Abstract
Tripartite motif-containing 27 (TRIM27) belongs to the tripartite motif (TRIM) protein family and is involved in various malignant tumor processes. However, the function and mechanism of TRIM27 in colorectal cancer (CRC) remains to be elucidated. In the present study, the expression of TRIM27 was analyzed in CRC tissues and adjacent normal tissues by reverse transcription-quantitative polymerase chain reaction and immunohistochemistry. LoVo and HCT116 cell lines were then selected to further investigate the function of TRIM27 in the proliferation, invasion and metastasis of CRC in vitro and in vivo. Finally, the potential mechanism underlying the effects of TRIM27 in CRC was examined by western blotting. The results showed that TRIM27 was upregulated in CRC tissues, and the expression level of TRIM27 was significantly associated with tumor invasion, metastasis and prognosis. Following TRIM27 inhibition and overexpression in CRC cells, it was found that TRIM27 promoted cell proliferation, possibly via the inhibition of apoptosis and cell cycle regulation. TRIM27 also facilitated invasion and metastasis. Finally, it was observed that TRIM27 promoted epithelial-mesenchymal transition and activated phosphorylated AKT serine/threonine kinase in CRC cells. These results suggested that TRIM27 is an oncogenic protein in the progression of CRC, and may represent a novel target for CRC detection and therapy.
Keywords: colorectal cancer, tripartite motif-containing 27, phosphorylated AKT serine/threonine kinase, epithelial-mesenchymal transition, oncogene
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in men and the second most commonly diagnosed cancer in women worldwide (1). According to cancer statistics in China, the incidence and mortality rates of CRC continue to increase (2). Tumor stage is the most important prognostic factor in CRC. For example, the 5-year survival rate of patients diagnosed with CRC in the USA in 2001–2007 was 90.1% for patients with CRC at the localized stage, but 11.7% for patients with distant tumor spread (3). However, only 40% of patients diagnosed with CRC were detected at the localized stage (4). Therefore, to improve the early diagnosis of patients with CRC, understanding the pathogenesis of this disease is important.
The tripartite motif (TRIM) protein family comprises proteins consisting of three domains, a RING finger protein domain, a B-box type I domain, and a B-box type II domain, followed by a coiled-coil region, and a highly variable C-terminal region (5). To date, >70 TRIM genes have been recognized in the human genome (6–8). As a member of the TRIM family, TRIM27 (also known as RFP) inherited the basic structure of this family. TRIM27 was first identified as a gene involved in the generation of the RET transforming gene activated by DNA rearrangement (9–11). In the majority of human tissues, TRIM27 has been reported to be detectable (12). The role of TRIM27 in cancer has received increased attention. It has been reported that TRIM27 can act as an oncogene in ovarian cancer, endometrial cancer, breast cancer and lung cancer (13–16). In addition, TRIM27 is important in promoting anticancer drug resistance in specific tumors (17,18). These studies suggested that TRIM27 may have an oncogenic role in various types of tumor. In CRC, TRIM27 has been reported to be upregulated and can predict chemotherapy resistance (18,19). However, the exact role of TRIM27 in the progression of CRC remains to be fully elucidated.
Epithelial-mesenchymal transition (EMT) is a developmental process that promotes invasion and metastasis in various types of tumor (20). During EMT, E-cadherin and N-cadherin are the most commonly detected epithelial and mesenchymal markers, respectively. Vimentin, as a major member of the intermediate filament, is expressed in almost all mesenchymal cells, and is important in maintaining cell integrity and resisting external injury (21). The multi-step process of EMT involves multiple regulatory mechanisms, including the activation of phosphorylated AKT serine/threonine kinase (p-AKT) (22). However, until now, the association between TRIM27 and EMT in CRC has not been investigated.
The aim of the present study was to analyze the expression of TRIM27 in CRC tissues and adjacent normal tissues. The study also aimed to further investigate the biological role of TRIM27 in CRC cells in vivo and in vitro via the inhibition and overexpression of TRIM27. Finally, the potential mechanism underlying the effects of TRIM27 on the progression of CRC was examined.
Materials and methods
Patients and tissue specimens
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (Nanjing, China; Ethics no. 2010-SR-091.A1). In total, 80 pairs of human CRC tissues and adjacent normal tissues were collected from patients with CRC, who had signed an informed consent form, between 2010 and 2012 at the First Affiliated Hospital of Nanjing Medical University. All the patients were aged between 27 and 88 years old (average, 61.6 years old), and none of the patients had a history of radiotherapy or chemotherapy prior to surgery. All samples were immediately preserved in liquid nitrogen within 5 min following resection and then placed at −70°C for long-term preservation. The tumor-node-metastasis stage was determined based on the National Comprehensive Cancer Network (23).
CRC cell lines and culture conditions
All CRC cell lines (LoVo, HCT116, SW480, DLD-1 and HT29) and normal epithelial colon cells (NCM460) were purchased from the American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (both from Winsent, Inc., St. Bruno, QC, Canada), 100 U/ml of penicillin and 100 µg/ml of streptomycin in a humid incubator (stabilized at 5% CO2 and 37°C)
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR analysis)
Total RNA was extracted from CRC tissues and cells using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s protocol. The cDNA was produced from the RNA by performing reverse transcription using a PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). The RT-qPCR experiment was performed in a 20 µl volume consisting of 2 µl cDNA, 1.2 µl primers, 6.8 µl dH2O and 10 µl SYBR, using a SYBR-Green PCR kit (Roche Diagnostics, Indianapolis, IN, USA). The final reaction was performed in a StepOnePlus Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) and comprised: Hot-start DNA polymerase activation to 95°C for 10 min; 40 cycles of 95°C for 15 sec and 60°C for 1 min; followed by one cycle of melt curve analysis at 95°C for 15 sec, 60°C for 1 min, and 95°C for 15 sec. The specific primers were as follows: TRIM27, forward, 5′-AGCCCATGATGCTCGACTG-3′ and reverse, 5′-GGGCACGACACGTTAGTCT-3′; GAPDH, forward, 5′-AGAAGGCTG GGGCTCATTTG-3′ and reverse, 5′-AGGGGCCATCCACAGTCTTC-3′. TRIM27 expression was normalized to GAPDH and relative expression levels were calculated using the 2−ΔΔCq method (24).
Immunohistochemistry (IHC)
To detect the level of TRIM27 in CRC, 50 CRC tissues and 25 adjacent normal tissues were evaluated using IHC. The formalin-fixed and paraffin-embedded tumor and normal tissue sections (4-µm) were deparaffinized in xylene and rehydrated in different concentrations of alcohol and distilled water, followed by microwave antigen retrieval. The sections were washed in phosphate-buffered saline (PBS) three times and were placed in 3% H2O2 for 20 min in the dark, followed by three washes with PBS. The tissues were then soaked in 5% BSA (Servicebio, Wuhan, China) for 30 min. Finally, the tissues were reacted with anti-TRIM27 antibodies (diluted 1:1,000, polyclonal, rabbit, cat. no. ab78393; Abcam, Cambridge, MA, USA) overnight at 4°C, followed by incubation with anti-rabbit antibodies (horseradish peroxidase-tagged, diluted 1:1,000, polyclonal, cat. no. ab6721; Abcam) at room temperature for 50 min, and with the color agent diaminobenzidine. The nuclei were counterstained with hematoxylin, and the sections were dehydrated using different grades of ethyl alcohol and xylene. The sections were then viewed under an inverted microscope (NIKON ECLIPSE TI-SR; Nikon Corporation, Tokyo, Japan). Based on the staining intensity, the level of TRIM27 was graded as 0 (no staining), 1 (+), 2 (++), and 3 (+++). According to the proportion of TRIM27-positive cells, the scores were as follows: 0 (negative), 1 (<30%), 2 (31–60%), 3 (>60%). The total score was calculated as the intensity score plus the positive rate score. Scores ≥4 were regarded as a high level of TRIM27, and scores <4 were regarded as a low level of TRIM27.
Knockdown and overexpression of TRIM27
Small interference RNA (siRNA) targeting TRIM27 and a negative control sequence (NC) were designed by GenePharma Corporation (Shanghai, China). The target sequences are shown in Table I (13). The TRIM27 inhibitor lentivirus [short hairpin (sh)TRIM27] in vivo experiment was designed based on the sequence of siRNA918, with the specific sequence: 5′-GCAGCTGATATCACTCCTTA-3′. For the overexpression of TRIM27, a plasmid expressing TRIM27 was designed by GeneCopoeia, Inc. (Rockville, ME, USA). The above siRNAs and plasmid were transfected into cells using Lipofectamine® 3000, according to the manufacturer’s protocol (Invitrogen; Thermo Fisher Scientific, Inc.).
Table I.
Sequences of small interference RNAs designed for TRIM27 knockdown.
| Name | Sequence |
|---|---|
| TRIM27-homo-1251 | F: 5′-GCAGUCAGAUAUGGAGAAATT-3′ |
| R: 5′-UUUCUCCAUAUCUGACUGCTT-3′ | |
| TRIM27-homo-1578 | F: 5′-GGCAGUGUCUUUGUGGUAUTT-3′ |
| R: 5′-AUACCACAAAGACACUGCCTT-3′ | |
| TRIM27-homo-918 | F: 5′-GCAGCUGUAUCACUCCUUATT-3′ |
| R: 5′-UAAGGAGUGAUACAGCUGCTT-3′ | |
| GAPDH-negative control | F: 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| R: 5′-ACGUGACACGUUCGGAGAATT-3′ | |
| GAPDH-positive control | F: 5′-GUAUGACAACAGCCUCAAGTT-3′ |
| R: 5′-CUUGAGGCUGUUGUCAUACTT-3′ |
TRIM27, tripartite motif-containing 27; F, forward; R, reverse.
Cell viability assay
To investigate the effect of TRIM27 on the proliferation of CRC cells, 2×103 cells per well were cultured in 96-well plates with each well containing 100 µl of medium. Cell viability was detected using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kummamoto, Japan) assay according to the manufacturer’s protocol. After 24, 48, 72 and 96 h, 10 µl of CCK-8 assay reagent was added to each well mixed with 90 µl of serum-free medium. The absorbance was measured 2 h later using a microplate reader at a test wavelength of 450 nm and a reference wavelength of 630 nm.
Plate colony formation assay
The cells (500 per well) were cultured in 6-well plates to investigate the effect of TRIM27 on the efficiency of colony formation in CRC cells. After 7 days, each well was washed with PBS three times at room temperature. The cells were then fixed in each well using ethyl alcohol for 30 sec and stained for 20 min using crystal violet dye. Following washing with PBS, colonies (≥50 cells/colony) in each well were manually counted and images were captured using a digital camera (Canon DS126211; Canon, Tokyo, Japan).
Wound-healing assay
The CRC cells (4×105) were seeded in 6-well plates and cultured until they reached a density of 90%. A 200-µl micropipette tip was then used make a wound in the cells and the medium was replaced with serum-free medium. Electron microscopy was used to observe the shape of wound at 0 and 24 h.
Transwell assay
Cell migration and invasion were assayed in a 24-well plate with polycarbonate sterile chambers (8-µm filters; BD Biosciences, Franklin Lakes, NJ, USA) with or without Matrigel coating. The CRC cells (2×104) were cultured with 100 µl of serum-free DMEM in the upper chamber and 600 µl of DMEM + 10% serum in the lower chamber. After 24 h, the lower chamber was washed twice with PBS and crystal violet dye was added to the lower chamber incubated for 20 min. The lower chamber was washed with PBS three times, following which a cotton bud was used to remove cells and medium from the upper chambers. The migrated or invaded cells in the lower chambers were observed under an electron microscope.
Cell apoptosis analysis
Following 48 h of the siRNA or plasmid transfection, the cells were collected and washed with cold PBS. The cells were then mixed with an Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences) in flow cytometry tubes, following the manufacturer’s protocol. The number of apoptotic cells was analyzed using flow cytometry (BD Biosciences).
Cell cycle analysis
At 48 h following siRNA or plasmid transfection, the transfected CRC cells were collected in PBS. Following washing twice with PBS, the cells were fixed with 75% ethyl alcohol at 4°C overnight. The following day, the separated ethyl alcohol was removed, and the cells were fixed with 500 µl of propidium iodide (PI) staining solution and incubated for 30 min in the dark at room temperature. The analysis of the cell cycle was performed using fluorescence-activated cell sorting with a FACSCalibur flow cytometer with CellQuest software (version 3.0; BD Biosciences).
Western blot analysis
Protein was extracted from the CRC cells using a Radioimmunoprecipitation Assay kit (Beyotime Institute of Biotechnology, Shanghai, China) according to the manufacturer’s protocol. The protein concentration, which determined the quantity loaded for SDS-PAGE, was determined using the Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology). The proteins (40 µg) were separated using 10% SDS-PAGE in running buffer and transferred onto polyvinylidene fluoride membranes (EMD Millipore, Bedford, MA, USA) in transfer buffer. The membranes were then blocked in 5% non-fat milk for >2 h and incubated with specific primary antibodies at 4°C overnight. Following washing with TBST three times (10 min each time), the membranes were incubated with secondary antibodies (anti-rabbit or anti-mouse) at room temperature for 2 h. The immunoreactive protein bands were visualized using ECL Plus (EMD Millipore) with a bio-imaging system. The specific primary and secondary antibodies were as follows: TRIM27 (diluted 1:1,000, polyclonal, rabbit, cat. no. ab78393), E-cadherin (diluted 1:500, monoclonal, mouse, cat. no. ab1416), N-cadherin (diluted 1:1,000, poly-clonal, rabbit, cat. no. ab18203), vimentin (diluted 1:2,000, monoclonal, rabbit, cat. no. ab92547), AKT (diluted 1:500, polyclonal, rabbit, cat. no. ab8805), p-AKT (diluted 1:500, polyclonal, rabbit, cat. no. ab38449) (all from Abcam), anti-rabbit secondary antibodies (diluted 1:5,000, polyclonal, cat. no. GAB007), anti-mouse secondary antibodies (diluted 1:5,000, polyclonal, cat. no. GAM007) (both from Hangzhou Multi Sciences Biotech Co., Ltd., Hangzhou, China). GAPDH (diluted 1:5,000, monoclonal, mouse; Abcam) was used as an internal control.
Tumor xenograft in a nude mouse model
The animal experiment was approved by Animal Ethics Committee of Nanjing Medical University. A total of 20 male mice (age, 3–4 weeks; weight, 13–15 g) were purchased from the Animal Center of Nanjing Medical University after signing the animal-raising agreement. The mice were maintained under the following conditions: Room temperature, 20–26°C and 12-h light/dark cycle. The mice received 5 g food and 100 ml water per 100 g body weight per day. To detect whether TRIM27 can affect tumor growth in mice, 2×106 of differently treated LoVo cells mixed with 200 µl of PBS were subcutaneously injected into the anesthetized mice. Each mouse was randomly injected with cells treated with shTRIM27 or NC in their right and left armpits. At 4 weeks post-injection, all mice were sacrificed and the tumor tissues were surgically removed. To further investigate the role of TRIM27 in tumor metastasis in vivo, 2×106 LoVo cells suspended in 200 µl of PBS were injected into the mouse tail vein. The liver tissues were removed after 7 weeks and stained with hematoxylin and eosin.
Statistical analysis
The results are expressed as the mean ± standard deviation. The mRNA expression was analyzed using unpaired t-tests. IHC and the clinical features were analyzed using Pearson χ2 tests. One-way analysis of variance and the least-significant difference post hoc test were the main statistical methods used to compare datasets containing multiple groups. Cumulative survival analysis was assessed using the Kaplan-Meier method. Independent prognostic factors were identified using univariate and multivariate Cox proportional hazard regression models. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using SPSS (version 15.0; SPSS, Inc., Chicago, IL, USA) and GraphPad Prism5 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
TRIM27 is upregulated in CRC tissues and correlated with clinicopathological features
To reveal the role of TRIM27 in the progression of CRC, the mRNA expression of TRIM27 was first detected in 80 pairs of CRC tissues and adjacent normal tissues using RT-qPCR analysis. It was found that the expression of TRIM27 was significantly upregulated in CRC tissues (Fig. 1A). Then we detected the presence of TRIM27 using IHC in 50 CRC tissues and 25 adjacent normal tissues. IHC also showed that the protein level of TRIM27 was significantly upregulated in CRC tissues (Fig. 1B and C; Table II). The patients were next divided into two groups based on the average TRIM27 levels, and the association between levels of TRIM27 and the clinicopathological features was analyzed. It was found that a higher expression of TRIM27 was associated with deeper invasion (P=0.005), increased lymph node metastasis (P=0.014), more advanced tumor stage (P=0.004), and increased liver metastasis (P=0.043) (Table III). However, no significant association was found for sex, age, CEA, or location. Kaplan-Meier curves were used to analyze the effect of TRIM27 on overall survival (OS) in patients with CRC. The median follow-up time was 60 months, and the result showed that higher mRNA expression of TRIM27 predicted poorer prognosis in patients diagnosed with CRC (Fig. 1D). Multivariate analysis further suggested that TRIM27 was an independent prognostic factor for CRC (Table IV).
Figure 1.
Expression of TRIM27 is upregulated in human CRC tissues and predicts poor prognosis. (A) mRNA expression levels of TRIM27 in CRC and normal tissues (n=80) were detected by reverse transcription-quantitative polymerase chain reaction. mRNA levels of TRIM27 were normalized to GAPDH. (B) Immunohistochemical results of TRIM27 in CRC tissues and normal adjacent tissues. (C) Percentage of specimens exhibiting strong or weak expression of TRIM27 in immunohistochemistry. (D) Kaplan-Meier survival analysis of 80 patients with CRC based on the expression level of TRIM27. ***P<0.001. TRIM27, tripartite motif-containing 27; CRC, colorectal cancer.
Table II.
Analysis of the expression of tripartite motif-containing 27 in colorectal cancer tissues and adjacent normal tissues by immunohistochemistry.
| Specimen | Total score | Number (%) | P-value |
|---|---|---|---|
| Carcinoma | 0–3 | 8 (10.7) | <0.001 |
| 4–6 | 42 (56.0) | ||
| Normal | 0–3 | 20 (26.7) | |
| 4–6 | 5 (6.6) |
Table III.
mRNA expression of TRIM27 in colorectal carcinoma and adjacent normal tissues.
| Clinical feature | n (%) |
TRIM27 expression
|
P-value | |
|---|---|---|---|---|
| High, n (%) | Low, n (%) | |||
| Sex | 40 | 40 | ||
| Male | 52 (65.0) | 25 (31.3) | 27 (33.8) | 0.639 |
| Female | 28 (35.0) | 15 (18.7) | 13 (16.2) | |
| Age (years) | ||||
| >60 | 36 (45.0) | 17 (21.3) | 19 (23.8) | 0.653 |
| ≤60 | 44 (55.0) | 23 (28.7) | 21 (26.2) | |
| Depth of invasion | ||||
| T1/T2 | 21 (26.3) | 5 (6.3) | 16 (20.0) | 0.005b |
| T3/T4 | 59 (73.7) | 35 (43.7) | 24 (30.0) | |
| Lymph node metastasis | ||||
| Absent | 39 (48.8) | 14 (17.5) | 25 (31.3) | 0.014a |
| Present | 41 (51.2) | 26 (32.5) | 15 (18.7) | |
| Tumor stage | ||||
| I/II | 37 (46.3) | 12 (15.0) | 25 (31.2) | 0.004b |
| III/IV | 43 (53.7) | 28 (35.0) | 15 (18.8) | |
| Liver metastasis | ||||
| Yes | 10 (12.5) | 8 (10.0) | 2 (2.5) | 0.043a |
| No | 70 (87.5) | 32 (40.0) | 38 (47.5) | |
| Location | ||||
| Rectum | 35 (43.8) | 19 (23.8) | 16 (20.0) | 0.499 |
| Colon | 45 (56.2) | 21 (26.2) | 24 (30.0) | |
| CEA (ng/ml) | ||||
| <4.7 | 31 (38.8) | 13 (16.2) | 18 (22.5) | 0.250 |
| >4.7 | 49 (61.2) | 27 (33.8) | 22 (27.5) | |
P<0.05;
P<0.01. TRIM27, tripartite motif-containing 27.
Table IV.
Univariate and multivariate analysis of the association of prognosis with clinicopathologic parameters and expression of TRIM27 in colorectal cancer.
| Variable | Univariable analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Sex (male vs. female) | 0.83 (0.38–1.82) | 0.642 | – | NA |
| Age (>60 vs. ≤60 years) | 0.92 (0.42–1.99) | 0.823 | – | NA |
| Depth of invasion (T3/T4 vs. T1/T2) | 1.50 (1.04–2.16) | 0.029a | 2.48 (0.86–7.15) | 0.093 |
| Lymph node metastasis (present vs. absent) | 3.18 (1.23–8.22) | 0.017a | 2.56 (1.04–6.28) | 0.040a |
| Liver metastasis (present vs. absent) | 3.28 (1.28–8.41) | 0.013a | 2.95 (1.19–7.31) | 0.020a |
| Location (rectum vs. colon) | 1.31 (0.61–2.82) | 0.495 | – | NA |
| CEA (>4.7 vs. <4.7) | 0.68 (0.32–1.42) | 0.303 | – | NA |
| TRIM27 expression (high vs. low) | 2.63 (1.14–6.05) | 0.023a | 2.52 (1.02–6.25) | 0.046a |
P<0.05. HR, hazard ratio; CI confidence interval, NA not adopted. TRIM27, tripartite motif-containing 27.
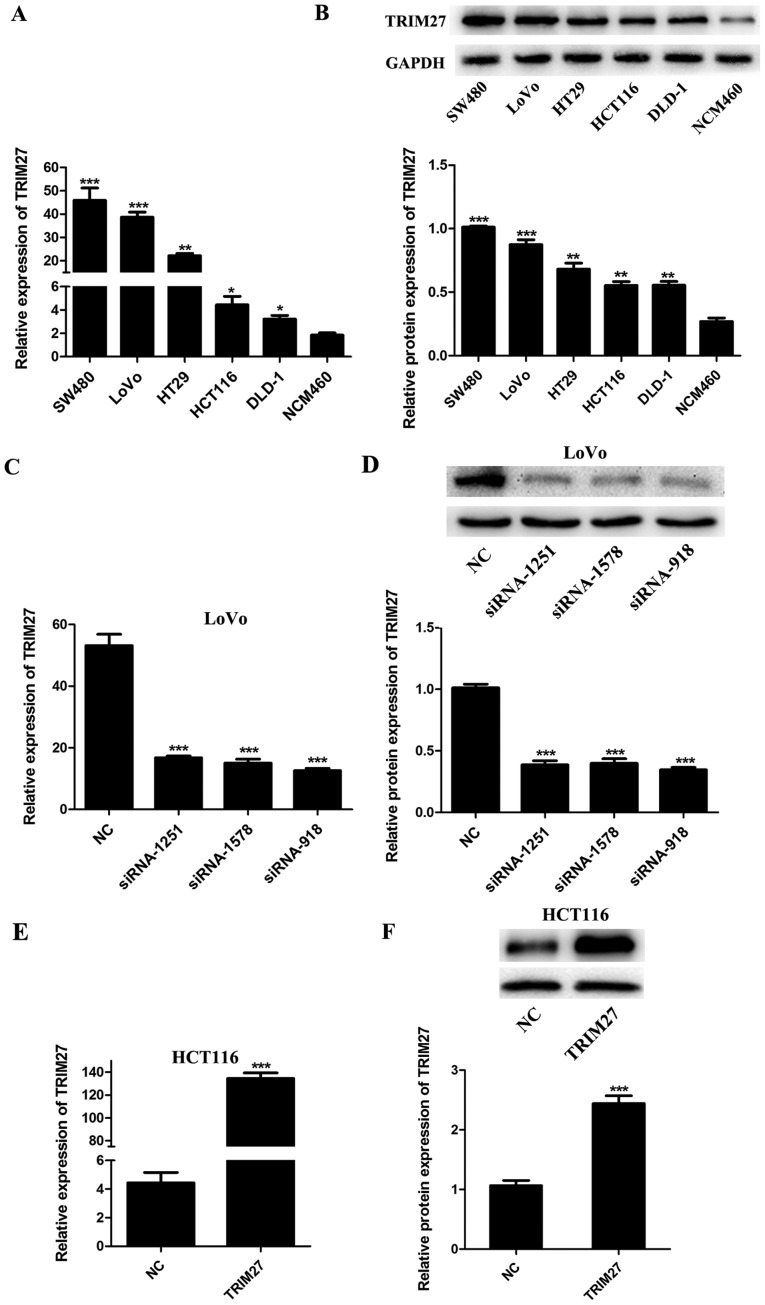
Expression of TRIM27 is upregulated in CRC cells and can be regulated by siRNAs and plasmids
To evaluate the expression level of TRIM27 in CRC cells, five CRC cell lines (SW480, LoVo, HT29, HCT116 and DLD-1) and the NCM460 normal colon epithelial cell line were detected by RT-qPCR and western blot analyses. It was found that the mRNA and protein levels of TRIM27 were upregulated in the CRC cell lines compared with those in the NCM460 cells (Fig. 2A and B). To knock down or overexpress TRIM27, the LoVo and HCT116 cells were transfected with a TRIM27 inhibitor and TRIM27-overexpressing plasmid, respectively. According to the results of RT-qPCR and western blot analysis, the expression of TRIM27 was significantly inhibited by siRNA-918, siRNA-1251 and siRNA-1578 in LoVo cells (Fig. 2C and D), and was overexpressed by trans-fection with the TRIM27-overexpressing plasmid in HCT116 cells (Fig. 2E and F). To further investigate the role of TRIM27 in CRC, the TRIM27-overexpressing plasmid and siRNA-918 were selected to perform further in vitro experiments.
Figure 2.
TRIM27 is upregulated in CRC cell lines and can be regulated by siRNA and plasmid transfection. (A) mRNA expression of TRIM27 in different CRC cell lines compared with NCM460 normal colon epithelial cells. (B) Relative protein levels of TRIM27 in different CRC cell lines compared with NCM460 normal colon epithelial cells. (C) Relative mRNA expression of TRIM27 in LoVo cells following siRNA transfection. (D) Relative protein levels of TRIM27 in LoVo cells following siRNA transfection. (E) Relative mRNA expression of TRIM27 in HCT116 cells following plasmid transfection. (F) Relative protein levels of TRIM27 in HCT116 cells following plasmid transfection. Data are presented as the mean ± standard deviation from three independent experiments; *P<0.05, **P<0.01 and ***P<0.001, compared with controls. TRIM27, tripartite motif-containing 27; CRC, colorectal cancer; NC, negative control sequence; siRNA, small interference RNA.
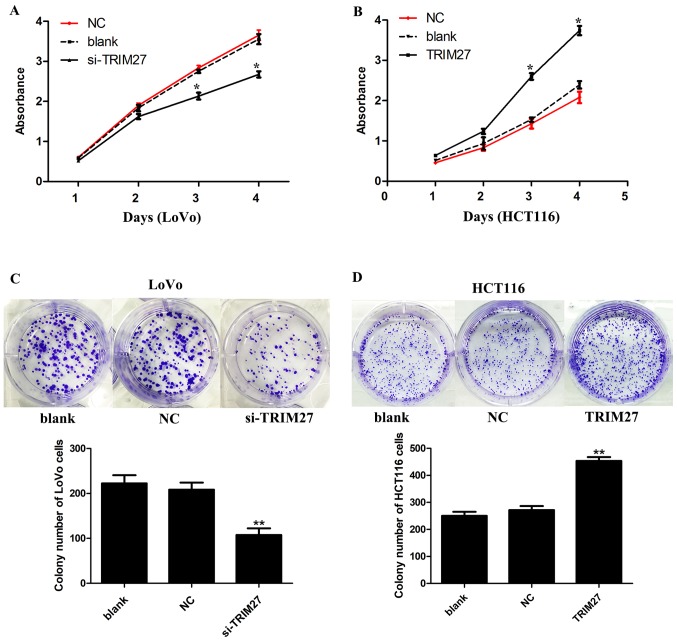
TRIM27 significantly increases CRC cell proliferation and colony formation in LoVo and HCT116 cells
CCK-8 and colony formation assays were used to detect the effect of TRIM27 on the proliferation of CRC cells. It was found that the knockdown of TRIM27 significantly inhibited cell proliferation in LoVo cells (Fig. 3A). By contrast, cell proliferation was promoted in HCT116 cells overexpressing TRIM27 (Fig. 3B). Consistently, in the colony formation assay, it was found that LoVo cells transfected with the siRNA showed reduced formation of colonies (Fig. 3C), whereas HCT116 cells transfected with the TRIM27-overexpressing plasmid showed the opposite result (Fig. 3D).
Figure 3.
TRIM27 promotes proliferation and colony formation in colorectal cancer cells. The cell counting kit-8 assay detected the cell viability of (A) LoVo and (B) HCT116 cells following TRIM27 knockdown or overexpression. Colony-forming abilities of (C) LoVo and (D) HCT116 cells were detected following TRIM27 knockdown or overexpression. Data are presented as the mean ± standard deviation from three independent experiments; *P<0.05 and **P<0.01, compared with controls. TRIM27, tripartite motif-containing 27; si-TRIM27, small interference RNA targeting TRIM27; NC, negative control sequence.
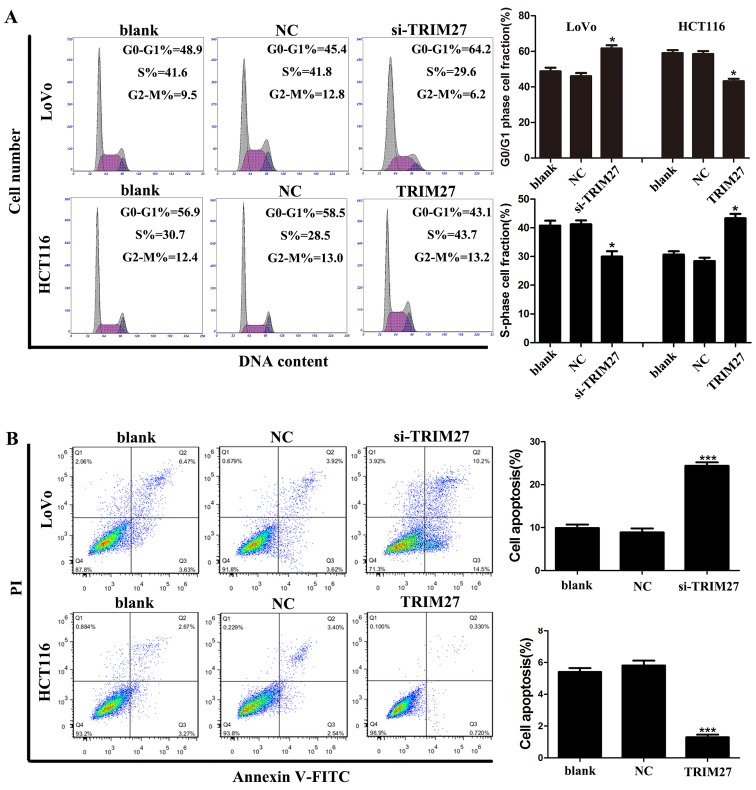
TRIM27 promotes apoptosis resistance and cell cycle G0-G1/S transition in CRC cells
Flow cytometry was used to investigate whether TRIM27 regulates apoptosis and the cell cycle of CRC cells. It was found that the LoVo cells transfected with the siRNA showed a significant increase in the percentage of cells in the G0-G1 phase and a reduced percentage of cells in the S phase, whereas HCT116 cells transfected with the TRIM27-overexpressing plasmid showed the reverse result (Fig. 4A). Similarly, LoVo cells transfected with the siRNA showed a significant increase in the percentage of apoptotic cells, whereas HCT116 cells transfected with the TRIM27-overexpressing plasmid showed fewer apoptotic cells (Fig. 4B). Taken together, these results suggested that TRIM27 promoted the proliferation of CRC cells by promoting apoptosis resistance and cell cycle transition in the G0-G1/S phase.
Figure 4.
TRIM27 induces cell apoptosis resistance and accelerated cell cycle in colorectal cancer cells. (A) Cell cycles of LoVo and HCT116 cells were detected using flow cytometry, and the comparative analysis of cell numbers in the G0/G1 and S phase are shown. (B) Total cell apoptosis of LoVo and HCT116 cells was detected by flow cytometry, and the comparative analysis of apoptotic cells is shown. Data are presented as the mean ± standard deviation from three independent experiments; *P<0.05 and ***P<0.001, compared with controls. TRIM27, tripartite motif-containing 27; si-TRIM27, small interference RNA targeting TRIM27; NC, negative control sequence; PI, propidium iodide.
Regulation of TRIM27 influences the invasion and metastasis of CRC cells in vitro
Wound-healing and Transwell assays were used to detect the metastasis and invasiveness in LoVo and HCT116 cells. In the wound-healing assay, it was found that the wound area was wider in LoVo cells transfected with siRNA (Fig. 5A), but narrower in HCT116 cells transfected with the TRIM27-overexpressing plasmid (Fig. 5B). In the Transwell assay, the numbers of cells that traversed the Transwell and Matrigel were significantly reduced by TRIM27 knockdown in the LoVo cells, whereas the overexpression of TRIM27 in HCT116 cells increased the number of cells that traversed the Transwell and Matrigel (Fig. 5C–F). These results suggested that TRIM27 promoted the invasion and metastasis of CRC cells.
Figure 5.
TRIM27 facilitates the migration and invasion of colorectal cancer cells. A wound-healing assay was performed in (A) LoVo and (B) HCT116 cells following TRIM27 knockdown and overexpression. Quantitative results in the wound-healing assay are shown. A Transwell assay was performed in (C) LoVo and (D) HCT116 cells following TRIM27 knockdown and overexpression. Magnification, ×200. (E) Numbers of migrated LoVo and HCT116 cells through the membrane. (F) Numbers of invaded LoVo and HCT116 cells through the Matrigel. Each assay was performed three times. Data are presented as the mean ± standard deviation; **P<0.01, compared with controls. TRIM27, tripartite motif-containing 27; si-TRIM27, small interference RNA targeting TRIM27; NC, negative control sequence.
Downregulation of TRIM27 significantly suppresses tumor proliferation and metastasis in nude mice
To determine whether TRIM27 affects tumor growth in vivo, tumor xenografting was performed in nude mice. At 4 weeks post-implantation, the tumor size (Fig. 6A and B) and weight (Fig. 6C) was significantly decreased in the group treated with shTRIM27 compared with the untreated control. To further investigate whether TRIM27 affects tumor metastasis in vivo, a tail vein metastatic assay was performed in nude mice using LoVo cells transfected with shTRIM27. At 7 weeks post-injection, the results of hematoxylin and eosin staining showed that the knockdown of TRIM27 correlated closely with reduced liver metastasis (Fig. 6D and E). These in vivo results further confirmed that TRIM27 promoted proliferation and metastasis in CRC.
Figure 6.
Knockdown of TRIM27 inhibits the proliferation and metastasis of colorectal cancer cells in vivo. (A) Nude mice 4 weeks following tumor implantation in the shTRIM27 group (left armpit) and control group (right armpit). (B) Representative images of isolated tumors in the control group (first row) and shTRIM27-treated group (second row). (C) Average weights of the isolated tumors in the two groups. (D) Representative images of hematoxylin and eosin staining results of the liver tissues from mice in the shTRIM27-treated group and control group. Magnification, ×200. (E) Comparative analysis of the number of liver metastases in mice. **P<0.01 and ***P<0.001. TRIM27, tripartite motif-containing 27; shTRIM27, short hairpin RNA targeting TRIM27; NC, negative control sequence.
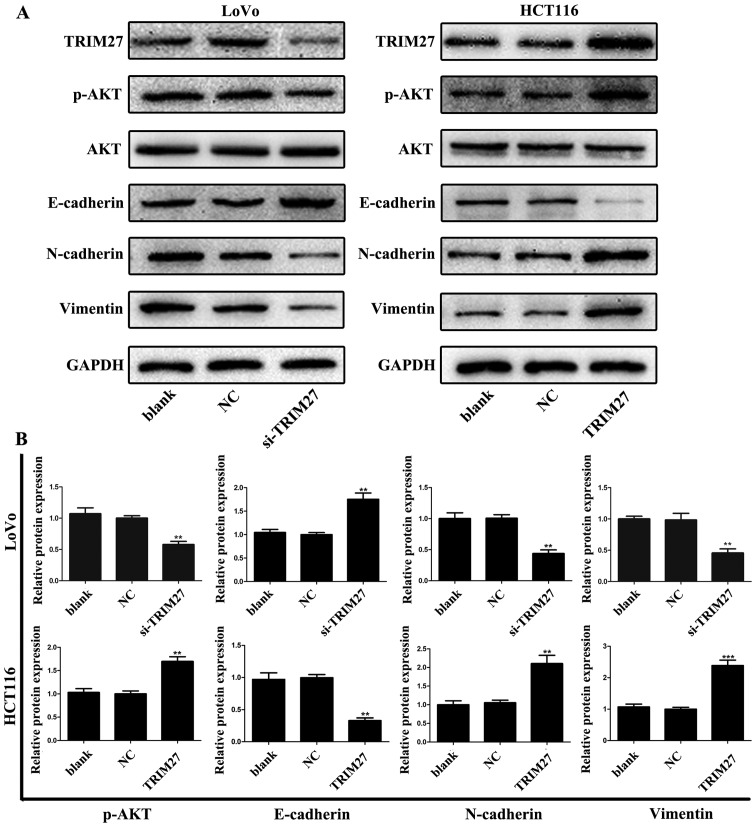
TRIM27 promotes the process of EMT and activation of p-AKT in CRC cells
To further detect whether TRIM27 was associated with the EMT process, the levels of EMT-associated proteins in LoVo and HCT116 cells were evaluated using western blot analysis. Following TRIM27 knockdown, it was found that the level of the epithelial marker E-cadherin increased, and the level of the mesenchymal marker N-cadherin decreased. The opposite effect was observed in HCT116 cells transfected with the TRIM27-overexpressing plasmid (Fig. 7). Further investigation revealed that the knockdown of TRIM27 decreased the level of vimentin in LoVo cells; however, the level of vimentin increased in HCT116 cells following the overexpression of TRIM27 (Fig. 7). These results suggested that TRIM27 promoted EMT in CRC cells. Additionally, the levels of p-AKT and AKT in LoVo and HCT116 cells were detected. It was found that the level of p-AKT increased when TRIM27 was overexpressed, but decreased when TRIM27 was knocked down. However, no significant changes occurred in the expression of AKT, suggesting that TRIM27 promoted the activation of p-AKT (Fig. 7).
Figure 7.
TRIM27 promotes the activation of p-AKT and the epiethlium-mesenchymal transition process in colorectal cancer cells. (A) Expression levels of p-AKT, AKT, E-cadherin, N-cadherin and vimentin in LoVo and HCT116 cells following TRIM27 knockdown and overexpression were detected using western blot analysis. (B) Relative protein levels of p-AKT, E-cadherin, N-cadherin and vimentin were quantified in LoVo and HCT116 cells. Data are presented as the mean ± standard deviation from three independent experiments; **P<0.01 and ***P<0.001, compared with controls. TRIM27, tripartite motif-containing 27; si-TRIM27, small interference RNA target TRIM27; p-, phosphorylated; NC, negative control sequence.
Discussion
The TRIM protein family has been recognized to comprise regulatory proteins in a variety of tumors, some of which have been reported to be associated with CRC (25–28). TRIM27, which belongs to the TRIM family, has been reported to be involved in various tumor processes. For example, in ovarian cancer, Ma et al showed that TRIM27 knockdown induced cell cycle arrest and apoptosis by upregulating the expression of p-p38 and downregulating the level of p-AKT (13). In endometrial cancer, Tsukamoto et al and Tezel et al revealed that TRIM27 knockdown significantly impaired cancer cell migration and invasion, with concomitant decreases in levels of integrin b1 and a2 (14,29). In lung cancer, Iwakoshi et al found that TRIM27 was involved in mutated epidermal growth factor receptor (EGFR) signaling and can be a prognostic factor for lung cancer with EGFR mutations (16). All of the above studies suggested that TRIM27 acts as an oncogene in facilitating tumor proliferation and metastasis in various tumors.
The first step of the present study involved determining whether TRIM27 was upregulated in CRC tissues, with further analysis to determine whether it was associated with patient prognosis. The results suggested that TRIM27 was significantly upregulated in CRC tissues, and a higher expression level of TRIM27 predicted a poorer prognosis, which was in accordance with the conclusion of Kato et al and Zoumpoulidou et al (18,19). The present study further showed that a higher expression level of TRIM27 predicted deeper invasion, increased lymph node metastasis, higher tumor stage, and increased distant metastases in CRC tissues. This suggested that TRIM27 is an oncogene in CRC. To test this hypothesis, the expression of TRIM27 was detected in CRC cells, and the results showed that TRIM27 was upregulated in CRC cells. Further investigation revealed that TRIM27 significantly promoted cell proliferation in vivo and in vitro, possibly by reducing cell apoptosis and promoting cell cycle G1/S transition. In addition, the results of wound-healing and Transwell assays showed that TRIM27 promoted invasion and metastasis, which was further supported by the results of tail vein metastasis assays in mice. Therefore, it was concluded that TRIM27 has an oncogenic role by promoting the proliferation, invasion, and metastasis of CRC cells.
EMT is a transformation in which epithelial cells break down cell-cell and cell-extracellular matrix connections (30). This process is considered to be closely associated with tumor invasion and metastasis in various types of cancer, and the overall process is always accompanied by decreased expression of epithelial markers and increased expression of mesenchymal markers (31,32). Although an association between EMT and TRIM27 has not been reported until now, other TRIM family members, including TRIM14, TRIM16 and TRIM59 have been reported to regulate EMT (28,33,34). Therefore, the present study assessed a series of proteins associated with EMT using western blot analysis. The results suggested that TRIM27 significantly promoted EMT, which verified the above hypothesis. The activation of AKT has been reported to transduce signals to regulate multiple biological processes, including cellular proliferation, survival, growth, angiogenesis, migration and EMT, in various types of cancer (35). In addition, Ma et al revealed that TRIM27 knockdown inhibited the expression of p-AKT in ovarian cancer (13). This suggested that TRIM27 may also be associated with the expression of p-AKT in CRC, and the results of the present study supported this. Therefore, it was hypothesized that TRIM27-mediated EMT may be promoted by the activation of p-AKT. However, the exact association between TRIM27, EMT, and p-AKT in promoting the proliferation, invasion and metastasis of CRC remains to be elucidated. Further in-depth investigations are required, with a focus on evaluating the role of TRIM27 in CRC.
In conclusion, the present study demonstrated that TRIM27 was significantly upregulated in CRC tissues, which indicated the TRIM27 acts as an oncogenic protein, and its expression level predicts poor prognosis in patients with CRC. Furthermore, TRIM27 promoted proliferation, invasion and metastasis, possibly by promoting EMT and the activation of p-AKT in CRC cells. Therefore, TRIM27 represents a potential prognostic and therapeutic target in CRC. Further investigations are required to determine the detailed mechanisms underlying the effects of TRIM27 in CRC.
Acknowledgments
The authors would like to thank Dr Hao Fan and Dr Yuanguangyan Zhang (the First Affiliated Hospital of Nanjing Medical University, Jiangsu, China) for providing language and technology support.
Funding
This study was supported in part by the Jiangsu Key Medical Discipline (General Surgery) (grant no. ZDXKA2016005).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
YS, ZF, YZ and YF conceived and designed the study. YZ, DJ, SW and WQ performed the experiments and acquired data. YF and CZ carried out the patient follow-up. DJ, QW, BJ and ZZ analyzed and interpreted the data. YZ, BJ and CZ performed the statistical analysis. YZ and YF drafted and edited the manuscript. All authors have given final approval of the version to be published.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (ethics no. 2010-SR-091.A1).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 5.Micale L, Chaignat E, Fusco C, Reymond A, Merla G. The tripartite motif: Structure and function. Adv Exp Med Biol. 2012;770:11–25. doi: 10.1007/978-1-4614-5398-7_2. [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto K, Nakamura N, Kashiwagi M, Honda S, Kato A, Hasegawa S, Takei Y, Hirose S. RING finger, B-box, and coiled-coil (RBCC) protein expression in branchial epithelial cells of Japanese eel, Anguilla japonica. Eur J Biochem. 2002;269:6152–6161. doi: 10.1046/j.1432-1033.2002.03332.x. [DOI] [PubMed] [Google Scholar]
- 7.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. BioEssays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 8.Nisole S, Stoye JP, Saïb A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell. 1985;42:581–588. doi: 10.1016/0092-8674(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi M, Cooper GM. ret transforming gene encodes a fusion protein homologous to tyrosine kinases. Mol Cell Biol. 1987;7:1378–1385. doi: 10.1128/MCB.7.4.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi M, Inaguma Y, Hiai H, Hirose F. Developmentally regulated expression of a human ‘finger’-containing gene encoded by the 5′ half of the ret transforming gene. Mol Cell Biol. 1988;8:1853–1856. doi: 10.1128/MCB.8.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y, Wei Z, Bast RC, Jr, Wang Z, Li Y, Gao M, Liu Y, Wang X, Guo C, Zhang L, et al. Downregulation of TRIM27 expression inhibits the proliferation of ovarian cancer cells in vitro and in vivo. Lab Invest. 2016;96:37–48. doi: 10.1038/labinvest.2015.132. [DOI] [PubMed] [Google Scholar]
- 14.Tsukamoto H, Kato T, Enomoto A, Nakamura N, Shimono Y, Jijiwa M, Asai N, Murakumo Y, Shibata K, Kikkawa F. Expression of Ret finger protein correlates with outcomes in endometrial cancer. Cancer Sci. 2009;100:1895–1901. doi: 10.1111/j.1349-7006.2009.01278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tezel GG, Uner A, Yildiz I, Guler G, Takahashi M. RET finger protein expression in invasive breast carcinoma: Relationship between RFP and ErbB2 expression. Pathol Res Pract. 2009;205:403–408. doi: 10.1016/j.prp.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Iwakoshi A, Murakumo Y, Kato T, Kitamura A, Mii S, Saito S, Yatabe Y, Takahashi M. RET finger protein expression is associated with prognosis in lung cancer with epidermal growth factor receptor mutations. Pathol Int. 2012;62:324–330. doi: 10.1111/j.1440-1827.2012.02797.x. [DOI] [PubMed] [Google Scholar]
- 17.Horio M, Kato T, Mii S, Enomoto A, Asai M, Asai N, Murakumo Y, Shibata K, Kikkawa F, Takahashi M. Expression of RET finger protein predicts chemoresistance in epithelial ovarian cancer. Cancer Med. 2012;1:218–229. doi: 10.1002/cam4.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato T, Shimono Y, Hasegawa M, Jijiwa M, Enomoto A, Asai N, Murakumo Y, Takahashi M. Characterization of the HDAC1 complex that regulates the sensitivity of cancer cells to oxidative stress. Cancer Res. 2009;69:3597–3604. doi: 10.1158/0008-5472.CAN-08-4368. [DOI] [PubMed] [Google Scholar]
- 19.Zoumpoulidou G, Broceño C, Li H, Bird D, Thomas G, Mittnacht S. Role of the tripartite motif protein 27 in cancer development. J Natl Cancer Inst. 2012;104:941–952. doi: 10.1093/jnci/djs224. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Li W. Epithelial-mesenchymal transition in human cancer: Comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther. 2015;150:33–46. doi: 10.1016/j.pharmthera.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adhes Migr. 2015;9:317–324. doi: 10.1080/19336918.2015.1016686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benson AB, III, Venook AP, Bekaii-Saab T, Chan E, Chen YJ, Cooper HS, Engstrom PF, Enzinger PC, Fenton MJ, Fuchs CS, et al. National Comprehensive Cancer Network: Colon cancer, version 3.2014. J Natl Compr Canc Netw. 2014;12:1028–1059. doi: 10.6004/jnccn.2014.0099. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Xu W, Xu B, Yao Y, Yu X, Cao H, Zhang J, Liu J, Sheng H. RNA interference against TRIM29 inhibits migration and invasion of colorectal cancer cells. Oncol Rep. 2016;36:1411–1418. doi: 10.3892/or.2016.4941. [DOI] [PubMed] [Google Scholar]
- 26.Tan Z, Liu X, Yu E, Wang H, Tang L, Wang H, Fu C. Lentivirus-mediated RNA interference of tripartite motif 68 inhibits the proliferation of colorectal cancer cell lines SW1116 and HCT116 in vitro. Oncol Lett. 2017;13:2649–2655. doi: 10.3892/ol.2017.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee OH, Lee J, Lee KH, Woo YM, Kang JH, Yoon HG, Bae SK, Songyang Z, Oh SH, Choi Y. Role of the focal adhesion protein TRIM15 in colon cancer development. Biochim Biophys Acta. 18532015:409–421. doi: 10.1016/j.bbamcr.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y, Ji B, Feng Y, Zhang Y, Ji D, Zhu C, Wang S, Zhang C, Zhang D, Sun Y. TRIM59 facilitates the proliferation of colorectal cancer and promotes metastasis via the PI3K/AKT pathway. Oncol Rep. 2017;38:43–52. doi: 10.3892/or.2017.5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tezel GG, Ordulu Z, Hımmetoğlu C, Usubütün A. The selective expression of ret finger protein in endometrial cancer: Can RFP be a marker of serous carcinomas. Turk Patoloji Derg. 2012;28:213–219. doi: 10.5146/tjpath.2012.01127. [DOI] [PubMed] [Google Scholar]
- 30.Radisky DC, LaBarge MA. Epithelial-mesenchymal transition and the stem cell phenotype. Cell Stem Cell. 2008;2:511–512. doi: 10.1016/j.stem.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Guo Xu G, Xu Y, Wang D, Shen Y, Wang Y, Lv F, Song Y, Jiang F, Zhang DY. TRIM14 regulates cell proliferation and invasion in osteosarcoma via promotion of the AKT signaling pathway. Sci Rep. 2017;7:42411. doi: 10.1038/srep42411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan H, Qi J, Chu G, Liu Z. Tripartite motif 16 inhibits the migration and invasion in ovarian cancer cells. Oncol Res. 2017;25:551–558. doi: 10.3727/096504016X14758370595285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suman S, Kurisetty V, Das TP, Vadodkar A, Ramos G, Lakshmanaswamy R, Damodaran C. Activation of AKT signaling promotes epithelial-mesenchymal transition and tumor growth in colorectal cancer cells. Mol Carcinog. 2014;53(Suppl 1):E151–E160. doi: 10.1002/mc.22076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.