Abstract
A new C11-norisoprenoid derivative, sargassumone (1), has been isolated from Sargassum naozhouense together with six known norisoprenoids and a highly oxygenated cyclopentene: (2R,6S,8S,9S)-hexahydro-2,9-dihydroxy-4,4,8-trimethyl-6-acetyloxy-3(2H)-benzofuranone (2), (6S,8S,9R)-hexahydro-6,9-dihydroxy-4,4,8-trimethyl-2(2H)-benzofuranone (3), (6S,8S,9R)-hexahydro-6,9-dihydroxy-4,4,8-trimethyl-2(2H)-benzofuranone (4), loliolide (5), (+)-epiloliolide (6), spheciospongones A (7), and (+)-kjellmanianone (8). Compound 1 was identified on the basis of nuclear magnetic resonance (NMR) and mass spectrometry (MS) analysis, and the absolute stereochemistry was defined by NOESY spectroscopy, minimizing energy calculation, and circular dichroism (CD) spectra. The known compounds 2–8, isolated from S. naozhouense for the first time, were identified by comparison of their physical and spectroscopic data with those reported in the literature. Compound 6 was tested for its inhibitory activity against protein tyrosine phosphatase 1B (PTP1B), antioxidant activity with 1,1-diphyl-2-picrylhydrazyl (DPPH) free radicals, and antimicrobial activity against resistant clinical isolates of Candida albicans, methicillin-resistant Staphylococcus aureus (MRSA), and Escherichia coli.
Keywords: brown alga, Sargassum naozhouense, norisoprenoids, cyclopentene
1. Introduction
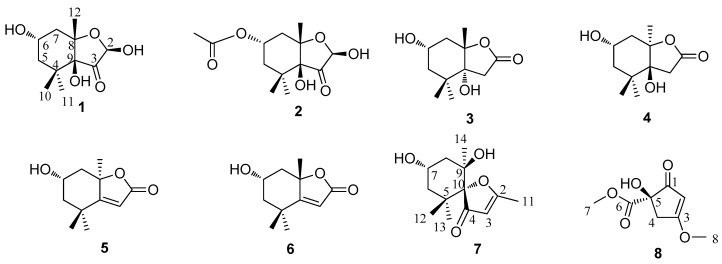
Sargassum naozhouense Tseng et Lu—one species of the Sargassum genus (phylum Phaeophyta, class Cyclosporeae, order Fucales, family Sargassaceae), called “black vegetable”—is distributed widely in the coastal region of the Leizhou Peninsula of China, and can be cultivated commercially under the northern South China Sea’s high-temperature condition [1,2,3,4]. It has a series of pharmaceutical functions in Chinese folk medicine, such as treating infections, laryngitis, dieresis, and other ailments [3]. However, there have been few studies of the chemical constituents that reported primarily carbohydrates, proteins, minerals, dietary fiber, sulfated polysaccharide, and phlorotanns from the brown seaweed S. naozhouense [3,5,6]. In order to fully utilize this species and obtain new bioactive compounds from seaweeds being able to produce a great variety of secondary metabolites characterized by a broad spectrum of biological activities, we investigated the brown alga S. naozhouense. Previous chemical research on this species has resulted in the isolation of 1-O-hexadecanoyl glycerol, pheophytin a, β-sitosterol, mannitol, uracil, thymine, p-hydroxybenzoic acid, 3,4-dihydroxybenzoic acid, 4-hydroxyphthalide, and 2′-deoxythmidine [2]. Now, in our further chemical investigation on the EtOH extract of S. naozhouense, a new C11-norisoprenoid derivative (namely sargassumone (1)), and six known norisoprenoids (2–7), together with a known highly oxygenated cyclopentenone (8) (Figure 1) were obtained. Compounds 2–8 were also isolated from S. naozhouense for the first time.
Figure 1.
Chemical structures of compounds 1–8 isolated from S. naozhouense.
In addition, brown algae present anti-diabetic, antioxidant, and antimicrobial effects [7,8,9], and so compound 6 was tested for its inhibitory activity against protein tyrosine phosphatase 1B (PTP1B), used as a potential therapeutic drug target in the treatment of type 2 diabetes, antioxidant activity with 1,1-diphyl-2-picrylhydrazyl (DPPH) free radicals, and antimicrobial activities against resistant clinical isolates of Candida albicans, methicillin-resistant Staphylococcus aureus (MRSA), and Escherichia coli.
2. Results and Discussion
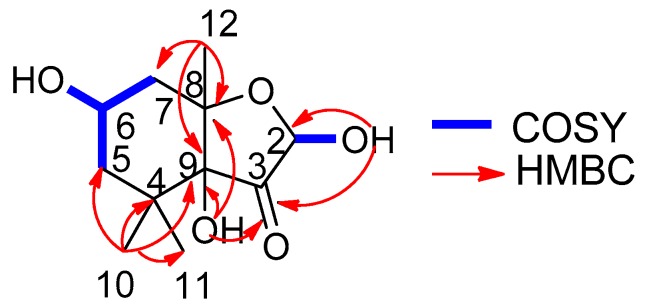
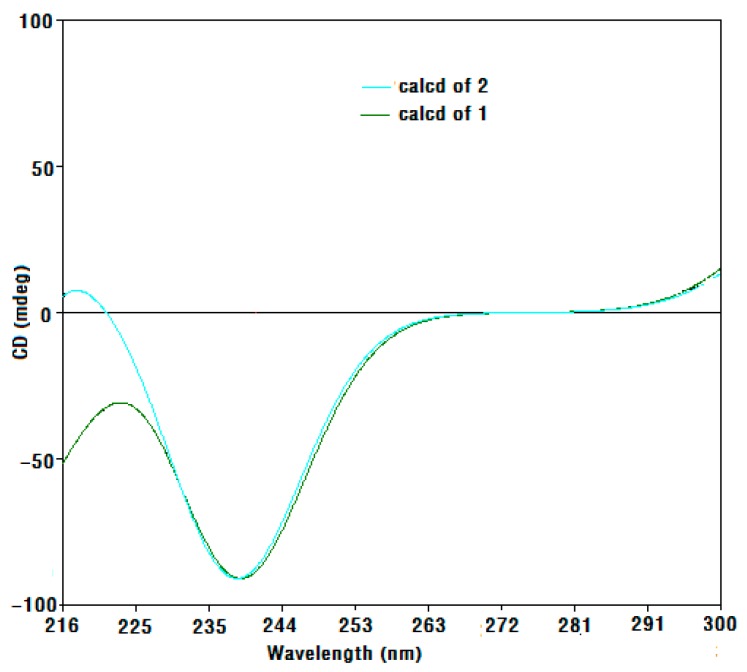
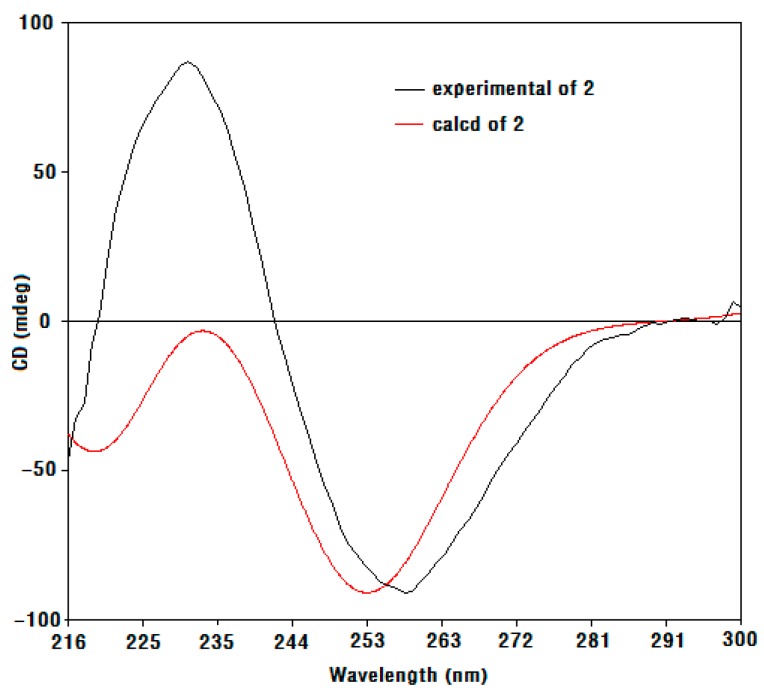
Compound 1 was obtained as a colorless oil, and its molecular formula was determined as C11H18O5 based on electron impact ionization source-mass spectrum (EI-MS) at m/z 212 [M − H2O]+ and NMR spectra data (see supplementary materials). The 1H-NMR spectra showed the resonances of three methyl singlets at δ 0.86 (3H, s), 0.95 (3H, s), and 1.14 (3H, s), four aliphatic protons at the range of δ 1.27–2.07 attributable to two methylene groups, two hydroxymethine protons at δ 3.79 (1H, m) and 4.93 (1H, d, J = 6.0 Hz), and three exchangeable protons at δ 5.36 (9-OH, s), 4.55 (6-OH, d, J = 4.5 Hz), and 7.03 (2-OH, d, J = 6.0 Hz). The 13C-NMR (Table 1), distortionless enchancement by polarization transfer (DEPT), and HMQC spectra revealed the presence of eleven carbons—namely, three methyl carbons, two methylene carbons, two oxymethine carbons, three quaternary carbons (two bearing oxygen), and one ketone carbon, which accounted for the three degrees of unsaturation. The 1H-1H COSY correlation (Figure 2) from H2-5 to H-6 and H-6 to H2-7, which established a moiety of CH2CHOHCH2, in conjunction with the key HMBC correlations from H3-10 to C-4, C-5, C-9, and C-11, H3-12 to C-7, C-8, and C-9, 9-OH to C-3, C-8, and C-9, and 2-OH to C-2 and C-3 (Figure 2), disclosed the structure of hexahydro-2,6,9-trihydyoxy-4,4,8-trimethyl-3(2H)-benzofuranone, which was the deacetyl derivative of compound 2 isolated from the brown alga Undaria pinnatifida as a new compound. The NOESY correlations (Figure 3) of H-6/Ha-7, Ha-7/H3-12, H3-12/2-OH and 9-OH, 9-OH/H3-10 (δ 0.86, s), and H-2/H3-11 (δ 0.95, s) clarified H-6, Ha-7, H3-12, 2-OH, 9-OH, and H3-10 (δ 0.86, s) being on the same side in opposite to H-2 and H3-11 (δ 0.95, s), which were similar to those of compound 2 whose stereochemistry was 2R, 6S, 8S, 9S [10], and so the stereochemistry of 1 may be 2R, 6S, 8S, 9S. By using a Gaussian-03 package (B3LYP/6-31 G(d) level) for minimizing energy calculation, molecular modeling of 1 suggested its most stable conformation was 2R, 6S, 8S, and 9S, which was in agreement with the NOESY results. Furthermore, the calculated circular dichroism (CD) curve of compound 1 was similar to those of compound 2 (Figure 4 and Figure 5), which indicated the same absolute configuration between 1 and 2. Thus, the structure of 1 was determined to be (2R,6S,8S,9S)-hexahydro-2,6,9-trihydroxy-4,4,8-trimethyl-3(2H)-benzofuranone, named sargassumon, which is the first example from natural sources to our knowledge.
Table 1.
1H- (500 MHz) and 13C-NMR (125 MHz) data of compound 1 (in DMSO-d6, δ in ppm, J in Hz).
| Position | δC | δH | HMBC (HC) |
|---|---|---|---|
| 2 | 92.6 | 4.93 d (6.0) | 84.6 |
| 3 | 214.8 | ||
| 4 | 37.1 | ||
| 5 | 44.4 | 1.36 m | |
| 6 | 61.6 | 3.79 m | |
| 7 | 43.1 | 2.07 dd (4.5, 4.5) | 44.4, 61.6, 80.6, 84.6 |
| 8 | 84.6 | ||
| 9 | 80.6 | ||
| 10 | 26.1 | 0.86 s | 25.7, 37.1, 44.4, 80.6 |
| 11 | 25.7 | 0.95 s | 26.1, 37.1, 44.4, 80.6 |
| 12 | 27.6 | 1.14 s | 43.1, 80.6, 84.6 |
| 2-OH | 7.03 d (6.0) | 92.6, 214.8 | |
| 6-OH | 4.55 d (4.5) | 44.4 | |
| 9-OH | 5.36 s | 37.1, 80.6, 84.6, 214.8 |
Figure 2.
The key HMBC and COSY correlation of compound 1.
Figure 3.
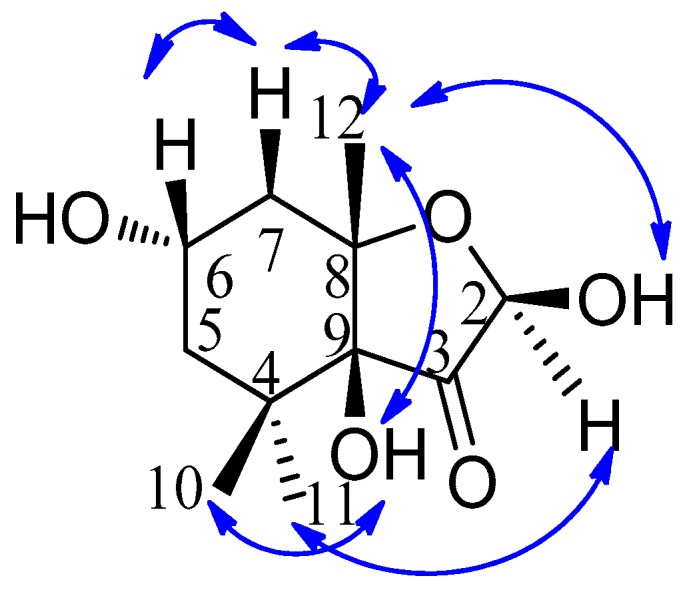
The NOE correlation of compound 1.
Figure 4.
Calculated circular dichroism (CD) spectra of compounds 1 and 2.
Figure 5.
Calculated and experimental CD spectra of compound 2.
Compounds 2–8 were identified as (2R,6S,8S,9S)-hexahydro-2,9-dihydroxy-4,4,8-trimethyl-6-acetyloxy-3(2H)-benzofuranone (2) [10], (6S,8S,9R)-hexahydro-6,9-dihydroxy-4,4,8-trimethyl-2(2H)-benzofuranone (3) [10], (6S,8S,9R)-hexahydro-6,9-dihydroxy-4,4,8-trimethyl-2(2H)-benzofuranone (4) [10], loliolide (5) [10,11], (+)-epiloliolide (6) [10,11,12], spheciospongones A (7) [13], and (+)-kjellmanianone (8) [14], respectively, by comparison of their physical and spectroscopic data with those in the literature.
Compound 6 was evaluated for its inhibitory activity as inhibitor of PTP1B in vitro, and the results showed it showed no or weak activity against PTP1B with IC50 value of over 50 μmol/L in Table 2. Meanwhile, compound 6 was also evaluated for its antioxidant activity with DPPH free radicals, and the results showed certain antioxidant activity with IC50 value of 16.65–17.35 mM in Table 3. In addition, compound 6 was assayed for its antimicrobial activity by the disc diffusion method in vitro. The results of inhibition zones showed that compound 6 exhibited antimicrobial activity against tested resistant strains such as Candida albicans, MRSA, and Escherichia coli, with inhibition zones ranging from 6.30 to 8.33 mm at a concentration of 4.08 µM in Table 4, which were lower than those of tetracycline used as a positive control at the same concentration. Interestingly, compound 6 exhibited better antimicrobial activities against tested resistant isolates than those of penicillin used as another positive control at the same concentration, which showed no or very weak activity. Furthermore, compound 6 showed antimicrobial activity against the standard strain (ATCC 29213, a non-antibiotic-resistant strain), with the inhibition zone of 7.10–7.90 mm at a concentration of 4.08 µM.
Table 2.
Protein tyrosine phosphatase 1B (PTP1B) inhibitory activity of compound 6.
| Compounds | Inhibitory Activity (IC50, μmol/L) # |
|---|---|
| 6 | >50 |
| Oleanolic acid | 1.9 ± 0.3 |
# IC50 values were determined by regression analysis and expressed as mean ± SD of three replicates.
Table 3.
1,1-Diphyl-2-picrylhydrazyl (DPPH) free radical scavenging activity of compound 6 #.
| Compounds | IC50 (mM) |
|---|---|
| 6 | 17 ± 0.35 |
| Vitamin C | 0.11 ± 0.01 |
# mean ± SD of three replicates.
Table 4.
Antimicrobial activity of compound 6.
| Strains | Zone of Inhibition (mm) # | ||
|---|---|---|---|
| 6 | Tetracycline | Penicillin | |
| Candida albicans 615 | 6.50 ± 0.20 | 10.50 ± 0.23 | – |
| MRSA 315 | 8.20 ± 0.11 | 11.00 ± 0.10 | – |
| Escherichia coli 234 | 7.00 ± 0.20 | 12.00 ± 0.06 | – |
| Staphyloccocus aureus 29213 | 7.50 ± 0.40 | 9.50 ± 0.52 | 11.00 ± 0.05 |
#: mean ± SD of three replicates; –: no or very weak activity. MRSA: methicillin-resistant Staphylococcus aureus.
3. Materials and Methods
3.1. General
Optical rotations were measured with a Perkin-Elmer model 341 polarimeter (Perkin Elmer Corporation, Boston, MA, USA). NMR spectra were recorded on a Bruker Avance 500 NMR spectrometer (Bruker Corporation, Faellanden, Switzerland). HR-ESI-MS spectra were recorded on a Bruker maXis impact mass spectrometer (Bruker Corporation, Bremen, Germany). EI-MS data were measured on a Thermo Scientific DSQ mass spectrometer (Thermo Fisher Scientific Corporation, New York, NY, USA). CD spectra were recorded on a Chirascan CD spectropolarimeter (Applied Photophysics Ltd., Surrey, UK). Column chromatography was performed on silica gel (100–200 and 200–300 mesh, Yantai Jiangyou Silica Gel Development Co., Ltd., Yantai, China) and Sephadex LH-20 (Pharmacia, Uppsala, Sweden). Thin-layer chromatography (TLC) was performed on pre-coated silica gel GF254 plates (Yantai Jiangyou Silica Gel Development Co., Ltd., Yantai, China), visualization under UV light, or by heating after spraying with 5% H2SO4–EtOH (v/v). Organic solvents for column chromatography were either spectral grade or analytical reagents and obtained from Guangzhou Dongju Test Instrument Co., Ltd. (Guangzhou, China).
Protein tyrosine phosphatase 1B (PTP1B), p-nitrophenyl phosphate disodium (pNPP), 3-(N-morpholino)-propanesulfonic acid (MOPS), and dimethyl sulphoxide (DMSO) were obtained from the National Center for Drug Screening (Shanghai, China). The clinical resistant strains (Candida albicans 615, MRSA 315, and Escherichia coli 234) were isolated from phlegm samples provided by Guangdong General Hospital (Guangdong, China). The standard strain (Staphyloccocus aureus ATCC 29213) was provided by the RNAM Center for Marine Microbiology, South China Sea Institute of Oceanology (Guangzhou, China).
3.2. Plant Materials
The brown seaweed S. naozhouense was collected in July 2012 from Leizhou Peninsula of Guangdong Province, China. The specimens were identified by Professor Enyi Xie of the College of Fisheries, Guangdong Ocean University. A voucher specimen (No. 20120711) was deposited at the CAS Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences.
3.3. Extraction and Isolation
The dried seaweeds (3 kg) were chopped and extracted with 75% EtOH at room temperature for 3 × 7 d. Removal of the solvent in vacuo afforded a syrupy residue (120 g), which was suspended in H2O followed by successive partition with petroleum ether, trichloromethane, ethyl acetate, and n-butanol. The trichloromethane extract (5.56 g) was subjected to column chromatography (CC) over silica gel (200–300 mesh) eluting with gradient CHCl3/acetone (100:0–0:100) to afford four fractions, Frs. 1–4. Fr.1 (100 mg) was subjected to CC eluting with gradient CHCl3/MeOH (25:1–10:1) to obtain two subfractions (Frs. 1–1–1–2). Frs. 1–1 was purified by Sephadex LH-20 to yield 1 (1 mg). Frs. 1–2 was directly purified by recrystallization to yield 2 (6 mg). Frs. 2 (200 mg) was further subjected to silica gel column chromatography eluting with CHCl3/MeOH (25:1) to yield 3 (20 mg) and 4 (2 mg). Fr. 3 (200 mg) was subjected to CC eluting with CHCl3/MeOH (20:1) to yield 5 (1 mg), 6 (50 mg), and 7 (1 mg). Fr. 4 was purified by Sephadex LH-20 to yield 8 (1 mg).
Compound 1: Colorless oil, [α +8.4° (c 0.018, CHCl3). EI-MS (m/z): 212 [M − H2O]+, 169, 156, 112, 101, 95, 83, 55; 1H-NMR (DMSO-d6, 500 MHz), see Table 1; 13C-NMR (DMSO-d6, 125 MHz), see Table 1.
Compound 2: Colorless oil, [α +16.7° (c 0.003, CHCl3). CD (CHCl3) Δε258nm −0.023, Δε231nm +0.023; 1H-NMR (CDCl3, 500 MHz) δ: 5.28 (1H, d, J = 4.6 Hz, H-2), 5.17 (1H, m, H-6), 3.14 (1H, d, J = 4.6 Hz, 2-OH), 2.65 (1H, s, 9-OH), 2.42 (1H, dd, J = 14.2, 4.6 Hz, Ha-7), 1.60 (2H, m, Hb-7, 5), 1.54 (1H, dd, J = 14.2, 11.0 Hz, Ha-5), 1.31 (3H, s, H-11), 1.15 (3H, s, H-12), 1.01 (3H, s, H-10); 13C-NMR (CDCl3, 125 MHz) δ: 93.1 (C-2), 214.1 (C-3), 37.8 (C-4), 40.4 (C-5), 66.9 (C-6), 39.2 (C-7), 85.6 (C-8), 81.3 (C-9), 25.6 (C-10), 27.4 (C-11), 25.5 (C-12), 170.5 (C-13), 21.3 (C-14).
Compound 3: Colorless oil, [α +8.4° (c 0.01, CHCl3). 1H-NMR (CD3OD, 500 MHz) δ: 3.86 (1H, m, H-6), 3.18 (1H, d, J = 15.5 Hz, Ha-3), 2.36 (1H, d, J = 17.5 Hz, Hb-3), 2.31 (1H, dt, J = 13.0, 4.0 Hz, Ha-7), 1.78 (1H, dt, J = 13.0,3.0 Hz, Hb-7), 1.58 (1H, d, J = 12.5 Hz, Ha-5), 1.50 (1H, d, J = 12.5 Hz, Hb-5), 1.56 (3H, s, H-12), 1.08 (3H, s, H-11), 1.00 (3H, s, H-10); 13C-NMR (CD3OD, 125 MHz) δ: 177.4 (C-2), 42.4 (C-3), 38.1 (C-4), 48.0 (C-5), 64.3 (C-6), 47.3 (C-7), 91.1 (C-8), 82.1 (C-9), 21.3 (C-10), 23.8 (C-11), 27.4 (C-12).
Compound 4: Colorless oil, [α −31.9° (c 0.0033, CHCl3). 1H-NMR (DMSO-d6, 500 MHz) δ: 3.00 (1H, d, J = 17.5 Hz, Ha-3), 2.25 (1H, d, J = 17.0 Hz, Hb-3), 2.00 (1H, dt, J = 14.0, 7.0 Hz, Ha-7), 1.78 (1H, dt, J = 14.0, 5.0 Hz, Hb-7), 1.58 (2H, m, H-5), 1.48 (3H, s, H-12), 1.10 (3H, s, H-11), 0.85 (3H, s, H-10); 13C-NMR (DMSO-d6, 125 MHz) δ: 174.7 (C-2), 40.8 (C-3), 36.8 (C-4), 43.3 (C-5), 63.9 (C-6), 42.7 (C-7), 88.6 (C-8), 79.8 (C-9), 22.5 (C-10), 25.3 (C-11), 27.1 (C-12).
Compound 5: Colorless needles, [α −21.3° (c 0.003, CHCl3). 1H-NMR (CDCl3, 500 MHz) δ: 5.68 (1H, s, H-3), 4.33 (1H, t, J = 3.2 Hz, H-6), 2.47 (1H, d, J = 14.0 Hz, Ha-7), 1.99 (1H, d, J = 14.5 Hz, Ha-5), 1.76 (1H, d, J = 3.5 Hz, Hb-7), 1.54 (1H, dd, J = 14.5, 3.5 Hz, Hb-5), 1.78 (3H, s, H-12), 1.46 (3H, s, H-11), 1.26 (3H, s, H-10); 13C-NMR (CDCl3, 125 MHz) δ: 182.5 (C-2), 112.9 (C-3), 35.9 (C-4), 47.4 (C-5), 66.8 (C-6), 45.7 (C-7), 86.7 (C-8), 171.6 (C-9), 26.5 (C-10), 27.0 (C-11), 30.7 (C-12).
Compound 6: Colorless needles, [α +42.9° (c 0.058, CHCl3). 1H-NMR (CDCl3, 500 MHz) δ: 5.65 (1H, s, H-3), 4.10 (1H, m, H-6), 2.50 (1H, ddd, J = 12.0, 3.5, 1.5 Hz, Ha-7), 2.01 (1H, ddd, J = 13.0, 4.0, 2.0 Hz, Ha-5), 1.54(3H, s, H-12), 1.48 (1H, t, J = 12.0 Hz, Hb-7), 1.31(1H, d, J = 12.0 Hz, Hb-5), 1.26 (3H, s, H-11), 1.22 (3H, s, H-10); 13C-NMR (CDCl3, 125 MHz) δ: 181.4 (C-2), 112.9 (C-3), 35.0 (C-4), 49.6 (C-5), 64.6 (C-6), 47.8 (C-7), 86.8 (C-8), 171.8 (C-9), 25.4 (C-10), 25.0 (C-11), 29.8 (C-12).
Compound 7: Colorless oil, HR-ESI-MS (m/z): 263.1799 [M + Na]+. CD (MeOH) Δε265nm −1.022; 1H-NMR (CDCl3, 500 MHz) δ: 5.58 (1H, s, H-3), 4.26 (1H, m, H-7), 2.07 (3H, s, H-11), 2.05 (1H, m, Ha-8), 1.60~1.68 (2H, m, H-6), 1.51~1.58 (1H, m, Hb-8), 1.36 (3H, s, H-13), 1.02 (3H, s, H-14), 0.82 (3H, s, H-12); 13C-NMR (CDCl3, 125 MHz) δ: 190.6 (C-2), 107.3 (C-3), 207.6 (C-4), 23.1 (C-5), 45.9 (C-6), 64.3 (C-7), 44.8 (C-8), 75.5 (C-9), 92.5 (C-10), 16.6 (C-11), 26.3 (C-12), 22.2 (C-13), 25.8 (C-14).
Compound 8: Colorless crystals, [α +1.6° (c 1.80, CHCl3). 1H-NMR (CDCl3, 500 MHz) δ: 5.34 (1H, s, H-2), 3.94 (3H, s, H-7), 3.80 (3H, s, H-8), 3.20 (1H, d, J = 17.5 Hz, Ha-4), 2.76 (1H, d, J = 17.5 Hz, Hb-4).
3.4. Bioactivity Assay
3.4.1. PTP1B Inhibition Assay
PTP1B (protein tyrosine phosphatase 1B) was expressed and purified in E. coli, and the enzymatic activities were measured at 30 °C by monitoring the hydrolysis of pNPP [15]. The enzymatic activities of PTP1B catalytic domain were also determined at 30 °C by monitoring the hydrolysis of pNPP. Dephosphorylation of pNPP generates p-nitrophenol, which can be monitored spectrophotometrically at 405 nm. In a 100-μL assay mixture containing 50 mM MOPS, pH 6.5, 2 mM pNPP, and recombinant enzymes, PTP1B activities were continuously monitored and the initial rate of the hydrolysis was determined from the early linear region of the enzymatic reaction kinetic curve as described by Zhang et al. [15].
The inhibitory activity of Compound 6 was tested against PTP1B by the colorimetric assay described by Zhang et al. Briefly, the compound was solubilized in DMSO at 20 μg/mL, and then added to a 96-well clear polystyrene plate (Corning, Action, MA, USA) with oleanolic acid (12.5 mM) as the positive inhibition. After adding an assay mixture (88 μL), 10 μL 300 nM GST-PTP1B was added to initiate the reaction. Then, the catalysis of pNPP was continuously monitored on SpectraMax 340 microplate reader (Molecular Devices, Sunnyvale, CA, USA) at 405 nm for 2 min at 30 °C in a final 100 μL volume containing 50 mM MOPS, pH 6.5, 2 mM pNPP, 30 nM PTP1B, and 2% DMSO. The IC50 value was calculated from the non-linear curve fitting percent inhibition (% inhibition) to inhibitor concentration [I] by the following equation, where k is the Hill coefficient:
| % Inhibition = 100/{(1 + (IC50)/[I])k}. | (1) |
3.4.2. DPPH Free Radical Scavenging Assay
The free radical scavenging potential of compound 6 was evaluated using the DPPH free radical scavenging assay described by Sharma and Bhat [16]. A total of 200 μL of the reaction mixture in a 96-well plate was composed of 100 μL of 0.1 mM DPPH (Sigma Aldrich, St. Louis, MO, USA) in methanol and 100 μL of different concentrations of test compound. The reaction mixture was then shaken well and incubated for 30 min in darkness at 37 °C. Meanwhile, a sample composed of 100 μL of methanol with 100 μL of 0.1 mM DPPH was taken as a control, while vitamin C (Shanxi Yishengtang Pharmaceutical Co., Ltd., Tongchuan, China) was taken as a reference material. The absorbance of reaction solution was measured at 517 nm with a plate reader (Thermo Multiskan MK3 spectrophotometer, Thermo Fisher Scientific, Waltham, MA, USA), and the percentage of scavenging capacity (%) was calculated by the following equation, where AControl stands for the absorbance of the control and ATreatment stands the absorbance of the treatment:
| Scavenging capacity (%) = (1 − ATreatment/AControl) × 100. | (2) |
The DPPH IC50 value is the concentration required to scavenge DPPH radical by 50%.
3.4.3. Antimicrobial Activity
Antimicrobial test for compound 6 was carried out against resistant microbes such as Candida albicans, MRSA, and Escherichia coli by disc diffusion method [17]. Briefly, a Petri dish was prepared with a base layer of Muller Hinton (MH) agar (10 mL), and then 0.1 mL culture with the concentration of 105 CFU/mL was uniformly distributed on to MH agar plates. Paper discs with the diameter of 5 mm, made and sterilized, were placed on the surface of MH agar plates at a distance of 2 cm. Drugs with the concentration of 800 μg/mL were added on each disc and incubated at 37 °C or 20 °C for 8–10 h. After incubation, the diameters of the inhibition zones were measured.
3.5. Statistical Analysis
For the bioactivity assay, all determinations were triplicates, and data were expressed as mean and standard deviation (SD). Analysis of variance was performed, and the mean separation was done by the least significant difference (LSD) (p ≤ 0.05) using SPSS 18.0 software for Windows (SPSS Inc., Chicago, IL, USA).
4. Conclusions
The phytochemical investigation of S. naozhouense afforded a new C11-norisoprenoid derivative and six known norisoprenoids, as well as a known cyclopentenone derivative. All isolates were isolated from S. naozhouense for the first time. Compound 6 showed a marginal inhibitory activity against PTP1B, certain antioxidant activity against DPPH radicals, and antimicrobial activities against some resistant microbes such as Candida albicans, MRSA, and Escherichia coli.
Acknowledgments
This study was supported by grants from the Natural Science Foundation of Guangdong Province (No. 2014A030307036), Natural Science Foundation of Zhanjiang Normal University (No. 2L1312), and Zhanjiang City-Science and Technology Program (Nos. 2013A01024 and 2016A03025).
Supplementary Materials
The following are available online.
Author Contributions
Yan Peng contributed in performing the experiment and writing the manuscript. Ri-Ming Huang and Xiu-Ping Lin analyzed the data. Yong-Hong Liu conceived and designed the format of the manuscript. All the authors contributed in critical reading and discussion on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Sample of the compound 6 is available from the authors.
References
- 1.Huang Z.G. Marine Species and Their Distributions in China’s Seas. 1st ed. China Ocean Press; Beijing, China: 1994. pp. 216–221. [Google Scholar]
- 2.Peng Y., Gao W.H., Lin X.P., Yan T., Liu Y.H. Chemical constituents of seaweed Sargassum naozhouense. J. Chin. Med. Mater. 2014;37:2210–2212. doi: 10.13863/j.issn1001-4454.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 3.Peng Y., Xie E., Zheng K., Fredimoses M., Yang X., Zhou X., Wang Y., Yang B., Lin X., Liu J., et al. Nutritional and chemical composition and antiviral activity of cultivated seaweed Sargassum naozhouense Tseng et Lu. Mar. Drugs. 2013;11:20–32. doi: 10.3390/md11010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie E.Y., Liu B.C., Jia C., Chen X.L., Yang B. Artificial seed production and cultivation of the edible brown alga Sargassum naozhouense Tseng et Lu. J. Appl. Phycol. 2013;25:513–522. doi: 10.1007/s10811-012-9885-2. [DOI] [Google Scholar]
- 5.Lu H., Chen X., Ou X., Mao J., Ji H. Anticoagulant activity of phlorotannins from S. naozhouense. Nat. Prod. Res. Dev. 2013;25:249–252. doi: 10.16333/j.1001-6880.2013.02.026. [DOI] [Google Scholar]
- 6.Feng M., Li Y., Cui L., Liu Y. The protective effect of phlorotannins from Sargassum naozhouense on bone loss in ovariectomized mice with hyperlipidemia. Chin. J. Osteoporos. 2015;21:586–591. doi: 10.3969/j.issn.1006-7108.2015.05.014. [DOI] [Google Scholar]
- 7.Sharifuddin Y., Chin Y.X., Lim P.E., Phang S.M. Potential Bioactive compounds from seaweed for diabetes management. Mar. Drugs. 2015;13:5447–5491. doi: 10.3390/md13085447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heo S.J., Park E.J., Lee K.W., Jeon Y.J. Antioxidant activities of enzymatic extracts from brown seaweeds. Bioresour. Technol. 2005;96:1613–1623. doi: 10.1016/j.biortech.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Shanmughapriya S., Manilal A., Sujith S., Selvin J., Seghal Kiran G., Natarajaseenivasan K. Antimicrobial activity of seaweeds extracts against multiresistant pathogens. Ann. Microbiol. 2008;58:535–541. doi: 10.1007/BF03175554. [DOI] [Google Scholar]
- 10.Kimura J., Maki N. New loliolide derivatives from the brown alga Undaria pinnatifida. J. Nat. Prod. 2002;65:57–58. doi: 10.1021/np0103057. [DOI] [PubMed] [Google Scholar]
- 11.Chávez J.P., Santos I.D.D., Cruz F.G., David J.M., Yang S.W., Cordell G.A. A quinoline alkaloid from Acanthosyris Paulo-alvinii. Phytochemistry. 1997;46:967–968. doi: 10.1016/S0031-9422(97)00363-4. [DOI] [Google Scholar]
- 12.Park K.E., Kim Y.A., Jung H.A., Lee H.J., Ahn J.W., Lee B.J., Seo Y.W. Three norisoprenoids from the brown alga Sargassum thunbergii. J. Korean Chem. Soc. 2004;48:394–398. doi: 10.5012/jkcs.2004.48.4.394. [DOI] [Google Scholar]
- 13.Liu D., Xu M.J., Wu L.J., Deng Z.W., Lin W.H. Norisoprenoids from the marine sponge Spheciospongia sp. J. Asian Nat. Prod. Res. 2009;11:811–816. doi: 10.1080/10286020903058941. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama M., Fukuoka Y., Nozaki H., Matsuo A., Hayashi S. Structure of (+)-kjellmanianone, a highly oxygenated cyclopentenone from the marin alga, Sargassum kjellmanianum. Chem. Lett. 1980;9:1243–1246. doi: 10.1246/cl.1980.1243. [DOI] [Google Scholar]
- 15.Zhang W., Hong D., Zhou Y.Y., Zhang Y.N., Shen Q., Li J.Y., Hu L.H., Li J. Ursolic acid and its derivative inhibit protein tyrosine phosphatase 1B, enhancing insulin receptor phosphorylation and stimulation glucose uptake. Biochim. Biophys. Acta. 2006;1970:1505–1512. doi: 10.1016/j.bbagen.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Sharma O.P., Bhat T.K. DPPH antioxidant assay revisited. Food Chem. 2009;113:1202–1205. doi: 10.1016/j.foodchem.2008.08.008. [DOI] [Google Scholar]
- 17.Santhosh S.K., Venugopal A., Radhakrishnan M.C. Study on the phytochemical, antibacterial and antioxidant activities of Simarouba glauca. Indian J. Biol. Sci. 2016;2:119–124. doi: 10.22205/sijbs/2016/v2/i1/100358. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.