Abstract
Gpi8p and Gaa1p are essential components of the GPI transamidase that adds glycosylphosphatidylinositols (GPIs) to newly synthesized proteins. After solubilization in 1.5% digitonin and separation by blue native PAGE, Gpi8p is found in 430–650-kDa protein complexes. These complexes can be affinity purified and are shown to consist of Gaa1p, Gpi8p, and Gpi16p (YHR188c). Gpi16p is an essential N-glycosylated transmembrane glycoprotein. Its bulk resides on the lumenal side of the ER, and it has a single C-terminal transmembrane domain and a small C-terminal, cytosolic extension with an ER retrieval motif. Depletion of Gpi16p results in the accumulation of the complete GPI lipid CP2 and of unprocessed GPI precursor proteins. Gpi8p and Gpi16p are unstable if either of them is removed by depletion. Similarly, when Gpi8p is overexpressed, it largely remains outside the 430–650-kDa transamidase complex and is unstable. Overexpression of Gpi8p cannot compensate for the lack of Gpi16p. Homologues of Gpi16p are found in all eucaryotes. The transamidase complex is not associated with the Sec61p complex and oligosaccharyltransferase complex required for ER insertion and N-glycosylation of GPI proteins, respectively. When GPI precursor proteins or GPI lipids are depleted, the transamidase complex remains intact.
INTRODUCTION
Many glycoproteins of lower and higher eucaryotes are attached to the plasma membrane by means of a glycosylphosphatidylinositol (GPI) (McConville and Ferguson, 1993; Kinoshita and Inoue, 2000). GPI anchoring of proteins is essential for the growth of Saccharomyces cerevisiae (Leidich et al., 1994; Schönbächler et al., 1995). S. cerevisiae contains ∼70 open reading frames predicting GPI proteins, and many of these have been found to be cell wall glycoproteins (Caro et al., 1997; Hamada et al., 1998).
Precursors of GPI-anchored proteins have a classic signal sequence for import into the ER at their N-terminus and a GPI-anchoring signal at their C-terminus; the C-terminal signal is necessary and sufficient to direct GPI addition (Caras et al., 1987). The C-terminal GPI-anchoring signal is recognized and removed by a GPI transamidase, which replaces it by a preformed GPI. GPI-anchoring signals are composed of a C-terminal hydrophobic domain, which is separated by a short hydrophilic spacer from the cleavage/attachment site (ω site) (Moran and Caras, 1991; Gerber et al., 1992; Nuoffer et al., 1993).
The transfer of preformed GPIs onto proteins has been studied in microsomal translation/translocation/GPI-anchoring systems (Ramalingam et al., 1996; Sharma et al., 1999) or translocation/GPI-anchoring systems (Doering and Schekman, 1997) in several organisms, and these studies allowed a preliminary biochemical characterization of the GPI transfer reaction. The GPI transferase is believed to act as a transamidase, i.e., to jointly remove the GPI-anchoring signal and transfer the preformed GPI (Ramalingam et al., 1996; Sharma et al., 1999). Genetic approaches have identified genes required for the addition of GPI anchors. Transamidase-deficient cells are expected to accumulate complete GPIs as well as GPI precursor proteins retaining the GPI-anchoring signal. This phenotype is exhibited by two yeast mutants, gaa1 and gpi8 (Hamburger et al., 1995; Benghezal et al., 1996). GAA1 is essential and encodes a 70-kDa ER protein with a large, hydrophilic, lumenal domain, followed by several transmembrane domains (TMDs) and a cytosolic ER retrieval signal on its extreme C-terminus. GPI8 is also an essential gene and encodes a type I ER membrane protein with a single TMD. Gpi8p has 25–28% homology to a family of cysteine proteinases, one of which is able to act as a transamidase (Benghezal et al., 1996). The Cys and His residues predicted to be active sites by sequence comparison with caspases indeed are essential, and their mutation to Ala yields nonfunctional GPI8 alleles (Chen et al., 1998; Meyer et al., 2000; Ohishi et al., 2000). Recent evidence shows that Gpi8p may be part of a larger protein complex. First, the human Gpi8p is efficiently coimmunoprecipitated with human Gaa1p when tagged forms are coexpressed in CHO cells, and even truncated versions of Gpi8p lacking the C-terminal TMD still can be coprecipitated with Gaa1p (Ohishi et al., 2000). Second, the overexpression in wild-type (wt) cells of any GPI8 allele that is mutated in one of the active site residues leads to cell growth arrest and causes the accumulation of unprocessed GPI lipids and protein precursors (Meyer et al., 2000). Here we isolate the complex and analyze its components.
MATERIALS AND METHODS
Strains, Growth Conditions, and Materials
Yeast strains were S. cerevisiae W303–1B (MATα ade2–1 can1–100 ura3–1 leu2–3,112 trp1–1 his3–11,15), FBY525 (MATa ade2–1 ura3–1 leu2–3,112 trp1–1 his3–11,15 lys- Δgpi8::kanMX containing plasmid YEpGPI8), FBY164 (MATα ade2–1 leu2–3,112 his3–11,15 gpi8::kanMX2 ura3–1::URA3-GAL1,10UAS-GPI8), RH932 (MATa gaa1 leu2 ura3 bar1–1) (Hamburger et al., 1995), 521–17A-H42 (MATa mcd4 trp1–289 leu2 ura3–52) (Packeiser et al., 1999), CWH4 (MATα gpi1 ura3–52 lys2) (this mutant was isolated as cwh4 by Frans Klis; it is allelic to gpi1; I. Imhof, unpublished data), FBY656 (MATa ade2–1 ura3–1 leu2–3,112 trp1–1 his3–11,15 lys- GPI8Δ::kanMX2 containing YCplac22-GST-GPI8), FBY733b (MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YHR188c::kanMX4 containing plasmid pYES2-GPI16), FBY735 (MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YHR188c::kanMX4 containing plasmid YCplac111-GAL1UAS-GPI16), Y22882 (MATa/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YHR188c::kanMX4/YHR188c, derived from BY4743, obtained from EUROSCARF), WADE060–01A(A) (MATa ura3–1 his3–11 leu2–3,112 trp1Δ2 ade2–1 can1–100 YJR015w(-11, 1503)::kanMX4 in W303 background), and FBY91 (sec18 gpi8–1 leu2). Yeast strains were cultured as previously described (Benghezal et al., 1996). SDaa medium is SD medium containing 20–60 mg/l of each of the 20 amino acids, SGaa is the same but with galactose used instead of glucose. Antibodies against Gpi16p were obtained by immunizing rabbits with an octameric peptide corresponding to residues 255–281 of Gpi16p. Antibodies were used after affinity purification on the same peptide. All peptides were synthesized by Alta Bioscience (University of Birmingham, UK). Oligonucleotide synthesis and DNA-sequencing services were provided by Microsynth (Balgach, Switzerland).
Construction of Vectors
Plasmid pYES-GPI16:
The open reading frame of YHR188c was amplified by PCR with the use of a forward primer that introduces a KpnI and a SalI at the 5′ end (5′-acgttgagctggatccggtaccgtcgacatgatcctcacactggcctatttcatgctg-3′) and a reverse primer introducing a XhoI site at the 3′ end (5′-acgttgagctctcgagttagtctgttttagtcttttttttccctag-3′). The PCR fragment was digested with KpnI/XhoI and was inserted into the multiple cloning site (MCS) of the pYES vector (Invitrogen, Carlsbad, CA), which was opened with KpnI/XhoI, thus yielding the plasmid pYES-GPI16.
YCplac111-GAL1UAS-GPI16:
The GAL1 promoter, i.e., nucleotides −453 to −1 of GAL1 of S. cerevisiae was amplified by PCR with the use of a forward primer that introduces a SphI site at the 5′ end (5′-acgttgagctgcatgcacggattagaagccgccgag-3′) and a reverse primer introducing a SalI site at the 3′ end (5′-acgttgagctgtcgacggatccggggttttttctcc-3′). The open reading frame plus the transcription-terminating region of YHR188c was amplified by PCR with the use of a forward primer introducing a SalI site at the 5′ end (5′-acgttgagctggatccggtaccgtcgacatgatcctcacactggcctatttcatgctg-3′) and a reverse primer introducing a KpnI site at the 3′ end (5′-acgttgagctggtacccgatatatttcaattacgacttgttaataaaag-3′). The GAL1 promoter fragment and the YHR188c fragment were digested with SphI/SalI, and SalI/KpnI, respectively, and both fragments were inserted into the MCS of YCplac111, which was opened with SphI/KpnI, thus yielding YCplac111-GAL1-GPI16.
YCplac22-GST-GPI8: harbors GPI8 under the control of the physiological GPI8 promoter containing the gene for glutathione-s-transferase (GST) inserted behind the N-terminal signal sequence of Gpi8p. For its construction, a BsiWI restriction site was introduced at nucleotide 72 of the open reading frame of GPI8.
PCR fragment 1 (primers G8NterF 5′-gagcaacaaatggaattagcc-3′ and G8BsiWINR 5′-agcatccgtacggttatttgc-3′) was digested with XhoI and BsiWI, and PCR fragment 2 (G8BsiWINF 5′-gcaaataaccgtacggatgct-3′ and G8NterR 5′-ggaatacattgtgtttgcctg-3′) were digested with BsiWI and BsmI and were ligated into pBF53 (Meyer et al., 2000), which was digested with XhoI and BsmI. GST (EC 2.5.1.18) from Schistosoma japonicum was amplified with the primers GSTBsiWIF 5′-ggctcgtacgatgtcccctatactaggt-3′ and GSTBsiWIR 5′-ggctcgtacgatccgattttggaggatg-3′, each of which contained a BsiWI restriction site. The GST fragment was digested with BsiWI and was introduced into the BsiWI site in GPI8.
Preparation of Microsomal Membranes and Solubilization of Membrane Proteins
Microsomal membranes were prepared as previously described (Reiss et al., 1997) with the following modifications: cells were grown at 37°C, and membranes were frozen at a higher concentration (0.2 ml membrane buffer was added to the membrane pellet from 1′000 OD600 units of cells). After thawing, 300 μl TM buffer (50 mM Tris-HCl pH7.4, 0.2 M mannitol, 0.1 M NaCl, 1 mM MgCl2, 1 mM CaCl2, 1 mM MnCl2 containing 1 mM DTT, and 1 mM PMSF plus the protease inhibitor mix described for membrane buffer) were added to every 100 μl of membrane suspension. DNA was digested with 0.2 mg/ml DNAse I (3000 U/mg; FLUKA, Buchs, Switzerland) for 45min at 25°C with shaking. Glycerol was adjusted to 10%, and the protein concentration was determined and diluted to 7 μg/μl with GTM-buffer (TM buffer with 10% glycerol). The solubilization of membrane proteins was achieved by adding digitonin and 6-aminocaproic acid to final concentrations of 1.5% and 620 mM, respectively. After incubating for 45 min at 4°C with shaking, insoluble material was removed by centrifugation for 30 min at 40,000 rpm (100,000 × g) at 4°C in a TFT 80.4 fixed angle rotor (Kontron, Munich, Germany). The concentration of solubilized proteins was determined and samples were snap frozen in liquid nitrogen and stored at −80°C.
Blue Native PAGE
Buffers and gel composition were used as previously described (Schägger and von Jagow, 1991; Schägger, 1995), but Tris-HCl, adjusted to pH 7.5 at 4°C, was used instead of Bistris. Solubilized proteins were adjusted to 1 μg/μl with GTM buffer containing 525 mM 6-aminocaproic acid, 1.5% digitonin, 1 mM DTT, 1 mM PMSF, and the described protease inhibitor mix. A 0.15 volume of sample buffer was added, samples were mixed gently and were loaded onto 5–15% or 5–12% polyacrylamide gradient gels. The electrophoresis was carried out at 4°C; after one third of the running time, the cathode buffer with SERVA-Blue was removed and replaced by a cathode buffer without dye. Western blots were revealed with the use of the chemiluminescence ECL kit from Amersham Pharmacia (Uppsala, Sweden).
Affinity Chromatography of GST-Gpi8p
All procedures were carried out at 4°C. Microsomes from 4000 OD600 units of FBY656 cells were prepared and solubilized as for blue native gel electrophoresis but without the use of 6-amino-caproic acid. The lysate was diluted to 10 ml with TM buffer and was mixed with 1 ml glutathione-Sepharose 4B. Beads were incubated overnight at 4°C on a wheel. Beads were sedimented by letting them stand for 1 h at 4°C, and the supernatant was carefully removed (unbound fraction, see Figure 2). The glutathione-Sepharose was washed three times, each time adding 10 ml TM buffer + 0.3% digitonin, rotating the tube on the wheel for 15 min, and letting it stand for 1 h. The supernatants were carefully removed (washes 1 to 3, Figure 2). The bound protein was eluted by adding 1 ml TM buffer plus 0.3% digitonin plus 20 mM reduced glutathione (Sigma, St. Louis, MO), and gently rotating the tube for 30 min. The supernatant was decanted carefully (eluate E20) and was concentrated by ultrafiltration with the use of a Centricon 3KD centrifugation device (Millipore Corp., Bedford, MA) at 4°C, 7000 × g for 2 h. This elution process was repeated by adding 1 ml of TM buffer plus 0.3% digitonin plus 100 mM reduced glutathione, pH 8.0. The supernatant was decanted and concentrated (eluate E100).
Figure 2.
Affinity chromatography purification of a multiprotein complex containing the GST-tagged form of Gpi8p. (A) Membranes of FBY656 cells (gpi8Δ GST-Gpi8p) were isolated and solubilized either in the presence (+) or absence (-) of 6-aminocaproic acid (aca isol). The bulk of the lysate prepared without 6-aminocaproic acid was subjected to affinity chromatography on glutathione-Sepharose 4B. Solubilized proteins were prepared for electrophoresis in the loading buffer with (+) or without (-) 6-aminocaproic acid (aca migr). Aliquots from extracts (lanes 1–4) and from affinity-purified protein (lanes 5 and 6) were separated on a 5–12% blue native gel. Gpi8p was detected by Western blotting with anti-Gpi8p antibody. (B) Material bound to glutathione-Sepharose was eluted with the use of 20 mM, then 100 mM, reduced glutathione (eluates 20 and 100, respectively). The crude extract (lane 1), material not binding to the Sepharose (lane 2), washes 1 and 3 (lanes 3 and 4), and eluates (lanes 5 and 6) were loaded onto a 12% polyacrylamide gel for SDS-PAGE. The gel was stained with silver nitrate. (C) The presence of the GST-tagged form of Gpi8p in the eluates was confirmed by probing a Western blot of a parallel gel containing the same fractions with anti-Gpi8p antibodies. The amounts of material loaded onto the gels are indicated in OD600 equivalents.
Tryptic Digestion and Mass Spectrometry of Proteins
The preparative SDS-PAGE gel (Figure 2) was stained with silver nitrate, and the interesting bands were cut out and cut into small pieces with a scalpel. Then, 100 μl of 25 mM NH4HCO3 were added, gel pieces were agitated with a Vortex for 8 min, and the supernatant was discarded. Washing by agitation was reported using 100 μl of acetonitrile. Alternating washes with these two solvents were repeated two more times. Gel pieces were dried completely with a SpeedVac evaporator before reduction and alkylation. For this, the gel pieces were covered with 100 μl of 10 mM DTT in 25 mM NH4HCO3 and were left at 57°C for 1 h. The supernatant was removed, 100 μl of 55 mM iodoacetamide in 25 mM NH4HCO3 were added, and gel pieces were left in the dark at room temperature for 1 h. The supernatant was removed, and the gel pieces were washed three times with 100 μl of NH4HCO3 and three times with acetonitrile, as above. Gel pieces were dried completely in the SpeedVac evaporator before tryptic digestion. The dried gel volume was evaluated, and three volumes of freshly diluted trypsin (12.5 ng/μl) in 25 mM NH4HCO3 were added. The digestion was performed at 35°C overnight. Then, 5 μl of 25% H2O/70% acetonitrile/5% HCOOH were added, and the samples were sonicated for 5 min and centrifuged. The supernatant was recovered, and the elution of peptides from gel was repeated once using the same solvent. The volume of the supernatant was reduced under a N2 flow to 4 μl, 1 μl of H2O/5% HCOOH was added, and 0.5 μl was used for matrix-assisted laser-desorption time-of-flight (MALDI-TOF) analysis.
Mass measurements were carried out on a Bruker BIFLEX MALDI-TOF mass spectrometer equipped with SCOUT high-resolution optics with X-Y multisample probe and gridless reflector. This instrument was used at a maximum accelerating potential of 20 kV and was operated in reflector mode. Ionization was accomplished with a 337-nm beam from a nitrogen laser with a repetition rate of 3 Hz. The output signal from the detector was digitized at a sampling rate of 1 GHz. A saturated solution of α–cyano-4-hydroxycinnamic acid in acetone was used as a matrix. A first layer of fine matrix crystals was obtained by the spreading and fast evaporation of 0.5 μl of matrix solution. Subsequently, a droplet of 0.5 μl of aqueous HCOOH (5%) solution was deposited onto this fine layer of crystals. Thereafter, 0.5 μl of sample solution was added, and a second droplet of 0.2 μl of the matrix-saturated solution of 50% H2O/50% acetonitrile was added. The preparation was dried in a vacuum. The sample was washed one to three times by applying 1 μl of aqueous HCOOH (5%) solution on the target and then blowing it off after a few seconds. The calibration was performed in internal mode with the following four peptides: angiotensin (1046.542 Da); substance P (1347.736 Da); bombesin (1620.807 Da); and ACTH (2465.199 Da).
Other Methods
The membrane association, N-glycosylation, and membrane orientation of Gpi16p were investigated as described previously (Benghezal et al., 1996). Cells were labeled with [2-3H]-myo-inositol, and lipid extracts were analyzed by TLC as previously described (Meyer et al., 2000). All SDS-PAGE procedures were done under reducing conditions.
RESULTS
Gpi8p Is Stabilized by Its Integration into a High-Molecular-Weight Complex
To investigate the interaction of Gpi8p with potential partners under physiological conditions, we ran digitonin extracts of microsomes in blue native gel electrophoresis. This method allows the preservation of the tertiary and quaternary structures of protein complexes but, nevertheless, allows the separation of the complexes according to their molecular weight (Schägger and von Jagow, 1991; Schägger, 1995). Cells lacking the wt GPI8 but expressing wt Gpi8p under the control of a GAL1,10 promoter were grown on galactose and then shifted to glucose. A previous study (Meyer et al., 2000) demonstrated that under these conditions the very same cells massively overexpress Gpi8p on galactose and reach wt levels of Gpi8p after 4.5 h of growth on glucose. As shown in Figure 1, material reacting with the anti-Gpi8p antibody is found as a broad smear covering the whole range of molecular weights from the top of the stacking gel down to a major band corresponding to ∼65–85 kDa. (In SDS-PAGE, the three glycoforms of Gpi8p have apparent molecular masses of 50, 48, and 46 kDa (Benghezal et al., 1996).) After 4 h on glucose, the amount of Gpi8p is drastically reduced, and almost all Gpi8p is found within high-molecular-weight bands corresponding to apparent molecular masses of 430–650 kDa. The large smear observed in cells overexpressing Gpi8p may represent Gpi8p interacting with numerous other proteins and/or with itself. The result strongly suggests that under physiological conditions (Figure 1, lane 2) Gpi8p is part of a defined stoichiometric protein complex and does not exist as a monomer. We previously reported (Meyer et al., 2000) that in the very same type of experiment, where one goes from overexpression to depletion, turnover of Gpi8p is at least four times faster in the period when Gpi8p is above physiological levels than afterward. The data in Figure 1 demonstrate that it is the Gpi8p fraction, which resides outside the high-molecular-weight complexes, that turns over rapidly, whereas the Gpi8p within these complexes turns over more slowly.
Figure 1.
Blue native PAGE of microsomal membrane proteins during Gpi8p depletion. FBY164 (gpi8::kanMX2 ura3–1:: URA3-GAL1,10-GPI8) cells were grown overnight at 24°C in SGaa medium, were shifted to SDaa medium, and were incubated for 0, 4, 7, and 23 h at 37°C. After incubation, the cells were harvested, microsomes were prepared, and membrane proteins were solubilized in digitonin and processed for blue native PAGE. Solubilized proteins (40 μg per lane) were loaded on a 5–15% polyacrylamide gradient gel. Proteins were transferred onto a PVDF membrane and probed with antibodies against Gpi8p. Marker proteins were bovine serum albumin (monomeric and dimeric form, 66 and 132 kDa, respectively), apoferritin (443 kDa), and thyroglobulin (660 kDa).
The Purified GPI Transamidase Complex Contains Gaa1p, Gpi8p, and a Novel Component, Gpi16p
The GPI transamidase complex was isolated by affinity chromatography over glutathione-Sepharose from the digitonin extract of microsomes prepared from cells containing GST-tagged Gpi8p rather than wt Gpi8p. The original protocol of blue native gel electrophoresis utilizes high concentrations of the detergent-like molecule 6-aminocaproic acid in addition to digitonin for solubilization and during electrophoresis. Our preliminary experiments showed that 6-aminocaproic acid increased the efficiency of extraction but that even without it the same 430–650-kDa complex was obtained. The same is shown in Figure 2A, lanes 1–4, although in this case the complex is bigger (490–780 kDa), since Gpi8p was tagged with GST. This suggested that the size of this complex is dictated by the inherent binding properties and interactions of its components rather than by the solubilization conditions. For large-scale purification of the complex, 6-aminocaproic acid was left out. The SDS-PAGE profile of the proteins present at the different purification steps is shown in Figure 2B. GST-Gpi8p was absorbed nearly quantitatively and could be eluted by free glutathione (Figure 2C, lanes 1, 2, 5, and 6). The complex was eluted in intact form, since when run side by side with the starting material in blue native gel electrophoresis, the purified complex had the same mobility as the complex present in the original microsomal extract (Figure 2A, lanes 1–4 vs. lanes 5 and 6). Thus, no major loss of subunits seems to occur during affinity chromatography. By silver nitrate staining it was revealed that the eluted complex contained only three distinct bands, among which GST-Gpi8p was detectable by Western blotting (Figure 2C, lanes 5 and 6). These bands were cut out and were further analyzed by tryptic digestion and MALDI-TOF mass spectrometry. As shown in Figure 3, only three yeast proteins could be identified by this procedure. Bands a and b of Figure 2B, lanes 5 and 6, which had been excised from the gel en bloc, contained tryptic fragments of GST-Gpi8p and YHR188c, and band c contained fragments of Gaa1p. Indeed, the genes for these three proteins predict translation products of 635, 610, and 614 amino acids. Gaa1p has been described to have an apparent mass of 70 kDa in SDS-PAGE (Hamburger et al., 1995).
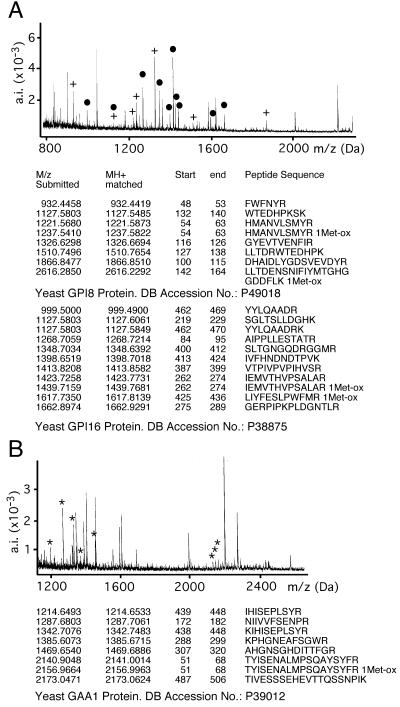
Figure 3.
Mass spectrometric identification of Gpi8p-associated proteins. (A) Spectrogram of peptide masses obtained from tryptic digestion of bands a and b (analyzed jointly) from the gel shown in Figure 2B, lanes 5 and 6. Peptide masses were used to query an amino acid sequence database (PROTEIN PROSPECTOR). Proteins were identified by correlating the measured peptide masses to a theoretical tryptic digest of all proteins present in the database. Yeast Gpi8p and Gpi16p were the only yeast proteins that were identified. Their peptides are marked with crosses (+, Gpi8p) and dots (●, Gpi16p). (B) Spectrogram of peptide masses obtained from band c (Figure 2B) identified Gaa1p (*). The exact tryptic peptides allowing identification are listed. a.i. = absolute intensity.
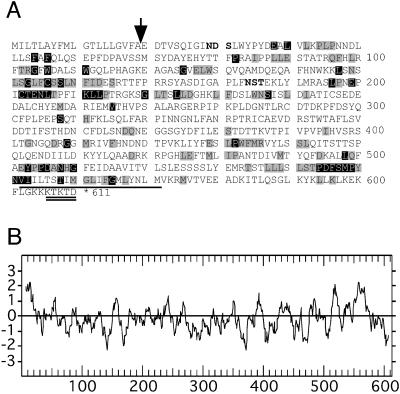
The sequence and the hydrophobicity plot of YHR188c, henceforth named GPI16, are shown in Figure 4. Two versions of Gpi16p differing by the presence or absence of eight hydrophobic residues at the N-terminus are currently found in genomic databases, the shorter version being proposed based on the sequence submission of C. Marci to the EMBL Data Library in 1994 (entry S46687), and the longer one based on the complete sequence of chromosome VIII submitted by Johnston et al. (1994). Both versions predict a type I membrane protein with an N-terminal signal sequence for insertion into the ER, a C-terminal TMD, and an ER retrieval motif at the end of the cytosolic C-terminus. Gpi16p is a protein of unknown function that has homologues in humans, flies, worms, plants, and other fungi. These organisms contain proteins of similar size (531–639 amino acids) and a similar hydropathy profile. When comparing the proteins over their entire length, these proteins (specified in the legend of Figure 4A) exhibit 24–32% identities and 43–49% similarities to GPI16. Identities are concentrated into several motifs appearing in yeast Gpi16p at residues 147–163, 196–213, 498–510, and 544–565 (Figure 4A). Only the yeast members have a C-terminal ER retrieval motif, but the proteins of other species may be retained in the ER by other signals or by interaction with Gaa1p or Gpi8p.
Figure 4.
Sequence of S. cerevisiae Gpi16p. (A) Potential N-glycosylation sites are shown in boldface, the C-terminal TMD is underlined, and the ER retrieval domain doubly underlined. The most likely cleavage site of the signal peptidase is indicated by a vertical arrow. The Gpi16p homologues of Schizosaccharomyces pombe (emb CAA22348.1 ), Drosophila melanogaster (gb AAF46367.1 ), Homo sapiens (CGI-06 protein, XM 009533.2), Caenorhabditis elegans (emb CAA96629.1), and Arabidopsis thaliana (gb AAF20232.1 ) were aligned usig the CLUSTALW multiple sequence alignments program 1.81. Only the yeast Gpi16p sequence is shown, but identities and conservative substitutions are represented by black and gray boxes. (B) A hydropathy plot, drawn according to Kyte and Doolittle, is shown in which the hydrophobic sequences get a positive score.
Gpi16p Is Essential for GPI Anchoring
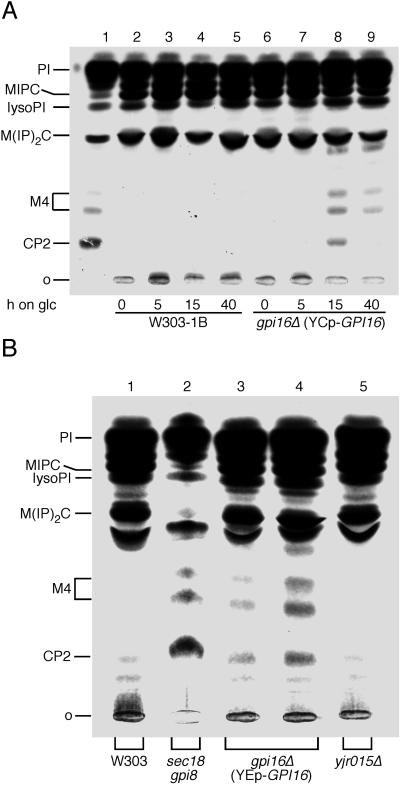
YHR188c has been found to be an essential gene. We therefore generated haploid gpi16Δ strains that contain the open reading frame of GPI16 behind the GAL1 promoter, either on a multicopy or a centromeric vector. Western blot analysis with the use of a rabbit antibody to an immunogenic peptide of Gpi16p demonstrated that Gpi16p could be significantly depleted when cells were shifted from a galactose- to a glucose-containing medium (see below). When Gpi16p-depleted cells were labeled with [3H]-myo-inositol, cells accumulated the GPI precursor lipids CP2 and two species of M4 (Figure 5A, lane 8, and B, lanes 3 and 4). These rather mature GPI precursor lipids are not normally present in the lipid extract of wt cells (Figure 5A, lanes 2–5, B, lane 1), but only appear if the GPI transamidation reaction is blocked, e.g., in gpi8 (Figure 5A, lane 1, B, lane 2). When Gpi16p is depleted for longer periods, the cells start to grow faster again and the accumulation of GPI lipids is less pronounced (Figure 5A, lane 9), indicating that cells may undergo some genetic or metabolic adaptation, possibly by increasing the number of plasmids carrying GPI16.
Figure 5.
Cells depleted of Gpi16p accumulate complete GPI lipids. (A) W303 and FBY735 (gpi16Δ YCplac111-GAL1-GPI16) were grown to exponential phase at 30°C in SGaa. The expression of Gpi16p was subsequently blocked by shifting cells to glucose medium (SDaa). Cultures were continued at 30°C for 0, 5, 15, or 40 h and were periodically diluted with fresh medium to keep the cell concentration in the range of 0.5–2.5 OD600. The sec18 gpi8 cells (lane 1) were grown at 24°C and were preincubated for 5 min at 37°C. Aliquots of cells were radiolabeled at 37°C with [2-3H]-myo-inositol for 80 min, and lipid extracts were analyzed by TLC. (The sec18 gpi8 double mutant was chosen as a positive control, since it accumulates more CP2 at 37°C than gpi8.) (B) FBY733b cells (gpi16Δ pYES2-GPI16) were grown to exponential phase at 30°C in SGaa medium and were shifted to SDaa for 2 h (lane 3) or 15 h (lane 4). W303 cells and yjr015Δ cells were grown at 30°C. Aliquots of cells were radiolabeled and processed as above.
The accumulation of immature ER forms of GPI proteins such as Gas1p (Nuoffer et al., 1991) or Cwp1p (Shimoi et al., 1995) is a symptom of a delay or deficiency in the GPI anchor addition to newly synthesized proteins (Doering and Schekman, 1996). Western blotting shows an accumulation of the immature 105-kDa form of Gas1p and the immature 45-kDa form of Cwp1p after 5 and 15 h of depletion of Gpi16p (Figure 6A), whereas there was no accumulation of immature carboxypeptidase Y (CPY) (not shown). In pulse-chase experiments, it appears that the maturation of Gas1p from the 105-kDa immature form to the 125-kDa mature form is severely delayed when Gpi16p is depleted, whereas the maturation of CPY is not significantly retarded (Figure 6B). Thus, it appears that the depletion of Gpi16p induces a specific defect in (with regard to) the attachment of GPI lipids to newly made GPI proteins and by consequence, delays their transport to the Golgi.
Figure 6.
Depletion of Gpi16p blocks maturation of Gas1p and Cwp1p. (A) W303–1B and FBY735 cells (gpi16Δ YCp-GAL1-GPI16) were grown to exponential phase at 30°C in SGaa medium and were shifted to glucose for 0, 5, and 15 h in order to block the expression of Gpi16p. Proteins were extracted, separated by SDS-PAGE, Western blotted, and probed with anti-Gas1p and anti-Cwp1p antibodies. The relative amounts of the immature 105-kDa form of Gas1p as well as of the immature 45-kDa form of Cwp1p were determined by densitometry and are given as the percentage of total Gas1p or Cwp1p in the extracts. m = mature; i = immature. (B) FBY735 cells were grown to exponential phase at 30°C in selective SGaa medium, were washed twice in distilled water, and were resuspended in SDaa medium. After 0 or 15 h of culture, cells were pulse-labeled with [35S]-methionine and [35S]-cysteine for 10 min and were chased for the indicated times. Gas1p was immunoprecipitated from the lysates and analyzed by SDS-PAGE and fluorography.
A systematic two-hybrid screen had detected a significant interaction of Gpi16p with YJR015w (Uetz et al., 2000), a nonessential protein of 510 amino acids having several TMDs. As seen in Figure 5B, lane 5, yjr015wΔ cells did not accumulate any GPI precursor lipids. Further studies are required to definitely rule out a role for this protein in GPI anchor addition.
Gpi16p Is an Integral Membrane Glycoprotein of the ER
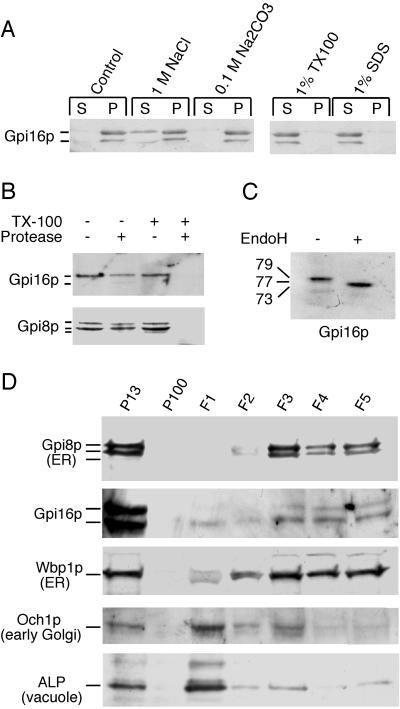
High salt or high pH treatment of microsomal membranes cannot remove Gpi16p from the membranes (Figure 7A), but the protein is rendered soluble by Triton X-100 at 4°C. Protease protection experiments on microsomes from wt cells indicate that Gpi16p is protease-resistant unless detergent is added in the same way as with Gpi8p (Figure 7B). The treatment of the cell lysate with endoglycosidase H shifted the molecular mass of Gpi16p by ∼6 kDa (Figure 7C), thus indicating that probably both of the two potential N-glycosylation sites are utilized. Subcellular fractionation indicates that Gpi16p is mainly present in fractions that contain the ER markers Gpi8p and Wbp1p, and is absent from lighter fractions that are enriched in the Golgi marker Och1p and the vacuolar alkaline phosphatase (ALP) (Figure 7D). These data indicate that Gpi16p is an integral ER membrane glycoprotein, with the bulk of the protein being oriented toward the lumen of the ER. It probably gets directed into the ER by its N-terminal signal sequence.
Figure 7.
Gpi16p is a lumenally oriented, integral membrane glycoprotein of the ER. (A) Exponentially growing W303–1B were broken with glass beads and cell walls were removed. Aliquots of the lysate (equivalent to 20 OD600) were incubated for 30 min at 0°C with NaCl, Na2CO3, Triton X-100, or SDS. Subsequently, membranes were sedimented by ultracentrifugation, proteins of supernatants (S) and pellets (P) were precipitated with TCA, were processed for SDS-PAGE and Western blotting, and were probed with affinity-purified rabbit anti-Gpi16p antibodies. (B) Microsomes from W303–1B were digested with 0.1 mg/ml proteinase K at 0°C for 30 min in the presence or absence of 0.5% of Triton X-100. Samples were resuspended in sample buffer, processed for Western blotting, and probed with anti-Gpi16p or anti-Gpi8p antibodies. (C) A microsomal pellet from W303–1B cells was solubilized in 1% SDS and was incubated with or without 1 mU/OD600 endoglycosidase H for 20 h at 37°C. The samples were processed as above. (D) Cellular localization of Gpi16p was determined by subcellular fractionation. For this experiment, a slightly modified version of a published procedure (Gaynor et al., 1994) was used. Spheroplasts of W303–1B at 67 OD600/ml were osmotically lysed in a syringe, the pellet P13 obtained through centrifugation at 13,000 × g for 15 min was further resolved by sedimenting 100-OD600 equivalents through a step gradient of 1.6 ml 1.2 M sucrose to >1.6 ml 1.5 M sucrose, yielding 5 0.7-ml fractions termed F1 to F5. Supernatant S13 was further centrifuged at 100,000 × g for 1 h to obtain pellet P100. Equivalent aliquots of all fractions were processed for SDS-PAGE and Western blotting. On prolonged incubation of microsomes, variable amounts of a 69-kDa degradation product of Gpi16p was generated.
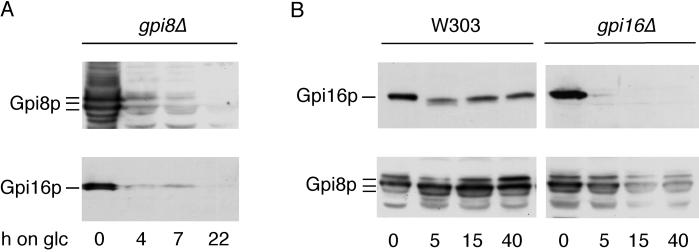
Gpi8p and Gpi16p Are Mutually Stabilizing Each Other
Figure 1 suggests that Gpi8p, if not integrated into the transamidase complex, is unstable. Since Gpi16p is a major component of this complex, we expect that the depletion of Gpi16p would lead to a depletion of Gpi8p. As shown in Figure 8B, this is indeed the case. When interpreting this figure, one has to keep in mind that at time 0 the cells strongly overexpress Gpi16p but contain physiological levels of Gpi8p. Thus, even after 40 h of depletion, the remaining Gpi16 and Gpi8p may still be present at a physiological ratio. Previous experiments have shown that during 24 h on glucose the Gpi8p content in the gpi8Δ cells used in Figure 8A is dropping from 1140% to <6% of the physiological amount of Gpi8p (Meyer et al., 2000). Figure 8A demonstrates that the depletion of Gpi8p in these cells leads to a concomitant loss of Gpi16p. This suggests that Gpi16p, in a manner similar to that of Gpi8p, is unstable unless integrated into the transamidase complex. The experiment shown in Figure 8B raised the possibility that the GPI-anchoring deficiency observed in Gpi16p-depleted cells may be due to the concomitant depletion of Gpi8p, which is believed to be the catalytic subunit of the transamidase complex. Thus, it was conceivable that Gpi16p affected the GPI anchor addition solely through the stabilization of Gpi8p. However, as shown in Figure 9, the overexpression of Gpi8p could not rescue the growth phenotype of Gpi16p-depleted cells. The same result also was obtained in liquid cultures (not shown). Moreover, the same cells used for the blotting experiment of Figure 9B were also metabolically labeled with [3H]-myo-inositol, and their lipid extracts were analyzed by TLC as described in Figure 5. This showed that the accumulation of the abnormal GPI lipids CP2 and M4 caused by the depletion of Gpi16p (Figure 5) could not be hindered or attenuated by the overexpression of Gpi8p (not shown). These data suggest that Gpi16p, besides stabilizing Gpi8p, also has a more direct effect on the GPI anchor addition.
Figure 8.
Gpi8p and Gpi16p are mutually stabilizing each other. FBY164 (gpi8::kanMX2 ura3–1:: URA3-GAL1,10-GPI8), W303, and FBY735 (YHR188c::kanMX4 containing plasmid YCplac111-GAL1-GPI16) were grown to exponential phase at 30°C in galactose medium (SGaa). The expression of Gpi8p (FBY164) or Gpi16p (FBY735) was subsequently blocked by shifting cells to glucose medium (SDaa). Cultures were continued at 30°C for 0, 4, 7, and 22 h (FBY164) or for 0, 5, 15, and 40 h (W303 and FBY735). Proteins were extracted, separated by SDS-PAGE, Western blotted, and probed with anti-Gpi8p (A) and anti-Gpi16p antibodies (B); membranes were stripped and probed again with anti-Gpi16p (A) and anti-Gpi8p (B) antibodies.
Figure 9.
Overexpression of Gpi8p cannot suppress the growth defect of Gpi16p-depleted cells. (A) W303–1B (wt), FBY735 (Δgpi16/YCplac111-GAL1-GPI16), FBY735 transfected either with empty YEplac195, or with YEplac195 harboring GPI8 placed under the control of the promoter of CUP1 (YEplac195-CUP1-GPI8) were grown to exponential phase at 30°C in galactose-containing medium. To deplete them of Gpi16p and to induce Gpi8p, 3-μl aliquots of cells at concentrations shown at the top (cells/ml) were inoculated onto plates containing glucose plus 100 μM Cu2+. The plates were incubated for 35 h at 30°C before being photographed. (B) W303 (lanes 1 and 5), FBY735 (lanes 2 and 6), FBY 735 transfected with YEplac195-CUP1-GPI8 (lanes 3 and 7), and FBY735 transfected with the empty vector YEplac195 (lanes 4 and 8) were grown at 30°C for 15 h in liquid culture with the use of either galactose medium or, alternatively, glucose medium supplemented with 100 μM Cu2+. Proteins were extracted, separated by SDS-PAGE, Western blotted, and probed with anti-Gpi8p antibodies.
The GPI Transamidase Complex Is not Associated with Sec61p or Wbp1p
The major components of the 430–650-kDa transamidase complex are Gaa1p, Gpi8p, and Gpi16p, but it seemed conceivable that before blue native gel electrophoresis, in the digitonin extract, this complex was associated with other protein complexes involved in the processing of GPI proteins. Some kind of association of the transamidase complex with the ER translocation pore is suggested by the fact that all current in vitro assays for the GPI anchor addition are combined translocation/GPI attachment assays, and all our attempts to assay the transamidase with artificial peptides that do not need to be translocated have failed so far (P. Fraering, unpublished data). Therefore, we tried to evaluate whether the transamidase complex is associated with the ER translocation pore and the oligosaccharyltransferase complex. The latter may be in close vicinity to the translocation pore because proteins can be cotranslationally N-glycosylated. The yeast translocation complex is composed of two subcomplexes, the Sec61 complex, consisting of Sec61p (41 kDa), Sss1p (9.1 kDa), and Sbh1p (8.7 kDa), and the Sec62/63 complex, consisting of Sec62p (30 kDa), Sec63 (73 kDa), Sec71p (31 kDa), and Sec72p (23 kDa). The Sec61 complex is competent for the cotranslational translocation of proteins, whereas the presence of both subcomplexes is required for posttranslational translocation (Ng et al., 1996). The oligosaccharyltransferase activity depends on Ost1p (64 kDa), Wbp1p (45 kDa), Swp1p (30 kDa), Ost2p (16 kDa), Ost3p (34 kDa), Stt3p (78 kDa), Ost4p (3.4 kDa), Ost5p (9.5 kDa), and Ost6p (37.3 kDa) (Knauer and Lehle, 1999). The sum of these subunits amounts to 317.6 kDa. In blue native gel electrophoresis, the oligosaccharyltransferase migrated as a 240-kDa complex that, as determined by Western blot analysis, contained all of these subunits (Knauer and Lehle, 1999). Each individual complex may, however, be composed of only a subset of components since some subunits may be redundant. As shown in Figure 10, when analyzed by blue native gel electrophoresis of digitonin extracts, Sec61p and Wbp1p do not exist in free form but are present mainly in high-molecular-weight complexes of 180, 310, and 470 kDa (Sec61p) and 440 kDa (Wbp1p), respectively. In this experiment, the cells contained GST-tagged Gpi8p rather than wt Gpi8p, so that the GPI transamidase complex could be isolated by affinity chromatography over glutathione-Sepharose. The purified transamidase complex contained neither Sec61p nor Wbp1p (lanes 5 and 6). Overall, the data indicate that an association of the GPI transamidase complex with these other complexes either does not exist or does not survive digitonin extraction and affinity purification. The lower mass estimation for the oligosaccharyltransferase complex obtained by Knauer and Lehle (1999) may be due to the fact that they used Nikkol plus 6-aminocaproic acid as detergents, whereas we used digitonin. Incidentally the data show that 6-aminocaproic acid is not absolutely required for solubilization and blue native gel electrophoresis of all three complexes since complexes of the same size are solubilized by digitonin alone. However, it is evident that the addition of 6-aminocaproic acid can increase the efficiency of solubilization, especially for the Sec61 complex.
Figure 10.
Association of the transamidase complex with other multimeric ER complexes. The membrane of the Western blot shown in Figure 2A was stripped and probed again with anti-Sec61p antibodies, was stripped again, and finally was probed with anti-Wbp1p antibodies. The top panel is the same as in Figure 2A.
The GPI High-Molecular-Weight Complex Persists in the Absence of GPI Lipids and Precursor Proteins
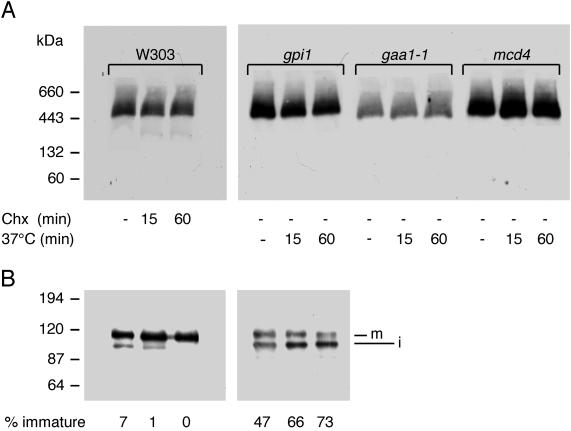
Although a stable interaction of Gpi8p and Gaa1p has been demonstrated by coimmunoprecipitation (Ohishi et al., 2000), it appeared possible that proteins in the GPI transamidase would not remain firmly associated throughout the catalytic cycle but that the normal workings of this complex required that some subunits dissociate at a certain stage. In fact, if overexpression is used to demonstrate the interaction, a large proportion of overexpressed proteins may never be engaged in GPI anchoring, and this could lead to the artificial perpetuation of a normally transient interaction. Blue native gel electrophoresis allowed detection of the transamidase complex also in normal wt cells (Figure 11A, lane 1). The following experiments were designed to probe the stability of this physiological transamidase complex under conditions where its substrates are depleted. As shown in Figure 11, Gpi8p remained completely confined within the complexes of 430–650 kDa when prepro forms of GPI proteins were depleted with the use of cycloheximide, a treatment that was effective since it led to the complete disappearance of the immature 105-kDa form of the GPI protein Gas1p (Figure 11B). The transamidase complex also persisted after a temperature shift of mutants that, upon a shift to 37°C, block the biosynthesis of GPI lipids at very early stages (gpi1 and mcd4) or interrupt the transfer of GPI lipids onto proteins (gaa1) (Hamburger et al., 1995; Leidich and Orlean, 1996; Gaynor et al., 1999; Packeiser et al., 1999). For gpi1, the efficiency of the block was assessed by following the gradual accumulation of the immature Gas1p (Figure 2B). In an independent experiment, we more carefully quantitated the amount of Gpi8p in the extracts shown in Figure 11 by SDS-PAGE/Western blotting, whereby the amount of protein loaded onto gels was verified by Coomassie blue staining of gels and blots (not shown). This demonstrated that the Gpi8p content of cells remains constant during 1 h of substrate deprivation, as was done here. Moreover, all cells contain similar amounts of Gpi8p. In Figure 11A, less Gpi8p is detected in gaa1 simply because less protein was loaded onto the gel in this mutant.
Figure 11.
Stability of the transamidase complex in the absence of GPI protein and GPI lipid precursors. (A) W303 cells were grown to exponential phase at 37°C in YPD medium. Cycloheximide (Chx; 100 μg/ml) was added 0, 15, or 60 min before protein extraction. Thermosensitive mutant cells gpi1, gaa1–1, and mcd4 were grown to exponential phase at 24°C in YPD medium. The cells were left at 24°C (-) or were shifted to 37°C for 15 or 60 min. Then, microsomal membranes were prepared and proteins were processed for blue native PAGE. Five μg per lane of solubilized proteins was loaded on a 5–15% gradient gel. Proteins were transferred onto a PVDF membrane and were probed with antibodies against Gpi8p. (B) Five micrograms of the same proteins from W303 and gpi1 were separated on SDS-PAGE (7.5%). Mature (m) and immature (i) forms of Gas1p were detected by Western blotting with the use of rabbit anti-Gas1p antibodies. The percentage of immature Gas1p, as quantitated by densitometry, is indicated below. Similarly to the original gpi1 isolate and to gpi1Δ cells (Leidich et al., 1994; Leidich and Orlean, 1996), the gpi1 mutant used here neither grows nor incorporates [2-3H]-myo-inositol into proteins at 37°C, whereas at 24°C GPI anchoring is only partially deficient. This partial deficiency explains the abnormally high percentage of immature Gas1p in gpi1 already at 24°C.
DISCUSSION
Biosynthesis of secretory proteins requires processing events such as insertion into the ER, removal of the signal peptide, and N-glycosylation, events that are operated by heteromultimeric complexes. The existence of a similar complex for the GPI anchor addition has been postulated before based on genetic experiments showing that the overexpression of active-site mutant alleles of Gpi8p such as the C199A allele blocks GPI addition by the wt Gpi8p that is also present in the same cell (Meyer et al., 2000). This dominant-negative effect could be overcome by the concomitant overexpression of wt Gpi8p, suggesting that the C199A and wt forms of Gpi8p are competing for other subunits that are required for activity (Meyer et al., 2000). On the other hand, the dominant-negative effect was not alleviated by the overexpression of Gaa1p, suggesting that other protein components beyond Gaa1p may be required for the formation of a functional complex (Meyer et al., 2000).
Here we indeed isolate a further component, Gpi16p, which is stably associated with Gaa1p and Gpi8p. The functional consequences of its depletion suggest that GPI16 is essential for yeast because it is required for GPI anchoring of proteins. Under depletion the complete precursor CP2 accumulates. This demonstrates unequivocally that Gpi16p is not required for the biosynthesis of GPI lipids but rather for their attachment to GPI proteins.
Although the functional importance of Gpi16p, as well as that of Gaa1p and Gpi8p, for the attachment of GPI lipids to newly made GPI proteins in the ER is beyond doubt, much remains to be learned about the exact function of these proteins. The homology of Gpi8p with the C13 thiol protease family suggests that Gpi8p is the subunit that proteolytically cleaves the proform of GPI proteins at the ω site. The other two subunits do not contain any known sequence motif that could give a hint with regard to their particular function. Also, Gaa1p, Gpi8p, and Gpi16p do not share any obvious sequence motif among each other. Gaa1p and Gpi16p may be required for binding of the protein and lipid substrates. As long as the membrane orientation of the GPI biosynthetic pathway is not established beyond doubt, it also remains possible, that they are involved in flipping complete GPI lipids from the cytosolic to the lumenal face of the ER. However, the recently identified ER membrane proteins PIG-B (Gpi10p) and PIG-M, which are involved in earlier steps of GPI biosynthesis, have their likely catalytic residues on the lumenal side of the ER membrane, suggesting that later steps of GPI biosynthesis occur on the lumenal side of the ER and that there is no need to flip complete precursors (Takahashi et al., 1996; Maeda et al., 2001). Finally, it also seems to be conceivable that some subunit would operate ER remodeling steps, by which the primary lipid moiety of GPI proteins is exchanged or modified. Indeed, soon after the addition to proteins all GPI anchors get remodeled in the ER, either in the sense that a C26 replaces a C16 fatty acid on the diacylglycerol type lipid moiety or in the sense that the primary diacylglycerol moiety is replaced by a ceramide (Conzelmann et al., 1992; Sipos et al., 1997).
Blue native gel electrophoresis allows a rapid assessment of the state of the GPI transamidase complex under various conditions. It was used to demonstrate that the digitonin-soluble transamidase complex can have various sizes (430–650 kDa) and that the complex remains intact during affinity chromatography (Figure 2A). The subunits that we identify in the complex are Gpi8p (50 kDa), Gaa1p (70 kDa), and Gpi16p (79 kDa), and together they add up to ∼200 kDa. It is possible that the 430–650-kDa complex contains more then one copy of some of these three proteins and/or contains yet other nonidentified subunits that were not stained by silver nitrate (Figure 2B). Blue native gel electrophoresis also shows that the complex is present under conditions in which proteins are expressed at physiological levels. To us, it seemed important to show that the complex is present under physiological conditions. A previous study demonstrated by coimmunoprecipitation that hGpi8p and hGaa1p interact stably when they are strongly overexpressed (Ohishi et al., 2000). In the situation of overexpression, the ER folding machinery may get overwhelmed, and incompletely folded proteins may accumulate and aggregate in a nonspecific way through hydrophobic interactions. (Microsomal ALDH, which was used as a specificity control, does not really rule out this possibility since it is mainly cytosolic and exposes only four amino acids on the lumenal side of the ER (Masaki et al., 1994), whereas the major extramembranous parts of Gaa1p and Gpi8p are lumenal.) However, our data clearly indicate that the complex exists under physiological conditions, not only because of data obtained by blue native gel electrophoresis, but also because the complex could be purified from cells, in which its constitutent proteins were under the control of their physiological promoters: GPI16 and GAA1 were transcribed from their normal genomic gene; GPI8 was deleted but present on a centromeric vector as a GST-tagged recombinant protein. Furthermore, blue native gel electrophoresis showed that Gpi8p is stabilized by the incorporation into the transamidase complex (Figure 1).
Previous experiments have suggested that the transamidase complex is not completely static in as much as the induction of dominant-negative alleles for 1 h completely blocked the addition of GPIs to proteins, whereas the half-life of Gpi8p amounts to 9 h. This suggested that newly made dominant-negative Gpi8p alleles get access to the preexisting complexes, implying that this complex is dynamic (Meyer et al., 2000). Also, soluble forms of Gpi8p could be added to ER membranes stripped of their endogenous Gpi8p and reconstitute transamidase activity (Sharma et al., 2000). Our present experiments (Figure 11) suggest that the complex is relatively stable even in the absence of GPI lipids or GPI precursor proteins, although it still remains possible that the depletion of these substrates was incomplete and that the complete absence of substrate would dissociate the complex. Indeed, the complex was stable not only during digitonin extraction and blue native gel electrophoresis, but also during the purification of the Gaa1p/GST-Gpi8p/Gpi16p complex. But even if we accept that the complex is stable in the absence of substrates, this does not exclude the possibility that the complex partially or totally disassembles each time it has to release products. If such were the case, our data support the view that the subunits can reassemble without previous binding of substrates. Clearly, further studies are required to solve this issue.
ACKNOWLEDGMENTS
We are grateful to Michael Wacker for introducing I.I. to blue native gel electrophoresis, to Frans Klis and Howard Riezman for the gift of reagents, to Dr. T. Suply for his scientific input, and to Anne-Lise Maury and Anne Schneider for technical assistance. This work was supported by grant No. 3100–032515 from the Swiss National foundation.
Abbreviations used:
- GPI
glycosylphosphatidylinositol
- GST
glutathione-s-transferase
- TMD
transmembrane domain
- wt
wild-type
REFERENCES
- Benghezal M, Benachour A, Rusconi S, Aebi M, Conzelmann A. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 1996;15:6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Caras IW, Weddell GN, Davitz MA, Nussenzweig V, Martin DWJ. Signal for attachment of a phospholipid membrane anchor in decay accelerating factor. Science. 1987;238:1280–1283. doi: 10.1126/science.2446389. [DOI] [PubMed] [Google Scholar]
- Caro LH, Tettelin H, Vossen JH, Ram AF, van den Ende H, Klis FM. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1477–1489. doi: 10.1002/(SICI)1097-0061(199712)13:15<1477::AID-YEA184>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Chen JM, Rawlings ND, Stevens RA, Barrett AJ. Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases. FEBS Lett. 1998;441:361–365. doi: 10.1016/s0014-5793(98)01574-9. [DOI] [PubMed] [Google Scholar]
- Conzelmann A, Puoti A, Lester RL, Desponds C. Two different types of lipid moieties are present in glycophosphoinositol-anchored membrane proteins of Saccharomyces cerevisiae. EMBO J. 1992;11:457–466. doi: 10.1002/j.1460-2075.1992.tb05075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TL, Schekman R. GPI anchor attachment is required for Gas1p transport from the endoplasmic reticulum in COP II vesicles. EMBO J. 1996;15:182–191. [PMC free article] [PubMed] [Google Scholar]
- Doering TL, Schekman R. Glycosyl-phosphatidylinositol anchor attachment in a yeast in vitro system. Biochem J. 1997;328:669–675. doi: 10.1042/bj3280669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, te Heesen S, Graham TR, Aebi M, Emr SD. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Mondesert G, Grimme SJ, Reed SI, Orlean P, Emr SD. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol Biol Cell. 1999;10:627–648. doi: 10.1091/mbc.10.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber LD, Kodukula K, Udenfriend S. Phosphatidylinositol glycan (PI-G) anchored membrane proteins: amino acid requirements adjacent to the site of cleavage and PI-G attachment in the COOH-terminal signal peptide. J Biol Chem. 1992;267:12168–12173. [PubMed] [Google Scholar]
- Hamada K, Fukuchi S, Arisawa M, Baba M, Kitada K. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol Gen Genet. 1998;258:53–59. doi: 10.1007/s004380050706. [DOI] [PubMed] [Google Scholar]
- Hamburger D, Egerton M, Riezman H. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J Cell Biol. 1995;129:629–639. doi: 10.1083/jcb.129.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, et al. Complete nucleotide sequence of Saccharomyces cerevisiae chromosome VIII. Science. 1994;265:2077–2082. doi: 10.1126/science.8091229. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Inoue N. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr Opin Chem Biol. 2000;4:632–638. doi: 10.1016/s1367-5931(00)00151-4. [DOI] [PubMed] [Google Scholar]
- Knauer R, Lehle L. The oligosaccharyltransferase complex from Saccharomyces cerevisiae: isolation of the OST6 gene, its synthetic interaction with OST3, and analysis of the native complex. J Biol Chem. 1999;274:17249–17256. doi: 10.1074/jbc.274.24.17249. [DOI] [PubMed] [Google Scholar]
- Leidich SD, Drapp DA, Orlean P. A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J Biol Chem. 1994;269:10193–1016. [PubMed] [Google Scholar]
- Leidich SD, Orlean P. Gpi1, a Saccharomyces cerevisiae protein that participates in the first step in glycosylphosphatidylinositol anchor synthesis. J Biol Chem. 1996;271:27829–27837. doi: 10.1074/jbc.271.44.27829. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Watanabe R, Harris CL, Hong Y, Ohishi K, Kinoshita K, Kinoshita T. PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 2001;20:250–261. doi: 10.1093/emboj/20.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Tashiro Y. Microsomal aldehyde dehydrogenase is localized to the endoplasmic reticulum via its carboxyl-terminal 35 amino acids. J Cell Biol. 1994;126:1407–1420. doi: 10.1083/jcb.126.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville MJ, Ferguson MA. The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem J. 1993;294:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U, Benghezal M, Imhof I, Conzelmann A. Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry. 2000;39:3461–3471. doi: 10.1021/bi992186o. [DOI] [PubMed] [Google Scholar]
- Moran P, Caras IW. Fusion of sequence elements from non-anchored proteins to generate a fully functional signal for glycophosphatidylinositol membrane anchor attachment. J Cell Biol. 1991;115:1595–1600. doi: 10.1083/jcb.115.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C, Jeno P, Conzelmann A, Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol Cell Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C, Horvath A, Riezman H. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J Biol Chem. 1993;268:10558–10563. [PubMed] [Google Scholar]
- Ohishi K, Inoue N, Maeda Y, Takeda J, Riezman H, Kinoshita T. Gaa1p and gpi8p are components of a glycosylphosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol Biol Cell. 2000;11:1523–1533. doi: 10.1091/mbc.11.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packeiser AN, Urakov VN, Polyakova YA, Shimanova NI, Shcherbukhin VD, Smirnov VN, Ter-Avanesyan MD. A novel vacuolar protein encoded by SSU21/MCD4 is involved in cell wall integrity in yeast. Yeast. 1999;15:1485–1501. doi: 10.1002/(SICI)1097-0061(199910)15:14<1485::AID-YEA477>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Ramalingam S, Maxwell SE, Medof ME, Chen R, Gerber LD, Udenfriend S. COOH-terminal processing of nascent polypeptides by the glycosylphosphatidylinositol transamidase in the presence of hydrazine is governed by the same parameters as glycosylphosphatidylinositol addition. Proc Natl Acad Sci USA. 1996;93:7528–7533. doi: 10.1073/pnas.93.15.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss G, te Heesen S, Gilmore R, Zufferey R, Aebi M. A specific screen for oligosaccharyltransferase mutations identifies the 9 kDa OST5 protein required for optimal activity in vivo and in vitro. EMBO J. 1997;16:1164–1172. doi: 10.1093/emboj/16.6.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schägger H. Native electrophoresis for isolation of mitochondrial oxidative phosphorylation protein complexes. Methods Enzymol. 1995;260:190–202. doi: 10.1016/0076-6879(95)60137-6. [DOI] [PubMed] [Google Scholar]
- Schönbächler M, Horvath A, Fassler J, Riezman H. The yeast spt14 gene is homologous to the human PIG-A gene and is required for GPI anchor synthesis. EMBO J. 1995;14:1637–1645. doi: 10.1002/j.1460-2075.1995.tb07152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DK, Vidugiriene J, Bangs JD, Menon AK. A cell-free assay for glycosylphosphatidylinositol anchoring in African trypanosomes: demonstration of a transamidation reaction mechanism. J Biol Chem. 1999;274:16479–16486. doi: 10.1074/jbc.274.23.16479. [DOI] [PubMed] [Google Scholar]
- Sharma DK, Hilley JD, Bangs JD, Coombs GH, Mottram JC, Menon AK. Soluble GPI8 restores glycosylphosphatidylinositol anchoring in a trypanosome cell-free system depleted of lumenal endoplasmic reticulum proteins. Biochem J. 2000;351:717–722. [PMC free article] [PubMed] [Google Scholar]
- Shimoi H, Iimura Y, Obata T. Molecular cloning of CWP1: a gene encoding a Saccharomyces cerevisiae cell wall protein solubilized with Rarobacter faecitabidus protease I. J Biochem (Tokyo) 1995;118:302–11. doi: 10.1093/oxfordjournals.jbchem.a124907. [DOI] [PubMed] [Google Scholar]
- Sipos G, Reggiori F, Vionnet C, Conzelmann A. Alternative lipid remodelling pathways for glycosylphosphatidylinositol membrane anchors in Saccharomyces cerevisiae. EMBO J. 1997;16:3494–3505. doi: 10.1093/emboj/16.12.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Inoue N, Ohishi K, Maeda Y, Nakamura N, Endo Y, Fujita T, Takeda J, Kinoshita T. PIG-B, a membrane protein of the endoplasmic reticulum with a large lumenal domain, is involved in transferring the third mannose of the GPI anchor. EMBO J. 1996;15:4254–4261. [PMC free article] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–67. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]