Abstract
Rosa chinensis var. spontanea, an endemic and endangered plant of China, is one of the key ancestors of modern roses and a source for famous traditional Chinese medicines against female diseases, such as irregular menses and dysmenorrhea. In this study, the complete chloroplast (cp) genome of R. chinensis var. spontanea was sequenced, analyzed, and compared to congeneric species. The cp genome of R. chinensis var. spontanea is a typical quadripartite circular molecule of 156,590 bp in length, including one large single copy (LSC) region of 85,910 bp and one small single copy (SSC) region of 18,762 bp, separated by two inverted repeat (IR) regions of 25,959 bp. The GC content of the whole genome is 37.2%, while that of LSC, SSC, and IR is 42.8%, 35.2% and 31.2%, respectively. The genome encodes 129 genes, including 84 protein-coding genes (PCGs), 37 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. Seventeen genes in the IR regions were found to be duplicated. Thirty-three forward and five inverted repeats were detected in the cp genome of R. chinensis var. spontanea. The genome is rich in SSRs. In total, 85 SSRs were detected. A genome comparison revealed that IR contraction might be the reason for the relatively smaller cp genome size of R. chinensis var. spontanea compared to other congeneric species. Sequence analysis revealed that the LSC and SSC regions were more divergent than the IR regions within the genus Rosa and that a higher divergence occurred in non-coding regions than in coding regions. A phylogenetic analysis showed that the sampled species of the genus Rosa formed a monophyletic clade and that R. chinensis var. spontanea shared a more recent ancestor with R. lichiangensis of the section Synstylae than with R. odorata var. gigantea of the section Chinenses. This information will be useful for the conservation genetics of R. chinensis var. spontanea and for the phylogenetic study of the genus Rosa, and it might also facilitate the genetics and breeding of modern roses.
Keywords: Rosa chinensis var. spontanea, chloroplast genome, repeats, SSRs, genome comparison, phylogeny
1. Introduction
Molecular data have suggested that Rosa chinensis Jacq. var. spontanea (Rehder et. Wilson) Yü et Ku is the maternal parent of R. chinensis and the possible paternal parent of R. odorata (Andrews) Sweet [1], which gave characters of recurrent flowering, tea scent, and multiple floral colors to modern roses [2,3]. As one of the key ancestors of modern roses, R. chinensis var. spontanea is not only a precious germplasm resource for improving modern roses but also valuable plant material for the genetic research of recurrent flowering and the study of the biosynthesis of flower scent components. Furthermore, with the effect of “promoting blood circulation for removing blood stasis” and “subdhing swelling and detoxicating”, R. chinensis var. spontanea is also a source for famous traditional Chinese medicines that treat female diseases such as irregular menses and dysmenorrhea [4].
Rosa chinensis var. spontanea originates from China and is endemic to the Hubei, Sichuan, Chongqing, and Guizhou provinces [5,6]. It has been overharvested by local people and pharmaceutical companies because of its medicinal usefulness and has become rare in its wild habitats. It was uncertain whether it still existed as a wild-living species because investigators failed to collect samples of this species in the field [1]. It has been listed as an endangered (EN) species in a recent biodiversity report [7]. Fortunately, during systematic and integrative field investigations focusing on this species, we recently found several populations in the wild.
It is important to mention that little information is available about R. chinensis var. spontanea except the fact that it is a diploid plant [8] and that it emits 1,3,5-trimethoxybenzene, together with methyleugenol and isomethyleugenol, as minor floral scent compounds [9], resulting from O-methytransferas genes [10]. The chloroplasts (cps) play important functional roles in the photosynthesis, biosynthesis, and metabolism of starch and fatty acids throughout the plant’s life cycle [11]. Typically, the cp genomes of angiosperm are circulars, with a characteristic quadripartite structure that is comprised of two inverted repeats (IRs) and two single copy regions: a large single copy region (LSC) and a small single copy region (SSC). The genetic composition of cp genome in angiosperm is more or less conserved, containing 110 to 120 genes including protein-coding genes (PCGs), transfer RNA (tRNA) genes and ribosomal RNA (rRNA) genes. In spite of the generally high conservation of gene order and gene content, cp genomes in angiosperm have undergone size changes, structure rearrangement, contraction and expansion of IRs, and even pseudogenization due to adaptations, even within genera, to the host plants’ environments [12].
Here we report the sequence and structural analyses of the complete cp genome of R. chinensis var. spontanea, including analyses of the repeats and SSRs. Furthermore, we carried out comparative sequence analysis studies of cp sequences in the genus Rosa. This information will be useful for the conservation genetics of R. chinensis var. spontanea, as well as for the phylogenetic study of the genus Rosa. It might also benefit the genetics and breeding of modern roses.
2. Results and Discussion
2.1. Characteristics of Chloroplast Genome of R. chinensis var. spontanea
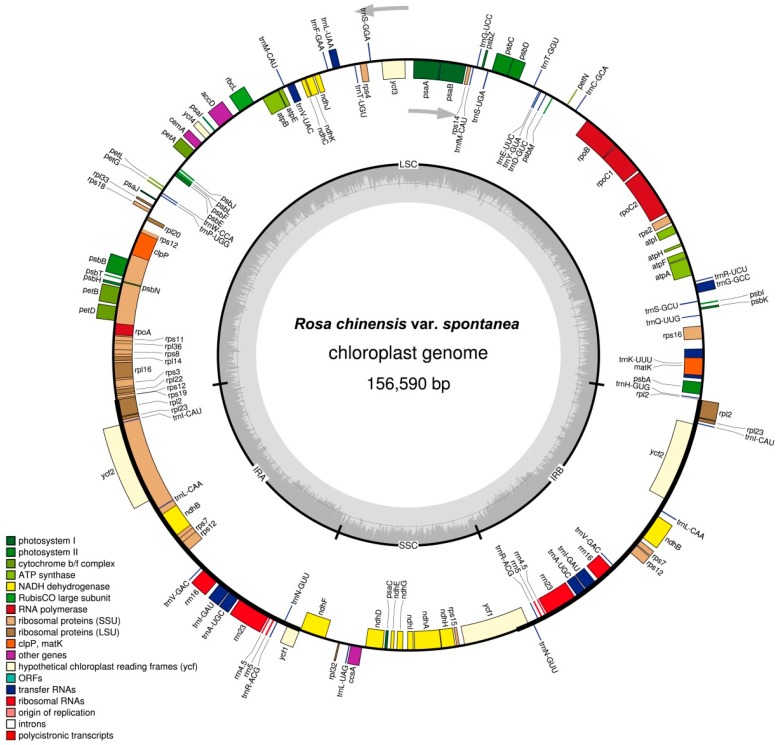
The complete cp genome of R. chinensis var. spontanea represents a typical quadripartite circular molecule that is 156,590 bp in length. It is composed by a LSC region of 85,910 bp and a SSC region of 18,762 bp, separated by two IR regions of 25,959 bp (Table 1 and Figure 1). The GC content of the total cp DNA sequence is 37.2%, similar to that of R. odorata (Andr.) Sweet var. gigantea (Crép) Rehd. et Wils.(KF753637) [13], R. praelucens Byhouwer (MG450565) [14] and R. roxburghii Tratt. (KX768420). The GC content of the IR regions is 42.8%, while the LSC and SSC regions exhibit lower GC content (35.2% and 31.2%, respectively) (Table 1). The complete cp genome includes 57.8% coding sequences (50.2% PCGs, 1.8% tRNAs, and 5.8% rRNAs) and 42.2% non-coding sequences (11.8% introns and 30.4% intergenic spacers). Among PCGs, the AT content of the first, second, and third positions is 54.7%, 62.5%, and 69.7%, respectively (Table 1). This kind of bias towards a higher AT content at the third position of the codons is used to discriminate cp DNA from nuclear and mitochondrial DNA [15] and has been widely reported in other plant cp genomes [16,17,18].
Table 1.
Base composition in the chloroplast genome of Rosa chinensis var. spontanea.
| Region | A | T (U) | G | C | Length | |
|---|---|---|---|---|---|---|
| LSC | 31.7 | 33.1 | 17.2 | 18.0 | 85,910 | |
| SSC | 34.4 | 34.3 | 15.1 | 16.3 | 18,762 | |
| IRB | 28.7 | 28.5 | 22.2 | 20.6 | 25,959 | |
| IRA | 28.7 | 28.5 | 22.2 | 20.6 | 25,959 | |
| Total | 31.0 | 31.8 | 18.6 | 18.7 | 156,590 | |
| PCGs | 30.6 | 31.4 | 20.3 | 17.7 | 79,773 | |
| 1st position | 30.7 | 24 | 26.9 | 18.7 | 26,591 | |
| 2nd position | 29.5 | 33 | 17.7 | 20.2 | 26,591 | |
| 3rd position | 31.7 | 38 | 16.4 | 14.1 | 26,591 |
PCGs: protein-coding genes.
Figure 1.
Chloroplast genome map of Rosa chinensis var. spontanea. Genes inside the circle are transcribed clockwise, and those outside are counterclockwise. Genes of different functions are color-coded. The darker gray in the inner circle shows the GC content, while the lighter gray shows the AT content.
The cp genome of R. chinensis var. spontanea contains 129 genes, including 84 PCGs, 37 tRNAs, and eight rRNAs (Table S1). Six PCGs (ndhB, rpl2, rpl23, rps7, rps12 and ycf2), four rRNAs (rrn16, rrn23, rrn 4.5 and rrn5) and seven tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) within the IR regions are completely duplicated. The LSC region contains 62 PCGs and 22 tRNAs. The SSC region contains one tRNA and 12 PCGs. Additionally, 14 genes, namely trnK-UUU, rps16, trnG-GCC, rpoC1, trnL-UAA, trnV-UAC, petB, rpl16, rp12, ndhB, trnI-GAU, trnA-UGC, ndhA, and petD, contain one intron, whereas the ycf3, rps12 and clpP genes contain two introns. Despite that, there are 17–20 group II introns within tRNA and protein-coding genes in land plant cp genomes [19], so far only the intron of trnL has been characterized as a group I intron in chloroplasts [20]. Thus, all these introns of R. chinensis var. spontanea, except the trnL-UAA intron, might be group II introns. The rps12 gene is trans-spliced in the cp genome of R. chinensis var. spontanea. C-terminal exon 2 and 3 of rps12 are located in the IR regions. Exon 1 is 28,259 bp downstream of the nearest copy of exons 2 and 3 while 72,017 bp away from the distal copy of exons 2 and 3 (Table S1). The trnK-UUU gene had the largest intron with a 2498 bp length, in which the matK gene was located. The matK gene encodes MatK, the maturase which is derived from reverse transcriptase and has been proved to be an essential splice factor for both the group I and group II introns [20,21].
Based on the sequences of PCGs and tRNAs, the frequency of codon usage of the cp genome of R. chinensis var. spontanea was estimated (Table 2). In total, 27,525 codons were found in all the coding sequences. Among these, leucine is the most frequent amino acid, representing 10.4% (2,871) of the total codons, while cysteine is the least frequent one with 1.2% (320) of the codons. A- and U-ending codons are common. Except for trnL-CAA, trnS-GGA and a stop codon (UAG), all types of preferred synonymous codons (RSCU > 1) ended with A or U.
Table 2.
Condon-anticodon recognition patterns and codon usage of the Rosa chinensis var. spontanea chloroplast genome.
| Amino Acid | Codon | Count | RSCU | tRNA | Amino Acid | Codon | Count | RSCU | tRNA |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 1015 | 1.3 | Ser | UCU | 580 | 1.62 | ||
| Phe | UUC | 545 | 0.7 | trnF-GAA | Ser | UCC | 370 | 1.03 | trnS-GGA |
| Leu | UUA | 897 | 1.87 | Ser | UCA | 406 | 1.13 | trnS-UGA | |
| Leu | UUG | 580 | 1.21 | trnL-CAA | Ser | UCG | 222 | 0.62 | |
| Leu | CUU | 595 | 1.24 | Pro | CCU | 424 | 1.45 | ||
| Leu | CUC | 217 | 0.45 | Pro | CCC | 241 | 0.82 | ||
| Leu | CUA | 380 | 0.79 | Pro | CCA | 320 | 1.09 | trnP-UGG | |
| Leu | CUG | 202 | 0.42 | Pro | CCG | 187 | 0.64 | ||
| Ile | AUU | 1136 | 1.48 | Thr | ACU | 542 | 1.55 | ||
| Ile | AUC | 451 | 0.59 | trnI-CAU | Thr | ACC | 269 | 0.77 | trnT-GGU |
| Ile | AUA | 716 | 0.93 | Thr | ACA | 418 | 1.19 | trnT-UGU | |
| Met | AUG | 635 | 1 | trnM-CAU | Thr | ACG | 171 | 0.49 | |
| Val | GUU | 550 | 1.44 | Ala | GCU | 645 | 1.76 | ||
| Val | GUC | 193 | 0.5 | trnV-GAC | Ala | GCC | 244 | 0.67 | |
| Val | GUA | 567 | 1.48 | Ala | GCA | 391 | 1.07 | ||
| Val | GUG | 223 | 0.58 | Ala | GCG | 183 | 0.5 | ||
| Tyr | UAU | 798 | 1.6 | Cys | UGU | 237 | 1.48 | ||
| Tyr | UAC | 198 | 0.4 | trnY-GUA | Cys | UGC | 83 | 0.52 | trnC-GCA |
| stop | UAA | 59 | 1.38 | stop | UGA | 21 | 0.49 | ||
| stop | UAG | 48 | 1.13 | Trp | UGG | 484 | 1 | trnW-CCA | |
| His | CAU | 476 | 1.49 | Arg | CGU | 362 | 1.28 | trnR-ACG | |
| His | CAC | 161 | 0.51 | trnH-GUG | Arg | CGC | 120 | 0.43 | |
| Gln | CAA | 734 | 1.51 | trnQ-UUG | Arg | CGA | 385 | 1.36 | |
| Gln | CAG | 236 | 0.49 | Arg | CGG | 144 | 0.51 | ||
| Asn | AAU | 1003 | 1.52 | Ser | AGU | 420 | 1.17 | ||
| Asn | AAC | 317 | 0.48 | Ser | AGC | 156 | 0.43 | trnS-GCU | |
| Lys | AAA | 1082 | 1.48 | Arg | AGA | 488 | 1.73 | trnR-UCU | |
| Lys | AAG | 385 | 0.52 | Arg | AGG | 194 | 0.69 | ||
| Asp | GAU | 890 | 1.62 | Gly | GGU | 612 | 1.3 | ||
| Asp | GAC | 207 | 0.38 | trnD-GUC | Gly | GGC | 209 | 0.44 | trnG-GCC |
| Glu | GAA | 1052 | 1.46 | trnE-UUC | Gly | GGA | 694 | 1.48 | |
| Glu | GAG | 390 | 0.54 | Gly | GGG | 365 | 0.78 |
RSCU: Relative Synonymous Codon Usage.
2.2. Repeat and SSR Analysis
For the repeat structure analysis, 33 forward and five inverted repeats with a minimal repeat size of 20 bp were detected in the cp genome of R. chinensis var. spontanea (Table 3). Most of these repeats are between 20 and 30 bp. The longest forward repeat is 41 bp in length, located in the intergenic region between the genes psbE and petL. Most of the repeats were found in the LSC region. Among them, repeat No. 5 is related to trnS-GCU and trnS-UGA (Table 3). Repeat No. 7 is related to trnG-GCU and trnG-UCC. Repeat No. 13 is associated with psa genes. Six forward repeats were located in IR regions, including two repeats associated with ycf2 genes and one repeat related to the ndhB gene. In addition, there were several repeat pairs with either repeated sequence located in a distinct region, e.g., each of the two sequences of repeat No. 16, 25, and 26 are located in the gene introns of LSC and SSC, respectively.
Table 3.
Repeat sequences in the Rosa chinensis var. spontanea chloroplast genome.
| ID | Repeat Start 1 | Type | Size (bp) | Repeat Start 2 | Mismatch (bp) | E-Value | Gene | Region |
|---|---|---|---|---|---|---|---|---|
| 1 | 4426 | F | 29 | 45,071 | −2 | 8.74 × 10−5 | IGS; ycf3(intron) | LSC |
| 2 | 4427 | F | 30 | 4428 | −3 | 6.56 × 10−4 | IGS | LSC |
| 3 | 4428 | F | 28 | 45,072 | −3 | 8.47 × 10−3 | IGS | LSC |
| 4 | 4432 | F | 26 | 45,072 | −2 | 4.48 × 10−3 | IGS | LSC |
| 5 | 8329 | F | 29 | 36,077 | −2 | 8.74 × 10−5 | trnS-GCU; trnS-UGA | LSC |
| 6 | 8873 | F | 20 | 8895 | 0 | 6.27 × 10−3 | IGS | LSC |
| 7 | 9804 | F | 27 | 37,135 | −1 | 3.10 × 10−5 | trnG-GCU; trnG-UCC | LSC |
| 8 | 13,510 | F | 20 | 89,606 | 0 | 6.27 × 10−3 | IGS; ycf2 | LSC; IRa |
| 9 | 14,236 | F | 20 | 29,560 | 0 | 6.27 × 10−3 | IGS | LSC |
| 10 | 27,619 | F | 24 | 27,643 | 0 | 2.45 × 10−5 | IGS | LSC |
| 11 | 29,555 | F | 24 | 29,556 | −1 | 1.76 × 10−3 | IGS | LSC |
| 12 | 33,157 | F | 20 | 33,177 | 0 | 6.27 × 10−3 | IGS | LSC |
| 13 | 39,390 | F | 30 | 41,614 | −3 | 6.56 × 10−4 | psaB; psaA | LSC |
| 14 | 42,625 | F | 25 | 147,248 | −1 | 4.59 × 10−4 | IGS | LSC; IRb |
| 15 | 44,406 | F | 39 | 100,262 | 0 | 2.28 × 10−14 | ycf3(intron); IGS | LSC; IRa |
| 16 | 44,406 | F | 38 | 122,332 | 0 | 9.13 × 10−14 | ycf3(intron); ndhA(intron) | LSC; SSC |
| 17 | 45,075 | F | 24 | 142,008 | −1 | 1.76 × 10−3 | ycf3(intron); IGS | LSC; IRb |
| 18 | 47,622 | F | 25 | 47,645 | 0 | 6.13 × 10−6 | IGS | LSC |
| 19 | 58,656 | F | 34 | 58,687 | 0 | 2.34 × 10−11 | IGS | LSC |
| 20 | 66,712 | F | 41 | 66,752 | 0 | 1.43 × 10−15 | IGS | LSC |
| 21 | 66,939 | F | 20 | 66,958 | 0 | 6.27 × 10−3 | IGS | LSC |
| 22 | 68,033 | F | 21 | 68,052 | 0 | 1.57 × 10−3 | IGS | LSC |
| 23 | 71,232 | F | 20 | 84,928 | 0 | 6.27 × 10−3 | IGS | LSC |
| 24 | 80,953 | F | 27 | 80,966 | −2 | 1.21 × 10−3 | IGS | LSC |
| 25 | 83,166 | F | 29 | 122,320 | −3 | 2.36 × 10−3 | rpl16(intron); ndhA(intron) | LSC;SSC |
| 26 | 83,172 | F | 28 | 122,326 | −3 | 8.47 × 10−3 | rpl16(intron); ndhA(intron) | LSC;SSC |
| 27 | 90,610 | F | 29 | 90,631 | −2 | 8.74 × 10−5 | ycf2 | IRa |
| 28 | 97,630 | F | 31 | 144,839 | −3 | 1.81 × 10−4 | ndhB(intron) | IRa; IRb |
| 29 | 100,260 | F | 40 | 122,330 | 0 | 5.70 × 10−15 | IGS; ndhA(intron) | IRa; SSC |
| 30 | 101,012 | F | 23 | 101,033 | 0 | 9.80 × 10−5 | IGS | IRa |
| 31 | 141,437 | F | 30 | 141,458 | −2 | 2.34 × 10−5 | IGS | IRb |
| 32 | 141,444 | F | 23 | 141,465 | 0 | 9.80 × 10−5 | IGS | IRb |
| 33 | 151,840 | F | 29 | 151,861 | −2 | 8.74 × 10−5 | ycf2 | IRb |
| 34 | 6406 | I | 20 | 71,231 | 0 | 6.27 × 10−3 | IGS | LSC |
| 35 | 6408 | I | 24 | 71,228 | −1 | 1.76 × 10−3 | IGS | LSC |
| 36 | 8622 | I | 26 | 45,073 | −2 | 4.48 × 10−3 | IGS | LSC |
| 37 | 8625 | I | 23 | 45,077 | −1 | 6.76 × 10−3 | IGS | LSC |
| 38 | 71,232 | I | 20 | 84,930 | 0 | 6.27 × 10−3 | IGS | LSC |
F: Forward; I: Inverted; IGS: intergenic space.
As chloroplast-specific SSRs are uniparentally inherited and exhibit a high level of intraspecific polymorphism, they are widely used in population genetics, species identification, evolutionary processes research of wild plants [22,23], and as markers for linkage map construction and the breeding of crop plants [24,25]. In total, 85 SSRs were identified in the cp genome of R. chinensis var. spontanea, most of which were detected in the LSC region (Table 4). Among them, 55 (64.7%) are mononucleotide SSRs, ten (11.8%) are dinucleotide SSRs, seven (8.2%) are trinucleotide SSRs, 10 (11.8%) are tetranucleotide SSRs, one (1.2%) is a pentanucleotide SSR, and two (2.4%) are hexanucleotide SSRs. Only 22 SSRs are located in genes and the others are in the intergenic regions. Fifty two (94.5%) of the mononucleotide SSRs belong to the A/T type, which is consistent with the hypothesis that cp SSRs are generally composed of short polyadenine (poly A) or polythymine (poly T) repeats and rarely contain tandem guanine (G) or cytosine (C) repeats. These cp SSR markers can be used in the conservation genetics of R. chinensis var. spontanea, as well as and in both the linkage map construction and molecular-marker-assisted selection of modern roses.
Table 4.
Simple sequence repeats (SSRs) in the Rosa chinensis var. spontanea chloroplast genome.
| ID | Repeat Motif | Length (bp) | Start | End | Region | Gene | ID | Repeat Motif | Length (bp) | Start | End | Region | Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | (A)11 | 11 | 279 | 289 | LSC | 44 | (TTTA)4 | 12 | 50,468 | 50,479 | LSC | ||
| 2 | (T)11 | 11 | 4108 | 4118 | LSC | 45 | (TA)5 | 10 | 52,742 | 52,751 | LSC | ||
| 3 | (A)19 | 19 | 4428 | 4446 | LSC | 46 | (T)10 | 10 | 55,811 | 55,820 | LSC | atpB | |
| 4 | (A)10 | 10 | 4449 | 4458 | LSC | 47 | (AAAT)3 | 12 | 55,911 | 55,922 | LSC | ||
| 5 | (A)10 | 10 | 4887 | 4896 | LSC | 48 | (TAAT)3 | 12 | 58,366 | 58,377 | LSC | ||
| 6 | (T)10 | 10 | 5023 | 5032 | LSC | 49 | (T)14 | 14 | 60,810 | 60,823 | LSC | ||
| 7 | (TATAT)3 | 15 | 6102 | 6116 | LSC | rps16 | 50 | (TC)5 | 10 | 62,280 | 62,289 | LSC | cemA |
| 8 | (T)17 | 17 | 6407 | 6423 | LSC | 51 | (T)11 | 11 | 64,513 | 64,523 | LSC | ||
| 9 | (AATA)3 | 12 | 6525 | 6536 | LSC | 52 | (T)10 | 10 | 69,689 | 69,698 | LSC | ||
| 10 | (AG)5 | 10 | 6755 | 6764 | LSC | 53 | (A)16 | 16 | 69,739 | 69,754 | LSC | ||
| 11 | (A)11 | 11 | 6945 | 6955 | LSC | 54 | (T)18 | 18 | 71,235 | 71,252 | LSC | ||
| 12 | (TAA)4 | 12 | 8257 | 8268 | LSC | 55 | (T)15 | 15 | 71,933 | 71,947 | LSC | clpP | |
| 13 | (A)10 | 10 | 8639 | 8648 | LSC | 56 | (A)10 | 10 | 72,733 | 72,742 | LSC | clpP | |
| 14 | (AT)6 | 12 | 10,093 | 10,104 | LSC | 57 | (AT)6 | 12 | 73,632 | 73,643 | LSC | ||
| 15 | (TAT)4 | 12 | 10,343 | 10,354 | LSC | 58 | (A)12 | 12 | 79,231 | 79,242 | LSC | ||
| 16 | (T)11 | 11 | 12,157 | 12,167 | LSC | 59 | (A)14 | 14 | 79,393 | 79,406 | LSC | ||
| 17 | (T)10 | 10 | 12,915 | 12,924 | LSC | 60 | (T)10 | 10 | 79,429 | 79,438 | LSC | rpoA | |
| 18 | (A)10 | 10 | 13,184 | 13,193 | LSC | 61 | (ATGT)3 | 12 | 79,529 | 79,540 | LSC | rpoA | |
| 19 | (C)10 | 10 | 14,237 | 14,246 | LSC | 62 | (T)11 | 11 | 81,586 | 81,596 | LSC | ||
| 20 | (T)10 | 10 | 14,247 | 14,256 | LSC | 63 | (A)10 | 10 | 82,641 | 82,650 | LSC | ||
| 21 | (T)11 | 11 | 18,361 | 18,371 | LSC | rpoC2 | 64 | (A)12 | 12 | 83,422 | 83,433 | LSC | rpl16 |
| 22 | (TA)5 | 10 | 19,730 | 19,739 | LSC | rpoC2 | 65 | (A)11 | 11 | 83,498 | 83,508 | LSC | rpl16 |
| 23 | (T)10 | 10 | 26,080 | 26,089 | LSC | rpoB | 66 | (T)18 | 18 | 84,931 | 84,948 | LSC | |
| 24 | (T)12 | 12 | 28,925 | 28,936 | LSC | 67 | (TAT)4 | 12 | 86,619 | 86,630 | IRa | rpl2 | |
| 25 | (C)15 | 15 | 29,556 | 29,570 | LSC | 68 | (TAGAAG)3 | 18 | 93,987 | 94,004 | IRa | ycf2 | |
| 26 | (T)10 | 10 | 29,571 | 29,580 | LSC | 69 | (T)11 | 11 | 101,618 | 101,628 | IRa | ||
| 27 | (AAT)4 | 12 | 30,504 | 30,515 | LSC | 70 | (AGGT)3 | 12 | 107,843 | 107,854 | IRa | rrn23 | |
| 28 | (T)14 | 14 | 30,519 | 30,532 | LSC | 71 | (TATT)3 | 12 | 110,028 | 110,039 | IRa | ||
| 29 | (A)10 | 10 | 30,666 | 30,675 | LSC | 72 | (TGT)4 | 12 | 111,869 | 111,880 | SSC | ||
| 30 | (TA)5 | 10 | 36,313 | 36,322 | LSC | 73 | (T)10 | 10 | 115,507 | 115,516 | SSC | ||
| 31 | (T)11 | 11 | 36,473 | 36,483 | LSC | 74 | (TAA)4 | 12 | 115,558 | 115,569 | SSC | ||
| 32 | (AT)5 | 12 | 37,070 | 37,079 | LSC | 75 | (A)13 | 13 | 115,612 | 115,624 | SSC | ||
| 33 | (C)13 | 13 | 37,303 | 37,315 | LSC | 76 | (T)10 | 10 | 120,845 | 120,854 | SSC | ||
| 34 | (A)11 | 11 | 37,316 | 37,326 | LSC | 77 | (AT)7 | 14 | 121,678 | 121,691 | SSC | ||
| 35 | (AT)5 | 10 | 43,682 | 43,691 | LSC | ycf3 | 78 | (A)16 | 16 | 122,551 | 122,566 | SSC | ndhA |
| 36 | (A)15 | 15 | 45,073 | 45,087 | LSC | ycf3 | 79 | (T)15 | 15 | 122,804 | 122,818 | SSC | ndhA |
| 37 | (A)10 | 10 | 45,392 | 45,401 | LSC | 80 | (T)10 | 10 | 129,830 | 129,839 | SSC | ycf1 | |
| 38 | (T)10 | 10 | 45,931 | 45,940 | LSC | 81 | (ATAA)3 | 12 | 132,463 | 132,474 | IRb | ||
| 39 | (A)11 | 11 | 47,296 | 47,306 | LSC | 82 | (CTAC)3 | 12 | 134,645 | 134,656 | IRb | rrn23 | |
| 40 | (TAAT)3 | 12 | 48,112 | 48,123 | LSC | 83 | (A)11 | 11 | 140,873 | 140,883 | IRb | ||
| 41 | (T)14 | 14 | 48,306 | 48,319 | LSC | 84 | (CTTCTA)3 | 18 | 148,497 | 148,514 | IRb | ycf2 | |
| 42 | (A)12 | 12 | 48,420 | 48,431 | LSC | 85 | (ATA)4 | 12 | 155,871 | 155,882 | IRb | ||
| 43 | (TA)5 | 10 | 48,500 | 48,509 | LSC |
2.3. Comparative Analysis of the Chloroplast Genomes of the Genus Rosa
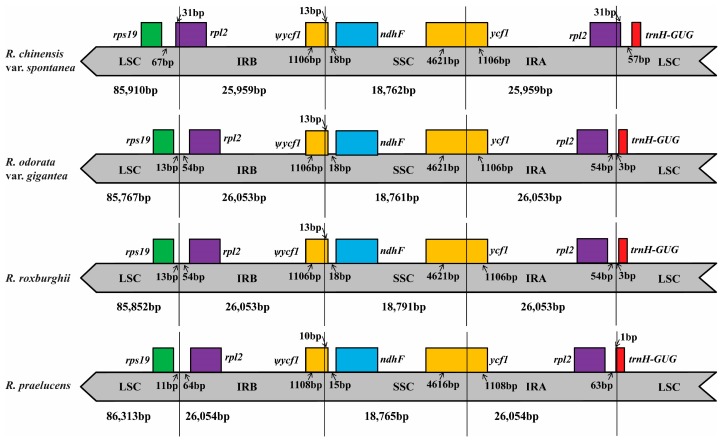
The complete cp genome sequence of R. chinensis var. spontanea was compared to that of R. odorata var. gigantea [13], R. roxburghii (KX768420) and R. praelucens (MG450565) [14]. Rosa chinensis var. spontanea has the smallest cp genome with the smallest IR region (25,959 bp), while R. praelucens has the largest cp genome with the largest LSC, at 86,313 bp (Table S2). No significant differences were found in the sequence lengths of SSC among the four species. The main reason for the length differences in cp genomes of different rose species is the size variation of the LSC and IR regions (Table S2).
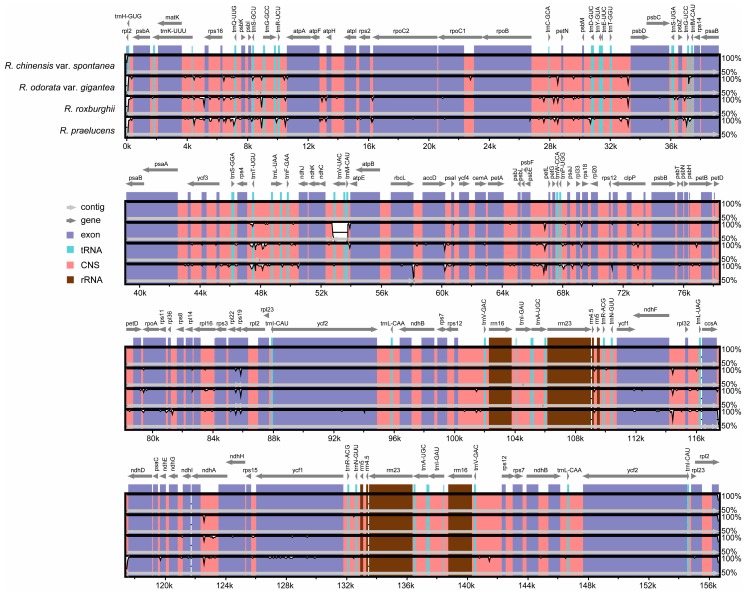
Sequence comparisons revealed that the LSC and the SSC regions were more divergent than the IR regions, and that higher divergence could be found in non-coding regions than in coding regions (Figure 2). Significant variations could be found in coding regions of some genes including rps19 and ycf1. The highest divergence in non-coding regions was found in the intergenic regions of the trnK-rps16, rps16-trnQ, trnS-trnG, trnR-atpA, atpF-atpH, rps2-rpoC2, rpoB-trnC, trnC-petN, trnT-psbD, psbZ-trnG, rps4-trnT, psbE-petL, trnP-psaJ, ndhF-rpl32, and ccsA-ndhD. The introns of rpl2, rps16, ndhA, trnV, clpP, and ndhA were relatively highly divergent, too. These results might indicate that these regions evolve rapidly in the genus Rosa, as well as in other Rosaceae plants [26,27].
Figure 2.
Complete chloroplast genome comparison of four rose species using the chloroplast genome of R. chinensis var. spontanea as a reference. The grey arrows and thick black lines above the alignment indicate the gene orientation. The y-axis represents the identity from 50% to 100%.
2.4. IR Contraction in the Chloroplast Genome of R. chinensis var. spontanea
Although IRs are the most conserved regions of the cp genomes, contraction and expansion at the borders of IR regions are common evolutionary events, and are hypothesized to be the main reason for size differences between cp genomes [28]. Detailed comparisons of the IR-SSC and IR-LSC boundaries among the cp genomes of the above four rose species were presented in Figure 3. IR regions are relatively highly conserved in the genus Rosa, but compared to other congeneric species, some position changes occurred in the IR/LSC regions of R. chinensis var. Spontanea. The rpl2 gene in the cp genome shifted by 31 bp from IRb to LSC at the LSC/IRb border, and that gene also shifted by 31 bp from IRa to LSC at the IRa/LSC border, indicating the IR contraction in the cp genome of this species. This contraction is mainly caused by the fragment deletions in the intergenic regions of the rps12-trnV, rrn4.5-rrn5, and trnR-trnN genes, and leads to the relatively smaller size of its IR regions and consequently a smaller size of the cp genome (Figure 3, Table S2).
Figure 3.
Comparison of the LSC, SSC and IR regions in chloroplast genomes of four species. Ψ: pseudogenes, →: distance from the edge.
Generally, the IRa/LSC border is located between the rpl2 and trnH genes in the rose family with rpl2 in IRa and trnH in LSC [27], like in R. roxburghii and R. odorata var. gigantea. The trnH gene of R. praelucens extends only one bp from LSC to IRa, but its LSC region was much larger than that of other species (Table S2). One 505 bp insertion in the intergenic region between the genes psbM and trnD was detected according to the result of the MAFFT alignment. This large insertion leads to the largest LSC region of R. praelucens and thus the largest cp genome among these four rose species. The extraction and contraction of the IR region at the IR-SSC boundaries among these species were not significant. Accordingly, the extension and contraction of IR regions at the IR/LSC borders, along with the large insertion/deletion in the LSC region, might be the main reason for the cp genome size variation in the genus Rosa.
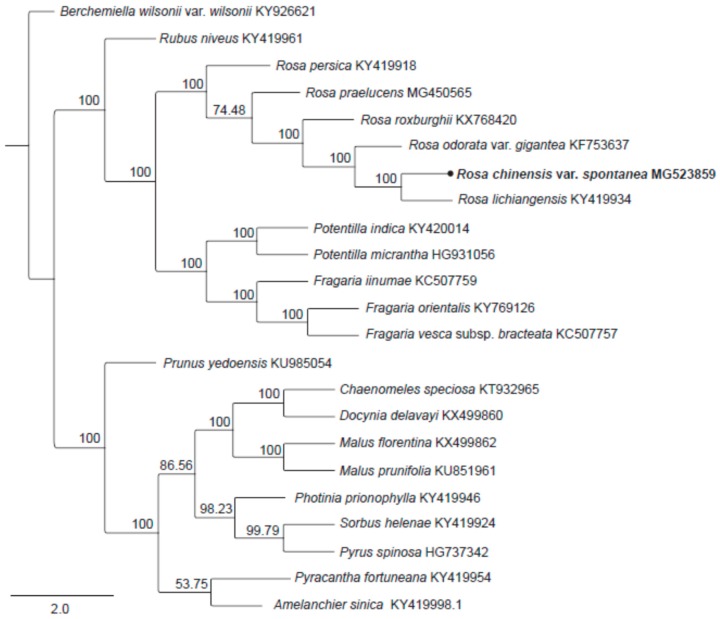
2.5. Phylogenetic Analysis
There have been many attempts to reconstruct the phylogeny of the genus Rosa. Most of them suggested that the extant classification system was artificial [29,30] and that interspecies relationships within the genus remained ambiguous. The specific relationships within the sections Chinenses and Synstylae were still obscure due to limited sampling, low genetic variation of molecular markers, and complex evolutionary histories [31]. The availability of the complete cp genomes will provide additional informative data for the reconstruction of a robust phylogenetic model for the rose species. The phylogenetic tree (Figure 4) based on the LSC, SSC and one-IR regions in the cp genomes of 22 species from Rosaceae showed that species from Rosaceae were monophyletic and that the intra-family relationships were almost in compliance with that found by Zhang et al. [32]. Species from the genus Rosa formed a monophyletic clade with 100% support. The representative of the subgenus Hulthemia, R. persica Michx. [33,34], was a sister to the clade composed by the other five rose species, supporting the subgenus position of Hulthemia. In the subgenus Rosa, R. chinensis var. spontanea from section Chinenses was sister to R. lichiangensis from section Synstylae, and then clustered with another species from section Chinenses, R. odorata var. gigantean, confirming that R. sections Chinenses and Synstylae, defined in the traditional taxonomic system, shared a more recent ancestor and could be merged as one section in the genus Rosa [30].
Figure 4.
Phylogeny of 22 species within Rosaceae based on the ML analysis of the chloroplast genome’s LSC, SSC, and one-IR regions with Berchemiella wilsonii (Rhamnaceae) as the outgroup. The position of R. chinensis var. spontanea is shown in block letters.
3. Materials and Methods
3.1. DNA Sequencing and Chloroplast Genome Assembly
Dry leaves of R. chinensis var. spontanea collected from Yichang of Hubei (111°10′ E, 30°47′ N, 400 m) were used to extract the total genomic DNA. A shotgun library was prepared and sequenced using the Illumina Hiseq 2000 (Illumina, CA, USA) at Novogene (Beijing, China). Approximately 3.68 Gb raw data of 150 bp paired-end reads were generated. The raw reads were filtered to obtain high-quality clean reads by using NGS QC Toolkit v2.3.3 with default parameters [35]. The cp genome was de novo assembled using the GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle).
3.2. Gene Annotation and Sequence Analysis
The genome was automatically annotated by using the CpGAVAS pipeline [36]. The annotation was adjusted and confirmed by Geneious 8.1 [37]. Sequence data was deposited into GenBank under the accession number MG523859. The circular cp map of R. chinensis var. spontanea was generated by OGDRAW [38]. Codon usage analysis, calculation of relative synonymous codon usage values (RSCU), and measurement of AT content were carried out by using MEGA 6.06 [39].
3.3. Genome Comparison
MUMer [40] was used to perform pairwise sequence alignments of cp genomes. The mVISTA [41] program was applied to compare the complete cp genome of R. chinensis var. spontanea to the other published cp genomes of its congeneric species, i.e., R. odorata var. gigantea, R. roxburghii and R. praelucens, using the shuffle-LAGAN mode [42] and using the annotation of R. chinensis var. spontanea as reference.
3.4. Repeats and Simple Sequence Repeats (SSRs)
REPuter [43] was used to find forward and inverted tandem repeats ≥ 20 bp with a minimum alignment score and maximum period size of 100 and 500, respectively. The minimum identity of repeats was limited to 85% (Hamming distance of 3). IMEx [44] was used to identify SSRs with the minimum repeat number set to 10, 5, 4, 3, 3 and 3 for mono-, di-, tri-, tetra-, penta- and hexanucleotides, respectively.
3.5. Phylogenetic Analysis
To identify the phylogenetic position of R. chinensis var. spontanea in Rosa, 21 published cp genomes of Rosaceae were used to construct a phylogeny tree, using Berchemiella wilsonii (C. K. Schneid.) Nakai (Rhamnaceae) as the outgroup. The LSC, SSC, and one-IR regions of the total 23 cp genomes were all aligned using MAFFT 7.308 [45]. The maximum likelihood (ML) tree was reconstructed by RAxML 8.2.11 [46] with the nucleotide substitution model of GTR + Gamma; node support was conducted by means of a bootstrap analysis with 1000 replicates.
4. Conclusions
In this study, we report and analyze the first complete cp genome of R. chinensis var. spontanea, one of the key ancestors of modern roses and a source for famous traditional Chinese medicines against female diseases. Compared to the cp genomes of other rose species, the cp genome of R. chinensis var. spontanea is the smallest, most likely due to the contraction of IR regions by 31 bps on each IR/LSC border. The cp genome of R. chinensis var. spontanea is rich in SSRs, which are valuable sources for developing new molecular markers. Our phylogenetic analysis showed that sampled species of the genus Rosa formed a monophyletic clade. Rosa chinensis var. spontanea shared a more recent ancestor with R. lichiangensis of the section Synstylae than with R. odorata var. gigantea of the section Chinenses. This supported the hypothesis that, in the traditional taxonomic system, Rosa sections Chinenses and Synstylae were closely related and could be merged to a single section within the genus Rosa. This information will be useful for the conservation genetics of R. chinensis var. spontanea and the phylogenetic study of genus Rosa, and might also facilitate the genetics and breeding of modern roses.
Acknowledgments
This study was supported by the National Natural Scientific Foundation of China (Grant 31760087), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDPB0201), and the Academic and Technical Talent Training Project of Yunnan Province, China (Grant 2013HB092).
Supplementary Materials
Supplementary materials are available online.
Author Contributions
Hong-Ying Jian and Shu-Dong Zhang conceived and designed the research framework; Hong-Ying Jian wrote the paper; Shu-Dong Zhang and Yong-Hong Zhang assembled and annotated the genome; Hong-Ying Jian, Yong-Hong Zhang, and Hui-Jun Yan analyzed the data; Hong-Ying Jian, Hui-Jun Yan, and Xian-Qin Qiu performed the experiments. Hong-Ying Jian, Yong-Hong Zhang, Qi-Gang Wang, Shu-Bin Li, and Shu-Dong Zhang collected the samples and made revisions to the final manuscript. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Sequence data of Rosa chinensis var. spontanea has been deposited into GenBank and are available from the authors.
References
- 1.Meng J., Fougère-Danezan M., Zhang L.B., Li D.Z., Yi T.S. Untangling the hybrid origin of the Chinese tea roses: Evidence from DNA sequences of single-copy nuclear and chloroplast genes. Plant Syst. Evol. 2011;297:157–170. doi: 10.1007/s00606-011-0504-5. [DOI] [Google Scholar]
- 2.Wylie A. The history of garden roses. J. R. Hortic. Soc. 1954;79:555–571. [Google Scholar]
- 3.Rix M. Rosa chinensis f. spontanea. Curtis’s Bot. Mag. 2005;22:214–219. doi: 10.1111/j.1355-4905.2005.00494.x. [DOI] [Google Scholar]
- 4.Ye J.Q. Modern Practical Herb. China Press of Traditional Chinese Medicine; Beijing, China: 2015. pp. 129–130. [Google Scholar]
- 5.Ku T.C. Rosa. In: Editorial Board of the Flora Republicae Popularis Sinicae, editor. Flora Reipublicae Popularis Sinicae. Volume 37. Science Press; Beijing, China: 1985. pp. 360–455. [Google Scholar]
- 6.Ku T.C., Robertson K.R. Rosa (Rosaceae) In: Wu Z.Y., Raven P.H., Hong D.Y., editors. Flora of China. Volume 9. Science Press; Beijing, China: Missouri Botanical Garden Press; St. Louis, MO, USA: 2003. pp. 339–381. [Google Scholar]
- 7.Qin H.N., Yang Y., Dong S.Y., He Q., Jia Y., Zhao L.N., Yu S.X., Liu H.Y., Liu B., Yan Y.H., et al. Threatened species list of China’s higher plants. Biodivers. Sci. 2017;25:696–744. doi: 10.17520/biods.2017144. [DOI] [Google Scholar]
- 8.Akasaka M., Ueda Y., Koba T. Karyotype analyses of five wild rose species belonging to septet A by fluorescence in situ hybridization. Chromosome Sci. 2002;6:17–26. [Google Scholar]
- 9.Yomogida K. Scent of modern roses. Kouryo. 1992;175:65–89. [Google Scholar]
- 10.Wu S., Watanabe N., Mita S., Ueda Y., Shibuya M., Ebizuka Y. Two O-methytransferases isolated from flower petals of Rosa chinensis var. spontanea involved in scent biosynthesis. J. Biosci. Bioeng. 2003;96:119–128. doi: 10.1016/S1389-1723(03)90113-7. [DOI] [PubMed] [Google Scholar]
- 11.Wicke S., Schneeweiss G.M., de Pamphilis C.W., Muller K.F., Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniell H., Lin C.S., Yu M., Chang W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J.B., Li D.Z., Li H.T. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol. Ecol. Resour. 2014;14:1024–1031. doi: 10.1111/1755-0998.12251. [DOI] [PubMed] [Google Scholar]
- 14.Jian H.Y., Zhang S.D., Zhang T., Qiu X.Q., Yan H.J., Wang Q.G., Tang K.X. Characterization of the complete chloroplast genome of a critically endangered decaploid rose species, Rosa praelucens (Rosaceae) Conserv. Genet. Resour. 2017 doi: 10.1007/s12686-017-0946-3. [DOI] [Google Scholar]
- 15.Clegg M.T., Gaut B.S., Learn G.H., Morton B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA. 1994;91:6795–6801. doi: 10.1073/pnas.91.15.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen X.F., Wu M.L., Liao B.S., Liu Z.X., Bai R., Xiao S.M., Li X.W., Zhang B.L., Xu J., Chen S.L. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 2017;22:1330. doi: 10.3390/molecules22081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang B., Li X., Qian J., Wang L., Ma L., Tian X., Wang Y. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii. Using the PacBio RS II platform. Molecules. 2016;21:1029. doi: 10.3390/molecules21081029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He L., Qian J., Sun Z.Y., Xu X.L., Chen S.L. Complete chloroplast genome of medicinal plant Lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules. 2017;22:249. doi: 10.3390/molecules22020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniell H., Wurdack K.J., Kanagaraj A., Lee S.B., Saski C., Jansen R.K. The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II Intron. Theor. Appl. Genet. 2008;116:723–737. doi: 10.1007/s00122-007-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C.H., Zhu H.T., Xing Y., Tan J.J., Chen X.H., Zhang J.J., Peng H.F., Xie Q.J., Zhang Z.M. Albino leaf 2 is involved in the splicing of chloroplast group I and II Introns in rice. J. Exp. Bot. 2016 doi: 10.1093/jxb/erw296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel J., Hübschmann T., Börner T., Hess W.R. Splicing and intron-internal RNA editing of trnK-matK transcript in barley plastids: Support for MatK as an essential splice factor 1. J. Mol. Biol. 1997;270:179–187. doi: 10.1006/jmbi.1997.1115. [DOI] [PubMed] [Google Scholar]
- 22.Provan J. Novel chloroplast microsatellites reveal cytoplasmic variation in Arabidopsis thaliana. Mol. Ecol. 2000;9:2183–2185. doi: 10.1046/j.1365-294X.2000.105316.x. [DOI] [PubMed] [Google Scholar]
- 23.Flannery M.L., Mitchell F.J., Coyne S., Kavanagh T.A., Burke J.I., Salamin N., Dowding P., Hodkinson T.R. Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theor. Appl. Genet. 2006;113:1221–1231. doi: 10.1007/s00122-006-0377-0. [DOI] [PubMed] [Google Scholar]
- 24.Powell W., Morgante M., McDevitt R., Vendramin G.G., Rafalski J.A. Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. USA. 1995;92:7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue J., Wang S., Zhou S.L. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae) Am. J. Bot. 2012;99:240–244. doi: 10.3732/ajb.1100547. [DOI] [PubMed] [Google Scholar]
- 26.Shen L., Guan Q., Amin A., Wei Z., Li M., Li X., Lin Z., Tian J. Complete plastid genome of Eriobotrya japonica (Thunb.) Lindl and comparative analysis in Rosaceae. SpringerPlus. 2016;5:2036. doi: 10.1186/s40064-016-3702-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng H., Li J.F., Zhang H., Cai B.H., Gao Z.H., Qiao Y.S., Mi L. The complete chloroplast genome sequence of strawberry (Fragaria × ananassa Duch.) and comparison with related species of Rosaceae. PeerJ. 2017;5:e3919. doi: 10.7717/peerj.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raubeson L.A., Peery R., Chumley T.W., Dziubek C., Fourcade H.M., Boorem J.L., Jansen R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007;8:174–201. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruneau A., Starr J.R., Joly S. Phylogenetic relationships in the genus Rosa: New evidence from chloroplast DNA sequences and an appraisal of current knowledge. Syst. Bot. 2007;32:366–378. doi: 10.1600/036364407781179653. [DOI] [Google Scholar]
- 30.Fougère-Danezan M., Joly S., Bruneau A., Gao X.F., Zhang L.B. Phylogeny and biogeography of wild roses with specific attention to polyploids. Ann. Bot. 2015;115:275–291. doi: 10.1093/aob/mcu245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z.M., Gao X.F., Fougère-Danezan M. Phylogeny of Rosa sections Chinenses and Synstylae (Rosaceae) based on chloroplast and nuclear markers. Mol. Phylogenet. Evol. 2016;87:50–64. doi: 10.1016/j.ympev.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S.D., Jin J.J., Chen S.Y., Chase M.W., Sotis D.E., Li H.T., Yang J.B., Li D.Z., Yi T.S. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 2017;214:1355–1367. doi: 10.1111/nph.14461. [DOI] [PubMed] [Google Scholar]
- 33.Rehder A. Manual of Cultivated Trees and Shrubs Hardy in North America Exclusive of the Subtropical and Warmed Temperate Regions. Macmillan; New York, NY, USA: 1940. [Google Scholar]
- 34.Wissemann V. Conventional taxonomy (wild roses) In: Roberts A.V., Debener T., Gudin S., editors. Encyclopedia of Rose Science. Volume 1. Elsevier; Amsterdam, The Netherlands: 2003. pp. 111–117. [Google Scholar]
- 35.Patel R.K., Jain M. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE. 2017;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C., Shi L., Zhu Y., Chen H., Zhang J., Lin X., Guan X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012;13:715. doi: 10.1186/1471-2164-13-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohse M., Drechsel O., Kahlau S., Bock R. Organellar Genome-DRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:575–581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2733. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 42.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mudunuri S.B., Nagarajaram H.A. IMEx: Imperfect Microsatellite Extractor. Bioinformatics. 2007;23:1181–1187. doi: 10.1093/bioinformatics/btm097. [DOI] [PubMed] [Google Scholar]
- 44.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.