Abstract
We have developed a method that allows quantitative amplification of single-stranded DNA (QAOS) in a sample that is primarily double-stranded DNA (dsDNA). Single-stranded DNA (ssDNA) is first captured by annealing a tagging primer at low temperature. Primer extension follows to create a novel, ssDNA-dependent, tagged molecule that can be detected by PCR. Using QAOS levels of between 0.2 and 100% ssDNA can be accurately quantified. We have used QAOS to characterise ssDNA levels at three loci near the right telomere of chromosome V in budding yeast cdc13-1 mutants. Our results confirm and extend previous studies which demonstrate that when Cdc13p, a telomere-binding protein, is disabled, loci close to the telomere become single stranded whereas centromere proximal sequences do not. In contrast to an earlier model, our new results are consistent with a model in which a RAD24-dependent, 5′ to 3′ exonuclease moves from the telomere toward the centromere in cdc13-1 mutants. QAOS has been adapted, using degenerate tagging primers, to preferentially amplify all ssDNA sequences within samples that are primarily dsDNA. This approach may be useful for identifying ssDNA sequences associated with physiological or pathological states in other organisms.
INTRODUCTION
Single-stranded DNA (ssDNA) is an important intermediate in the processes of DNA replication, recombination and repair. It is also an important mediator of DNA damage-dependent cell cycle arrest in a range of organisms. In Escherichia coli, double strand breaks (DSBs) must be processed by the RecBC helicase/exonuclease and pyrimidine dimers must be replicated to generate ssDNA, to activate RecA, and induce the SOS response and cell cycle arrest (1). In budding yeast, meiotic cell DSBs need to be processed to ssDNA in order to signal arrest (2,3). In mammalian cells, a p53-dependent DNA damage sensor also responds to ssDNA, but not to nicked or covalently closed circular DNA (4). ssDNA has also been associated with DNA replication (5–7), transcription (8), trans-lesion synthesis (9), apoptosis (10,11), DNA repair (12), mitotic and meiotic recombination (13,14) and telomeres (15–18).
In order to better understand the role of ssDNA in cells it is important to identify and quantify sequences that are single stranded under differing physiological or pathological conditions. Several methods exist to detect ssDNA that is generated in vivo. Sucrose gradient (5), monoclonal antibody (11,12), hydroxyapapatite chromatography (8), filter-binding (17,19), in-gel hybridisation (15,20), two-dimensional agarose gel electrophoresis (17) and restriction enzyme digestion (21) assays have all been used to detect ssDNA. Previously we used a filter-binding assay, developed by Garvik et al. (17), to detect ssDNA accumulating at telomeres in synchronous cultures of yeast cells that contained a defective Cdc13p, telomere-binding protein (19,22). In this paper we describe a novel, simple and robust method that allows detection and quantification of ssDNA. Using quantitative amplification of ssDNA (QAOS) we have shown that different loci become single stranded at different rates when Cdc13-1p is inactivated and that checkpoint mutations affect these rates. In addition, we have adapted QAOS, using degenerate primers, to preferentially amplify small amounts of undefined ssDNA present in samples that are primarily double stranded.
MATERIALS AND METHODS
Reagents
All oligonucleotides were purchased from Sigma-Genosys, except for Taqman probes, which were purchased from Applied Biosystems. The oligonucleotides are described in Table 1 and are given the prefix M (for Manchester). PCR reagents except water and oligonucleotides were from TaKaRa Shuzo Company Ltd and Qiagen. Chemicals were purchased from Sigma Chemical Company. DNA purification kits were purchased from Qiagen and Amersham Pharmacia.
Table 1. Oligonucleotides used in this study.
| M |
Gene |
Sequence |
Tail Tm (°C) |
Coordinates on chromosome V |
Type |
| 7 |
|
GATCTCGAGCTCGATATCGGATCCATT |
|
|
Tag |
| 8 |
YER186C |
GTATGTGGGATATTCTCTCCTTATTGCGA |
|
562155–562183 |
Reverse |
| 10 |
YER186C |
GATCTCGAGCTCGATATCGGATCcatttacggcgga |
42 |
562405–562393 |
Tagging |
| 12 |
YER186C |
GATCTCGAGCTCGATATCGGATCCattctctcctta |
17 |
562166–562178 |
Tagging |
| 20 |
Random |
GATCTCGAGCTCGATATCGGATCCATTnnnnnnnnn |
|
|
Tagging |
| 21 |
Random |
GATCTCGAGCTCGATATCGGATCCATTnnnnnnnnnnnn |
|
|
Tagging |
| 22 |
Random |
GATCTCGAGCTCGATATCGGATCCATTnnnnnn |
|
|
Tagging |
| 57 |
YER186C |
GATCTCGAGCTCGATATCGGATCcatttacggcgga |
42 |
562405–562393 |
Tagging |
| 96 |
YER186C |
AATCTCGCCTAACAAAAAAAGGCTTCTTAGTG |
|
562332–562363 |
Reverse |
| 97 |
|
GATCTCGAGCTCGATATCGGATCCATT |
|
|
Tag |
| 174 |
PAC2 |
GATCTCGAGCTCGATATCGGATCCattgagctatga |
18 |
165689–165700 |
Tagging |
| 178 |
PAC2 |
GATCTCGAGCTCGATATCGGATCCatttcatacgac |
18 |
165745–165734 |
Tagging |
| 188 |
YER186C |
GCAAGTAGGAAGCATCCCTTCAAGTCATT |
|
562431–562403 |
Reverse |
| 234 |
|
AAGGAGCGCAGCGCCTGTACCA |
|
|
Tag |
| 235 |
|
TGCCCTCGCATCGCTCTCACA |
|
|
Tag |
| 239 |
|
AACCAGCGCAGCGGCATGTGT |
|
|
Tag |
| 256 |
YER186C |
AGGCAAATAACGGCAAGCCCTCTCC |
|
562368–562392 |
Probe |
| 264 |
YER188W |
TCATGCCGTTCAAATTCTGAGGGTTCT |
|
568402–568376 |
Reverse |
| 265 |
YER188W |
AACGTACAGGTTACGATCGCGTCATTTTA |
|
568296–568324 |
Reverse |
| 266 |
YER188W |
TGCCCTCGCATCGCTCTCacaggttacgatc |
26 |
568302–568314 |
Tagging |
| 267 |
YER188W |
AAGGAGCGCAGCGCCTGTAccatagcgtgat |
28 |
568374–568363 |
Tagging |
| 271 |
PDA1 |
AAGGAGCGCAGCGCCTGTAccatctatctgttg |
28 |
547144–547157 |
Tagging |
| 272 |
PDA1 |
AACCAGCGCAGCGGCATGtgtgatgatggaa |
28.5 |
547215–547203 |
Tagging |
| 280 |
YER188W |
TAGCCGTTATCATCGGGCCCAAAACCGTATTCATTG |
|
568360–568325 |
Probe |
| 281 |
PDA1 |
TGGCATTCTCGATACCGACAGCAATGGCCTC |
|
547193–547163 |
Probe |
| 287 |
PDA1 |
TGGAGATGGCTTGTGACGCCTTG |
|
547092–547114 |
Reverse |
| 288 |
PDA1 |
ACGGCTTTCACTGAGGCACCTCTC |
|
547272–547249 |
Reverse |
| 310 |
PAC2 |
AATAACGAATTGAGCTATGACACCAA |
|
165681–165706 |
Reverse |
| 322 |
PAC2 |
AGCTTACTCATATCGATTTCATACGACTT |
|
165760–165732 |
Reverse |
| 326 | PAC2 | CTGCCGCGTTGGTCAAGCCTCAT | 165708–165730 | Probe |
The sequences of the oligonucleotides used in this study. The oligonucleotides are classified as tags, tagging primers, probes and reverse primers (see Fig. 1A). In the case of the tagging primers the sequence that anneals with the locus detected is lower case, the sequence analogous to the tag is upper case, and the overlap is bold and lower case. The annealing temperature of the tail is shown in the ‘Tail Tm’ column. The coordinates refer to the position of the relevant oligonucleotide sequence on chromosome V of yeast and are taken from the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces)
Preparation of control templates and standards
To mimic the concentration of a single copy gene in the yeast genome, plasmid pHR85-31 (pDL339, 5.7 kb), containing the yER186C gene (19), was cut to completion with restriction enzymes and serially diluted in 10 ng/µl calf thymus DNA (Sigma Chemical Company D8661) in TE (10 mM Tris–HCl, pH 8.0, 1 mM EDTA). EcoRI-digested pHR85-31 was diluted to 2.5 pg/µl, and SalI-digested pHR85-31 was diluted to 5 pg/µl. Thus, at 1:4000 (EcoRI) or 1:2000 (SalI) dilutions the plasmid was at a similar concentration to a 5.7 kb gene in the 13 469 kb yeast genome (1:2350). After dilution, a fraction of each sample was boiled at 96°C for 7 min, then rapidly chilled on ice. To create standard series, boiled and non-boiled DNA samples were mixed together, e.g. 3.2% ssDNA was prepared by mixing 96.8 µl of non-boiled DNA with 3.2 µl of boiled DNA. For global QAOS the target ssDNA was pHR85-31 cut with SalI, heated at 95°C for 5 min and then chilled on ice and the double-stranded DNA (dsDNA) was λ DNA purchased from New England Biolabs (NEB No301-3s). Sonication was used to prepare low molecular weight samples of pHR85-31 and λ. DNA was sonicated until the average length was 400–500 bp, purified on Amersham GFX gel purification columns and concentration determined by absorbance at 260 nM.
Preparation of yeast genomic DNA
Yeast DNA was prepared from yeast strains containing bar1Δ cdc13-1 cdc15-2 mutations (19,22). Some of the strains carried additional checkpoint mutations indicated in parentheses: DLY408 (RAD+), DLY409 (rad9Δ), DLY410 (rad24Δ), DLY411 (rad9Δ rad24Δ). The cells were released from G1 arrest at 23–36°C, exactly as previously described (19). A Qiagen, rather than CsCl, based method was used to purify DNA (23,24). Quantity (40, 20 and 4 ng) and ssDNA (51.2, 3.2 and 0.2% ssDNA) standards were prepared from DNA isolated from alpha factor arrested strain DLY62 (RAD+ bar1).
QAOS followed by agarose gel electrophoresis
Four microlitres (40 ng) of DNA was added to 40 µl of a PCR master mix comprising (final concentrations in parentheses): water, ExTaq PCR buffer (1×, 2 mM MgCl2), dNTPs (250 µM), M10 (3.3 nM), M7 and M8 (330 nM each), ExTaq DNA polymerase (0.025 U/µl). To create the boiled DNA control (Fig. 1C), 20 µl of each mix was removed to a separate tube, and heated at 95°C, for 5 min, before cooling rapidly on ice. All samples were then subject to the following PCR protocol. Step 1: 1 cycle; 40°C for 5 min, ramp to 72°C, at 2°C/min. Step 2: 1 cycle; 95°C, for 5 min. Step 3: 35 cycles; 94°C for 30 s, 67°C for 30 s, 72°C for 60 s. Step 4: 1 cycle; 72°C, for 10 min. After the PCR, 6× loading buffer was added and 15 µl of each sample was run at 120 V on a 1% agarose gel in 0.5× TBE, containing 0.5 µg/ml ethidium bromide. After electrophoresis the gel was exposed to UV and photographed.
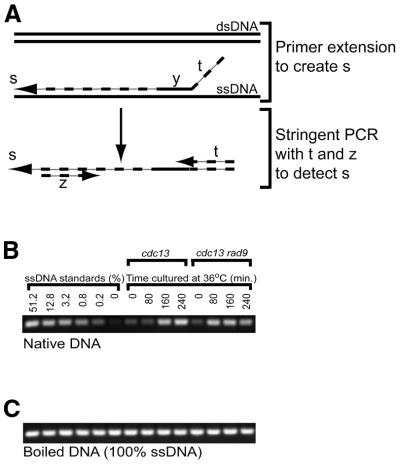
Figure 1.
(A) Schematic outline of QAOS. A tagging primer, ty, containing a tag t at its 5′ end, binds to ssDNA, at low temperature, via the y region at its 3′ end. A round of primer extension creates a novel molecule s, that contains the t sequence at its 5′ end. s is detected by stringent PCR using primers t and z (a reverse primer) at temperatures which allow binding of primers t and z, but which are too high to allow binding of the y sequences to ssDNA generated during each cycle of PCR. (B) Application of QAOS to native samples. An ethidium bromide stained agarose gel shows the amount of YER186C PCR product obtained following the application of QAOS to samples containing various amounts of ssDNA. The left half of the gel shows the amount of PCR product obtained from a 4-fold dilution series of ssDNA. The right half of the gel shows the amount of PCR product obtained when QAOS was applied to DNA purified from yeast cells that accumulate ssDNA in vivo. The yeast contained cdc13 (DLY408) or cdc13 rad9 (DLY409) mutations and had been cultured at the non-permissive temperature of 36°C, for 0, 80, 160 and 240 min, before their DNA was extracted. (C) Application of QAOS to boiled (100% ssDNA) samples. Samples are identical to those in (B) except they were boiled before QAOS was performed.
Real-time analysis of QAOS products
The template was pHR85-31 linearised with SalI at 5 pg/µl in 10 ng/µl calf thymus DNA. PCR was carried out in two stages. In the first stage 2 µl (10 pg pHR85-31, 20 ng total) of DNA was mixed with 10 µl of a master mix comprising (final concentration in parentheses): water, ExTaq PCR buffer (1×), dNTPs (250 µM), M10 (3.3 nM) and ExTaq DNA polymerase (0.025 U/µl) and the samples subjected to: 1 cycle; 40°C for 5 min ramp to 72°C, at 2°C per min. Samples were then stored at –20°C before being thawed and 2 µl of the first stage material added to 21 µl of a second master mix comprising (final concentrations in brackets): PCR Buffer A (1× PE Biosystems), MgCl2 (3.5 mM), dNTPs (250 µM), M96 and M97 (300 nM), M256 (200 nM) and Qiagen Hot Star Taq DNA polymerase (0.025 U/µl). Real-time PCR was performed in an ABI7700 with the following profile. Step 1: 1 cycle; 50°C, for 2 min. Step 2: 1 cycle; 95°C, for 10 min. Step 3: 40 cycles; 95°C for 15 s, 67°C for 60 s.
Real-time quantification of total genomic DNA
The concentration of DNA in yeast genomic DNA preparations was initially determined by absorbance at 260 nm, and generally adjusted to 2 ng/µl. To more accurately determine the amount of genomic DNA in each preparation the amount of DNA at the centromeric PAC2 locus was measured, in triplicate, by real-time quantitative PCR with reference to 40, 20 and 4 ng quantity standards. PAC2 lies close to the centromere on chromosome V and does not become detectably single stranded in cdc13-1 mutants and so it can be used as a ‘loading’ control. From these PAC2 measurements a correction factor was then calculated. For example, if the PAC2 measurements indicated that there were 12 rather than 20 ng of DNA in a particular preparation, the correction factor was 0.6.
To measure the levels of PAC2, 15 µl of PCR master mix was added to 10 µl of template to create solutions comprising (final concentrations in parentheses): water, ExTaq PCR Buffer (1×), dNTPs (200 µM), M310 and M322 (300 nM), M326 (200 nM) ExTaq polymerase (0.025 U/µl). PCR conditions were as follows. Step 1: 1 cycle; 40°C for 5 min, ramp to 72°C, at 2°C/min. Step 2: 1 cycle; 72°C, for 10 min. Step 3: 1 cycle; 94°C, for 5 min. Step 4: 40 cycles; 95°C for 15 s, 67°C for 60 s.
Real-time QAOS to measure the fraction of ssDNA at genomic loci
To measure ssDNA at genomic loci triplicate samples (each 10 µl, 20 ng) were frozen at –20°C until use. A sample (15 µl) of PCR master mix was added to create solutions that comprised (final concentrations in parentheses): water, ExTaq PCR Buffer (1×) dNTPs (200 µM), tagging primer (33 nM), tag (330 nM), reverse primer (330 nM), probe (200 nM) and ExTaq polymerase (0.025 U/µl). Primers and probes used are listed in Table 2 and the PCR protocol was as described for measuring the amount of PAC2 genomic DNA.
Table 2. Primers and probes used to detect ssDNA at genomic loci.
| Locus |
Strand |
Primers/probes |
| PAC2 | TG | M178, M97, M310, M326 |
| PAC2 | AC | M174, M97, M322, M326 |
| PDA1 | TG | M272, M239, M287, M281 |
| PDA1 | AC | M271, M234, M288, M281 |
| YER186C | TG | M57, M96, M97, M256 |
| YER186C | AC | M12, M97, M188, M256 |
| YER188W | TG | M267, M234, M265, M280 |
| YER188W | AC | M266, M235, M264, M280 |
Oligonucleotides used in real-time QAOS to detect ssDNA at the loci indicated. With the exception of M326, which was labelled with VIC at the 5′ end, all Taqman probes were 5′ labelled with FAM and 3′ labelled with TAMRA.
To construct a ssDNA standard curve 51.2, 3.2 and 0.2% ssDNA standards were created by mixing boiled and non-boiled DNA prepared from alpha factor arrested strain DLY62. The amount of ssDNA at any strand of any locus was calculated using these standards and adjusted by the PAC2 based correction factor before being plotted. In most cases the results plotted represent the mean values of the triplicate samples. However, if quantitative PCR showed that an individual differed from its duplicates >2-fold (one Ct value) it was ignored and the mean of the other two duplicates plotted.
Primer and probe design for locus-specific QAOS
Tagging primers were designed to contain tails 10–15 bp in length with Tm of between 16 and 45°C (determined by Primer Express software). A short overlapping sequence that was common to both the tag and the tail was always included but we have not rigorously tested to see if this overlap is important (see Table 1). Experimentation with tail length (and Tm) was necessary to determine which length best distinguishes ssDNA from dsDNA. The tag and reverse primers had a Tm of >55°C and the probes of >65°C.
Global QAOS
PCR was performed in two rounds. In the first round 20 ng of λ DNA (containing 0, 1 or 0.1 ng of boiled pHR85-31) was mixed and diluted to a total volume of 20 µl in a mix comprising: dNTPs (200 µM), a degenerate tagging primer [333 nM: M20 t(n)9, M21 t(n)12 or M22 t(n)6], ExTaq buffer (1×) and ExTaq polymerase (0.025 U/µl). PCR was performed as follows. Step 1: 2 cycles; 30°C for 30 min, ramp to 94°C, at 2°C/min. Samples were then placed on ice. Five microlitres (5 ng) was removed and added a different master mix comprising (final concentrations in parentheses, ignoring any components from the first round): dNTPs (200 µM), M7 (2.7 µM), Qiagen Hot Star buffer (1×), MgCl2 (additional 0.5 mM) Qiagen Hot Star DNA polymerase (0.025 U/µl). Step 1: 1 cycle; 95°C for 15 min. Step 2: 35 cycles; 94°C for 30 s, 67°C 45 s. Step 3: 1 cycles; 72°C for 10 min. After PCR agarose gel electrophoresis loading buffer was added and aliquots of each reaction examined by agarose gel electrophoresis and Southern blotting. For Southern blotting a single agarose gel was sandwiched between two MSI Magna Nylon Membranes. After blotting the membranes were hybridised with fluorescene labelled, random primed (Amersham kit RPN 3511) probes derived from pHR85-31 or λ DNA. Hybridisation was detected using the Amersham Gene Images detection kit (RPN3510), and exposure to X-ray film. Quantification of the autoradiographs was performed using a BioRad GS700 scanning densitometer.
RESULTS
Locus-specific QAOS
Figure 1A shows a schematic outline of locus directed QAOS. Figure 1B shows the application of QAOS to the detection of ssDNA at the yeast YER186C locus, which in its natural context lies 15 kb from the telomere of chromosome V (see Fig. 3). The application of QAOS to a standard series created by mixing defined amounts of ssDNA and dsDNA illustrates that the amount of PCR product correlated well with the amount of ssDNA initially present and that QAOS can discriminate between 0.2 and 0% ssDNA samples (Fig. 1B). In Figure 1C QAOS was applied to identical samples to those shown in Figure 1B with the exception that the samples were first boiled. A comparison of Figure 1B and C shows that although similar amounts of DNA were present in all samples QAOS was capable of preferentially amplifying YER186C when single stranded.
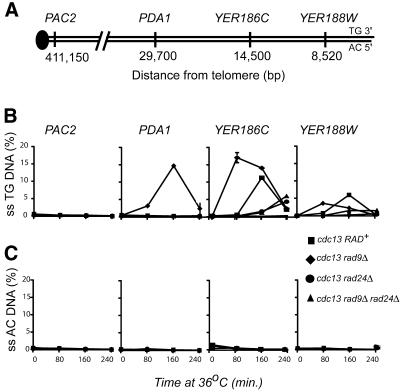
Figure 3.
Detection of ssDNA at telomeric loci in cdc13-1 mutants. (A) The location of four loci along the right arm of chromosome V. The 3′ strand of the telomere consists of TG repeats and the 5′ strand of AC repeats. (B) Appearance of ssDNA on the TG strand in cdc13-1mutants. QAOS was used to measure ssDNA levels in synchronous cultures of cdc13-1 mutants. QAOS primers detected ssDNA on the strand that ends with the 3′, TG, sequences at the telomere. The yeast strains analysed were DLY408 cdc13-1 RAD+ (squares), DLY409 cdc13-1 rad9Δ (diamonds), DLY410 cdc13-1 rad24Δ (circles) and DLY411 cdc13-1 rad9Δ rad24Δ (triangles). (C) Appearance of ssDNA on the AC strand in cdc13-1 mutants. QAOS primers were chosen to detect ssDNA on the DNA strand that ends with the 5′, AC, sequences at the telomere. The samples and symbols are the same as in (B).
QAOS is capable of detecting ssDNA generated in vivo. Previously we showed, using a filter-binding assay, that cdc13-1 mutants accumulate high levels of ssDNA at YER186C, 160 min after incubation at their non-permissive temperature (36°C) and that cdc13-1 rad9Δ double mutants accumulated ssDNA earlier, by 80 min (22). The data in the right lanes of Figure 1B are in complete agreement with our previous findings and show that QAOS is capable of measuring ssDNA produced at YER186C in vivo, in budding yeast cdc13 mutants.
Despite the promising results shown in Figure 1B it is difficult to determine by agarose gel electrophoresis precisely how much ssDNA was present in each sample at the start of the experiment (e.g. compare the signal produced for samples that contained 12.8 and 0.8% ssDNA, Fig. 1B). To more accurately measure the amount of ssDNA in a sample we applied real-time quantitative PCR to QAOS.
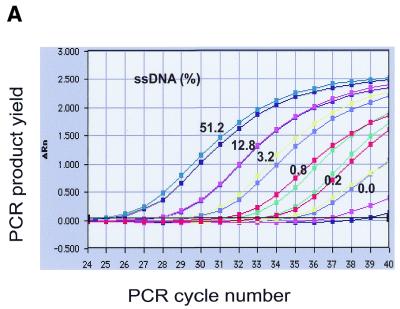
The application of real-time QAOS to samples that contained defined amounts of ssDNA shows that real-time QAOS can discriminate between samples in the range 0.2–51.2% ssDNA. Figure 2A shows that duplicate samples behave very similarly when subject to real-time QAOS and that there is a dose-dependent relationship between the initial ssDNA concentration and the time (PCR cycle number) at which measurable amplification was initiated. The samples in 4-fold standard series of ssDNA reached similar fluorescence values (e.g. a ΔRn of 1) at approximately two cycle intervals, which matched their theoretical behaviour based on a 2-fold increase in the amount of product each cycle.
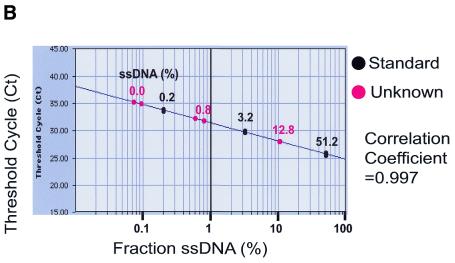
Figure 2.
Real-time QAOS. (A) Amplification plots of real-time QAOS samples. A duplicate 4-fold dilution series, as described in Figure 1B, was subject to real-time QAOS. The products of PCR are measured as an increase in fluorescence (ΔRn) and are plotted versus PCR cycle number. Duplicate samples are labelled together, the two lines without labels at the right side of the graph contained no target DNA and the small amount of signal is probably due to contamination. (B) Construction of a standard curve to measure the fraction of ssDNA. The samples in (A) have been classified as standards (51.2, 3.2 or 0.2% ssDNA, black) or unknowns (12.8, 0.8 and 0% ssDNA, red). The threshold cycle, the point at which the fluorescence (ΔRn) reached a fixed value for each sample, is plotted versus the fraction of ssDNA that was present initially. A regression line has been drawn through the standards (correlation coefficient 0.997) and can be used to calculate the fraction of ssDNA in the unknown samples (drawn as red dots on the regression line).
To directly assess the ability of real-time QAOS to measure the amount of ssDNA a series of samples containing known amounts of ssDNA were analysed. In Figure 2B samples containing 0.2, 3.2 and 51.2% ssDNA were used as standards while samples containing 0, 0.8 and 12.8% ssDNA were classified as ‘unknowns’. The correlation coefficient of the standard curve was 0.997 and the 0.8 and 12.8% ssDNA samples lie extremely close to their predicted positions on the standard curve. The negative control samples (0% ssDNA) were estimated to have <0.1% ssDNA indicating that the limit of detection of real-time QAOS is ∼0.1% ssDNA.
To assess the utility of QAOS we used it to measure the kinetics of ssDNA appearance at four loci on the right arm of chromosome V in yeast containing a cdc13-1 mutation plus additional rad9Δ and rad24Δ deletions (Fig. 3). In all strains we were unable to detect ssDNA on either strand at the centromeric PAC2 locus. At the PDA1 locus, ∼30 kb from the telomere, we could detect ssDNA in the cdc13-1 rad9Δ strain, but not in cdc13-1 RAD+, cdc13-1 rad24Δ and cdc13-1 rad9Δ rad24Δ strains. At PDA1 and at all loci where ssDNA was measured, noticeable levels were only detected on the strand that ends with the a 3′ end and TG repeats at the telomere, and not on the opposite (5′ AC repeat) strand. At PDA1 levels of ssDNA rose from 3% at 80 min, peaked at ∼15% at 160 min and declined to 3% at 240 min. At YER186C, ssDNA was first detected at 80 min in cdc13-1 rad9Δ mutants but by 240 min was detectable in all yeast strains. At YER186C ssDNA levels peaked in the range of 10–20%. At YER188W, like at YER186C, cdc13-1 rad9Δ mutants accumulated ssDNA earlier than cdc13-1 RAD+ cells, which in turn accumulated ssDNA earlier than cdc13-1 rad24Δ and cdc13-1 rad9Δ rad24Δ strains. The levels of ssDNA at YER188W peaked at ∼7%. We conclude that real-time QAOS is a useful method to measure ssDNA generated at the telomeres of budding yeast cdc13-1 mutants.
Global QAOS
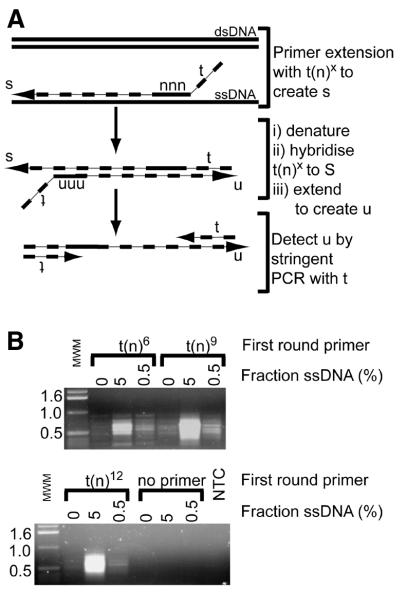
In order to identify ssDNA in samples that contain primarily dsDNA we have applied a similar approach to the one used to detect ssDNA at specific loci. An outline of global QAOS, which combines elements of degenerate oligonucleotide primer PCR (DOP-PCR) (25), with two rounds of low temperature polymerisation, is shown in Figure 4A. Figure 4B shows that when degenerate tagging primers, containing 6, 9 or 12 random nucleotides, were used for global QAOS and applied to samples containing 0, 0.5 and 5% ssDNA the amount of PCR product strongly correlated with the amount of ssDNA present at the start of the experiment. The size of the PCR products was in the range of 400–800 bp.
Figure 4.
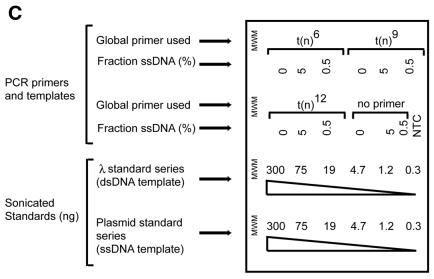
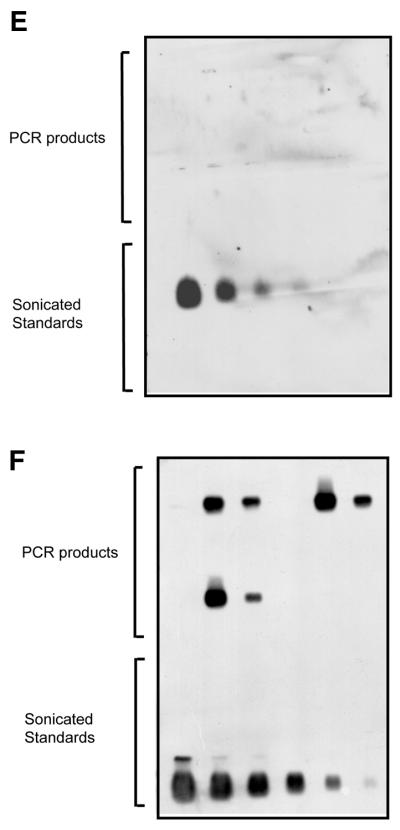
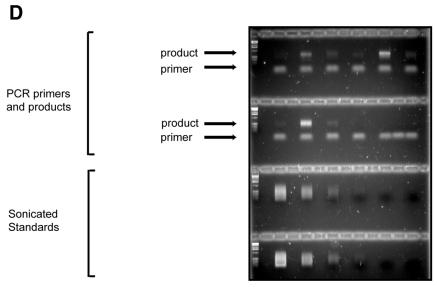
Global QAOS. (A) Outline of global QAOS. A random tagging primer t(n) x, containing a tag t, and a random nucleic acid-binding sequence with x random nucleotides, anneals to ssDNA. The first round of primer extension creates a library of molecule s. The mixture is then denatured, and cooled to allow the primer t(n)x to anneal to the library of molecule s. A second round of primer extension creates a library of molecules u. The library u is amplified by stringent PCR using the primer t. (B) Detection of ssDNA-dependent PCR products. Samples containing 0, 5 and 0.5% ssDNA were mixed with random tagging primers containing 6 = t(n)6, 9 = t(n)9 or 12 = t(n)12 random nucleotides at the 3′ end. A no template control (NTC) and no first round primer controls were also subject to global QAOS. After PCR the samples were analysed by agarose gel electrophoresis, along with molecular weight markers (MWM). The sizes of the major molecular weight markers are indicated on the left in kilobases. (C) Key of agarose gel examined in (D–F). The same set of samples described in (B), and a 4-fold dilution series of sonicated DNA standards were separated on an agarose gel. The λ standard series contained sonicated λ DNA (the dsDNA initially present in all tubes). The plasmid standard series contained sonicated, double-stranded, plasmid DNA (that had been present, in single stranded form, in the 5 and 0.5% ssDNA samples). (D) Agarose gel stained with ethidium bromide after electrophoresis. The samples are as described in (C). The low molecular weight bands present in the top two rows are PCR primers. The bands visible above the primers are the ssDNA-dependent PCR product. (E) Southern blot hybridised to λ DNA (dsDNA) probe. The gel was blotted on to a nylon membrane and hybridised with labelled λ DNA. (F)Southern blot hybridised to plasmid DNA (ssDNA) probe. The gel in (D) was blotted on to a nylon membrane and hybrised with labelled plasmid DNA.
To confirm that the PCR products were derived from ssDNA we performed Southern blot analyses with probes from the plasmid pHR85-31 (ssDNA in the starting samples) and λ DNA (dsDNA in the starting samples). Hybridisation results clearly show that there was preferential amplification of the ssDNA (Fig. 4C–F). Quantification of the autoradiograms allowed us to estimate that ssDNA was amplified 100–500-fold, whereas there was no detectable amplification of dsDNA (Table 3).
Table 3. Amplification of ssDNA by global QAOS.
| Primer | Fraction ssDNA (%) | ssDNA amplification | dsDNA amplification | ||||
| |
|
Input (ng) |
Yield (ng) |
Amplification (fold) |
Input (ng) |
Yield (ng) |
Amplification (fold) |
| T(n)6 | 5 | 0.021 | 2 | 95 | 0.42 | <4.7 | <11 |
| T(n)6 | 0.5 | 0.0021 | 0.8 | 381 | 0.42 | <4.7 | <11 |
| T(n)9 | 5 | 0.021 | 3.5 | 167 | 0.42 | <4.7 | <11 |
| T(n)9 | 0.5 | 0.0021 | 0.7 | 333 | 0.42 | <4.7 | <11 |
| T(n)12 | 5 | 0.021 | 10.2 | 486 | 0.42 | <4.7 | <11 |
| T(n)12 | 0.5 | 0.0021 | 0.7 | 333 | 0.42 | <4.7 | <11 |
The starting amount of ssDNA and dsDNA that was initially present in the PCR tubes, and would have been loaded on the agarose gel (Fig. 4D) is indicated in the ‘Input’ columns. The amount of DNA that was detected, by Southern blot hybridisation (Fig. 4E and F) and quantification by scanning densitometry is illustrated in the ‘Yield’ columns. The amplification value was calculated by dividing the yield by the input.
DISCUSSION
Locus-specific QAOS
QAOS is a rapid, sensitive and reproducible technique capable of detecting and quantifying ssDNA in samples that contain primarily dsDNA. We have used QAOS to detect and quantify ssDNA on each strand at four, single copy loci, on the right arm of chromosome V in yeast cdc13-1 mutants. QAOS is a considerable improvement on a filter-binding assay that was previously used to detect ssDNA in cdc13-1 mutants. The filter-binding assay was time consuming, required large amounts of highly purified DNA, radioactive RNA probes, a slot blot apparatus and numerous duplicate filters (19). In contrast QAOS uses considerably less DNA (20 compared with 500 ng), can be performed in 96 well format, and in combination with a real-time PCR machine can generate quantitative data very rapidly (3 h versus 3 days). We see no reason why QAOS should not be able to detect ssDNA at any 150 bp sequence in the yeast genome, or why it could not be used to detect ssDNA produced in other cell types.
Use of QAOS to examine effects of RAD9 and RAD24 in cdc13-1 mutants
Experiments using real-time QAOS have forced us to modify an earlier model of how checkpoint gene products affect DNA damage processing in vivo. Previously we proposed that Rad24p, Rad17p and Mec3p controlled the activity of a 3′ to 5′ exonuclease that degraded the strand that ended at the telomere with 5′ AC sequences, while Rad9p inhibited the exonuclease (22). The model was primarily based on the homology of Rad17p to 3′ to 5′ exonucleases, and the kinetics of ssDNA accumulation at YER186C in strains carrying combinations of cdc13-1, rad9Δ and rad24Δ mutations. By using real-time QAOS to analyse ssDNA levels at four loci on chromosome V in synchronous cultures of cdc13-1 RAD+ cdc13-1 rad9Δ, cdc13-1 rad24Δ and cdc13-1 rad9Δ rad24Δ strains we now show that the TG strand at telomeres becomes single stranded close to the telomere, earlier than at loci further from the telomere (Fig. 3). Therefore, this data is most consistent with the ssDNA being generated by a 5′ to 3′ exonuclease, moving from the telomere to the centromere, rather than a 3′ to 5′ exonuclease moving in the opposite direction, as previously proposed.
In agreement with previous data, analysis of ssDNA accumulation in cdc13-1 mutants by real-time QAOS confirms that (i) the only strand to become single stranded is the one that ends at the telomere with TG (3′) sequences (as opposed to AC 5′ sequences) (17,22), (ii) ssDNA is detected near telomeres (up to 30 kb from telomeres) but not at centromeres (17,22), (iii) cdc13-1 rad9Δ mutants accumulate ssDNA more rapidly than cdc13-1 RAD+ mutants (22), (iv) strains containing a rad24Δ accumulate ssDNA more slowly than cdc13-1 RAD+ e) significant levels of ssDNA in cdc13-1 rad24Δ strains are seen at the two most telomeric loci at 240 min (up to 6% at YER186C) (22).
The kinetics of appearance of ssDNA along chromosome V in cdc13-1 rad9Δ mutants allow us to make a crude estimation of the rate at which an exonuclease moves in the absence of RAD9. If ssDNA generated in cdc13-1 mutants is due a processive exonuclease then it causes a peak of ssDNA to appear at YER186C at 80 min, in these and earlier experiments (19,22), but at PDA1, 15 kb closer to the centromere, the peak of ssDNA is at 160 min. Therefore, in the absence of Rad9p an exonuclease moves at ∼200 bp/min (15 000/80). For comparison the Saccharomyces cerevisiae replication fork moves ∼20 times faster at 3700 bp/min (26). Additional experiments will be needed to allow a more accurate estimate of its speed in both the presence and absence of Rad9p.
In cdc13-1 rad9Δ and cdc13-1 RAD+ mutants the levels of ssDNA rise then fall at all telomeric loci examined [disappearance of YER186C ssDNA sequences was also observed using a filter-binding assay (19,22)]. This may be because the ssDNA is only transiently present in cdc13-1 mutants before it is converted back to dsDNA. Alternatively this phenomenon could be caused by loss of ssDNA sequences either inside the cell or during the DNA extraction process. As the peak levels of ssDNA in cdc13-1 mutants are in the order of 15–20% it is difficult to determine which is true. However, the fact that cdc13-1 RAD+ mutants maintain very high viability (more than cdc13-1 rad24Δ mutants) after incubation at 36°C for 4 h (22) strongly suggests that they are not losing telomeric DNA in vivo, and argues that the disappearance of ssDNA observed in these experiments is an artefact of the DNA extraction process.
Global QAOS
The degenerate oligonucleotide priming approach, global QAOS, offers the possibility of identifying as yet unknown ssDNA sequences. This may prove to be a useful method to identify ssDNA sequences associated with physiological or pathological processes. Given that the presence of ssDNA at telomeric sequences is ‘diagnostic’ of a cdc13-1 defect in yeast it is conceivable that ssDNA is also associated with pathological states in other eukaryotes, including humans. If true then the identification of these sequences may be of diagnostic value in human disease. We have demonstrated that starting with samples that contain 0.5% ssDNA it is possible to preferentially amplify the ssDNA sequences to the point in which the majority of DNA derived from sequences were originally single stranded (Fig. 4). Therefore, global QAOS offers the potential to first amplify, and then identify, sequences that are associated with disease states.
Localised regions of ssDNA are generated in yeast as part of normal physiological processes. Perhaps the best example of this is the transient appearance of ssDNA at the DSBs that initiate meiotic recombination (14). There is increasing evidence that DSBs initiate recombination not only in yeast but also in the majority of eukaryotes, including plants and mammals (27,28). If global QAOS was applied to DNA purified from cells undergoing the process of meiotic recombination it should preferentially amplify those regions that initiate the process of meiotic recombination. The amplified sequences could be used to identify the sites of meiotic recombination.
In conclusion, QAOS offers a powerful approach to amplify, identify and quantify ssDNA generated during normal physiological and pathological cellular processes. QAOS may prove to be a useful method to analyse the ssDNA intermediates that arise during DNA replication, recombination and repair.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by the award of a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science to D.L.
References
- 1.Sassanfar M. and Roberts,J.W. (1990) Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol., 212, 79–96. [DOI] [PubMed] [Google Scholar]
- 2.Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) DMC1: a meiosis-specific yeast homolog of E.coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- 3.Lydall D., Nikolsky,Y., Bishop,D.K. and Weinert,T. (1996) A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature, 383, 840–843. [DOI] [PubMed] [Google Scholar]
- 4.Huang L.C., Clarkin,K.C. and Wahl,G.M. (1996) Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc. Natl Acad. Sci. USA, 93, 4827–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okazaki R., Okazaki,T., Sakabe,K., Sugimoto,K. and Sugino,A. (1968) Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc. Natl Acad. Sci. USA, 59, 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henson P. (1978) The presence of single-stranded regions in mammalian DNA. J. Mol. Biol., 119, 487–506. [DOI] [PubMed] [Google Scholar]
- 7.Wanka F., Brouns,R.M., Aelen,J.M., Eygensteyn,A. and Eygensteyn,J. (1977) The origin of nascent single-stranded DNA extracted from mammalian cells. Nucleic Acids Res., 4, 2083–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tapiero H., Leibowitch,S.A., Shaool,D., Monier,M.N. and Harel,J. (1976) Isolation of single stranded DNA related to the transcriptional activity of animal cells. Nucleic Acids Res., 3, 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordeiro-Stone M., Makhov,A.M., Zaritskaya,L.S. and Griffith,J.D. (1999) Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol., 289, 1207–1218. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe I., Toyoda,M., Okuda,J., Tenjo,T., Tanaka,K., Yamamoto,T., Kawasaki,H., Sugiyama,T., Kawarada,Y. and Tanigawa,N. (1999) Detection of apoptotic cells in human colorectal cancer by two different in situ methods: antibody against single-stranded DNA and terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end-labeling (TUNEL) methods. Jpn J. Cancer Res., 90, 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankfurt O.S., Robb,J.A., Sugarbaker,E.V. and Villa,L. (1996) Monoclonal antibody to single-stranded DNA is a specific and sensitive cellular marker of apoptosis. Exp. Cell Res., 226, 387–397. [DOI] [PubMed] [Google Scholar]
- 12.Raderschall E., Golub,E.I. and Haaf,T. (1999) Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl Acad. Sci. USA, 96, 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugawara N. and Haber,J.E. (1992) Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol., 12, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun H., Treco,D. and Szostak,J.W. (1991) Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell, 64, 1155–1161. [DOI] [PubMed] [Google Scholar]
- 15.Dionne I. and Wellinger,R.J. (1996) Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl Acad. Sci. USA, 93, 13902–13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- 17.Garvik B., Carson,M. and Hartwell,L. (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol., 15, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polotnianka R.M., Li,J. and Lustig,A.J. (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol., 8, 831–834. [DOI] [PubMed] [Google Scholar]
- 19.Lydall D. and Weinert,T. (1997) Use of cdc13-1-induced DNA damage to study effects of checkpoint genes on DNA damage processing. Methods Enzymol., 283, 410–424. [DOI] [PubMed] [Google Scholar]
- 20.Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 21.White C.I. and Haber,J.E. (1990) Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J., 9, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lydall D. and Weinert,T. (1995) Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science, 270, 1488–1491. [DOI] [PubMed] [Google Scholar]
- 23.Grandin N., Reed,S.I. and Charbonneau,M. (1997) Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev., 11, 512–527. [DOI] [PubMed] [Google Scholar]
- 24.Wu J.R. and Gilbert,D.M. (1995) Rapid DNA preparation for 2D gel analysis of replication intermediates. Nucleic Acids Res., 23, 3997–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telenius H., Carter,N.P., Bebb,C.E., Nordenskjold,M., Ponder,B.A. and Tunnacliffe,A. (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics, 13, 718–725. [DOI] [PubMed] [Google Scholar]
- 26.Rivin C.J. and Fangman,W.L. (1980) Replication fork rate and origin activation during the S phase of Saccharomyces cerevisiae. J. Cell Biol., 85, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grelon M., Vezon,D., Gendrot,G. and Pelletier,G. (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J., 20, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahadevaiah S.K., Turner,J.M., Baudat,F., Rogakou,E.P., de Boer,P., Blanco-Rodriguez,J., Jasin,M., Keeney,S., Bonner,W.M. and Burgoyne,P.S. (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nature Genet., 27, 271–276. [DOI] [PubMed] [Google Scholar]