Abstract
R2R3 MYB transcription factors play key functions in the regulation of secondary metabolites. In the present study, a R2R3 MYB transcriptional factor NtMYB2 was identified from Chinese narcissus (Narcissus tazetta L. var. Chinensis Roem) and functionally characterized. NtMYB2 belongs to subgroup 4 of the R2R3 MYB transcription factor family that are related to repressor MYBs involved in the regulation of anthocyanin and flavonoids. Transient expression confirmed that NtMYB2 strongly reduced the red pigmentation induced by MYB- anthocyanin activators in agro-infiltrated tobacco leaves. Ectopic expression of NtMYB2 in tobacco significantly reduced the pigmentation and altered the floral phenotypes in transgenic tobacco flowers. Gene expression analysis suggested that NtMYB2 repressed the transcript levels of structural genes involved in anthocyanin biosynthesis pathway, especially the UFGT gene. NtMYB2 gene is expressed in all examined narcissus tissues; the levels of transcription in petals and corona is higher than other tissues and the transcription level at the bud stage was highest. These results show that NtMYB2 is involved in the regulation of anthocyanin biosynthesis pathway and may act as a repressor by down regulating the transcripts of key enzyme genes in Chinese narcissus.
Keywords: Chinese narcissus, R2R3 MYB, anthocyanin repressor, anthocyanin pathway, transcriptional regulation
1. Introduction
Chinese narcissus (Narcissus tazetta L. var. chinensis Roem) is a perennial bulbous plant which belongs to the Amaryllidaceae family. It has very popular attractive ornamental flowers with significant commercial and cultural value. It has been grown in China and East Asia from many years as a traditional and commercial flower. Narcissus has been reported as a triploid species, mostly propagated by a vegetative method, as genetically advanced improvement using traditional cross breeding is complicated [1]. Therefore, only a few cultivars have been used for commercial production in China and many agronomic characters have to be improved such as flower colors [2]. The flower color and pigmentation patterns are the very important consideration for consumer choice. In Chinese narcissus, the flower colors are white and yellow. The compositions of the flavonoid compounds in narcissus cultivars were analyzed, and found to be flavonols, with no anthocyanins detected [3]. Therefore, the investigation on the molecular mechanisms of why narcissus has no anthocyanins biosynthesis could provide benefit for developing new cultivars with diverse color patterns.
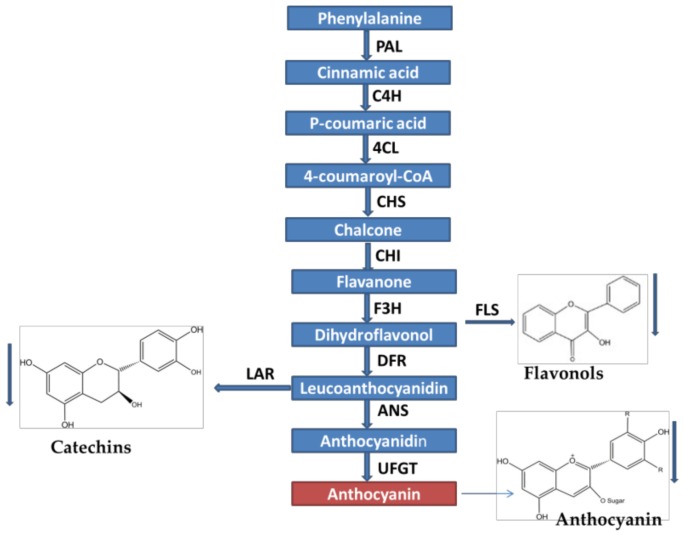
Flavonoids are a group of diverse secondary metabolites which are broadly distributed in plants consisting of the flavones, isoflavones, flavonols, anthocyanins and proanthocyanidins (PAs). Anthocyanins are reported as common natural plant pigments which contribute different colors such as blue, purple and red to the fruits, vegetables and flowers [4]. In plants, key genes encoding enzymes involved in anthocyanin pathway have been identified. Chalcone is considered as the general precursor of flavonoid compounds that comprise of coumaroyl-CoA and malonyl-CoA. Leucoanthocyanidins is derived from coumaroyl-CoA and malonyl-CoA by chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H) and dihydroflavonol reductase (DFR) [5]. Finally, anthocyanins and PAs products are obtained by the activity of anthocyanin synthase (ANS). Anthocyanidin reductase leads to the end product of catechin while epicatechin is derived from leucoanthocyanidin reductase, both are related to proanthocyanin biosynthesis pathway [6,7,8].
Flavonoid biosynthesis is regulated by several transcriptional factors, including R2R3 MYB, basic helix-loop-helix (bHLH) and WD40 repeat (WDR) proteins [9]. These TFs act together as a MYB-bHLH-WDR (MBW) complex to activate the structural genes which are involved in flavonoid biosynthesis pathway. These TFs have been functionally characterized in many plants including maize, Arabidopsis and soybean, strawberry, petunia, apple, grapevine and populous [10,11,12,13,14,15,16].
In plants, many R2R3 MYB transcriptional factors have been identified as activators of flavonoid biosynthesis, for example, PAP1 is involved with the modulation of anthocyanin and TT2 regulates the proanthocyanidin biosynthesis in Arabidopsis [17,18]. In grapevine, VvMYBA1 and VvMYBA2 are involved in the activation of anthocyanin biosynthesis by modulating UFGT gene expression [19]. VvMYBA1 is also involved in the regulation of PA production by stimulating LAR and ANR expression [20]. In apple, MdMYB1 is associated in the biosynthesis of anthocyanin in apple fruit skin [21].
R2R3 MYB proteins are more conserved in the N-terminal DNA binding domains while protein sequence at the C-terminus are divergent and believed to be responsible for several kinds of regulatory functions [22]. Based on specific conserved regions in the C-terminus, R2R3 (MYBs TFs) have been classified into 22 subgroups [23]. Subgroup 4 R2R3-MYB transcriptional factors function as negative regulators in phenylpropanoid and flavonoid biosynthesis. VvMYB4-like from grapevine, PhPH4, PhMYB27 from petunia, MdMYB16 from apple, GmMYB100 from soybean, PtrMYB57, PtrMYB182 from poplar, AtMYB60 from Arabidopsis, SsMYB3 from coleus reduces the accumulation of anthocyanin by suppressing the key anthocyanin biosynthesis pathway genes [16,24,25,26,27,28,29,30,31]. CmMYB1 from chrysanthemum, AtMYB7 from Arabidopsis, FaMYB1 from strawberry reduced the flavonol biosynthesis [10,32,33].
In addition, ectopic expression of AmMYB308 and AmMYB330 from Antirrhinum majus in tobacco plants negatively affects phenolic acids and lignin biosynthesis [34]. Overexpression of ZmMYB31 and ZmMYB42 in Arabidopsis reduced the content of lignin in Arabidopsis [35,36]. AtMYB4 and AtMYB32 are involved in the regulation of flavonol, sinapate esters and lignin biosynthesis [37,38]. In addition to R2R3 MYB repressors in Subgroup 4, single-repeat R3-MYB proteins also have been identified which are involved in the negative regulation of anthocyanin accumulation, including ETC1, AtMYBL2 and MYBx from Arabidopsis and Petunia [39,40]. AtMYBL2 has a C-terminal TLLLFR motif, which also contributes to suppressive activities [41].
A lack of anthocyanins may be due to inhibition of the anthocyanin biosynthesis pathway. Transcriptional factors involved in the downregulation of anthocyanin or flavonoid biosynthesis have not been identified in narcissus. The present investigation on molecular mechanisms controlling flower color in narcissus would provide beneficial breeding programs that look to develop new cultivars with diverse color patterns. In present study, a putative R2R3-MYB gene, NtMYB2 was cloned from Narcissus. It was functionally characterized in detail by bioinformatics analysis, transient co-transformation, qPCR analysis and ectopic expression in tobacco. The results suggest NtMYB2 may be a repressor of anthocyanin biosynthesis. The putative role of NtMYB2 will provides new approaches to understanding the regulator mechanisms of flavonoid biosynthesis in Narcissus spp.
2. Results
2.1. Cloning of NtMYB2
A partial sequence (769 bp) of a R2R3-MYB gene was obtained from transcriptome data and named NtMYB2. The 3′ end of NtMYB2 ORF was isolated by 3′ RACE-PCR with two gene specific primers GP1, GP2 (Table 1) and universal primer (UPM) and a 912 bp fragment was obtained. The full length of NtMYB2 ORF -was 756 bp encoding 251 amino acids residues with theoretical pI 7.95 and molecular weight of 28.3 kDa was obtained.
Table 1.
List of primers used for NtMYB2 isolation and characterization.
| Name | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Note |
|---|---|---|---|
| GSP1 | ATGGGTAAACTATCTTCGGC | UPM (provided by company) | 1st round of 3′-RACE |
| GSP2 | GGAAGGACAGACAACGAGAT | UPM (provided by company) | 2nd round of 3′-RACE |
| NtMYB2 | AATATGGGTAGGTCTCCTTGTTGTG | GATTACAGGACACGCAAAGTAAATCTA | Cloning of full length of ORF |
| pSAK-NtMYB2 | TGGATCCAAAGAATTCAATATGGGTAGGTCTCCTTGTTGTG | TACTCTCGAGAAGCTTGATTACAGGACACGCAAAGTAAATCTA | Vector construction |
| QRT | TGGGTAGGTCTCCTTGTTGTG | AAATTGCCTCTCTTGAGATCGG | qRT-PCR analysis |
2.2. Subcellular Localization
Insilico investigation for subcellular localization (Available online: http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) showed the existence of nuclear localization signal “RPDLKRGNFTEEEDDLIIKLHGM” starting from amino acid at 62th location, suggesting nuclear localization of NtMYB2.
2.3. Sequence Analysis and Phylogenetic Tree
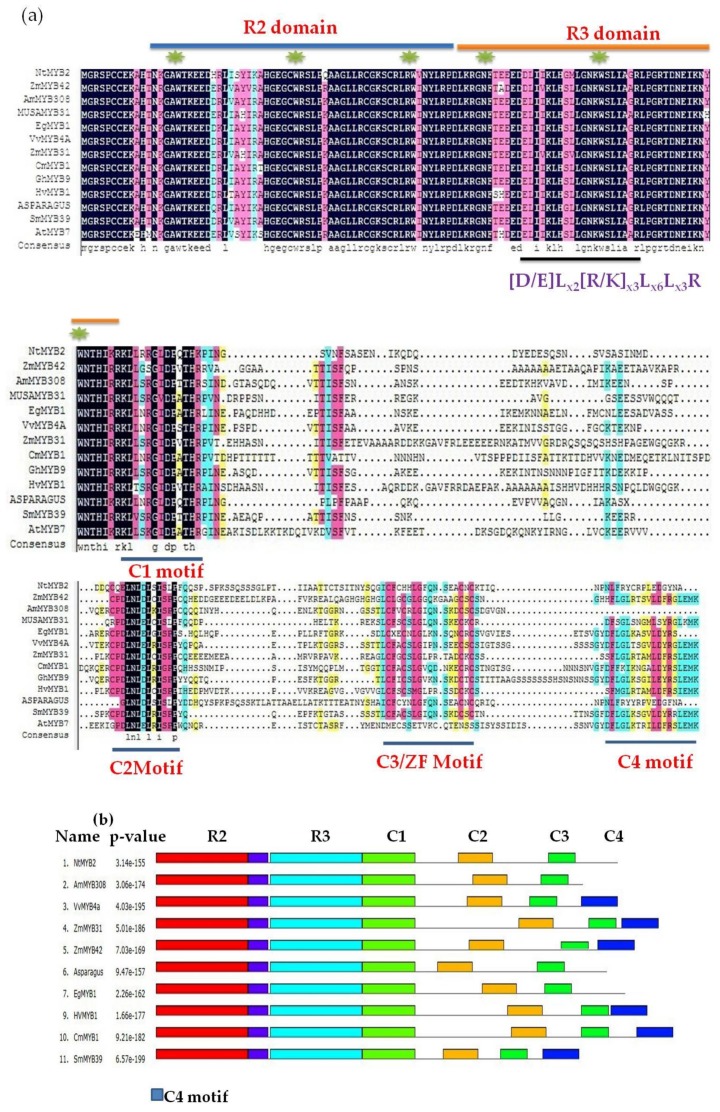
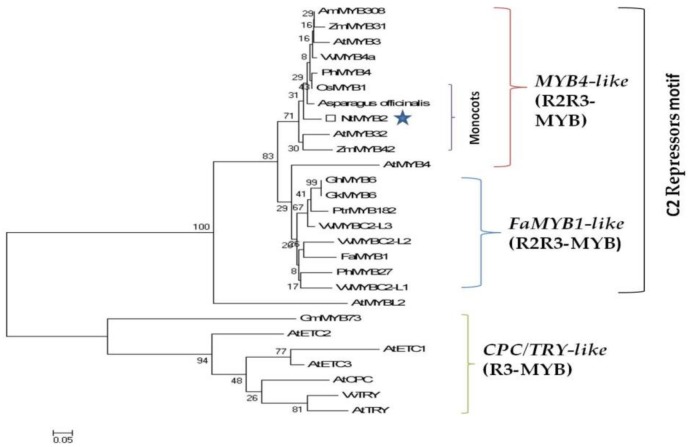
Sequence analysis showed that NtMYB2 contained conserved R2R3 domain at the N-terminal end. In addition, NtMYB2 protein has the conserved motif of [D/E]Lx2[R/K]x3Lx6Lx3R in the R3 domain accountable for interaction with R like bHLH protein. NtMYB2 protein also contained the conserved motif of L17x3L21x2R24 in the R3 domain (Figure 1a). Comparisons of NtMYB2 with other R2R3 MYB transcriptional factors indicated that NtMYB2 show high homology with AoMYB (73.05%, similarity) in asparagus, MUSAMYB31 (56.75% similarity) in banana, VvMYB4a (55.64%, similarity) in grapevine AmMYB308 (55.38%, similarity) in antirrhinum and PhMYB4 (54.75%, similarity) in Petunia. These genes belong to the subgroup 4 of R2R3 MYB transcriptional factors which consist of four conserved motifs at C-terminal region. These conserved motif are called C1 motif (KLIsrGIDPxT/SHRxI/L), C2 motif (pdLNLD/ELxiG/S), C3 or ZnF (zink-finger) like motif (CX1-2CX7-12CX2C) and C4 motif (FLGLx4-7V/LLD/GF/YR/Sx1LEMK) [12,42]. C2 repressor motif associated with EAR repressor domain [37,43]. The analysis of protein sequence revealed that NtMYB2 shows the LxLxL type EAR motif with others subgroup 4 R2R3 MYB transcriptional factors such as VvMYB4a and VvMYB4b, AtMYB7 and AtMYB32 which are involved in the suppression of flavonoids and lignin biosynthesis pathway [43] (Figure 1a). However, NtMYB2 lacks C4 motif unlike other members of subgroup 4 R2R3 MYB proteins (Figure 1b,c). The phylogenetic analysis indicated that R2R3 MYB TFs that consist of C2 repressor motif were divided into two clades such as AtMYB4-like and FaMYB1-like. Single- repeat R3-MYB protein containing repressor CPC/TRY like clade was separated. NtMYB2 was grouped within MYB4- like R2R3 MYB TFs clade (Figure 2), which is concerned for transcriptional repression including PhMYB4 from Petunia, VvMYB4a, from the grapevine, AmMYB308 from Antirrhinum, AtMYB32. AtMYB4, AtMYB3 from Arabidopsis, ZmMYB31 and ZmMYB42 from Maize, AoMYB from asparagus, OsMYB1 from rice. NtMYB2 also has the close relationship with other MYB TFs from monocot plants such as rice, maize and asparagus (Figure 2). On the basis of sequence alignments and phylogenetic analysis, NtMYB2 may act as a transcriptional repressor of flavonoids biosynthesis pathway.
Figure 1.
(a) Multi -alignment of the amino acid sequence of NtMYB2 with other repressor R2R3 MYB of subgroup 4. The R2 and R3 binding domain are indicated by blue and gray colored lines respectively on top. A black underline within the R3 domain shows the motif (consensus sequence [DE]Lx2[RK]x3Lx6Lx3R) interacting with the bHLH protein. The conserved amino acids, tryptophan (W) and phenylalanine (F) are shown by the green star. The C1 and C2 functional motifs are underlined in the blue line. The C3/ZF like and C4 motif are also underlined with blue color. Sequences utilized for alignment are: Narcissus tazetta L. var. chinensis NtMYB2 (KY860527.1). Antirrhinum majus AmMYB308 (P81393), Vitis vinifera VvMYB4a (XP_002278222), Gossypium hirsutum GhMYB9 (AAK19619), Hordeum vulgare HvMYB1 (P20026), Chrysanthemum morifolium CmMYB1 (AEO27497), Arabidopsis thaliana AtMYB7 (NP_179263), Salvia miltiorrhiza SmMYB39 (KC213793), Eucalyptus gunnii EgMYB1 (CAE09058.1), Asparagus officinalis (XP_020246745) and Zea mays ZmMYB42 (NP_001106009.2), ZmMYB31 (NP_001105949.2), (b) MEME motif logo with p–value, (c) MEME motif logos with bit scores.
Figure 2.
Phylogenetic tree analysis of MYB repressors based on amino acid sequences consisting R2R3 domain or R3 domains. Bar 0.05 substitutions per site. The accession numbers of MYB proteins used in tree building are: Zea mays ZmMYB42 (NP_001106009.2), Zea mays ZmMYB31 (NP_195225.1), Vitis vinifera VvMYB4a (ABL61515.1), Arabidopsis thaliana AtMYB32 (NP_195225.1), AtMYB3 (NP_564176.2), AtMYB4 (AAC83582.1), AtMYBL2 (AEE35154), Petunia hybrida PhMYB4 (ADX33331.1), PhMYB27 (AHX24372.1), Fragaria ananassa FaMYB1 (AAK84064.1), Vitis vinifera VvMYBC2-L1 (ABW34393.1), VvMYBC2-L2 (ACX50288.2), VvMYBC2-L3 (AIP98385.1), Antirrhinum majus AmMYB308 (P81393), Oryza sativa OsMYB1 (AK104457), Gossypium hirsutum GhMYB6 (AAN28286), Glycine max GmMYB73 (ABH02868), VvTRY (ABW34395), Arabidopsis thaliana AtTRY (Q8GV05), CPC (BAA21917), ETC1(NP_171645), (ETC2 AEC08385), ETC3 (AEE81974), Gossypioides kirkii GkMYB6 (AAN28289), Populus tremuloides PtrMYB182 (Potri.004G088100).
2.4. The Expression Pattern of NtMYB2 in Chinese Narcissus
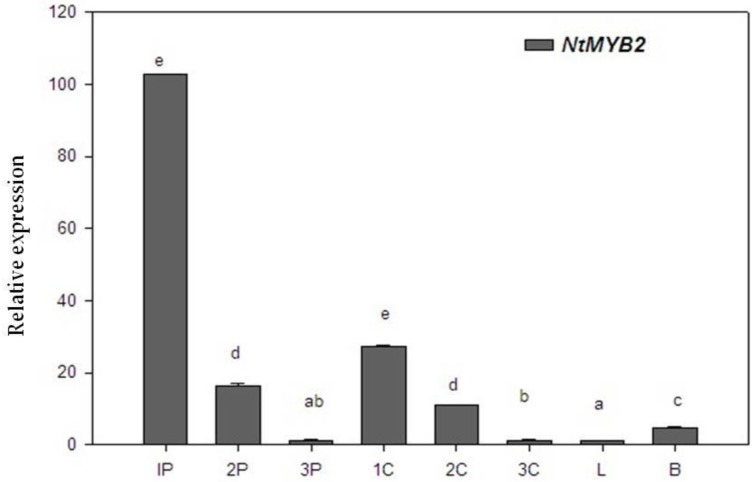
The expression profile of NtMYB2 was examined in different tissues of Chinese narcissus. Quantitative RT-PCR results indicated that NtMYB2 was expressed in all types of tissues examined. The expressions in petals and corona was significantly higher than in basal plates and leaves. Flower petals at the bud stage showed a relatively higher expression as compared to petals at half opened and full opened stage. The similar trend of expression levels was observed in corona (Figure 3).
Figure 3.
The expression pattern of NtMYB2 in Chinese Narcissus.
1P, 2P and 3P indicate the different developmental stages of petals at bud stage, half opened stage and full stage respectively. 1C, 2C and 3C denote for corona different developmental stages as bud stage, half opened stage and full opened stage. L and B represent for leaves and basal plates respectively. The bars indicate the standard error of three biological replicates. Letter represents a significant difference at the level of p < 0.05 using LSD statistical analysis.
2.5. Transient Expression of NtMYB2 in Tobacco
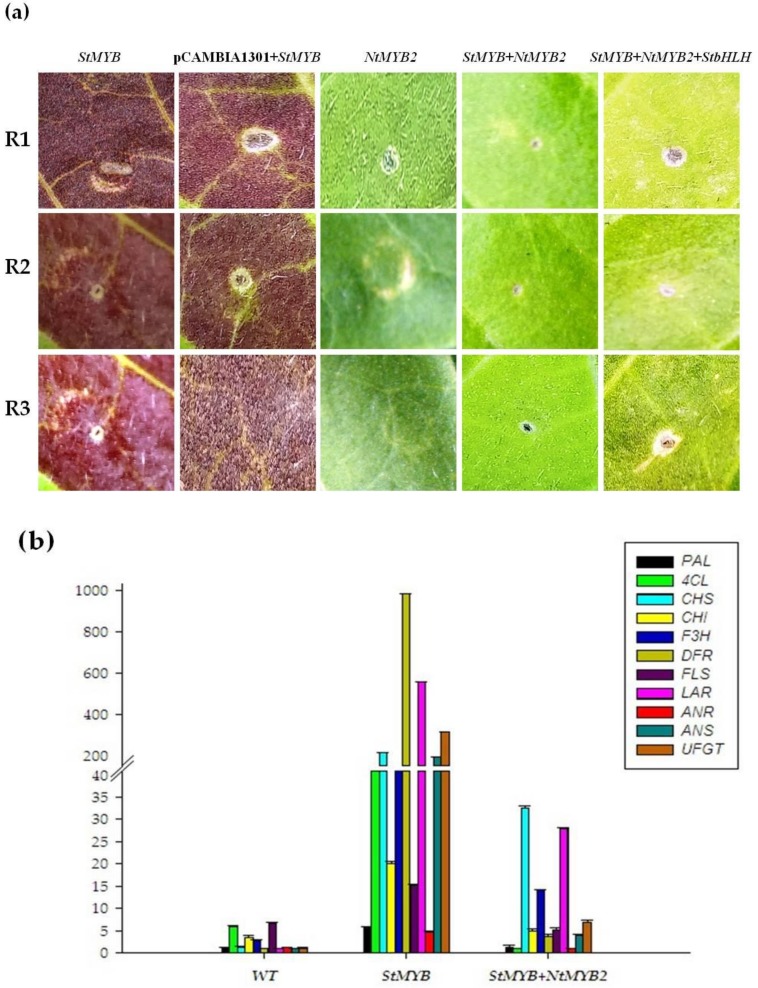
In order to functionally characterize NtMYB2, transient assays were carried out in Nicotiana tabacum through Agrobacterium infiltration method as previously report [44]. Agroinfiltration with different treatments were performed. Visible red pigmentation was observed with syringe-infiltrated StMYB (potato StAN1, positive control) [45]. Interestingly, red pigmentation disappeared in tobacco leaves co-infiltrated with StMYB and NtMYB2. No red pigmentation or anthocyanin patch was observed with NtMYB2 infiltrated by itself. There was also no observed induction of anthocyanin pigmentation in leaves co-infiltrated with 3 genes of NtMYB2, StMYB and StbHLH (Figure 4a).
Figure 4.
Transient transformation of Nicotiana tabacum leaves. (a) Color changes in agroinfiltrated leaves of Nicotiana tabacum. R1, R2 and R3 indicated three replicates. (b) Expression analysis of flavonoid biosynthesis pathway genes in agroinfiltrated leaves of Nicotiana tabacum by qPCR.
In order to verify the effect of NtMYB2 on the expression levels of the structural genes which are involved in the biosynthesis of flavonoid pathway in syringe infiltrated tobacco leaves, the expression of PAL, 4CL, CHS, CHI, F3H, FLS, DFR, ANR, LAR, ANS and UFGT were assessed by qRT-PCR. The expression levels of all these structural key genes except for PAL and ANR were significantly induced in the injected leaves with single StMYB and were significantly reduced in co-infiltrated leaves with NtMYB2 and StMYB (Figure 4b).
2.6. Stable Expression of NtMYB2 in Tobacco
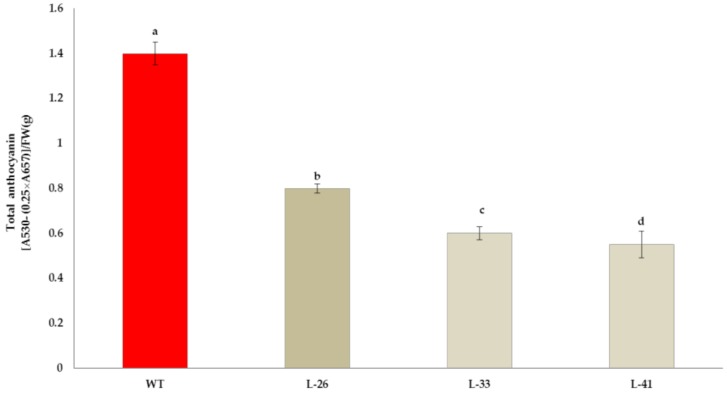
NtMYB2 under the expression of the 35S promoter was transformed into tobacco. The existence of introduced NtMYB2 was confirmed by PCR (Figure S1). Transgenic tobacco plants didn’t show any growth changes. The transgenic plant flowers showed visible phenotypic changes in petal pigmentation patterns; from light pink to almost white. In particular, the transgenic tobacco Line-41 (L-41) had a significant change of flower color. The L-26 showed very pale pink flowers with pale red veins and the edge of petal (Figure 5a). In addition to the flowers color, the length of pistil of transgenic flowers was elongated, compared to that of the WT flower. Furthermore, the flowers size was smaller as compared to the control. The stigma color of transgenic lines turned yellow and a green to slightly green color was noticed in the sepals of transgenic flowers (Figure 5b). The transgenic tobacco flowers at early developmental stage showed no pigmentation, while in wild type tobacco flowers visible pigmentation were observed (Figure 5c). These results showed that ectopic overexpression of NtMYB2 in tobacco leads to decrease the accumulation of anthocyanin to alter the flower color in transgenic flowers. Quantitative analysis confirmed the reduction of anthocyanin levels in the flowers of transgenic plants (Figure 6).
Figure 5.
Floral phenotypes of transgenic tobacco plants. Floral phenotypes of transgenic tobacco plants over expressing NtMYB2 gene. WT, wild type tabacco; L-26, L-33 and L-41, three transgenic tobacco lines. (a) Comparison of flower color in transgenic tobacco flowers and WT; (b) Comparison of flower size and pistil length in WT and transgenic tobacco; (c) Comparison of flower color of in transgenic tobacco flowers and WT at different flower stages. 1-bud stage, 2-half opened stage, 3-full open stage.
Figure 6.
Quantification of total anthocyanin. Quantification of total anthocyanin in flowers of NtMYB2 tobacco lines (L-26, L-33) and wild type (WT). The bars indicate the standard error of three biological replicates. Letter represents a significant difference at the level of p < 0.05 using LSD statistical analysis.
2.7. Overexpression of NtMYB2 in Tobacco Influences the Expression Levels of Flavonoid Biosynthetic Pathway Genes
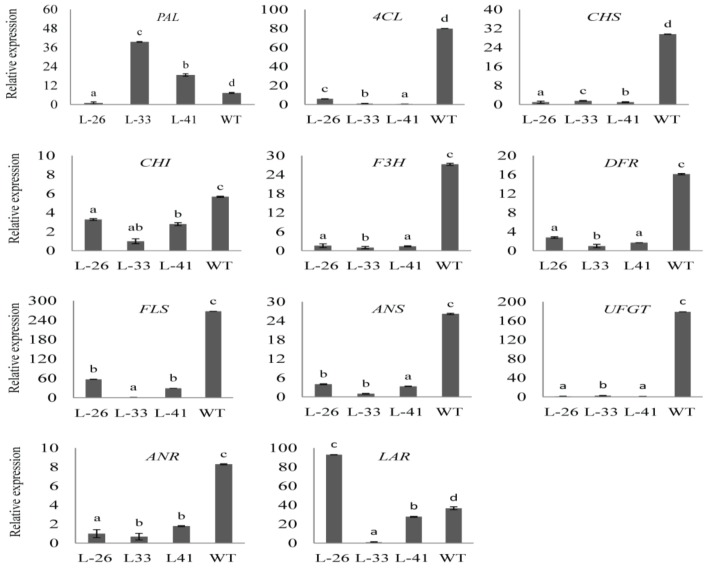
The effect of overexpression of NtMYB2 on the transcript levels of flavonoid biosynthetic pathway genes in transgenic tobacco flowers was assessed by qRT-PCR assay. The analysis of qPCR results indicated that tobacco anthocyanin biosynthetic genes including CHS, CHI, F3H, DFR, ANS, and UFGT were significantly down-regulated in NtMYB2-26, NtMYB2-33 and NtMYB2-41 overexpression lines compared to the control tobacco flowers (Figure 7). The transcript levels of FLS and ANR were also significantly down-regulated in three overexpression lines. The LAR expression was down-regulated in the NtMYB2-33 and NtMYB-41 overexpression lines but was up regulated in the NtMYB2-26 overexpression line as compared to the control. The transcript level of PAL which was involved in general phenylpropanoid pathway was up-regulated in transgenic tobacco Lines 33 and 41. These results showed that ectopic expression of NtMYB2 transcriptional factors significantly down-regulates the expression levels of main flavonoid biosynthetic genes in tobacco.
Figure 7.
Expression analysis of flavonoid biosynthesis pathway genes. Expression analysis of flavonoid biosynthesis pathway genes in flowers of NtMYB2 transgenic tobacco. L-26, L-33, L-41 means three transgenic line. The bars indicate the standard error of three biological replicates. Letters (a, b, c and d) represent the significant difference from wild type at the level of p < 0.05 using LSD test.
3. Discussion
R2R3-MYB transcriptional factors are a large gene-family in plants and have been reported to play key roles in the regulation of secondary metabolites. In the current study, we identified a novel R2 R3 MYB transcriptional factor from narcissus, termed as NtMYB2, which may repress anthocyanin biosynthesis in transgenic tobacco as well as in narcissus.
3.1. Bioinformatics Analysis Showed NtMYB2 Was a Transcriptional Factor
Flavonoid biosynthesis is regulated by MBW complexes, among which MYB proteins that consist of activators and repressors. The amino acid sequence of NtMYB2 showed high homology with other R2R3 -MYB proteins from plants, such as AtMYB7, AmMYB308, ZmMYB42, VvMYB4 and CmMYB1 [25,32,33,34,46]. These are implicated in the negative regulation of flavonoid biosynthesis. Further protein motif analysis indicated that NtMYB2 contained typically three conserved motifs as C1, C2 and C3 motif at C-terminal. Subgroup 4 R2R3-MYB was initially defined by the existence of C1, C2, and C3 motifs [12]. Members of subgroup 4 R2R3-MYB proteins have C2 repressor motif which is responsible for repression activity in flavonoid biosynthesis pathway genes [47,48,49,50]. The results of NtMYB2 protein analysis showed the sequence to be similar to PtrMYB57 and VvMYB4- like, both have C2 motif and are involved in the suppression of anthocyanin accumulation [25,28]. The presence of a conserved C2 motif sequence “pdLNLD/ELxiG/S” suggests that NtMYB2 may be act as a repressor of flavonoid biosynthesis. However, NtMYB2 lacks the C4 (dFLGL and LDF/YRxLEMK) motif and showed divergence in the C-terminus from other members of subgroup 4. The C4 motif is also absent in snapdragon AmMYB308, which is known as a flavonoid repressor [34]. In addition, we have found the existence of bHLH interaction residues that are present in R3 repeat of NtMYB2. Theses residues indicate the conserved region ([DE]Lx2[RK]x3Lx6Lx3R) interacting with the bHLH proteins as described in Arabidopsis [51]. NtMYB2 protein also contained the conserved motif of L17x3L21x2R24 in the R3 domain and supposed to stabilize the interaction of bHLH-MYB proteins [16]. Phylogenetic analysis revealed that NtMYB2 was grouped in MYB4-like clade with VvMYB4a, PhMYB4, and CmMYB1. In Subgroup 4, AtMYB4, AtMYB3 from Arabidopsis, VvMYB4a and VvMYB4b from grapevine and PtrMYB182, PtrMYB57 from poplar are associated with down-regulation of the anthocyanin and PA biosynthesis [12,37]. AtMYB7 in subgroup 4 acts as a repressor of flavonol biosynthesis [33]. The results of bioinformatics analysis showed that NtMYB2’s function may be similar with other MYBs in subgroup 4, acting as a transcriptional repressor. R2R3 MYB repressors may have two kinds, one of which functioned on MBW complexes such as FaMYB1 and the other directly binds on target genes such as AtMYB4 [27]. NtMYB2 is grouped with MYB4-like clade indicates this gene may directly bind on target genes consistent with the results of MdMYB16 from apple which directly binds on ANS and UFGT that inhibit the anthocyanin biosynthesis [27].
Effective accumulation of anthocyanin biosynthesis by the MdMYB10 is dependent on the co-expression of a specific bHLH protein. In Petunia, PhAn1, a bHLH transcriptional factor directly induces the expression of the biosynthetic gene DFR and is controlled by MYB factor PhAn2 [52]. This difference may be due to the characteristic of the exogenous MYBs and the interactions with endogenous tobacco bHLH factors. The previous study showed that the PamMYBA.1 and PamMYBA.2 regulate the anthocyanin biosynthesis in citrus and tobacco without requiring any further cofactors. While PamMYBA.3 required the additional cofactors (bHLH) for the induction of anthocyanin accumulation [53]. These results indicate that some MYB transcriptional factors need co-expression of the bHLH simultaneously to perform functional activates, while other MYB transcriptional factors alone are sufficient to induce or reduce the accumulation of anthocyanin.
3.2. NtMYB2 Reduced the Pigmentation in Tobacco Leaves with Transient Assay
StMYB is from potato, which is involved in positive regulation of DFR gene [45]. When StMYB was agro-infiltrated in tobacco leaves alone, it induced obvious red pigmentation on leaves (Figure 4). A significant loss of red pigmentation was observed in co-injected leaves with StMYB and NtMYB2 indicating NtMYB2 represses anthocyanin biosynthesis. qPCR analysis showed that in injected leaves co-infiltrated with NtMYB2 and StMYB, the transcript levels of key anthocyanin biosynthesis structural genes (CHS, CHI, F3H, DFR, ANS and UFGT) were significantly down-regulated. Previous study has shown that poplar PtrMYB57 reduced the transcript level of flavonoid biosynthetic genes including 4CL, CHS, DFR, ANS, ANR and LAR leading to the repressing of anthocyanins and PAs in tobacco leaves [28]. The evidence continued repression in the presence of StbHLH co-infiltration indicated that NtMYB2 may not be competing with for StbHLH with StMYB and may play a role by directly binding with the promoters of structural genes.
3.3. NtMYB2 Reduces the Flower Color of Transgenic Tobacco
Ectopic expression of NtMYB2 in tobacco repressed the pigmentation in petals of transgenic tobacco flowers. In addition, a change in stigma color was observed in some overexpression lines. Phenotypic changes were only observed in flowers, not in vegetative parts. This present study agrees with the results of VvMYB4-like transcriptional regulators [25]. Overexpression of VvMYB4-Like in tobacco lead to the loss of pigmentation in flowers due to the reduction in anthocyanin accumulation [25]. Lower anthocyanin levels in NtMYB2 overexpression lines correlated with the observed loss of pigmentation. PtrMYB57 (Subgroup 4) that reduced the contents of anthocyanin in transgenic poplar plants than wild type [28]. Overexpression of Coleus SsMYB3 significantly reduced the anthocyanin contents in all transgenic tobacco plants and also strongly decreased the color pigmentation of transgenic flowers than those of wild type [31]. In grape, VvMYBPA1 showed significantly lower amounts of anthocyanin and reduction of petal pigmentation in transgenic tobacco flowers as compared to flowers of wild type plants [54]. These support the hypothesis that NtMYB2 is an anthocyanin repressor.
In addition to a loss of flower pigmentation, differences in other morphological characters, such as smaller shorter flower were seen in NtMYB2 lines compared to wild type. Our findings are similar with previous studies, when AmMYB308 from Antirrhinum is over expressed in transgenic tobacco, flower size and the level of anthocyanin are reduced as compared to wild type [34]. Furthermore, the length of pistils of NtMYB2 overexpression lines were elongated compared to wild type, with stigmas being above the anther. Similar results have been reported for transgenic tobacco lines expressing the apple MdMYB3 gene [55]. How these MYB repressors induce other morphological character changes needs further investigation.
3.4. NtMYB2 Is Involved in Regulation of Anthocyanin/Flavonoid Biosynhesis Patway Genes in Tobacco
The change of anthocyanin biosynthesis in transgenic tobacco flowers implied that the expressions of flavonoid biosynthetic genes were affected by NtMYB2 overexpression (Figure 8). In transgenic tobacco flowers, all of anthocyanin pathway genes were significantly down-regulated, including CHS, CHI, F3H, DFR, ANS and UFGT. VvMYB4-like gene negatively regulates ANR and FLS gens in transgenic tobacco flowers [25]. Our results showed that NtMYB2 down-regulates both early and late key anthocyanin biosynthesis pathway genes. In addition, NtMYB2 down-regulated the proanthocyanidin (PA) biosynthetic pathway gene NtANR in tobacco flowers, while NtLAR showed variable expression levels in transgenic tobacco flowers. These results suggest NtMYB2 may negatively regulate the catechin biosynthesis pathway. Moreover, the expression level of NtFLS, involved in flavonol biosynthesis, also decreased in NtMYB2-overexpression tobacco flowers.
Figure 8.
General flavonoid biosynthesis pathway. Arrow indicating the decreasing flavonol, catechins and anthocyanin biosynthesis pathway in transgenic flower of tobacco lines.
The transcripts level of UFGT was significantly reduced in all NtMYB2 overexpression tobacco lines as compared to the control plants. In grapevine VvMYB4-like and VvMYB4A reduced the anthocyanin by down regulating the DFR, ANS and UFGT genes respectively [25,56]. Ectopic expression of SsMYB3 in tobacco significantly down-regulated the transcripts level of NtUFGT that leads to reduce the accumulation of anthocyanin [31]. Over expression of FaMYB1 in tobacco reduce the contents of anthocyanin by reducing the expression level of UFGT [10].
3.5. Expression Profiles of NtMYB2 in Narcissus
The results of qPCR analysis showed that NtMYB2 has expression in flowers, leaves and basal plates of narcissus. The transcript level of NtMYB2 was much higher in petals and corona as compared to vegetative tissues, indicating that NtMYB2 may play a repressive role in downregulating anthocyanin levels in flowers. This may be a key reason why no anthocyanin accumulates in flowers of narcissus. In addition, the expression of NtMYB2 is affected by different developmental stages of flowers. In ornamental peach flowers, PpMYB17 and PpMYB20 repressors were significantly expressed at bud stages, but remarkably decreased at full open stages and showed decreasing expression pattern during flower development [57].
4. Materials and Methods
4.1. Plant Materials
The flowers, basal plate and leaf of Chinese narcissus were used for the RNA extraction and further experiments. The seeds of Nicotiana tabacum plants were grown in pots contain compost mix (1:1 compost: perlite and 3:1 compost: perlite for seed and plants, respectively) and maintained in the glasshouse. The young leaves of tobacco were used for transient expression. The flowers of transgenic tobacco were used for the qPCR analysis and also used for extraction of total anthocyanin and total phenolics contents.
4.2. RNA Extraction
Total RNA was extracted using the Universal Plant total RNA extraction Kit (Bioteke Corporation, Beijing, China), following the manufacturer’s instructions. RNase free DNase I treatment (Invitrogen, Waltham, MA, USA) was performed to eliminate contaminant and residual genomic DNA. The integrity of extracted RNA was checked on agarose gels stained with ethidium bromide. The RNA concentration was calculated in an ND-1000 UV spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). The cDNA first strand was synthesized from 1 µg total RNA using SMARTer®RACE 5′/3′ kit (Clontech Laboratories, Inc., Palo Alto, CA, USA).
4.3. 3′RACE of NtMYB2
The full-length cDNA of NtMYB2 was obtained by extending the 3′end using SMARTer®RACE 5′/3′ kit (Clontech Laboratories, Inc.). The 3′end sequence was obtained by two rounds of PCR amplifications with gene specific primers and a universal primer (provided in the kit). Nested PCR involved the nested universal primer and the second gene specific primer. The first gene specific-primer was NtMYB2-GSPF1 5′-ATGGGTAAACTATCTTCGGC-3′ and the second gene-specific primer was NtMYB2-GSPF2 5′-GGAAGGACAGACAACGAGAT-3′. The total 50 µL PCR mixture (Ex Taq, Takara, Dalian, China) contained 5 µL buffer, 4 µL dNTPs, 1 µL DNA polymerase, 2 µL first strand cDNA, 0.5 µM each primer and 36 µL double distilled water. The PCR reaction was carried out as follows: pre-heating at 95 °C for 5 min, 34 cycles at 95 °C for 30 s, the annealing temperature was 57 °C for 1 min and 72 °C for 1 min, then an extension at 72 °C for 7 min. The PCR amplified fragment was purified and then cloned into pMD 18-T vector (Takara) and colonies were randomly selected and sent to the company for the sequencing. A pair of gene- specific primers designed from the putative 5′ and 3′ UTR sequences and were used to amplify the complete NtMYB2 open reading frame (ORF). The sequence of forward and reverse primers are NtMYB2 Forward: 5′-AATATGGGTAGGTCTCCTTGTTGTG-3′ and NtMYB2 reverse: 5GATTACAGGACACGCAAA GTAAATCTA-3′ respectively. The PCR conditions were same as above. The amplified PCR product was purified using a gel extraction Kit (Omega, Norcross, GA, USA). The PCR product cloned into pMD18-T (Vector Takara, Dalian, China). The cloning product was transformed into Escherichia coli which were cultured on LB plate containing ampicillin at a final concentration of 50 mg/mL. Resistant colonies were confirmed by PCR using gene specific primers and M13 Primers and sent to the company (Biosune, Shangai, China) for the sequencing on both strands.
4.4. Expression Vector Construction
The ORF of NtMYb2 was amplified and cloned into a plant expression vector pSAK277 containing the CaMV 35S promoter using In-Fusion®HD cloning kit (Clontech® Laboratories, Inc., Palo Alto, CA, USA). The complete coding sequence of NtMYB2 was amplified using a gene- specific forward primer with an EcoRI restriction site (TGGATCCAAAGAATTCAATATGGGTAGGTCTCCTTGT TGTG) and reverse gene-specific with primer with a HindIII restriction site (TACTCTCGAGAAGCTT GATTACAGGACACGCAAAGTAAATCTA). This overexpression constructed vector pSAK277-NtMYB2 was introduced into the Agrobacterium tumefaciens GV3101 stain using the freeze-thaw method.
4.5. Sequence Analysis of NtMYB2
The theoretical NtMYB2 coding sequence translation, isoelectric point (pI) and molecular weight of the NtMYB2 was found by using expasy server (www.ExPASy.org). R2R3 MYB transcriptional factors (TFs) which are close related to NtMYB2 used for building a phylogenetic tree. Phylogenetic trees were built using the neighbor joining tree method with partial deletion gap treatment. The bootstrap analysis with 1000 replicates was used to evaluate the tree nodes through MEGA5 software with default parameters. The sequences of amino acid of R2R3 MYB in other different plants were obtained from NCBI (Available online: www.ncbi.nlm.nih.gov/BLAST/) and TAIR database (Available online: https://www.arabidopsis.org). NtMYB2 sequence was aligned with other deduced the amino acid sequence of other species using DNAMAN7.0 (Lynnon Corporation, San Ramon, CA, USA).
4.6. The Expression Pattern of NtMYB2 in Different Tissues of Chinese Narcissus
The NtMYB2 gene expression in various tissues (Leaves, basal plate, flower petals and corona at different developmental stages) of Chinese narcissus was examined by real-time qRT-PCR. Total RNA was extracted using an Omega Total RNA Kit (Omega) from different tissues of Chinese Narcissus by following the instructions of plant extraction kit. The genomic DNA was removed from total RNA and cDNAs weresynthesized with the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Dalian, China). The synthesized cDNAs was diluted and used as template for qRT-PCR analysis. Quantitative Real time PCR was performed with a Light cyclar®480 real time PCR machine (Roche Diagnostics, Indianapolis, IN, USA) using the SYBR Green PCR master mix kit (Takara). The qRT-PCR condition was as follows: 95 °C for 30 s, 40 cycles of 95 °C for 3 s and 60 °C for 30 s and a default melt curve program. The relative expression level was estimated with the NtActin gene (Table S1) used as an internal standard. The comparative Ct method was carried out to estimate the gene expression level [58]. Three biological replicates and same technical replicate for each sample were carried out. Gene specific primers (Table 1) were used in Quantitative real time PCR analysis.
4.7. Transient Assays of NtMYB2 in Tobacco Leaves
The seeds of Nicotiana tabacum plants were grown in pots contain compost mix (1:1 compost: perlite and 3:1 compost: perlite for seed and plants respectively) and maintained in the glasshouse. The constructed pSAK277-NtMYB2, pSAK277-StMYB, pSAK277-StbHLH and pCAMBIA1301 vector was transformed into Agrobacterium tumifaciens stain GV3101 by electroporation. Potato StMYB (StAN1) and StbHLH were provided by Liu [45]. The Agrobacterium colonies were cultured in YEB media supplement with appropriate antibiotics and incubated at 28 °C at 190 rpm for overnight. The O.D.600 was adjusted at 0.6–0.8 and checked by Spectrophotometer. Agrobacterium culture was centrifuged for 3 min at 3000 rpm in a microfuge tube and supernatant were removed. The obtained pellet was resuspended in agroinfiltration medium (MS salts 4.32 mg/L; 10 mM MES; 20 g/L sucrose; 200 mM acetosyringone) and left for 2–4 h at room temperature. Agroinfiltration suspension was injected into abaxial side of the young leaves of Nicotiana tabacum. NtMYB2, StMYB, StMYB + NtMYB2 and StMYB + StbHLH + NtMYB2 were used as treatments StMYB + pCAMBIA1301 as control. Three biological replications are carried out in this transient assay. Agrobacterium tumefaciens infiltrated plants were keep at 25 °C in a growth chamber with a 10 h light and 14 h dark photoperiod for 4–6 days. After 5–6 days of agroinfiltration injection, change in color were observed in injected tobacco leaves. The photographs of infiltrated leaves were taken and the samples of injected leaves were collected and stored at −80 °C for further analysis and experiment.
4.8. Stable Transformation of Tobacco
Transformation of Nicotiana tabacum was carried out using the leaf disc method as previously described [59]. The positively transformed tobacco shoots were screened on MS medium using 100 mg/L kanamycin and 350 mg/L carbenicillin. Transgenic tobacco plants were confirmed by PCR assay from genomic DNA with the help of specific primers for NtMYB4. The generated kanamycin resistant tobacco plants were then transferred to a mixture of soil (1:1:1 of vermiculite, perlite and peat moss) for their adaptation under normal condition.
4.9. Extraction and Measurement of Total Anthocyanins and Phenolics
Total anthocyanin content was extracted and measured in overexpression NtMYB2 tobacco flowers according to the modified method reported previously [60]. The fresh flowers (0.5 g) were ground in liquid nitrogen and transferred into clean tube containing 6ml of methanol (1% HCl) and incubated overnight at 4 °C. Consequently, the tissues extracts were homogenized and centrifuged at 13,000 rpm (10 min, 4 °C). The absorbance of supernatants was determined at 530 nm and 657 nm. Total anthocyanin concentration was calculated as Q = (A530 − 0.25 × A657) × FW−l.
4.10. Statistical Analysis
Statistical analysis was carried out by one-way ANOVA. LSD values were calculated using p = 0.05 and used to compare treatment means.
5. Conclusions
In this study, a novel R2R3 MYB transcriptional factor, NtMYB2, with the potential to down-regulate the anthocyanin, flavonol and proanthocyanidin biosynthesis pathway, was identified. This MYB belongs to subgroup 4 R2R3 MYBs, which act as repressors of transcripts of key enzyme genes involved in flavonoid biosynthesis. Transcripts of NtMYB2 are detected in flowers, leaves and basal plates in Chinese narcissus. Transcripts of NtMYB2 are higher in flowers than other organs. Transient expression of NtMYB2 in tobacco leaves and ectopic expression of NtMYB2 in tobacco indicated that this MYB represses anthocyanin biosynthesis. The results from this study will be helpful to understand the regulating mechanism of anthocyanin and flavonoid biosynthesis in Narcissus and provide direction to improve the flower color and pattern in this species.
Acknowledgments
This work was supported by Fujian Natural Science Fund, China (No. 2016J01109) and Fujian Agriculture and Forestry University International Scientific and Technological Exchange and Cooperation Project, China (Kxb16013A). We thank Yuhui Liu from Gansu Agricultural University kindly providing StMYB and StbHLH genes.
Supplementary Materials
Supplementary materials can be found at.
Author Contributions
Lihui Zeng and Muhammad Anwar conceived the study. Muhammad Anwar, Lihui Zeng and Andrew C. Allan wrote the manuscript; Muhammad Anwar performed Lab, Field experiments and statistical analysis. Guiqing Wang, Jiacheng Wu, Saquib Waheed, Andrew C. Allan and Lihui Zeng discussed the results and commented on the main manuscript.
Conflicts of Interest
The authors declare no competing interests.
Footnotes
Sample Availability: Samples of the compounds are available from the authors.
References
- 1.Wu J., Chen L., Gu L., Wang Y., Tian H. Embryological studies on Narcissus tazetta var. Chinensis. J. Xiamen Univ. Nat. Sci. 2004;44:112–115. [Google Scholar]
- 2.Lu G., Zou Q., Guo D., Zhuang X., Yu X., Xiang X., Cao J. Agrobacterium tumefaciens-mediated transformation of Narcissus tazzeta var. Chinensis. Plant Cell Rep. 2007;26:1585–1593. doi: 10.1007/s00299-007-0382-z. [DOI] [PubMed] [Google Scholar]
- 3.Li X., Lu M., Tang D., Shi Y. Composition of carotenoids and flavonoids in narcissus cultivars and their relationship with flower color. PLoS ONE. 2015;10:e0142074. doi: 10.1371/journal.pone.0142074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies K.M., Albert N.W., Schwinn K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 2012;39:619–638. doi: 10.1071/FP12195. [DOI] [PubMed] [Google Scholar]
- 5.Marles M.S., Gruber M.Y., Scoles G.J., Muir A.D. Pigmentation in the developing seed coat and seedling leaves of brassica carinata is controlled at the dihydroflavonol reductase locus. Phytochemistry. 2003;62:663–672. doi: 10.1016/S0031-9422(02)00488-0. [DOI] [PubMed] [Google Scholar]
- 6.Abrahams S., Lee E., Walker A.R., Tanner G.J., Larkin P.J., Ashton A.R. The arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 2003;35:624–636. doi: 10.1046/j.1365-313X.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- 7.Xie D.-Y., Sharma S.B., Paiva N.L., Ferreira D., Dixon R.A. Role of anthocyanidin reductase, encoded by banyuls in plant flavonoid biosynthesis. Science. 2003;299:396–399. doi: 10.1126/science.1078540. [DOI] [PubMed] [Google Scholar]
- 8.Yuan L., Wang L., Han Z., Jiang Y., Zhao L., Liu H., Yang L., Luo K. Molecular cloning and characterization of PtrLAR3, a gene encoding leucoanthocyanidin reductase from populus trichocarpa, and its constitutive expression enhances fungal resistance in transgenic plants. J. Exp. Bot. 2012;63:2513–2524. doi: 10.1093/jxb/err425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koes R., Verweij W., Quattrocchio F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Aharoni A., De Vos C., Wein M., Sun Z., Greco R., Kroon A., Mol J.N., O’connell A.P. The strawberry famyb1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28:319–332. doi: 10.1046/j.1365-313X.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- 11.Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell Online. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavallini E., Matus J.T., Finezzo L., Zenoni S., Loyola R., Guzzo F., Schlechter R., Ageorges A., Arce-Johnson P., Tornielli G.B. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 2015;167:1448–1470. doi: 10.1104/pp.114.256172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colquhoun T.A., Kim J.Y., Wedde A.E., Levin L.A., Schmitt K.C., Schuurink R.C., Clark D.G. PhMYB4 fine-tunes the floral volatile signature of petunia× hybrida through PhC4h. J. Exp. Bot. 2010;62:1133–1143. doi: 10.1093/jxb/erq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espley R.V., Hellens R.P., Putterill J., Stevenson D.E., Kutty-Amma S., Allan A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho J.-S., Nguyen V.P., Jeon H.-W., Kim M.-H., Eom S.H., Lim Y.J., Kim W.-C., Park E.-J., Choi Y.-I., Ko J.-H. Overexpression of PtrMYB119, a R2R3-MYB transcription factor from populus trichocarpa, promotes anthocyanin production in hybrid poplar. Tree Physiol. 2016;36:1162–1176. doi: 10.1093/treephys/tpw046. [DOI] [PubMed] [Google Scholar]
- 16.Yan J., Wang B., Zhong Y., Yao L., Cheng L., Wu T. The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 2015;89:35–48. doi: 10.1007/s11103-015-0349-3. [DOI] [PubMed] [Google Scholar]
- 17.Nesi N., Jond C., Debeaujon I., Caboche M., Lepiniec L. The arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker A.R., Lee E., Bogs J., McDavid D.A., Thomas M.R., Robinson S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007;49:772–785. doi: 10.1111/j.1365-313X.2006.02997.x. [DOI] [PubMed] [Google Scholar]
- 20.Bogs J., Jaffé F.W., Takos A.M., Walker A.R., Robinson S.P. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007;143:1347–1361. doi: 10.1104/pp.106.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takos A.M., Jaffé F.W., Jacob S.R., Bogs J., Robinson S.P., Walker A.R. Light-induced expression of a myb gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prouse M.B., Campbell M.M. Interactions between the R2R3-Myb transcription factor, AtMYB61, and target DNA binding sites. PLoS ONE. 2013;8:e65132. doi: 10.1371/journal.pone.0065132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kranz H.D., Denekamp M., Greco R., Jin H., Leyva A., Meissner R.C., Petroni K., Urzainqui A., Bevan M., Martin C. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- 24.Kasajima I., Sasaki K. A chimeric repressor of petunia PH4 R2R3-MYB family transcription factor generates margined flowers in torenia. Plant Signal. Behav. 2016;11:e1177693. doi: 10.1080/15592324.2016.1177693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez-Díaz J.R., Pérez-Díaz J., Madrid-Espinoza J., González-Villanueva E., Moreno Y., Ruiz-Lara S. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 2016;90:63–76. doi: 10.1007/s11103-015-0394-y. [DOI] [PubMed] [Google Scholar]
- 26.Albert N.W., Lewis D.H., Zhang H., Schwinn K.E., Jameson P.E., Davies K.M. Members of an R2R3-MYB transcription factor family in petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011;65:771–784. doi: 10.1111/j.1365-313X.2010.04465.x. [DOI] [PubMed] [Google Scholar]
- 27.Xu H., Wang N., Liu J., Qu C., Wang Y., Jiang S., Lu N., Wang D., Zhang Z., Chen X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdBHLH33 genes. Plant Mol. Biol. 2017;94:149–165. doi: 10.1007/s11103-017-0601-0. [DOI] [PubMed] [Google Scholar]
- 28.Wan S., Li C., Ma X., Luo K. PtrMYb57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 2017;36:1263–1276. doi: 10.1007/s00299-017-2151-y. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida K., Ma D., Constabel C.P. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 2015;167:693–710. doi: 10.1104/pp.114.253674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J.-S., Kim J.-B., Cho K.-J., Cheon C.-I., Sung M.-K., Choung M.-G., Roh K.-H. Arabidopsis R2R3-myb transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa) Plant Cell Rep. 2008;27:985–994. doi: 10.1007/s00299-008-0521-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Q., Sui S., Lei X., Yang Z., Lu K., Liu G., Liu Y.-G., Li M. Ectopic expression of the coleus R2R3 MYB-type proanthocyanidin regulator gene ssmyb3 alters the flower color in transgenic tobacco. PLoS ONE. 2015;10:e0139392. doi: 10.1371/journal.pone.0139392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu L., Shan H., Chen S., Jiang J., Gu C., Zhou G., Chen Y., Song A., Chen F. The heterologous expression of the chrysanthemum R2R3-MYB transcription factor cmmyb1 alters lignin composition and represses flavonoid synthesis in Arabidopsis thaliana. PLoS ONE. 2013;8:e65680. doi: 10.1371/journal.pone.0065680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fornalé S., Lopez E., Salazar-Henao J.E., Fernández-Nohales P., Rigau J., Caparros-Ruiz D. AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:507–516. doi: 10.1093/pcp/pct187. [DOI] [PubMed] [Google Scholar]
- 34.Tamagnone L., Merida A., Parr A., Mackay S., Culianez-Macia F.A., Roberts K., Martin C. The ammyb308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fornale S., Sonbol F.M., Maes T., Capellades M., Puigdomenech P., Rigau J., Caparros-Ruiz D. Down-regulation of the maize and Arabidopsis thaliana caffeic acid o-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol. Biol. 2006;62:809–823. doi: 10.1007/s11103-006-9058-2. [DOI] [PubMed] [Google Scholar]
- 36.Fornale S., Shi X., Chai C., Encina A., Irar S., Capellades M., Fuguet E., Torres J.L., Rovira P., Puigdomenech P., et al. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. Cell Mol. Biol. 2010;64:633–644. doi: 10.1111/j.1365-313X.2010.04363.x. [DOI] [PubMed] [Google Scholar]
- 37.Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. Transcriptional repression by atmyb4 controls production of UV-protecting sunscreens in arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preston J., Wheeler J., Heazlewood J., Li S.F., Parish R.W. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2004;40:979–995. doi: 10.1111/j.1365-313X.2004.02280.x. [DOI] [PubMed] [Google Scholar]
- 39.Nemie-Feyissa D., Olafsdottir S.M., Heidari B., Lillo C. Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in arabidopsis leaves. Phytochemistry. 2014;98:34–40. doi: 10.1016/j.phytochem.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Zhu H.-F., Fitzsimmons K., Khandelwal A., Kranz R.G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in arabidopsis. Mol. Plant. 2009;2:790–802. doi: 10.1093/mp/ssp030. [DOI] [PubMed] [Google Scholar]
- 41.Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.M., Alboresi A., Weisshaar B., Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- 42.Shen H., He X., Poovaiah C.R., Wuddineh W.A., Ma J., Mann D.G., Wang H., Jackson L., Tang Y., Neal Stewart C. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 2012;193:121–136. doi: 10.1111/j.1469-8137.2011.03922.x. [DOI] [PubMed] [Google Scholar]
- 43.Kagale S., Rozwadowski K. Ear motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics. 2011;6:141–146. doi: 10.4161/epi.6.2.13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin-Wang K., Bolitho K., Grafton K., Kortstee A., Karunairetnam S., McGhie T.K., Espley R.V., Hellens R.P., Allan A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in rosaceae. BMC Plant Biol. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y., Lin-Wang K., Espley R.V., Wang L., Yang H., Yu B., Dare A., Varkonyi-Gasic E., Wang J., Zhang J. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016;67:2159–2176. doi: 10.1093/jxb/erw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi X. Ph.D. Thesis. University of Toledo; Toledo, OH, USA: 2011. Regulatory Functions of ZmMYB31 and ZmMYB42 in Maize Phenylpropanoid Pathway. [Google Scholar]
- 47.Huang Y.F., Vialet S., Guiraud J.L., Torregrosa L., Bertrand Y., Cheynier V., This P., Terrier N. A negative myb regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 2014;201:795–809. doi: 10.1111/nph.12557. [DOI] [PubMed] [Google Scholar]
- 48.Paolocci F., Robbins M.P., Passeri V., Hauck B., Morris P., Rubini A., Arcioni S., Damiani F. The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in lotus corniculatus leaves. J. Exp. Bot. 2010;62:1189–1200. doi: 10.1093/jxb/erq344. [DOI] [PubMed] [Google Scholar]
- 49.Kazan K. Negative regulation of defence and stress genes by ear-motif-containing repressors. Trends Plant Sci. 2006;11:109–112. doi: 10.1016/j.tplants.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 50.Sonbol F.-M., Fornalé S., Capellades M., Encina A., Tourino S., Torres J.-L., Rovira P., Ruel K., Puigdomenech P., Rigau J., et al. The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol. 2009;70:283. doi: 10.1007/s11103-009-9473-2. [DOI] [PubMed] [Google Scholar]
- 51.Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- 52.Spelt C., Quattrocchio F., Mol J., Koes R. Anthocyanin1 of petunia controls pigment synthesis, vacuolar PH, and seed coat development by genetically distinct mechanisms. Plant Cell. 2002;14:2121–2135. doi: 10.1105/tpc.003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dasgupta K., Thilmony R., Stover E., Oliveira M.L., Thomson J. Novel R2R3-MYB transcription factors from prunus americana regulate differential patterns of anthocyanin accumulation in tobacco and citrus. GM Crops Food. 2017;8:85–105. doi: 10.1080/21645698.2016.1267897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Passeri V., Martens S., Carvalho E., Bianchet C., Damiani F., Paolocci F. The R2R3MYB VvMYBPA1 from grape reprograms the phenylpropanoid pathway in tobacco flowers. Planta. 2017;246:185–199. doi: 10.1007/s00425-017-2667-y. [DOI] [PubMed] [Google Scholar]
- 55.Vimolmangkang S., Han Y., Wei G., Korban S.S. An apple myb transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013;13:176. doi: 10.1186/1471-2229-13-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matus J.T., Aquea F., Arce-Johnson P. Analysis of the grape myb R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across vitis and arabidopsis genomes. BMC Plant Biol. 2008;8:83. doi: 10.1186/1471-2229-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou H., Peng Q., Zhao J., Owiti A., Ren F., Liao L., Wang L., Deng X., Jiang Q., Han Y. Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 2016;7:1557. doi: 10.3389/fpls.2016.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 59.Horsch R., Fry J., Hoffman N., Eichholtz D., Rogers S.A., Fraley R. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1232. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- 60.Mehrtens F., Kranz H., Bednarek P., Weisshaar B. The arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.