Fahima T. Chromosomal location of a Triticum dicoccoides-derived powdery mildew resistance gene in common wheat by using microsatellite markers. Theor. Appl. Genet. 2003;106:341–345. doi: 10.1007/s00122-002-1022-1. [DOI] [PubMed] [Google Scholar]
- 58.Kroonenberg P.M.K. The TUCKALS line: A suite of programs for three-way data analysis. Comput. Stat. Data Anal. 1994;18:73–96. doi: 10.1016/0167-9473(94)90133-3. [DOI] [Google Scholar]
- 59.Gabriel K.R. The biplot graphical display of matrices with applicationto principal component analysis. Biometrika. 1971;58:453–467. doi: 10.1093/biomet/58.3.453. [DOI] [Google Scholar]
- 60.Zhang J.Y., Yuan Q.H., Zhang W.S., Lu T. Genomic DNA extraction and optimizing RAPD procedure of Lespedeza floribunda. Acta Agrestia Sin. 2004;12:219–223. [Google Scholar]
- 61.Williams W.M., Ansari H.A., Ellison N.W., Hussain S.W. Evidence of Three Subspecies in Trifolium nigrescens Viv. Ann. Bot. 2001;87:683–691. doi: 10.1006/anbo.2001.1399. [DOI] [Google Scholar]
- 62.Taberlet P., Gielly L., Pautou G., Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 63.Chen K., Abbott R.J., Milne R.I., Tian X.M., Liu J. Phylogeography of Pinus tabulaeformis Carr. (Pinaceae), a dominant species of coniferous forest in northern China. Mol. Ecol. 2008;17:4276–4288. doi: 10.1111/j.1365-294X.2008.03911.x. [DOI] [PubMed] [Google Scholar]
- 64.Larkin M., Blackshields G., Brown N., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 65.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ronquist F., Teslenko M., Mark P.V.D., Huelsenbeck J. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 68.Fu Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Excoffier L., Smouse P.E., Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Excoffier L., Laval G., Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2005;1:47–50. doi: 10.1177/117693430500100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.