Abstract
Chop is a ubiquitously expressed mammalian gene encoding a small nuclear protein related to the CCAAT/enhancer-binding protein (C/EBP) family of transcription factors. CHOP protein plays an important role in various cellular processes such as growth, differentiation and programmed cell death. CHOP expression is strongly increased in response to a large variety of stresses including perturbation of the endoplasmic reticulum function, DNA damage and nutrient deprivation. Multiple mechanisms including transcriptional and post-transcriptional controls are involved in the regulation of CHOP expression. We show here that the 5′UTR of the Chop transcript plays an important role in controlling the synthesis of CHOP protein. In particular, the 5′UTR contains a conserved uORF which encodes a 31 amino acid peptide that inhibits the expression of the downstream ORF. Mutational analysis of the 5′ leader region and peptide coding sequences suggests that the peptide itself inhibits expression of the downstream ORF. Such results suggest a role for uORF in limiting ribosomal access to downstream initiation sites. With respect to the importance of CHOP protein in the regulation of cellular functions, the mechanisms that regulate its basal level are of considerable interest.
INTRODUCTION
Chop is a ubiquitously expressed mammalian gene encoding a small nuclear protein related to the CCAAT/enhancer-binding protein (C/EBP) family of transcription factors. Members of the C/EBP family have been implicated in the regulation of processes relevant to energy metabolism (1), cellular proliferation, differentiation and expression of cell-type-specific genes (2–4). Over-expression of CHOP attenuates the process of adipocyte differentiation in 3T3-L1 cells (5), leads to cell cycle arrest (6) and can interfere with programmed cell death (7,8). Additional evidence for the importance of CHOP in cellular growth comes from the observation that, in a vast majority of cases of myxoid and liposarcoma, the Chop gene is structurally rearranged, giving rise to the constitutive expression of an altered, oncogenic form of the protein (9–14).
The Chop gene encodes a protein that acts as a transcriptional activator, essential for the expression of a set of genes (15). By forming heterodimers with other members of the C/EBP family, CHOP can influence gene expression as both a dominant-negative regulator of C/EBP and by redirecting CHOP–C/EBP heterodimers to other sequences (5,6,11,16,17). It has also been recently shown that CHOP can interact with members of the growth-promoting transcription factor family, including jun-d, c-jun and c-fos, to activate promoter elements in the somatostatin, jun-d and collagenase genes (18).
In a wide variety of cells, the Chop gene is induced by stress giving rise to DNA damage or decreased function of the endoplasmic reticulum (ER) such as genotoxic agents (19), oxidative or reductive stress (20,21) and glucose deprivation (22). In addition, Chop is responsive to amino acid deprivation through a pathway that is distinct from the ER stress-signaling cascade (23,24). In animals, Chop mRNA is increased during the acute phase response initiated by treatment with lipopolysaccharide (25) and during caloric restriction (26).
In every cell type tested, the basal expression level of CHOP is very low, almost undetectable. However, CHOP expression can be rapidly induced in response to different stimuli. Taking into consideration the important consequences of its expression, it is reasonable to expect that CHOP level is highly regulated. It has been previously shown that Chop induction involves both transcriptional and post-transcriptional mechanisms (23,27). These observations led us to investigate the potential role for translational control in determining CHOP expression level. In this study we report that an upstream open reading frame (uORF) located in the Chop 5′-untranslated region (5′UTR) inhibits the rate of CHOP translation. Analysis of the mechanisms by which the Chop uORF inhibits downstream translation demonstrates that the uORF encodes a functional polypeptide that inhibits translation of the downstream cistron.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were from Eurogentec (Belgium). When double-stranded oligonucleotides were required, equal numbers of moles of complementary strands were heated to 90°C for 1 min and annealed by slow cooling to room temperature.
Cell culture and treatment conditions
HeLa cells were cultured at 37°C in Dulbecco’s modified Eagle’s medium F12 (DMEM F12; Sigma) containing 10% decomplemented fetal bovine serum (FBS).
DNA transfection
Cells were plated on 60 mm plates and transfected by the calcium phosphate coprecipitation method as described previously (28). LUC plasmid (5 µg) was transfected into the cells along with 0.25 µg of pCMV–β-gal, a plasmid carrying the bacterial Lac-Z gene (encoding β-galactosidase) fused to the human cytomegalovirus immediate-early enhancer/promoter region, as an internal control. Cells were exposed to the precipitate for 16 h, washed twice in PBS, and then incubated with DMEM F12 containing 10% FBS. Forty-eight hours after transfection, cells were trypsinised and diluted in 1 ml of PBS. For each sample, 500 µl of cell suspension was used for luciferase assay and 500 µl was used for RNA isolation and transcripts quantification.
Luciferase assay
Trypsinised cells were pelleted by short centrifugation and the supernatant was discarded. Cells were then suspended in 150 µl of lysis buffer (Promega) and centrifuged at 13 000 g for 2 min. Twenty microliters of the supernatant were assayed for luciferase activity (PRODEMAT, Anduze, France). β-Galactosidase activity was measured as described previously (29). Relative luciferase activity was given as the ratio of relative light unit/ relative β-gal unit. All values are the means calculated from the results of at least three independent experiments.
RNA isolation and quantification of LUC and LacZ transcripts
RNA was prepared from a cells pellet using a QIAGEN RNeasy Mini Kit. After quantification of total RNA by measuring OD at 260 nm, 0.5 µg of RNA was treated with 1 U of DNase I (Gibco BRL). Reverse transcription using random primers (Hexanucleotides Mix; Boehringer Mannheim) was performed with Superscript RT II (Gibco BRL) according to the protocol of the manufacturer. The first strand obtained was quantified using a real time quantitative PCR system: TaqMan assay on ABI PRISM 7700 Sequence Detection System (SYBR Green I double-stranded DNA binding dye chemistry). The following oligonucleotides were used for the amplification of 103 and 105 bp fragments of the cDNAs corresponding to LUC or LacZ, respectively: LUC primers, 5′-AGG GAC AAG ACA ATT GCA CTG A-3′ and 5′-TGC GAG AAT CTC ACG CAG G-3′; LacZ primers, 5′-GCT GCA TAA ACC GAC TAC ACA AA-3′ and 5′-GCC GCA CAT CTG AAC TTC AG-3′. The samples were first heated at 50°C for 10 min and then at 95°C for 10 min and amplified during 40 cycles as follows: 95°C for 15 s, 60°C for 30 s, 72°C for 30 s. The relative LUC mRNA level was given as the ratio of LUC mRNA/LacZ mRNA.
In order to validate the techniques used in these experiments, we verified that the protein content encoded by one reporter gene is proportional to its mRNA content in the absence of translational regulation (results not shown). This experiment was performed following transfection of various amounts of pCMV–β-gal and pCMV–LUC plasmids. We can conclude that Luc and β-gal activities and the transcript contents are accurately measured.
Expression of tagged CHOP protein
The identical 9E10-tagged CHOP protein was expressed from recombinant retroviral vectors (pBABE.puro) that either lacked the 5′UTR of Chop or contained the 5′UTR of human Chop. Construction of the CHOP-expressing retrovirus was performed as described previously (5). NIH-3T3 cells infected with recombinant retrovirus were selected for puromycine resistance. CHOP protein levels in NIH-3T3 cells stably expressing equal amounts of the retroviral mRNA were measured by western immunoblotting using a rabbit polyclonal antiserum against CHOP.
Plasmid constructions
All constructs were generated by PCR using Pfu polymerase (Stratagene), forward and reverse primers containing appropriate restriction sites at their 5′ end. Amplified fragments were then cloned into the pGL3-basic reporter construct (Promega) using the corresponding restriction sites.
The CHOP–5′UTR–LUC construct (Fig. 1B) was generated by inserting the +6 to +170 PCR fragment of the 5′UTR of Chop (generated by using AflII-ended forward primer and HindIII-ended reverse primer) in between the AflII and HindIII restriction sites of the –649/+91CHOP–LUC construct (30), a plasmid containing the CHOP promoter upstream LUC. The CHOP–5′UTR(21)–LUC (Fig. 1B) construct was generated by substituting the +6 to +170 5′UTR fragment by a deleted fragment (+6 to +21) generated by PCR using AflII-ended forward and HindIII-ended reverse primers.
Figure 1.
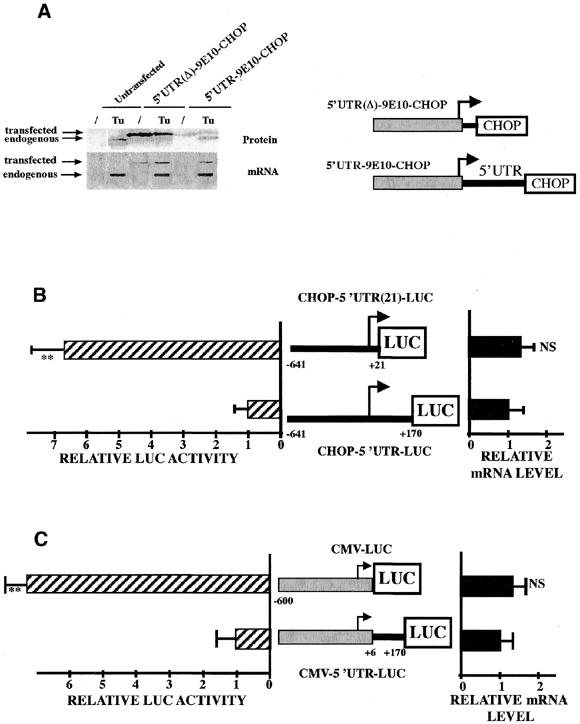
The Chop 5′UTR sequence represses translation of the downstream coding sequences. (A) NIH-3T3 cells were infected with recombinant retrovirus expressing, under the control of the CMV promoter, the tagged CHOP ORF that contained the 5′UTR of human Chop (5′UTR–9E10–CHOP, lane 3) or not [5′UTR(Δ)–9E10–CHOP; lane 2]. A control experiment was performed by infecting cells with an empty virus (Untransfected, lane 1). Cells were then incubated for 4 h with or without 2.5 µg/ml of tunicamycin. On the construct scheme, the arrow represents the transcription start site. Protein extracts were analyzed for the presence of CHOP protein and mRNA as described in Materials and Methods. (B) HeLa cells were transiently transfected with LUC constructs containing either the wild-type (5′UTR–LUC) or the deleted [5′UTR(21)–LUC] Chop 5′UTR downstream of the Chop promoter. Forty-eight hours after transfection, cells were harvested and analyzed for relative luciferase activities and relative LUC mRNA level as described in Materials and Methods. (C) HeLa cells were transiently transfected with constructs containing the CMV promoter driving a LUC construct containing or not containing the Chop 5′UTR. The CHOP 5′UTR was cloned +6 nt after the start site for transcription. The relative Luc mRNA content was not affected by the CHOP 5′UTR (t-test, n > 3, ** = P < 0.0001, NS = not significant).
The CMV–LUC construct (Figs 1C and 4) containing the human cytomegalovirus immediate-early enhancer/promoter region (CMV) (–600 to +64) was generated by PCR using XhoI-ended forward primer and HindIII-ended reverse primer. The amplified fragment was then cloned into pGL3 at the XhoI and HindIII restriction sites.
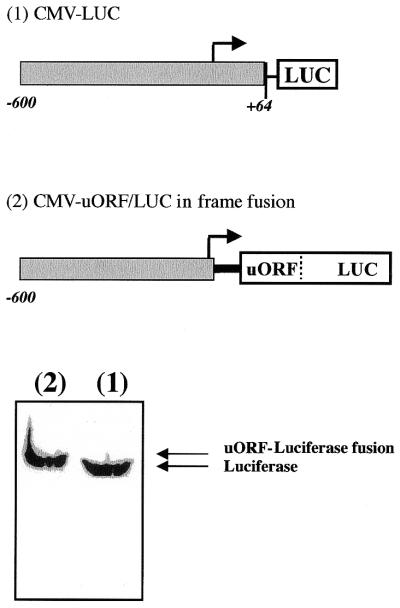
Figure 4.
Translation is initiated at the uORF AUG codon. HeLa cells were transiently transfected with LUC constructs containing either (1) LUC downstream of a CMV promoter or (2) a construct containing CHOP-uORF/LUC in-frame fusion downstream of the CMV promoter. Seventy-two hours after transfection, whole cell lysates were prepared and analyzed for the luciferase protein content by western blotting as described in Materials and Methods.
The CMV–5′UTR–LUC construct [uAUG(+) construct; Figs 1C, 3, 5(1), 6(5) and 7A(1)] containing the human CMV promoter (–600 to +6) and the 5′UTR of Chop (+6 to +170) was generated by cloning a PCR fragment of the CMV promoter using XhoI-ended forward primer and AflII-ended reverse primer in between the XhoI and AflII sites of CHOP–5′UTR–LUC.
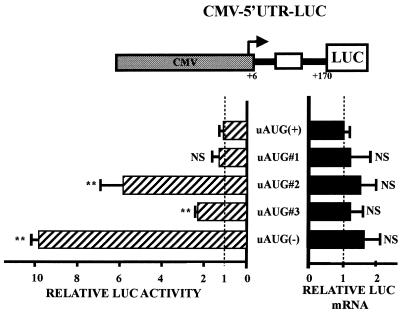
Figure 3.
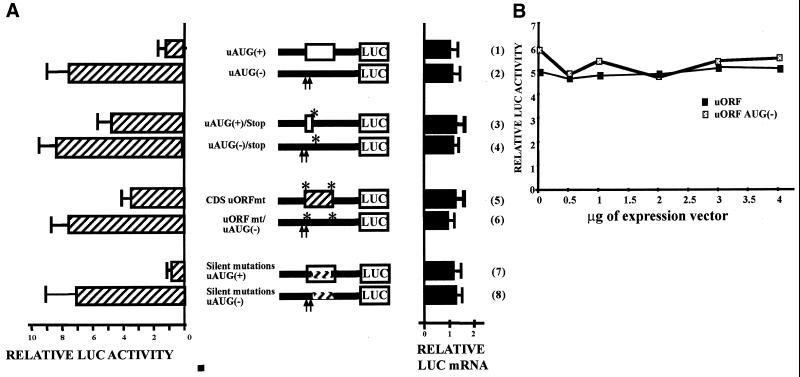
Mutations in the uAUGs abolish the repressor effect of the 5′UTR. HeLa cells were transiently transfected with LUC constructs containing the wild-type [uAUG(+)] or mutated Chop 5′UTR downstream of the CMV promoter. The AUGs shown in Figure 2 were mutated individually (uAUG#1, uAUG2, uAUG#3) or all together [uAUG(–)]. On the construct scheme, the arrow represents the transcription start site and the open box shows the uORF. Forty-eight hours after transfection cells were trypsinised, then half was proceeded for luciferase and β-galactosidase activity assays, and the second half was used for RNA extraction and transcript quantification. Relative LUC activities and mRNA levels were determined as the ratio LUC/βGAL as described in Materials and Methods. (LUC activity: t-test, n > 5, ** = P < 0.0001, NS = not significant; LUC mRNA: t-test, n = 3, NS = not significant).
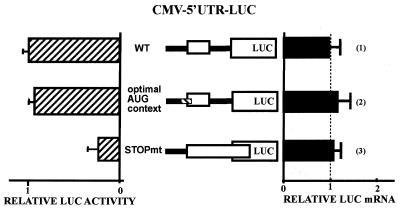
Figure 5.
Translational repression by the Chop uORF is impaired by elongated uORF. The constructs shown in this figure are derived from the CMV–5′UTR–LUC (1). (2) The uAUG#2 was mutated in an optimum consensus start site (gccgccaccAUGg). In the following construct, the STOPmt construct (3) was generated by mutation of the stop codon of the uORF and by the introduction of a 1 nt frame shift in order to generate an extended uORF that is not in-frame with the LUC ORF. The CHOP 5′UTR and the LUC coding sequence are presented for each construct and the uORF is boxed. Forty-eight hours after transfection, cells were harvested and analyzed for relative LUC activities and mRNA level as described in Materials and Methods.
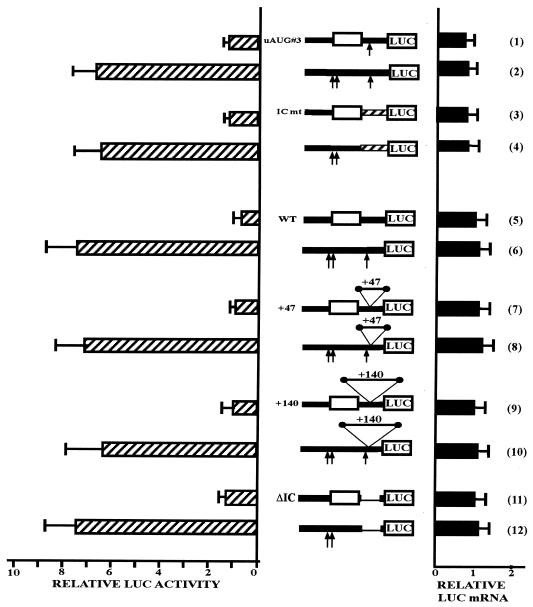
Figure 6.
Translational repression by the Chop uORF is not dependent on the intercistronic region. Expression constructs containing the 5′UTR either in a wild-type form (5) or mutated on the uAUGs (6) were used to mutate the intercistronic region. The arrows represent point mutation of the uAUGs. When present the uORF is boxed. These constructs were transfected in HeLa cells then, 48 h after transfection, cells were harvested. The relative LUC activities and mRNA levels were analyzed as described above. In plasmid ICmt, the CHOP intercistronic region was replaced by a LacZ sequence which does not contain any AUG [(3) and (4)]. The initiation context of the LUC AUG was not changed. Plasmids mutated on the AUG#3 are given as a control (1) (2). A spacer of 47 bp [(7) and (8)] or 140 bp [(9) and (10)] that does not contain AUG codon was introduced into the intercistronic region of the pCMV–5′UTR–LUC plasmid mutated or not on the uAUGs. In the constructs ΔIC, the intercistronic region was deleted [(11) and (12)]. The mutations do not affect the Kozak consensus sequence of the LUC AUG.
Figure 7.
Inhibition of translation by the uORF is dependent on its peptide sequence. (A) HeLa cells were transiently transfected with LUC constructs [derived from the CMV–5′UTR–LUC(1) (2)] mutated in the uORF sequence. Forty-eight hours after transfection cells, were harvested then relative LUC activities and mRNA levels were analyzed. Plasmids contain the 5′UTR either in a wild-type form or mutated on uAUG#1 and AUG#2. The arrows represent point mutation of the uAUGs. When present, the uORF is boxed. Point mutations of the uORF coding sequence are indicated by asterisks. In the first set of constructs a stop codon was introduced at the nucleotide +52, the UGG codon is replaced by the UGA codon (3) and (4) generating a three amino acid peptide. In the second set of constructs, the sequence of the synthesized peptide was mutated by introduction of a frame shift in the nucleotide sequence (5) and (6). One base was introduced after AUG#2 and one base was withdrawn just before the stop codon. The mutation does not affect the Kozak consensus sequence of AUG#2. The mutated uORF is boxed and hatched. In the last set of constructs, we have generated ‘silent mutation’ in the uORF coding sequence, which modify the mRNA sequence but not the amino acid sequence [we have mutated 13 nt located in the last part the uORF sequence; (7) and (8)]. (B) HeLa cells were transiently co-transfected with 0.5 µg of the reporter plasmid [CMV–5′UTR–uAUG(–)–LUC] and increasing amounts (0, 0.5, 1, 2, 3 and 4 µg) of a CHOP uORF expression vector encoding the uORF peptide (pCMV–uORF). In the control experiment, this expression vector has been mutated on AUG#1 and AUG#2 [pCMV–uORF–AUG(–)]. The total amount of expression vector was kept constant by addition of empty vector (pCMV). Forty-eight hours after transfection, cells were harvested and relative LUC activities were analyzed.
The CMV–5′UTR–LUC mutated constructs (Fig. 3) were generated by cloning mutated PCR fragments of the +6 to +170 5′UTR using wild-type or mutated AflII-ended forward primers and wild-type or mutated HindIII-ended reverse primer generating mutation of the first uAUG (uAUG#1 construct), the second uAUG (uAUG#2 construct), the third uAUG (uAUG#3 construct) and of the three uAUGs [uAUG(–) construct] in between the AflII and HindIII sites of CMV–5′UTR–LUC construct. For each AUG mutation, the A of the AUG was substituted by T.
The CMV–uORF/LUC (Fig. 4) in-frame fusion construct was generated by inserting a PCR fragment amplified from the CMV–5′UTR–LUC construct using the NcoI-ended primers, 5′-GCG CCA TGG TGA TGC GGT TTT GG-3′ (forward primer) and 5′-CGC CCA TGG CTG CAG TTG GAG CAG TCT GGA AAA GC-3′ (reverse primer where the stop codon of the 5′UTR is replaced by the NcoI site), in between a NcoI digested CMV–5′UTR–LUC construct. This cloning leads to a fusion between the stop codon of the Chop 5′UTR and the ATG codon of the LUC coding sequence.
The STOPmt construct [Fig. 5(3)] was generated by inserting an AflII/HindIII fragment in between the AflII and HindIII sites of CMV–5′UTR–LUC construct. This fragment was obtained by fusion of two PCR fragments: (i) a PCR fragment amplified from the CMV–5′UTR–LUC construct using the forward AflII-ended primer 5′-AGA CTT AAG TCT AAG GCA CTG AGC GTA TC-3′ and the reverse primer 5′-CAT CTG CTT TCT GGT GTG GTG ATG TAT G-3′ (containing the mutation of the stop codon of the uORF) and (ii) a PCR fragment amplified from the CMV–5′UTR–LUC construct using the forward primer 5′-CAT ACA TCA CCA CAC CAG AAA GCA GAT G-3′ and the reverse HindIII-ended primer 5′-GCC AAG CTT CAG TTG GAT CAG T-3′. This fragment contains a mutation of the stop codon of the uORF and introduction of a 1 nt frame shift in order to generate an extended uORF that is not in-frame with the LUC gene. The fusion of the two PCR fragments was obtained by PCR amplification with the first forward primer and the second reverse primer, using the two fragments already synthesized as a template.
The CMV–5′UTR–LUC construct containing an optimal context for ribosomal initiation at the AUG#2 [Fig. 5(2)] was generated by cloning mutated PCR fragments of the +6 to +170 5′UTR using a mutated AflII-ended forward primer (replacing the AUG#2 Kozak sequence by the following sequence: gccgccaccAUGg) and wild-type HindIII-ended reverse primers in between the AflII and HindIII sites of CMV–5′UTR–LUC construct.
The ICmt construct [Fig. 6(3) and (4)] wild-type or mutated on the uAUGs was generated by inserting an AflII/HindIII fragment in between the AflII and HindIII sites of CMV–5′UTR–LUC construct. This fragment was obtained by fusion of two PCR fragments: (i) a PCR fragment amplified from the CMV–5′UTR–LUC construct (wild-type or mutated on the uAUG#1 and #2) using the forward AflII-ended primer 5′-AGA CTT AAG TCT AAG GCA CTG AGC GTA TC-3′ and the reverse primer 5′-GCA GTT CAG GCC TCA GGT GTG GTG ATG TAT GAA G-3′ and (ii) a PCR fragment corresponding to an AUG-free LacZ sequence amplified from the LACZ gene using the forward primer 5′-CAT CAC CAC ACC TGA GGC CTG AAC TGC CAG CTG GCG CAG G-3′ and the reverse HindIII-ended primer 5′-CGC AAG CTT GCT CTG CTA CCT GCG CCA G-3′. The fusion of the two PCR fragments was obtained by PCR amplification with the first forward primer and the second reverse primer using the two fragments already synthesized as a template. In this construct a restriction site StuI was added just after the stop codon.
The +47 constructs [Fig. 6(7) and (8)], wild-type [uAUG(+)] or mutated on the uAUGs [uAUG(–)] were generated by inserting a double-strand 47-bp HindIII-ended fragment (5′-AAG CTT AAG GCA GAT CCC AGC GGT CAA AAC AGG CCA ATC CGC GCC GGA AGC TT-3′) in between the HindIII site of dephosphorylated CMV–5′UTR–LUC or CMV–5′UTR–uAUG(–)–LUC plasmids, respectively.
The +140 constructs [Fig. 6(9) and (10)], wild-type [uAUG(+)] or mutated on the uAUGs [uAUG(–)] were generated by inserting a 140-bp HindIII-ended fragment. This 140-bp fragment corresponds to an AUG-free PCR fragment of the LacZ gene generated using HindIII-ended forward primer (5′-GCG AAG CTT CCG GCG CGG ATT GGC CTG-3′) and HindIII-ended reverse primer (5′-GCG AAG CTT AAG GCA GAT CCC AGC GGT CAA AAC AGG-3′).
The ΔIC constructs [Fig. 6(11) and (12)] were generated from the ICmt constructs mutated or not on the uAUG. The lacZ sequence of the ICmt constructs contains a restriction site StuI that is located immediately after the CHOP uORF stop codon (see above). The fragment StuI–HindIII was removed from the ICmt constructs. The plasmids were then blunted and ligated.
The uAUG(+)/Stop and uAUG(1+2)/Stop constructs [Fig. 7(3) and (4)] were generated by inserting an AflII/HindIII fragment in between the AflII and HindIII sites of CMV–5′UTR–LUC construct. This fragment was obtained by fusion of two PCR fragments: (i) a PCR fragment amplified from the CMV–5′UTR–LUC [uAUG(+) or uAUG(–)] construct using the forward AflII-ended primer 5′-AGA CTT AAG TCT AAG GCA CTG AGC GTA TC-3′ and the reverse primer 5′-GGC TCT GTC GCT GTC ACC CGC TCA TC-3′ [replacing the UGG codon of the uORF positioned at +50 by a stop codon (UGA)] and (ii) a PCR fragment amplified from the CMV–5′UTR–LUC construct using the forward primer 5′-GAT GAG CGG GTG ACA GCG ACA GAG CC-3′ [replacing the UGG codon of the uORF positioned at +50 by a stop codon (UGA)] and the reverse HindIII-ended primer 5′-CGC AAG CTT GCA GTT GGA TCA GTC TGG A-3′. The fusion of the two PCR fragments was obtained by PCR amplification with the first forward primer and the second reverse primer using the two fragments already synthesized as a template.
The coding sequence (CDS) uORFmt construct [Fig. 7 (5) and (6)] was generated by cloning a PCR fragment of the +6 to +170 5′UTR containing a mutated uORF sequence. The mutation was obtained by adding one base after AUG#2 (ATG AGg CGG), whereas one base was withdrawn before the uORF stop codon (CCA CAC– TGA). The PCR was performed using a mutated AflII-ended forward primer (frame shift +1 nt) and a mutated HindIII-ended reverse primer (frame shift –1 nt), the fragment was then cloned in between the AflII and HindIII sites of CMV–5′UTR–LUC construct. For PCR the pCMV–5′UTR–LUC mutated or not on AUG#1 and AUG#2 were used as template (construct mutated or not on the AUG#1 and AUG#2).
The ‘silent mutation’ [Fig. 7A(7) and (8)] was generated by cloning a PCR fragment at the Bsu36I and HindIII sites into the CMV–5′UTR–LUC construct mutated or not on AUG#1 and #2 [Fig. 7A(1) and (2)]. The PCR fragment was amplified using the reverse HindIII-ended primer 5′-GCC AAG CTT CAG TTG GAT CAG T-3′ and the forward primer 5′-G AAC CTG AGG CGT GAA TGC TCT AGA CGA AAA TGC ATA TTT ATC CAC CAC CAC ACC TGA-3′. The mutated nucleotides are underlined, the bold nucleotides are not mutated in the mouse sequence. The CMV–5′UTR–LUC construct was used as template.
The uORF and uORF uAUG(–) expression vectors (Fig. 7B) were obtained by deletion of the luciferase coding region of the construct CMV–5′UTR–LUC and CMV–5′UTR–uAUG(–)–LUC, respectively (HindIII/XbaI fragment).
All the luciferase plasmid constructs were sequenced before utilization using a BigDye terminator cycle sequencing ready reaction kit (Perkin Elmer) and an ABI PRISM 310 Genetic Analyser (Perkin Elmer) according to the manufacturer’s instructions.
Western blot analysis
Transfected cells were harvested in 150 µl of lysis buffer (Tris 20 mM pH 6.9, NP-40 0.5%, PMSF 0.5 mM) and centrifuged at 13 000 g for 2 min. Five hundred micrograms (total protein) of each extract were applied on a mono Q HR 5/5 column (ion exchange chromatography; Amersham Pharmacia) equilibrated with a 30 mM Tris–HCl pH 8.0 buffer. After passage of unretarded proteins through the column, a linear gradient (20 ml total) was initiated from 0 to 0.5 M NaCl. The fractions containing LUC activity were eluted in the gradient at ∼0.2 M NaCl. After precipitation (5% TCA, 0.5% PTA) 20 µg of extracts were separated by SDS–PAGE on a 10% (w/v) polyacrylamide gel and electrophoretically transferred to a hybond C membrane in 25 mM Tris/190 mM glycine. Membranes were blocked for 1 h at room temperature with a 5% non-fat milk powder solution in TN buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl). The blots were then incubated with the anti-LUC antibody (1/500) in blocking solution overnight at 4°C, then washed four times in TN containing 0.05% Triton X-100, and incubated with an anti-rabbit antibody conjugated with alkaline phosphatase (1/5000) for 1 h at room temperature. The blots were then treated with the ECF western blotting reagent (Amersham) and visualized by a chemiofluorescence detection system (Molecular Dynamics).
RESULTS
The Chop 5′UTR sequence represses translation of downstream coding sequences
To determine whether the Chop 5′UTR can regulate translation of the Chop ORF, we constructed a retroviral vector expressing the tagged CHOP ORF containing or lacking the 5′UTR (Fig. 1A). After infection of NIH-3T3 cells, CHOP protein and mRNA accumulation was measured in control cells and in response to an agent (tunicamycin) that induces ER stress. As expected, both mRNA and protein content of the endogenous Chop gene are strongly induced in response to tunicamycin treatment. The construct [5′UTR(Δ)–9E10–CHOP], containing the Chop gene without 5′UTR, shows a high expression of the CHOP protein. In contrast, CHOP expression is strongly repressed when the 5′UTR is inserted between the transcription start site and the CHOP coding sequence (5′UTR–9E10–CHOP). Tunicamycin treatment does not affect CHOP expression in both constructs, containing or not containing the 5′UTR. In the same experiment, the mRNA content is not significantly affected by the Chop 5′UTR. Similar results were obtained using HeLa cells (results not shown). Taken together, these results demonstrate that Chop translation is strongly repressed by the leader region of the Chop transcript but this region is not involved in the regulation of Chop expression in response to stress.
We next investigated whether the Chop 5′UTR could repress the downstream ORF in a different context (Fig. 1B and C). Reporter gene LUC, driven either by the CMV or by the Chop promoter, was used in place of Chop and the constructs were transfected into HeLa cells. The AUG of the LUC gene is in an optimum context to be initiated by ribosomes. Our results show that the level of luciferase activity is repressed when the +21 to +170 fragment of the CHOP 5′UTR is present whereas the LUC mRNA level is not affected. For these experiments the LUC transcript was measured by quantitative reverse transcription PCR and we considered that the LUC activity was proportional to the LUC protein content (31).
The Chop 5′UTR contains a conserved uORF
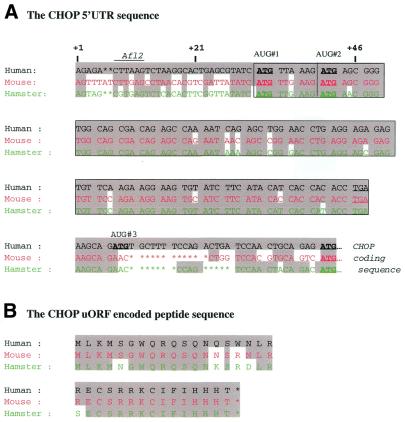
To better understand the mechanisms by which the Chop 5′UTR region can repress translation of the downstream ORF, its nucleotide sequence was analyzed. The alignment of the 5′UTR of human, mouse and hamster Chop transcripts reveals a high sequence homology (Fig. 2A), and the presence of three translation start sites (upstream AUG–uAUG) for the human sequence and two for the hamster and mouse sequences. The AUG#1 and AUG#2 are in-frame with the same stop codon (UGA) delineating an ORF of 34 and 31 amino acids, respectively. The amino acid sequences of the uORF peptides in hamster, mouse and humans also show a high degree of homology (Fig. 2B). It is noticeable that the human 5′UTR sequence also contains a third uAUG which delineates an ORF overlapping the Chop ORF. The conservation of the first uORF between species and its location in the transcript suggest that it is functionally important for the regulation of CHOP expression. These uAUGs, as well as the CHOP initiation codon, are found within a sequence context that allows translational initiation of the ribosome (32,33). Altogether, these observations led us to focus on the effects of the uAUGs in modulating the translation of downstream coding sequence.
Figure 2.
CHOP 5′UTR contains a conserved uORF. Sequence alignment of the 5′UTR region of the human, mouse and hamster Chop transcript. (A) Nucleotide sequence, (B) amino acid sequence. The uAUG codons and the stop codon are indicated. The conserved nucleotides or amino acids are shown and the nucleotide sequence of the uORF is boxed.
Mutations in the upstream AUGs abolish the repressor effect of the 5′UTR
To determine the importance of the uAUGs in regulating Chop translation, we constructed a series of plasmids (derived from pCMV–5′UTR–LUC) containing a point mutation for each uAUG. After transfection, a cellular extract was prepared and assayed for LUC activity and mRNA content (Fig. 3). The level of transcripts is very similar among all constructs, indicating that differences in LUC activity do not result from variation in mRNA accumulation. In contrast, LUC activity is affected by mutation of the uAUGs. Particularly, a mutation within the second or third uAUG (uAUG#2 or uAUG#3) causes a 6- and 2-fold increase of the LUC activity, respectively, whereas mutation of the uAUG#1 has no significant effect. When all the uAUGs are mutated [uAUG(–)], the repressing effect of the 5′UTR is absent since the LUC activity was equivalent to that measured in the absence of the Chop 5′UTR (Figs 3 and 1C). Compared to the wild-type leader sequence, mutation of the three uAUGs (the A of the AUG codon was mutated) leads to an ∼10-fold increase in LUC activity (similar results were obtained after mutation of the G of the AUG codon; results not shown). These results show that the repression of the LUC activity due to the presence of the Chop 5′UTR occurs at a translational level. In addition, the uAUG#2 is strongly involved in this repressive effect.
Translation initiation at the upstream ORF
The results described above suggest that Chop uORF is translated after initiation of the uAUG#2 codon. To test directly the possibility that the uORF can be translated, we generated a construct in which the stop codon of the uORF as well as the intercistronic region were deleted and the Chop uORF was fused in-frame to the luciferase ORF (pCMV–uORF/Luc in-frame fusion). In this experiment, LUC synthesis should only occur if ribosomes initiate translation at the uORF uAUGs codons. Transfected cell extracts were roughly purified and analyzed for LUC expression by western blotting using a specific antibody that recognizes luciferase protein. The immuno-blot presented in Figure 4 shows that the ORF–LUC fusion protein is expressed at a similar level as the LUC protein expressed from the pCMV–LUC vector. As expected, this protein has a molecular weight slightly higher than the LUC protein. The above data demonstrate that the uORF is efficiently translated.
Translation initiation at the downstream ORF
Because uAUG#2 is well initiated by ribosomes and because uAUG#2 cannot be removed following alternative splicing of the transcript or utilization of another transcriptional intiation site, we were interested in investigating the mechanism by which the downstream ORF is translated. One possibility is that translation occurs through ‘leaky scanning’ in which some ribosomes subunits do not initiate at the uAUG codon and translate the downstream ORF. Another possibility is that a few ribosomes that translate the uORF may re-initiate translation at the downstream AUG (34,35). To evaluate these possibilities, several sets of experiments were conducted.
First, we mutated the context of the uAUG#2 in a sequence (gccgccaccAUGg) known to yield maximum initiation frequency (36,37). Figure 5 shows that placing the uAUG in an optimal context does not significantly affect LUC expression. Therefore, the uAUG#2 codon is equally initiated in both a wild-type and optimally mutated context. These data reinforce the conclusion that the uAUG#2 is present within a context able to efficiently allow translation and thus uORF is efficiently translated.
To further evaluate the contribution of ‘leaky scanning’ to translation of the downstream ORF, we mutated the stop codon of the uORF and introduced a frame shift of 1 nt in order to generate an extended uORF that is not in-frame with the LUC gene [Fig. 2)]. This uORF overlaps with the LUC ORF and terminates at the next stop codon located 22 nt downstream of the LUC initiation codon. This mutation prevents the translation of the LUC ORF by reinitiation after translation of the CHOP uORF. The reduction of the luciferase activity observed suggests that ribosomal reinitiation could be responsible for a part of the translation of the downstream ORF (Fig. 5). However, because ribosomes translating the expanded uORF may interfere with initiation events at the overlapping LUC start codon, it is difficult to determine from this experiment the respective role of leaky scanning and reinitiation in translating LUC.
Effect of intercistronic spacing on translation of the downstream ORF
In a number of genes containing uORF (34,38) it has been shown that ribosomal re-initiation after the uORF stop codon is affected by both the sequence of nucleotides following the upstream termination codon and the distance between the uORF and the downstream start site. We introduced several mutations in the intercistronic region to understand its role in translation of the downstream ORF (Fig. 6). In a first set of experiments, the wild-type sequence was substituted by a LacZ sequence that does not contain an AUG [Fig. 6(3)]. Because substitution of the spacing region removed the uAUG#3, a construct containing a mutated 5′UTR on this uAUG#3 is presented as a control [Fig. 6(1)]. Our results show that mutation of the intercistronic region does not significantly affect the LUC activity [Fig. 6(3)]. Moreover, in order to detect unexpected uORF-independent effects of the mutations, the plasmids previously used were mutated on the uAUG#1+2 [Fig. 6(2) and (4)]. In this context, mutation of the intercistronic region does not affect expression of the downstream cistron.
In a second set of experiments, we modified the length of the intercistronic region. First, we inserted fragments of different lengths into the intercistronic region [Fig. 6(7)–(10)]. These spacers were PCR amplified from the Escherichia coli LacZ gene and did not contain an AUG start codon. Expansions of both 47 and 140 nt do not affect LUC activity in the uAUG(+) constructs [Fig. 6(7) and (9)]. Insertion of the spacer in the opposite orientation gives the same result (result not shown). Similarly, reducing the intercistronic region does not impair LUC translation [Fig. 6(11)]. In all constructs mutated within the uAUGs, modifying the length of the intercistronic region does not alter LUC activity significantly [Fig. 6(8), (10) and (12)].
Taken together, these results demonstrate that the sequence of the intercistronic region is not involved in controlling translation of the downstream ORF.
Mutations within the peptide sequence encoded by the uORF enhance translation of the downstream cistron
Inhibition by uORFs in few other eukaryotic genes is dependent upon the peptide-coding sequence of the uORF (34,39–42). To assess the importance of this peptide in the translation of the downstream ORF, the uORF sequence was mutated in the following ways: First, we introduced a stop codon in the uORF sequence [Fig. 7A(3)] in order to synthesize a three amino acid peptide after initiation of the translation at uAUG#2 (the UGG codon was mutated in UGA; Fig. 2). Secondly, we performed a frame shift mutation within the mRNA that generates a mutated sequence of the uORF-encoded peptide without affecting its length [Fig. 7A(5)]. Figure 7A shows that these different mutations of the uORF peptide sequence increase LUC activity by about ∼3–4-fold, but do not affect the mRNA content [Fig. 7A, compare (3) and (5) with (1)].
In order to measure the full de-repression of LUC activity, we constructed control plasmids, containing the same modification in the uORF sequence but mutated on uAUG#1 and uAUG#2 codons [Fig. 7A(2), (4) and (6)]. Elimination of the uORF start codons results in a higher LUC activity than that found after mutation of the peptide sequence of the uORF suggesting that mutations of the peptide sequence encoded by the uORF do not achieve the full de-repression of the LUC activity. From these experiments we can conclude that (i) initiation at the uAUG alone contributes, for a part, to the inhibition of the downstream ORF translation independently of peptide sequence and (ii) when the amino acid sequence of uORF is mutated, translation of the downstream ORF is increased.
The mutations used in these experiments have a drastic effect on the amino acid sequence, but correspond only to minor changes in the mRNA sequence. Nevertheless, we cannot exclude that the mRNA sequence of the 5′UTR by itself could be involved in the control of the residual expression of the downstream cistron. To investigate its role we performed silent mutations in the uORF coding sequence, which modify the mRNA but not the uORF protein sequence (13 nt located on the last part of the uORF sequence were mutated). Figure 7A(7) shows that silent mutations of the uORF sequence slightly decrease LUC activity (∼2-fold). These results demonstrate that the uORF mRNA sequence plays a role in the initiation of the downstream AUG. This mutation does not affect significantly the expression of the downstream cistron when uAUG#1+2+3 are mutated [Fig. 7A, compare (2) and (8)].
uORF-mediated translation inhibition is exerted in cis
We next addressed the possibility that the uORF encoded peptide inhibits translation of the downstream ORF in trans. The reporter plasmid mutated on the uAUGs [pCMV–5′UTR–LUC–uAUG(–)] was transfected together with increasing amounts of the expression vector, pCMV–uORF, which contains the uORF driven by the CMV promoter (Fig. 7B). A control experiment was performed by co-transfecting the reporter vector with an expression plasmid mutated on the uAUG codons [pCMV–uORF–uAUG(–)]. Transfection of increasing amounts of the uORF expression vector does not diminish LUC expression encoded by the reporter plasmid. Although we cannot directly assess synthesis of the uORF-encoded peptide in the cell, the high expression of the fusion construct shown in Figure 4 demonstrates that translation is initiated efficiently at the CHOP uORF. The inability of the uORF to repress translation when expressed from a separate transcript suggests that the uORF inhibits the expression of a downstream cistron via a cis-acting mechanism.
DISCUSSION
The expression of the CHOP is strongly enhanced in response to various stresses including perturbation of ER function and amino acid deprivation. In these situations, global protein synthesis is inhibited. We initiated this study on the CHOP 5′UTR in the hope of understanding how CHOP is translated while most other transcripts are not. We examined this question by comparing the relative translation levels of a CHOP mRNA expressed from a strong constitutive retroviral promoter in the presence or absence of ER stress. We found that the presence of this region suppressed translation in both stressed and unstressed cells. This is due to a set of three upstream AUGs (uAUG) that is present in the region from nucleotides +1 to +170 in the human 5′UTR. The first two uAUGs codons are in-frame and start an uORF localized in the 5′ leader. Our results show that the uAUG#2 is well initiated by ribosomes and strongly represses translation of the downstream ORF. The third uAUG codon of the human Chop 5′UTR, which is not present in the hamster and mouse 5′UTR, slightly inhibits initiation at the main AUG. Removal of all the uAUGs improves translational activity by ∼10-fold. Our results are consistent with a mechanism in which translation of the CHOP uORF cistron represses initiation of the downstream AUG codon. Several examples of genes containing inhibitory uORFs have been described (34). However, uORFs do not always inhibit translation. For instance, the 5′UTR of ornithine decarboxylase mRNA represses translation, but the uORFs present in its leader contribute poorly to its repression (43).
The current study shows that the mutation of two nucleotides, changing the sequence of the peptide encoded by the uORF, strongly increases translation of the downstream cistron. These data favor the hypothesis that uORF inhibits downstream translation by a mechanism that depends on the amino acid coding information. We cannot completely exclude that mutations used in these constructs could influence the initiation at the downstream codon. However, it is unlikely that point mutation strongly alters the RNA structure and thereby affects LUC expression. In addition, the results obtained using the ‘silent mutations’ show that the mRNA sequence encoded by the uORF can affect the initiation at the downstream cistron. Taken together, our results show that when mutated only on the mRNA sequence, the uORF region slightly decreases the LUC expression, whereas mutations on the amino acid sequence strongly enhanced LUC expression. Therefore, it is likely that the effects of the point mutation used to modify the amino acid sequence of the uORF (Fig. 7A) are due to the peptide sequence of the uORF and thus the inhibitory effect of the uORF is a ‘protein driven-mechanism’.
Other eukaryotic gene transcripts that contain uORF which inhibit downstream translation by a mechanism that depends on the amino acid coding information have been described. For example, in S-Adenosylmethionine decarboxylase (44), the β2 adrenergic receptor (45), the yeast CPA1 gene (46) or the cytomegalovirus gp48 (or CMV gp UL4) (47–49) genes, the peptide product of the uORF mediates repression of translation. For most of these genes the uORF peptide mediates inihibition in cis. Only the β2 adrenergic receptor uORF peptide inhibits translation in trans. However, this result has been shown only in cell-free extracts and at high peptide concentrations (45). The mechanisms involved to explain the inhibitory effects of the uORF peptide on translation are not understood, however several models can be proposed. For example, the peptide of the uORF is synthezised and has the ability to inhibit translation only at high concentrations in the local microenvironment (45). In addition, it is possible that the nascent peptide, still attached to the translational machinery, inhibits translation through interaction with the ribosome or a ribosome-associated translation factor. In the gp48 gene, it has been shown that the uORF peptide leads to ribosome arrest during termination of translation of uORF2 (39,47,49). Therefore, the uORF would be required to be part of the same transcript preceding the major cistron. Further investigations will be required to determine if a similar mechanism takes place within the Chop transcript.
Our data show that despite an efficient translation of the uORF, the main cistron remains slightly translated. Several mechanisms can account for the translation of the downstream cistron: internal ribosomal entry, ribosome re-initiation and ‘leaky scanning’. The efficiency of re-initiation after uORF translation can be affected by both the nature of the sequence surrounding the upstream termination codon and the distance between the uORF and the downstream start site (34,35,50). Substitution of the intercistronic sequence by a neutral LacZ sequence or altering the length of this sequence does not affect translation of the downstream ORF. Therefore, the intercistronic region does not contain sequence involved in the control of ribosome re-initiation or internal ribosomal entry site. However, ‘silent mutations’ of the uORF sequence slightly decrease the residual translation of the downstream cistron, suggesting that the mRNA sequence can play a role in the initiation of the downstream cistron. This conclusion is reinforced by the observation that elongating uORF (Fig. 5) partially inhibits the residual translation of LUC. However, the residual LUC activity that is measured when the uORF is expanded shows that ‘leaky scanning’ can also contribute for a part to the translation of the downstream cistron.
Taken together these data are consistent with the idea that the residual translation of the downstream ORF occurs in part by re-initiation of the ribosomes and in part by leaky scanning. Further work will be necessary to quantify the respective part of the both mechanisms in the translation of the CHOP ORF.
The observation that most of the mRNAs that initiate downstream of one or several uAUGs codons encode regulatory proteins leads to the suggestion that uAUGs may down-regulate the translation of proteins whose expression has to be finely controlled. Supporting evidence for this idea comes from the findings that (i) the oncogenic potential of c-mos (51,52) or c-lck (53) is increased by rearrangements that remove the uORF, and (ii) the repressive effect of the uORF can be regulated. For example, in the well-studied case of the Saccharomyces cerevisiae GCN4 gene, four uORFs regulate translation of GCN4 in response to amino acid availability by controlling which downstream AUG codons are utilized by the re-initiating ribosomes (54,55). Another example involves the uORF of the S-adenosylmethionine decarboxylase which by itself can provide polyamine regulation of translation of the downstream ORF (56). In addition, the inhibitory role of the upstream AUG can be affected by cell type. For instance, elements in the leader sequence of c-sis transcript inhibit its translation, but this repression is transiently relieved in differentiating megakaryocytes (57).
Given the importance of the CHOP protein in the regulation of numerous cellular functions, requirement for precise control of its expression may be necessary. We have described a new feature of CHOP expression that involves translational repression by a highly conserved uORF. An example of constitutive expression of an abnormal form of CHOP is the fusion of CHOP with TLS due to the chromosomal translocation found in a vast majority of human myxoid liposarcomas (10). The oncogenic potential of this fusion protein is due to the properties of the TLS part of the protein and to its constitutive expression (11). It is noticeable that the constitutive expression of this oncogenic form of CHOP could be partially explained by the absence of translational repression due to the deletion of the CHOP 5′UTR.
Acknowledgments
ACKNOWLEDGEMENTS
We thank C. Tassy and Dr A. Ouali (Station de Recherche sur la Viande, INRA de Theix, France) for the ion exchange chromatography, S. Lafarge and Prof. Y. J. Bignon for the mRNA quantification, Drs S. Mordier and E. Pinzar for critically reading the manuscript and for helpful discussion. This work was supported by grants from the Institut National de la Recherche Agronomique and the Fondation pour la Recherche Médicale. C.J. was awarded a DANONE research scholarship. F.U. is a recipient of the Uehara Memorial Foundation Fellowship.
References
- 1.McKnight S.L., Lane,M.D. and Gluecksohn-Waelsch,S. (1989) Is CCAAT/enhancer-binding protein a central regulator of energy metabolism? Genes Dev., 3, 2021–2024. [DOI] [PubMed] [Google Scholar]
- 2.Umek R.M., Friedman,A.D. and McKnight,S.L. (1991) CCAAT-enhancer binding protein: a component of a differentiation switch. Science, 251, 288–292. [DOI] [PubMed] [Google Scholar]
- 3.Cao Z., Umek,R.M. and McKnight,S.L. (1991) Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev., 5, 1538–1552. [DOI] [PubMed] [Google Scholar]
- 4.Birkenmeier E.H., Gwynn,B., Howard,S., Jerry,J., Gordon,J.I., Landschulz,W.H. and McKnight,S.L. (1989) Tissue-specific expression, developmental regulation and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev., 3, 1146–1156. [DOI] [PubMed] [Google Scholar]
- 5.Batchvarova N., Wang,X.Z. and Ron,D. (1995) Inhibition of adipogenesis by the stress-induced protein CHOP (Gadd153). EMBO J., 14, 4654–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barone M.V., Crozat,A., Tabaee,A., Philipson,L. and Ron,D. (1994) CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev., 8, 453–464. [DOI] [PubMed] [Google Scholar]
- 7.Friedman A.D. (1996) GADD153/CHOP, a DNA damage-inducible protein, reduced CAAT/enhancer binding protein activities and increased apoptosis in 32D c13 myeloid cells. Cancer Res., 56, 3250–3256. [PubMed] [Google Scholar]
- 8.Zinszner H., Kuroda,M., Wang,X., Batchvarova,N., Lightfoot,R.T., Remotti,H., Stevens,J.L. and Ron,D. (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev., 12, 982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aman P., Ron,D., Mandahl,N., Fioretos,T., Heim,S., Arheden,K., Willen,H., Rydholm,A. and Mitelman,F. (1992) Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11). Genes Chromosom. Cancer, 5, 278–285. [DOI] [PubMed] [Google Scholar]
- 10.Crozat A., Aman,P., Mandahl,N. and Ron,D. (1993) Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature, 363, 640–644. [DOI] [PubMed] [Google Scholar]
- 11.Zinszner H., Albalat,R. and Ron,D. (1994) A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev., 8, 2513–2526. [DOI] [PubMed] [Google Scholar]
- 12.Rabbitts T.H., Forster,A., Larson,R. and Nathan,P. (1993) Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nature Genet., 4, 175–180. [DOI] [PubMed] [Google Scholar]
- 13.Panagopoulos I., Hoglund,M., Mertens,F., Mandahl,N., Mitelman,F. and Aman,P. (1996) Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene, 12, 489–494. [PubMed] [Google Scholar]
- 14.Panagopoulos I., Aman,P., Mertens,F., Mandahl,N., Rydholm,A., Bauer,H.F. and Mitelman,F. (1996) Genomic PCR detects tumor cells in peripheral blood from patients with myxoid liposarcoma. Genes Chromosom. Cancer, 17, 102–107. [DOI] [PubMed] [Google Scholar]
- 15.Wang X.Z., Kuroda,M., Sok,J., Batchvarova,N., Kimmel,R., Chung,P., Zinszner,H. and Ron,D. (1998) Identification of novel stress-induced genes downstream of chop. EMBO J., 17, 3619–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ron D. and Habener,J.F. (1992) CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant- negative inhibitor of gene transcription. Genes Dev., 6, 439–453. [DOI] [PubMed] [Google Scholar]
- 17.Fawcett T.W., Eastman,H.B., Martindale,J.L. and Holbrook,N.J. (1996) Physical and functional association between GADD153 and CCAAT/enhancer- binding protein beta during cellular stress. J. Biol. Chem., 271, 14285–14289. [DOI] [PubMed] [Google Scholar]
- 18.Ubeda M., Vallejo,M. and Habener,J.F. (1999) CHOP enhancement of gene transcription by interactions with Jun/Fos AP- 1 complex proteins. Mol. Cell. Biol., 19, 7589–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luethy J.D. and Holbrook,N.J. (1992) Activation of the gadd153 promoter by genotoxic agents: a rapid and specific response to DNA damage. Cancer Res., 52, 5–10. [PubMed] [Google Scholar]
- 20.Guyton K.Z., Xu,Q. and Holbrook,N.J. (1996) Induction of the mammalian stress response gene GADD153 by oxidative stress: role of AP-1 element. Biochem. J., 314, 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halleck M.M., Holbrook,N.J., Skinner,J., Liu,H. and Stevens,J.L. (1997) The molecular response to reductive stress in LLC-PK1 renal epithelial cells: coordinate transcriptional regulation of gadd153 and grp78 genes by thiols. Cell Stress Chaperones, 2, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson S.G., Fawcett,T.W., Bartlett,J.D., Bernier,M. and Holbrook,N.J. (1993) Regulation of the C/EBP-related gene gadd153 by glucose deprivation. Mol. Cell. Biol., 13, 4736–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruhat A., Jousse,C., Wang,X.Z., Ron,D., Ferrara,M. and Fafournoux,P. (1997) Amino acid limitation induces expression of CHOP, a CCAAT/enhancer binding protein-related gene, at both transcriptional and post-transcriptional levels. J. Biol. Chem., 272, 17588–17593. [DOI] [PubMed] [Google Scholar]
- 24.Jousse C., Bruhat,A., Harding,H.P., Ferrara,M., Ron,D. and Fafournoux,P. (1999) Amino acid limitation regulates CHOP expression through a specific pathway independent of the unfolded protein response. FEBS Lett., 448, 211–216. [DOI] [PubMed] [Google Scholar]
- 25.Sylvester S.L., ap Rhys,C.M., Luethy-Martindale,J.D. and Holbrook,N.J. (1994) Induction of GADD153, a CCAAT/enhancer-binding protein (C/EBP)-related gene, during the acute phase response in rats. Evidence for the involvement of C/EBPs in regulating its expression. J. Biol. Chem., 269, 20119–20125. [PubMed] [Google Scholar]
- 26.Lee C.K., Klopp,R.G., Weindruch,R. and Prolla,T.A. (1999) Gene expression profile of aging and its retardation by caloric restriction. Science, 285, 1390–1393. [DOI] [PubMed] [Google Scholar]
- 27.Bartlett J.D., Luethy,J.D., Carlson,S.G., Sollott,S.J. and Holbrook,N.J. (1992) Calcium ionophore A23187 induces expression of the growth arrest and DNA damage inducible CCAAT/enhancer-binding protein (C/EBP)-related gene, gadd153. Ca2+ increases transcriptional activity and mRNA stability. J. Biol. Chem., 267, 20465–20470. [PubMed] [Google Scholar]
- 28.Davis L.G., Dibner,M.D. and Battey,J. (1986) Basics Methods in Molecular Biology. Elsevier Science Publishing Co., Inc., New York, NY.
- 29.Hall C.V., Jacob,P.E., Ringold,G.M. and Lee,F. (1983) Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J. Mol. Appl. Genet., 2, 101–109. [PubMed] [Google Scholar]
- 30.Bruhat A., Jousse,C., Carraro,V., Reimold,A.M., Ferrara,M. and Fafournoux,P. (2000) Amino acids control mammalian gene transcription: activating transcription factor 2 is essential for the amino acid responsiveness of the CHOP promoter. Mol. Cell. Biol., 20, 7192–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Wet J.R., Wood,K.V., DeLuca,M., Helinski,D.R. and Subramani,S. (1987) Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol., 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell, 44, 283–292. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. (1987) At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol., 196, 947–950. [DOI] [PubMed] [Google Scholar]
- 34.Geballe A.P. (1996) Translational control mediated by upstream AUG codons. In Hershey,I.W.B., Mathews,M.B. and Sonenberg,N. (eds) Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 173–197.
- 35.Morris D.R. and Geballe,A.P. (2000) Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol., 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. (1995) Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc. Natl Acad. Sci. USA, 92, 7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak M. (1989) Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol., 9, 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lincoln A.J., Monczak,Y., Williams,S.C. and Johnson,P.F. (1998) Inhibition of CCAAT/enhancer-binding protein alpha and beta translation by upstream open reading frames. J. Biol. Chem., 273, 9552–9560. [DOI] [PubMed] [Google Scholar]
- 39.Cao J. and Geballe,A.P. (1996) Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol., 16, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z. and Sachs,M.S. (1997) Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J. Biol. Chem., 272, 255–261. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds K., Zimmer,A.M. and Zimmer,A. (1996) Regulation of RAR beta 2 mRNA expression: evidence for an inhibitory peptide encoded in the 5′-untranslated region. J. Cell Biol., 134, 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Z. and Sachs,M.S. (1996) Role of an upstream open reading frame in mediating arginine-specific translational control in Neurospora crassa. J. Bacteriol ., 178, 2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manzella J.M. and Blackshear,P.J. (1990) Regulation of rat ornithine decarboxylase mRNA translation by its 5′-untranslated region. J. Biol. Chem., 265, 11817–11822. [PubMed] [Google Scholar]
- 44.Ruan H., Hill,J.R., Fatemie-Nainie,S. and Morris,D.R. (1994) Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Influence of the structure of the 5′ transcript leader on regulation by the upstream open reading frame. J. Biol. Chem., 269, 17905–17910. [PubMed] [Google Scholar]
- 45.Parola A.L. and Kobilka,B.K. (1994) The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J. Biol. Chem., 269, 4497–4505. [PubMed] [Google Scholar]
- 46.Delbecq P., Werner,M., Feller,A., Filipkowski,R.K., Messenguy,F. and Pierard,A. (1994) A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol. Cell. Biol., 14, 2378–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schleiss M.R., Degnin,C.R. and Geballe,A.P. (1991) Translational control of human cytomegalovirus gp48 expression. J. Virol., 65, 6782–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degnin C.R., Schleiss,M.R., Cao,J. and Geballe,A.P. (1993) Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J. Virol., 67, 5514–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao J. and Geballe,A.P. (1995) Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J. Virol., 69, 1030–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozak M. (1987) Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol., 7, 3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rechavi G., Givol,D. and Canaani,E. (1982) Activation of a cellular oncogene by DNA rearrangement: possible involvement of an IS-like element. Nature, 300, 607–611. [DOI] [PubMed] [Google Scholar]
- 52.Steel L.F., Telly,D.L., Leonard,J., Rice,B.A., Monks,B. and Sawicki,J.A. (1996) Elements in the murine c-mos messenger RNA 5′-untranslated region repress translation of downstream coding sequences. Cell Growth Differ., 7, 1415–1424. [PubMed] [Google Scholar]
- 53.Marth J.D., Overell,R.W., Meier,K.E., Krebs,E.G. and Perlmutter,R.M. (1988) Translational activation of the lck proto-oncogene. Nature, 332, 171––173.. [DOI] [PubMed] [Google Scholar]
- 54.Hinnebusch A.G. (1997) Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J. Biol. Chem., 272, 21661–21664. [DOI] [PubMed] [Google Scholar]
- 55.Hinnebusch A.G. (1994) Translational control of GCN4: an in vivo barometer of initiation-factor activity. Trends Biochem. Sci., 19, 409–414. [DOI] [PubMed] [Google Scholar]
- 56.Ruan H., Shantz,L.M., Pegg,A.E. and Morris,D.R. (1996) The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J. Biol. Chem., 271, 29576–29582. [DOI] [PubMed] [Google Scholar]
- 57.Bernstein J., Shefler,I. and Elroy-Stein,O. (1995) The translational repression mediated by the platelet-derived growth factor 2/c-sis mRNA leader is relieved during megakaryocytic differentiation. J. Biol. Chem., 270, 10559–10565. [DOI] [PubMed] [Google Scholar]