Abstract
In the present work, 15 new 1-(4-(1H-imidazol-1-yl)phenyl)-3-(4-substituedphenyl)prop-2-en-1-one derivatives (3a–3o) were synthesized to evaluate their antifungal activity. Structures of newly synthesized imidazole derivatives (3a–3o) were characterized by IR, 1H-NMR, 13C-NMR, and LCMSMS spectroscopic methods. The anticandidal activity of compounds (3a–3o) against C. albicans (ATCC 24433), C. krusei (ATCC 6258), C. parapsilosis (ATCC 22019), and C. glabrata (ATCC 90030) was elucidated according to the EUCAST definitive (EDef 7.1) method. Consistent with the activity studies, 3a–3d were found to be more potent derivatives with their MIC50 values (0.78 µg/mL–3.125 µg/mL) against Candida strains. Compound 3c indicated similar antifungal activity to ketoconazole against all Candida species and was evaluated as the most active derivative in the series. Effects of the most potent derivatives 3a–3d on ergosterol biosynthesis were observed by LC-MS-MS method, which is based on quantification of the ergosterol level in C. krusei. Moreover, these compounds were subjected to a cytotoxicity test for the preliminary toxicological profiles and were found as non-cytotoxic. Furthermore, docking studies for the most active derivative 3c were performed to evaluate its binding modes on lanosterol 14-α-demethylase. In addition to in vitro tests, docking studies also revealed that Compound 3c is a potential ergosterol biosynthesis inhibitor.
Keywords: imidazole, anticandidal activity, ergosterol inhibition, 14-alpha demethylase, docking study
1. Introduction
During the last few years, there has been an increased awareness of morbidity and mortality related to invasive and systemic fungal disease due to resistant fungi and immunocompromised infections such as AIDS. Candidiasis, aspergillosis, and cryptococcosis represent the three most common invasive fungal infections and have the highest mortality rates [1,2,3]. Cryptococcus, Trichosporon, Geotrichum, and Rhodotorula species also lead to diverse occurrence and restrict clinical therapy owing to the fungus variety. However, Candida species have been the primary reason of fungal infections [4,5]. Candida spp. are presently the third-to-fourth leading reason of bloodstream infections in the USA [6]. Candida is generally part of the normal flora of the mouth, vagina, skin, and intestinal tract and can exist in the oral cavity in 40–60% of the population without causing any problems [7,8]. If the environment of normal flora changes in a way that encourages fungal growth, Candida can increase and cause infections. Currently, various antifungal drugs have been used clinically in an attempt to reduce effects of fungal infections.
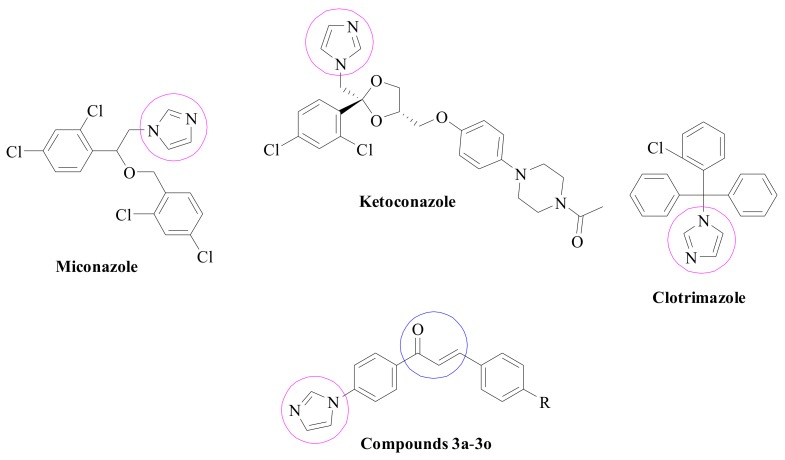
The “azoles” are class of antifungal agents whose molecules are based on a pharmacophore that inhibits the activity of fungal cytochrome P45014DM (also known as lanosterol 14-α-demethylase, Erg11p, Cyp51p, and Erg16p). Azoles are administered against lanosterol 14-α-demethylase in the ergosterol pathway. Antifungal azoles bind with nitrogen atoms to iron atoms of the heme group in the target protein and block fungal membrane ergosterol biosynthesis by inhibiting the demethylation of lanosterol to ergosterol and altering the fungal membrane structure and function [9,10,11]. The variability of azole nuclei is demonstrated in their present applications as pharmaceutical drugs used in the cure of bacterial, viral, and fungal infections, worm infestations, acid reflux, cancer, inflammations, and diabetes [12]. Imidazoles, the first group to be developed in azole antifungals also block the accumulation of methylated sterols, and disrupt the ergosterol biosynthesis, which is an essential component of the fungal cell wall. Moreover, some imidazole drugs, at high concentrations, could display a direct inhibitory effect on membranes, without intervene in sterols and sterol esters [13,14,15]. Ketoconazole, miconazole, and clotrimazole are some common drugs used for the treatment of patients affected by different Candida species [16,17,18]. Chemical structures of these antifungal agents are summarized in Figure 1 along with compounds targeted in the present study.
Figure 1.
Structures of some antifungal agents and synthesized compounds (3a–3o).
In other respects, it is well known that chalcones (1,3-diaryl-2-propen-1-one) plays an important role for anticandidal activity [19,20,21,22]. Their anticandidal action has been mainly related to the reactive enone moiety. As a Michael reaction acceptor, the enone unit binds thiol groups of certain proteins. Hence, most chalcones inhibit biosynthesis of the fungal cell wall and thus clarify their antifungal potential [23].
Knowledge of antifungal activities in both functional groups (azole and chalcone) has prompted curiosity about the antifungal effects of compounds containing these two groups. Therefore, within the scope of this study, a series of new imidazole–chalcone derivatives were synthesized and evaluated for their antifungal activities.
2. Results and Discussion
2.1. Chemistry
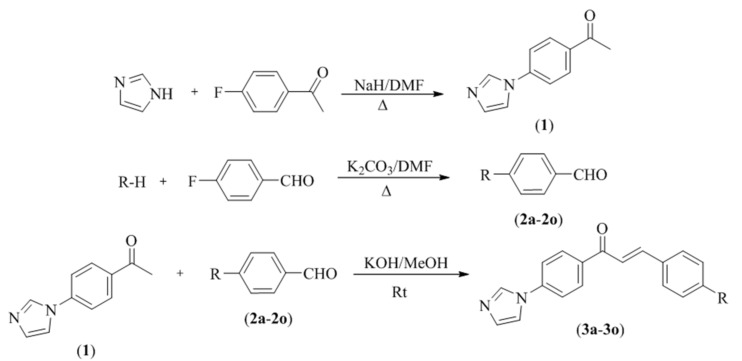
Compounds 3a–3o were synthesized as summarized in Scheme 1. Initially, 4′-(imidazol-1-yl)acetophenone (1) was obtained under reflux by a reaction of 1-(4-fluorophenyl)ethan-1-one and 1H-imidazole. Secondly, 4-fluorobenzaldehyde and appropriate proton donoring group were reacted in order to obtained 4-substitutedbenzaldehydes (2a–2o). In the last step, the target compounds (3a–3o) were synthesized by means of claisen schmidt condensation using 4′-(imidazol-1-yl)acetophenone (1) and appropriate 4-substituted benzaldehydes (2a–2o). IR, 1H-NMR, 13C-NMR, and LCMSMS spectroscopic methods were used to perform structure elucidations of the final compounds. In the IR spectrum, C=O and C=N bonds were observed at 1601–1657 cm−1 and 1296–1389 cm−1, respectively. 1,4-Disubstituted benzene bands were observed at 808–814 cm−1. In the 1H-NMR spectrum, 1,4-disubstituted benzene protons had doublet peaks between 6.58 and 8.30 ppm. Three imidazole hydrogens gave broad singlet or triplet peaks with small coupling constant values at 7.16–7.17 ppm, 7.90–7.91 ppm, and 8.44–8.47 ppm. Vinyl protons were observed as two doublet peaks with coupling constant values, approximately 15 Hz. In the 13C-NMR spectrum, aromatic peaks were gained between 113 and 161 ppm. Carbonyl carbon gave a peak over 187 ppm. Aliphatic carbons belonging to substituents were observed between 12 and 64 ppm. In the mass spectrum, all masses were matched with the expected M + H values
Scheme 1.
Synthesis way of the target compounds (3a–3o).
| Compounds | R |
|---|---|
| 3a | 4-methylphenoxy |
| 3b | 4-methylphenylthio |
| 3c | 4-methoxyphenoxy |
| 3d | 4-methoxyphenylthio |
| 3e | pyrrolidinyl |
| 3f | morpholinyl |
| 3g | piperidinyl |
| 3h | 3-methylpiperidinyl |
| 3i | 4-methylpiperidinyl |
| 3j | 3,5-dimethylpiperidinyl |
| 3k | 4-benzylpiperidinyl |
| 3l | 4-methylpiperazinyl |
| 3m | 4-ethylpiperazinyl |
| 3n | 4-(2-dimethylaminoethyl)piperazinyl |
| 3o | 4-(3-dimethylaminopropyl)piperazinyl |
2.2. Antifungal Activity
Synthesized compounds (3a–3o) were evaluated for anticandidal activity against C. albicans (ATCC 24433), C. krusei (ATCC 6258), C. parapsilosis (ATCC 22019), and C. glabrata (ATCC 90030). MIC50 values were determined via fluorometric measurements, using resazurin solution [24,25]. Ketoconazole and fluconazole were used as a standard drug in the activity test. Anticandidal activity results are presented in Table 1.
Table 1.
MIC50 (µg/mL) values of Compounds 3a–3o.
| Comp. | C. albicans | C. glabrata | C. krusei | C. parapsilosis |
|---|---|---|---|---|
| 3a | 3.125 | 3.125 | 0.78 | 0.78 |
| 3b | 3.125 | 3.125 | 0.78 | 0.78 |
| 3c | 1.56 | 0.78 | 0.78 | 0.78 |
| 3d | 1.56 | 0.78 | 1.56 | 3.125 |
| 3e | 50 | 12.50 | 12.50 | 12.50 |
| 3f | 50 | 12.50 | 12.50 | 12.50 |
| 3g | 50 | 12.50 | 25 | 12.50 |
| 3h | 50 | 12.50 | 25 | 12.50 |
| 3i | 50 | 12.50 | 12.50 | 12.50 |
| 3j | 50 | 12.50 | 25 | 25 |
| 3k | 50 | 25 | 25 | 12.50 |
| 3l | 50 | 12.50 | 25 | 12.50 |
| 3m | 50 | 12.50 | 12.50 | 25 |
| 3n | 50 | 12.50 | 12.50 | 12.50 |
| 3o | 50 | 12.50 | 12.50 | 12.50 |
| Ketoconazole | 0.78 | 1.56 | 1.56 | 1.56 |
| Fluconazole | 0.78 | 1.56 | 1.56 | 0.78 |
Consistent with the activity studies, Compounds 3a–3d, were found to be more potent derivatives with their MIC50 values (0.78–3.125 µg/mL) against Candida strains. Compounds 3a–3c showed higher antifungal activity against C. krusei with an MIC50 value of 0.78 µg/mL compared with standard drugs. Furthermore, MIC50 value of 0.78 µg/mL was recorded for Compounds 3c and 3d against C. glabrata. Compound 3c indicated similar antifungal activity to ketoconazole and fluconazole against all Candida species and was evaluated as the most active derivative in the series.
Antimicrobial activity results clearly indicated that variable groups at the 4th position of the phenyl moiety have an essential impact on antifungal activity. It was observed that the presence of more liphophilic phenoxy and phenylthio groups in Compounds 3a–3d have significantly enhanced anticandidal activity when compared with the compounds that carry cyclic secondary amines. It is known that lipophilicity is a key property that influences the ability of a drug to reach the target by transmembrane diffusion and to have a major effect on the biological activity [26]. Besides, augmented electron density may be another reason for better anticandidal activity due to the presence of electronically rich aromatic rings in Compounds 3a–3d. Thus, it may be suggested that increased lipophilic and/or electronic characters of Compounds 3a–3d caused an increase in anticandidal activity.
2.3. Quantification of the Ergosterol Level
Sterols are neutral lipids of eukaryotic cells, among which ergosterol is the main constituent of fungal membranes. Ergosterol has biological functions such as membrane fluidity, regulation, activity and distribution of integral membrane proteins, and control of the cell cycle [27,28]. The critical role of sterols in maintenance of cell membranes make ergosterol and its biosynthetic pathway essential for fungal growth, and a primary target for antifungal drugs to treat fungal infections [6]. From this point of view, we performed an LCMSMS (Shimadzu LCMS 8040, Kyoto, Japan) method for quantitative determination of ergosterol content of C. krusei as reported in our recent study [29]. Compounds 3a–3d, displaying the best anticandidal activity and low cytotoxic effect, and reference agents ketoconazole and fluconazole were tested at concentrations of 0.78–3.12 µg/mL. Ergosterol standard (Product No.: 45480, Sigma-Aldrich, Darmstadt, Germany) was used for the quantification of ergostrerol in samples, which were treated with reference agents and Compounds 3a–3d. Ergosterol quantity of the negative control was considered as 100%. Quadruplicate analyses were performed for all concentrations and the obtained data were expressed as mean ± standard deviation (SD) (Table 2).
Table 2.
Cytotoxic activity and ergosterol biosynthesis inhibition potency of Compounds 3a–3d against NIH/3T3 cell line and C. krusei, respectively.
| Compound | IC50 (µg/mL) | Inhibition of Ergosterol Biosynthesis (%) | ||
|---|---|---|---|---|
| 0.78 µg/mL | 1.56 µg/mL | 3.12 µg/mL | ||
| 3a | >500 | 66.19 ± 2.23 | 79.45 ± 3.16 | 86.47 ± 4.77 |
| 3b | >500 | 68.59 ± 1.98 | 81.62 ± 4.07 | 83.49 ± 3.18 |
| 3c | 436.04 ± 1.03 | 71.14 ± 4.9 | 78.16 ± 2.70 | 84.28 ± 4.65 |
| 3d | 387.64 ± 20.20 | 52.47 ± 1.83 | 67.14 ± 2.70 | 74.14 ± 2.21 |
| Ketoconazole | - | 60.99 ± 2.94 | 73.12 ± 4.16 | 84.56 ± 3.01 |
| Fluconazole | - | 61.74 ± 1.70 | 70.12 ± 3.22 | 82.13 ± 4.45 |
According to obtained results, the decrease in ergosterol level after administration of Compounds 3a–3d are noticeable when compared to the reference agents. Compounds 3a–3d and reference agents significantly decreased the level of ergosterol at all tested concentrations. Therefore, it can be interpreted that Compounds 3a–3d play a role in the ergosterol biosynthesis pathway.
2.4. Cytotoxicity Test
The main cause of failure in all stages of the new drug development process is toxicity. Early identification of toxicities of drug candidates is very important in terms of drug development studies [30]. Therefore, the MTT cell viability assay, which is suggested for cytotoxicity screening of drug candidates by ISO (10993-5, 2009) was performed [31]. Cytotoxicity of selected compounds (3a–3d), displaying strong anticandidal activity and good predicted pharmacokinetics, was determined against NIH/3T3 mouse embryonic fibroblast cell lines (ATCC CRL1658).
The cytotoxicity results of the tested compounds are presented in Table 2. IC50 of compounds against NIH/3T3 was much higher than their MIC50 values (0.78–3.125 µg/mL) against Candida strains. This finding shows that the antifungal activity of Compounds 3a–3d is not due to general toxicity, but can be ascribed to their selective action against Candida species. Thus, cytotoxicity test findings enhanced the importance of Compounds 3a–3d as anticandidal drug candidates.
2.5. Molecular Docking Studies
Docking studies were performed in order to gain more insight into the binding mode of the most active compound 3c to lanosterol 14-α-demethylase. Lanosterol 14-α-demethylase from Mycobacterium tuberculosis has high homology compared with lanosterol 14-α-demethylase from Candida species. It has been reported that these two enzymes have a high degree of similarity between the hydrophobic cavities of the catalytic site [29,32]. Therefore, docking studies were carried out using X-ray crystal structure of lanosterol 14-α-demethylase from Mycobacterium tuberculosis in complex with fluconazole (PDB ID: 1EA1) [33] obtained from the Protein Data Bank server (www.pdb.org).
As stated in the antifungal activity section, Compound 3c was found to be the most active derivative against C. albicans, C. glabrata, C. krusei, and C. parapsilosis with 1.56, 0.78, 0.78, and 0.78 µg/mL MIC values, respectively. Thus, the main purpose of docking studies was to investigate the possible interaction of this compound with lanosterol 14-α-demethylase enzyme.
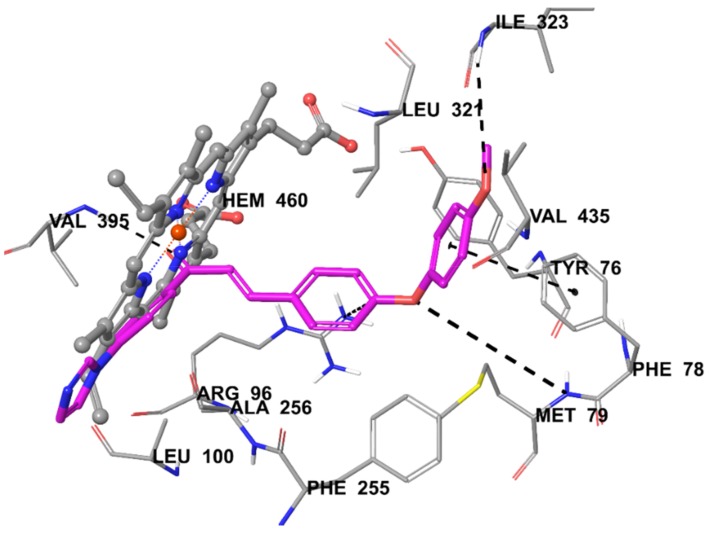
The docking pose on lanesterol 14-α-sterol demethylase reveals that the interactions between Compound 3c and HEM450 are very important in terms of binding to the active site of the enzyme (Figure 2). Imidazole and its neighboring phenyl established two π–π interactions. The other two π–π interactions were formed by other phenyl rings of the structure. Phenyl of benzylidene was in interaction with Arg96, while phenyl substituted with methoxy group created a π–π interaction with the phenyl of Phe78. Additionally, the docking pose showed that there were three hydrogen bonds providing polar interactions. Carbonyl of Compound 3c established a hydrogen bond with the amino of Val395. Oxygen atoms of phenoxy created this interaction with the amino of Met79, whereas oxygen atoms of the methoxy group formed with the amino of Ile323. These interactions explain the stronger anticandidal activity of 3c. It could be that C-4 of phenyl is very important in terms of binding to the enzyme active site and anticandidal activity. As a result, this additional interaction could explain the greater binding capability and stronger activity of Compound 3c compared with other compounds.
Figure 2.
The interacting mode of Compound 3c in the active region of 14-alpha-sterol demethylase. The inhibitor is colored with purple and HEM with grey.
3. Materials and Methods
3.1. Chemistry
All chemicals used in the syntheses were purchased either from Sigma-Aldrich Chemicals (Sigma-Aldrich Corp., St. Louis, MO, USA) or Merck Chemicals (Merck KGaA, Darmstadt, Germany). Melting points of the synthesized compounds were measured by MP90 digital melting point apparatus (Mettler Toledo, Columbus, OH, USA) and were presented as uncorrected. 1H-NMR and 13C-NMR spectra were recorded by a Bruker 300 MHz and 75 MHz digital FT-NMR spectrometer (Bruker Bioscience, Billerica, MA, USA) in DMSO-d6, respectively. In the NMR spectra, splitting patterns were designated as follows: s: singlet; d: doublet; t: triplet; m: multiplet. Coupling constants (J) were reported as Hertz. The IR spectra of the compounds were recorded using an IRAffinity-1S Fourier transform IR (FTIR) spectrometer (Shimadzu, Tokyo, Japan). LC-MS-MS studies were performed on a Schimadzu, 8040 LCMSMS spectrophotometer (Shimadzu, Tokyo, Japan). The purities of compounds were checked by TLC on silica gel 60 F254 (Merck KGaA, Darmstadt, Germany).
3.1.1. Synthesis of 4′-(Imidazol-1-yl)acetophenone (1)
4-Fluoro acetofenone (4.976 mL, 0.041 mol), imidazole (2.789 g, 0.041 mol), and sodium hydride (NaH) (1.080 g, 0.045 mol) in DMF were refluxed for 12 h. After cooling, the mixture was poured into the ice water, and the precipitated product was washed with water, dried, and recrystallized from ethanol.
3.1.2. Synthesis of Four Substituted Benzaldehydes (2a–2o)
A mixture of 4-fluoro benzaldehyde (0.259 mL, 0.002 mol), corresponding phenol, thiophenol, or amine (0.002 mol), and a catalytic quantity of potassium carbonate (K2CO3) was refluxed in DMF (20 mL) for 36 h. After completion of the reaction, the mixture was poured into ice water (50 mL), and the precipitated product was filtered, washed with deionized water, dried, and recrystallized from ethanol.
3.1.3. General Procedure for the Synthesis of Target Compounds (3a–3o)
1-(4-(1H-imidazol-1-yl)phenyl)ethan-1-one (1) (0.316 g, 0.0017 mol) and appropriate 4-substituted benzaldehydes (2a–2o) derivatives (0.0017 mol) in methanol were stirred for 10 h in the presence of potassium hydroxide. The precipitated product was washed with water, dried, and recrystallized from ethanol.
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(p-tolyloxy)phenyl)prop-2-en-1-one (3a): Yield: 79%, M.P. = 146–148 °C, FTIR (ATR, cm−1): 3128 (C-H), 1653 (C=O), 1373 (C=N), 814. 1H-NMR (300 MHz, DMSO-d6): 2.32 (3H, s, CH3), 7.01 (2H, d, J = 8.5 Hz, disubstituted CH), 7.02 (2H, d, J = 8.8 Hz, disubstituted CH), 7.17 (1H, s, imidazole CH), 7.25 (2H, d, J = 8.2 Hz, methylphenyl CH), 7.76 (1H, d, J = 15.6 Hz, -HC=CH-), 7.87–7.94 (6H, m, disubstituted CH, methylphenyl CH, imidazole CH, -HC=CH-), 8.00 (1H, d, J = 15.6 Hz, -HC=CH-), 8.30 (2H, d, J = 8.8 Hz, disubstituted CH), 8.47 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 20.8, 118.2, 118.3, 120.1, 120.3, 121.0, 129.8, 130.9, 131.1, 131.5, 134.0, 136.1, 136.2, 140.7, 144.1, 153.6, 160.1, 188.2. ESI-MS [M + H]+: 381.25 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(p-tolylthio)phenyl)prop-2-en-1-one (3b): Yield: 84%, M.P. = 168–169 °C, FTIR (ATR, cm−1): 3120 (C-H), 1651 (C=O), 1337 (C=N), 812. 1H-NMR (300 MHz, DMSO-d6): 2.35 (3H, s, CH3), 7.17 (1H, t, J = 1.1 Hz, imidazole CH), 7.21 (2H, d, J = 8.3 Hz, disubstituted CH), 7.29 (2H, d, J = 7.9 Hz, methylphenyl CH), 7.40 (2H, d, J = 8.3 Hz, disubstituted CH), 7.72 (1H, d, J = 15.6 Hz, -HC=CH-), 7.84–7.88 (3H, m, methylphenyl CH, imidazole CH), 7.89–7.98 (3H, m, disubstituted CH, -HC=CH-), 8.00 (1H, d, J = 15.6 Hz, -HC=CH-), 8.29 (2H, d, J = 8.8 Hz, disubstituted CH), 8.47 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 21.2, 118.3, 120.3, 122.0, 128.4, 128.9, 130.3, 130.9, 130.9, 131.0, 132.9, 133.8, 135.9, 136.2, 139.1, 140.7, 141.1, 143.9, 188.2. ESI-MS [M + H]+: 397.20 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(4-methoxyphenoxy)phenyl)prop-2-en-1-one (3c): Yield: 92%, M.P. = 168–170 °C, FTIR (ATR, cm−1): 3144 (C-H), 1653 (C=O), 1342 (C=N), 814. 1H-NMR (300 MHz, DMSO-d6): 3.78 (3H, s, OCH3), 6.98 (2H, d, J = 8.8 Hz, methoxyphenyl CH), 7.01 (2H, d, J = 9.2 Hz, disubstituted CH), 7.09 (2H, d, J = 9.2 Hz, disubstituted CH), 7.17 (1H, t, J = 1.1 Hz, imidazole CH), 7.75 (1H, d, J = 15.5 Hz, -HC=CH-), 7.87–7.93 (6H, m, methoxyphenyl CH, disubstituted CH, imidazole CH, -HC=CH-), 8.29 (2H, d, J = 8.8 Hz, disubstituted CH), 8.47 (1H, t, J = 1.1 Hz, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 55.9, 115.7, 117.5, 118.3, 120.3, 120.8, 121.8, 129.5, 130.9, 131.3, 131.5, 136.1, 136.2, 140.6, 144.2, 148.9, 156.6, 160.8, 188.2. ESI-MS [M + H]+: 397.25 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-((4-methoxyphenyl)thio)phenyl)prop-2-en-1-one (3d): Yield: 80%, M.P. = 177–179 °C, FTIR (ATR, cm−1): 3111 (C-H), 1657 (C=O), 1369 (C=N), 808. 1H-NMR (300 MHz, DMSO-d6): 3.81 (3H, s, OCH3), 7.06 (2H, d, J = 8.8 Hz, methoxyphenyl CH), 7.12 (2H, d, J = 8.4 Hz, disubstituted CH), 7.17 (1H, s, imidazole CH), 7.49 (2H, d, J = 8.8 Hz, methoxyphenyl CH), 7.70 (1H, d, J = 15.6 Hz, -HC=CH-), 7.81 (2H, d, J = 8.4 Hz, disubstituted CH), 7.87 (2H, d, J = 8.8 Hz, disubstituted CH), 7.91 (1H, t, J = 1.4 Hz, imidazole CH), 7.92 (1H, d, J = 15.6 Hz, -HC=CH-), 8.27 (2H, d, J = 8.8 Hz, disubstituted CH), 8.46 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 55.9, 116.1, 118.3, 120.3, 121.7, 121.8, 127.2, 130.2, 130.9, 132.5, 136.0, 136.2, 136.6, 140.7, 142.6, 143.9, 160.7, 188.2. ESI-MS [M + H]+: 413.20 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(pyrrolidin-1-yl)phenyl)prop-2-en-1-one (3e): Yield: 77%, M.P. = 186–188 °C, FTIR (ATR, cm−1): 3109 (C-H), 1655 (C=O), 1373 (C=N), 810. 1H-NMR (300 MHz, DMSO-d6): δ 1.94–1.98 (4H, m, pyrrolidine CH2), 3.28–3.32 (4H, m, pyrrolidine CH2), 6.58 (2H, d, J = 8.8 Hz, disubstituted CH), 7.16 (1H, s, imidazole CH), 7.63–7.73 (4H, m, disubstituted CH, -HC=CH-), 7.85 (2H, d, J = 8.7 Hz, disubstituted CH), 7.90 (1H, t, J = 1.2 Hz, imidazole CH), 8.25 (2H, d, J = 8.7 Hz, disubstituted CH), 8.44 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 25.4, 47.8, 112.2, 115.6, 118.3, 120.2, 121.9, 130.5, 130.8, 131.6, 136.2, 136.9, 140.2, 146.2, 149.9, 187.6. ESI-MS [M + H]+: 344.25 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-morpholinophenyl)prop-2-en-1-one (3f): Yield: 79%, M.P. = 214–215 °C, FTIR (ATR, cm−1): 3109 (C-H), 1653 (C=O), 1337 (C=N), 812. 1H-NMR (300 MHz, DMSO-d6): δ 3.27 (4H, t, J = 4.9 Hz, morpholine CH2), 3.75 (4H, t, J = 4.9 Hz, morpholine CH2), 7.00 (2H, d, J = 8.8 Hz, disubstituted CH), 7.16 (1H, t, J = 1.0 Hz, imidazole CH), 7.71 (2H, d, J = 15.4 Hz, -HC=CH-), 7.74–7.82 (3H, m, disubstituted CH, -HC=CH-), 7.86 (2H, d, J = 8.8 Hz, disubstituted CH), 7.90 (1H, t, J = 1.2 Hz, imidazole CH), 8.25 (2H, d, J = 8.7 Hz, disubstituted CH), 8.44 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 47.6, 63.4, 114.6, 117.9, 118.3, 120.2, 125.2, 130.7, 130.8, 131.1, 136.2, 136.6, 140.4, 145.3, 153.2, 187.9. ESI-MS [M + H]+: 360.25 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(piperidin-1-yl)phenyl)prop-2-en-1-one (3g): Y Yield: 73%, M.P. = 198–199 °C, FTIR (ATR, cm−1): 3078 (C-H), 1657 (C=O), 1389 (C=N), 812. 1H-NMR (300 MHz, DMSO-d6): δ 1.58 (6H, s, piperidine CH2), 3.32 (4H, s, piperidine CH2), 6.95 (2H, d, J = 8.9 Hz, disubstituted CH), 7.16 (1H, t, J = 1.0 Hz, imidazole CH), 7.71–7.82 (4H, m, disubstituted CH, -HC=CH-), 7.86 (2H, d, J = 8.7 Hz, disubstituted CH), 7.91 (1H, t, J = 1.3 Hz, imidazole CH), 8.26 (2H, d, J = 8.7 Hz, disubstituted CH), 8.45 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 24.4, 25.4, 48.5, 114.6, 117.2, 118.3, 120.2, 123.9, 130.7, 130.8, 131.3, 136.2, 136.7, 140.3, 145.5, 153.2, 187.8. ESI-MS [M + 2H]2+: 179.70 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(3-methylpiperidin-1-yl)phenyl)prop-2-en-1-one (3h): Yield: 80%, M.P. = 163–165 °C, FTIR (ATR, cm−1): 3107 (C-H), 1655 (C=O), 1339 (C=N), 812. 1H-NMR (300 MHz, DMSO-d6): 0.92 (3H, d, J = 6.6 Hz, CH3), 1.04–1.17 (1H, m, piperidine CH), 1.43–1.80 (5H, m, piperidine CH2), 2.73–2.82 (1H, m, piperidine CH2), 3.77–3.85 (2H, m, piperidine CH2), 6.96 (2H, d, J = 9.0 Hz, disubstituted CH), 7.16 (1H, s, imidazole CH), 7.71–7.73 (4H, m, disubstituted CH, -HC=CH-), 7.86 (2H, d, J = 8.8 Hz, disubstituted CH), 7.91 (1H, t, J = 1.2 Hz, imidazole CH), 8.26 (2H, d, J = 8.8 Hz, disubstituted CH), 8.45 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 19.6, 24.8, 30.6, 33.0, 47.9, 55.3, 114.6, 117.1, 118.3, 120.2, 123.8, 130.6, 130.8, 131.4, 136.2, 136.7, 140.3, 145.5, 152.9, 187.8. ESI-MS [M + 2H]2+: 186.70 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(4-methylpiperidin-1-yl)phenyl)prop-2-en-1-one (3i): Yield: 77%, M.P. = 189–191 °C, FTIR (ATR, cm−1): 3117 (C-H), 1657 (C=O), 1339 (C=N), 812. 1H-NMR (300 MHz, DMSO-d6): δ 0.92 (3H, d, J = 6.4 Hz, CH3), 1.10–1.24 (2H, m, piperidine CH2), 1.52–1.70 (3H, m, piperidine CH2), 2.76–2.85 (2H, m, piperidine CH2), 3.87–3.91 (2H, m, piperidine CH2), 6.96 (2H, d, J = 8.9 Hz, disubstituted CH), 7.16 (1H, t, J = 1.0 Hz, imidazole CH), 7.71–7.74 (4H, m, disubstituted CH, -HC=CH-), 7.86 (2H, d, J = 8.7 Hz, disubstituted CH), 7.91 (1H, t, J = 1.3 Hz, imidazole CH), 8.26 (2H, d, J = 8.7 Hz, disubstituted CH), 8.45 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 22.2, 30.8, 33.6, 47.9, 114.7, 117.2, 118.3, 120.2, 123.9, 130.7, 130.8, 131.3, 136.2, 136.7, 140.3, 145.5, 153.0, 187.8. ESI-MS [M + 2H]2+: 186.75 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(3,5-dimethylpiperidin-1-yl)phenyl)prop-2-en-1-one (3j): Yield: 81%, M.P. = 160–161 °C, FTIR (ATR, cm−1): 3035 (C-H), 1622 (C=O), 1335 (C=N), 815. 1H-NMR (300 MHz, DMSO-d6): δ 0.90 (6H, d, J = 6.5 Hz, CH3), 1.59–1.79 (4H, m, piperidine CH2), 2.28–2.36 (2H, m, piperidine CH2), 3.85–3.90 (2H, m, piperidine CH2), 6.97 (2H, d, J = 9.0 Hz, disubstituted CH), 7.16 (1H, t, J = 1.0 Hz, imidazole CH), 7.70–7.73 (4H, m, disubstituted CH, -HC=CH-), 7.86 (2H, d, J = 8.8 Hz, disubstituted CH), 7.91 (1H, t, J = 1.3 Hz, imidazole CH), 8.23 (2H, d, J = 8.8 Hz, disubstituted CH), 8.45 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 19.5, 30.5, 42.4, 54.8, 114.5, 117.1, 118.3, 120.2, 123.7, 130.6, 130.8, 131.4, 136.2, 136.7, 140.3, 145.5, 152.7, 187.9. ESI-MS [M + 2H]2+: 193.70 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(4-benzylpiperidin-1-yl)phenyl)prop-2-en-1-one (3k): Yield: 83%, M.P. = 178–180 °C, FTIR (ATR, cm−1): 3062 (C-H), 1601 (C=O), 1296 (C=N), 814. 1H-NMR (300 MHz, DMSO-d6): δ 1.17–1.31 (2H, m, piperidine CH2), 1.63–1.77 (3H, m, piperidine CH2), 2.54 (2H, d, J = 7.1 Hz, CH2), 2.73–2.81 (2H, m, piperidine CH2), 3.88–3.92 (2H, m, piperidine CH2), 6.96 (2H, d, J = 9.0 Hz, disubstituted CH), 7.16–7.21 (4H, m, monosubstituted benzene CH, imidazole CH), 7.27–7.29 (2H, m, monosubstituted benzene CH), 7.71–7.74 (4H, m, disubstituted CH, -HC=CH-), 7.86 (2H, d, J = 8.8 Hz, disubstituted CH), 7.91 (1H, t, J = 1.4 Hz, imidazole CH), 8.26 (2H, d, J = 8.8 Hz, disubstituted CH), 8.45 (1H, t, J = 1.0 Hz, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 31.5, 37.8, 42.7, 47.8, 114.7, 117.2, 118.3, 120.2, 123.9, 126.3, 128.6, 129.5, 130.7, 130.8, 131.3, 136.2, 136.7, 140.3, 140.6, 145.5, 152.9, 187.9. ESI-MS [M + 2H]2+: 224.75 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(4-methylpiperazin-1-yl)phenyl)prop-2-en-1-one (3l): Yield: 82%, M.P. = 205–207 °C, FTIR (ATR, cm−1): 3109 (C-H), 1655 (C=O), 1337 (C=N), 812. 1H-NMR (300 MHz, DMSO-d6): δ 2.22 (3H, s, CH3), 2.43 (4H, t, J = 5.0 Hz, piperazine CH2), 3.30 (4H, t, J = 5.0 Hz, piperazine CH2), 6.98 (2H, d, J = 8.9 Hz, disubstituted CH), 7.16 (1H, t, J = 1.2 Hz, imidazole CH), 7.72–7.76 (4H, m, disubstituted CH, -HC=CH-), 7.86 (2H, d, J = 8.8 Hz, disubstituted CH), 7.91 (1H, t, J = 1.3 Hz, imidazole CH), 8.27 (2H, d, J = 8.8 Hz, disubstituted CH), 8.45 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 46.2, 47.3, 54.8, 114.7, 117.7, 118.3, 120.2, 124.7, 130.7, 130.8, 131.2, 136.2, 136.6, 140.4, 145.3, 153.0, 187.9. ESI-MS [M + 2H]2+: 187.20 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(4-ethylpiperazin-1-yl)phenyl)prop-2-en-1-one (3m): Yield: 86%, M.P. = 188–189 °C, FTIR (ATR, cm−1): 3125 (C-H), 1653 (C=O), 1337 (C=N), 814. 1H-NMR (300 MHz, DMSO-d6): δ 1.03 (3H, t, J = 7.2 Hz, CH3), 2.37 (2H, q, J = 7.2 Hz, CH2), 2.48 (4H, t, J = 5.0 Hz, piperazine CH2), 3.30 (4H, t, J = 5.0 Hz, piperazine CH2), 6.99 (2H, d, J = 8.9 Hz, disubstituted CH), 7.17 (1H, s, imidazole CH), 7.73–7.77 (4H, m, disubstituted CH, -HC=CH-), 7.87 (2H, d, J = 8.8 Hz, disubstituted CH), 7.91 (1H, t, J = 1.3 Hz, imidazole CH), 8.28 (2H, d, J = 8.8 Hz, disubstituted CH), 8.46 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 12.4, 47.4, 52.1, 52.6, 114.6, 117.7, 118.3, 120.2, 124.7, 130.7, 130.8, 131.2, 136.2, 136.6, 140.4, 145.4, 153.1, 187.9. ESI-MS [M + 2H]2+: 194.20 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(4-(2-(dimethylamino)ethyl)piperazin-1-yl)phenyl)prop-2-en-1-one (3n): Yield: 78%, M.P. = 146–149 °C, FTIR (ATR, cm−1): 3036 (C-H), 1655 (C=O), 1300 (C=N), 812. 1H-NMR (300 MHz, DMSO-d6): δ 2.14 (6H, s, CH3), 2.33–2.44 (4H, m, CH2), 2.52 (4H, t, J = 4.7 Hz, piperazine CH2), 3.28 (4H, t, J = 4.7 Hz, piperazine CH2), 6.97 (2H, d, J = 8.9 Hz, disubstituted CH), 7.16 (1H, s, imidazole CH), 7.72–7.76 (4H, m, disubstituted CH, -HC=CH-), 7.86 (2H, d, J = 8.7 Hz, disubstituted CH), 7.91 (1H, t, J = 1.2 Hz, imidazole CH), 8.27 (2H, d, J = 8.7 Hz, disubstituted CH), 8.45 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 40.0, 47.4, 53.3, 56.3, 57.1, 114.6, 117.6, 118.3, 120.2, 124.7, 130.7, 130.8, 131.2, 136.2, 136.6, 140.4, 145.4, 153.0, 187.9. ESI-MS [M + H]+: 430.35 (100%).
1-(4-(1H-Imidazol-1-yl)phenyl)-3-(4-(4-(3-(dimethylamino)propyl)piperazin-1-yl)phenyl)prop-2-en-1-one (3o): Yield: 79%, M.P. = 138–141 °C, FTIR (ATR, cm−1): 3086 (C-H), 1655 (C=O), 1339 (C=N), 812. 1H-NMR (300 MHz, DMSO-d6): δ 1.56 (2H, p, J = 7.3 Hz, CH2), 2.10 (6H, s, CH3), 2.20 (2H, t, J = 7.3 Hz, CH2), 2.31 (2H, t, J = 7.3 Hz, CH2), 2.47 (4H, t, J = 4.8 Hz, piperazine CH2), 3.28 (4H, t, J = 4.8 Hz, piperazine CH2), 6.97 (2H, d, J = 8.9 Hz, disubstituted CH), 7.16 (1H, s, imidazole CH), 7.70-7.76 (4H, m, disubstituted CH, -HC=CH-), 7.86 (2H, d, J = 8.7 Hz, disubstituted CH), 7.91 (1H, t, J = 1.2 Hz, imidazole CH), 8.27 (2H, d, J = 8.7 Hz, disubstituted CH), 8.45 (1H, s, imidazole CH). 13C-NMR (75 MHz, DMSO-d6): δ = 24.9, 45.7, 47.3, 53.0, 56.5, 57.8, 113.7, 114.6, 118.2, 120.2, 124.6, 130.7, 130.8, 131.2, 136.2, 136.6, 140.4, 145.3, 153.0, 187.9. ESI-MS [M + H]+: 444.35 (100%).
3.2. Antifungal Activity
Microbiological studies were performed according to the EUCAST definitive (EDef 7.1) method [34] for C. albicans (ATCC 24433), C. krusei (ATCC 6258), C. parapsilosis (ATCC 22019), and C. glabrata (ATCC 90030). Fluconazole and ketoconazole were used as control drugs. Two MIC readings were carried out for each chemical agent. The yeasts were maintained in RPMI after overnight incubation at 37 °C. The inocula of test microorganisms was adjusted to match the turbidity of a MacFarland 0.5 standard tube as determined with a spectrophotometer, and the final inoculum size was 0.5–2.5 × 105 cfu/mL for the antifungal assay. Testing was carried out in RPMI at pH = 7, and the two-fold serial dilutions technique was applied. The last well on the microplates containing only inoculated broth was kept as a control, and the last well with no growth of microorganism was recorded to represent the MIC50 expressed in μg/mL. For the antifungal assays, the compounds were dissolved in DMSO. Further dilutions of the compounds and standard drugs in the test medium were prepared at the required quantities of 800, 400, 200, 100, 50, 25, 12.5, 6.25, 3.125, 1.5625, and 0.78 μg/mL concentrations RPMI. The completed plates were incubated for 24 h. At the end of the incubation, resazurin (20 µg/mL) was added into each well to control the growth in the wells. Completed plates were incubated for 2 h. MIC50 values were determined using a microplate reader at 590 nm excitation and 560 nm emission. Each experiment in the antimicrobial assays was replicated twice in order to define the MIC50 values given in Table 1.
3.3. Quantification of Ergosterol Level
Extraction of total sterols from C. krusei was performed as recorded by Breivik and Owades [35]. Quantification of ergosterol level in this extract was carried out in accordance with our recently described method [36].
3.4. Cytotoxicity Test
Cytotoxicity was tested using the NIH/3T3 mouse embryonic fibroblast cell line (ATCC® CRL-1658™, London, UK). NIH/3T3 cells were incubated according to the supplier’s recommendations. NIH/3T3 cells were seeded at 1 × 104 cells into each well of the 96-well plates. The MTT assay was performed as previously described [37,38]. The compounds were tested between 800 and 0.78 µg/mL concentrations. Inhibition % was calculated for each concentration according to the formula below, and IC50 values were determined by plotting a dose–response curve of inhibition % versus compound concentrations tested [39].
| % inhibition = 100 − (mean sample × 100/mean solvent). |
3.5. Molecular Docking Studies
A structure-based in silico procedure was applied to discover the binding modes of the most active compound 3c to 14-alpha-sterol demethylase enzyme active sites. The crystal structure of the enzyme (PDB ID: 1EA1) [33], which was crystallized with the reference drug (fluconazole) of antifungal activity assay, was retrieved from the Protein Data Bank server (www.pdb.org).
The structure of the ligand was built using the Schrödinger Maestro [40] interface and was then submitted to the Protein Preparation Wizard protocol of the Schrödinger Suite 2016 Update 2 [41]. The ligands were prepared by LigPrep 3.8 [42] to assign the protonation states at pH 7.4 ± 1.0 and the atom types correctly. Bond orders were assigned and hydrogen atoms were added to the structures. The grid generation was formed using Glide 7.1 [43], and docking runs were performed with the standard precision docking mode (SP).
4. Conclusions
In the present study, 15 new imidazole compounds that incorporate chalcone pharmacophores as antifungal agents were designed and synthesized. Activity studies revealed the potency of Compounds 3a–3d as antifungal agents. Furthermore, toxicological study enhanced the biological importance of these compounds. Results of the ergosterol level quantification assay revealed that the mechanism of action of the compounds is related to the inhibition of the biosynthesis of ergosterol, a vital sterol regulating membrane fluidity, plasma membrane biogenesis, and function. The docking studies clearly explained the molecular interacting mode of Compound 3c in the active region of 14-alpha-sterol demethylase. Consequently, all of this information may pave the way for medicinal chemists to synthesize similar compounds that have enhanced antifungal profiles.
Acknowledgments
This study was financially supported by Anadolu University Scientific Projects Fund, Project No.: 1705S183.
Supplementary Materials
The following are available online.
Author Contributions
Y.O. and Z.A.K. conceived and designed the experiments; D.O. and B.K.Ç. performed the synthesis; S.L. performed analysis studies; B.N.S. and U.A.C. performed activity tests; B.N.S. performed docking studies; Ö.A. performed the toxicity tests; D.O., B.K.Ç., B.N.S., S.L., U.A.Ç., Ö.A., Y.O. and Z.A.K. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds 3a–3o are available from the authors.
References
- 1.Tang H., Wub J., Zhang W., Zhao L., Zhang Y.H., Shen C.W. Design, Synthesis and Biological Evaluation of Novel Non-Azole Derivatives as Potential Antifungal Agents. Chin. Chem. Lett. 2015;26:1161–1164. doi: 10.1016/j.cclet.2015.04.030. [DOI] [Google Scholar]
- 2.Canuto M.M., Rodero F.G. Antifungal Drug Resistance to Azoles and Polyenes. Infect. Dis. 2002;2:550–563. doi: 10.1016/S1473-3099(02)00371-7. [DOI] [PubMed] [Google Scholar]
- 3.Jiang Z., Gu J., Wang C., Wang S., Liu N., Jiang Y., Dong G., Wang Y., Liu Y., Yao J., et al. Design, synthesis and antifungal activity of novel triazole derivatives containing substituted 1,2,3-triazole-piperdine side chains. Eur. J. Med. Chem. 2014;82:490–497. doi: 10.1016/j.ejmech.2014.05.079. [DOI] [PubMed] [Google Scholar]
- 4.Cast’on-Osorio J.J., Rivero A., Torre-Cisneros J. Epidemiology of Invasive Fungal Infection. Int. J. Antimicrob. Agents. 2008;32:103–109. doi: 10.1016/S0924-8579(08)70009-8. [DOI] [PubMed] [Google Scholar]
- 5.Marchetti O., Bille J., Fluckiger U., Eggimann P., Ruef C., Garbino J., Calandra T., Glauser M.P., Tauber M.G., Pittet D. Fungal Infection Network of Switzerland (FUNGINOS), Epidemiology of Candidemia in Swiss Tertiary Care Hospitals: Secular Trends, 1991–2000. Clin. Infect. Dis. 2003;38:311–320. doi: 10.1086/380637. [DOI] [PubMed] [Google Scholar]
- 6.Walsh T.J., Viviani M.A., Arathoon E., Chiou C., Ghannous M., Groll A.H., Odds F.C. New targets and delivery systems for antifungal therapy. Med. Mycol. 2000;38:335–347. doi: 10.1080/mmy.38.s1.335.347. [DOI] [PubMed] [Google Scholar]
- 7.Epstein J.B. Antifungal therapy in oropharyngeal mycotic infections. Oral Surg. Oral Med. Oral Pathol. 1990;69:32–41. doi: 10.1016/0030-4220(90)90265-T. [DOI] [PubMed] [Google Scholar]
- 8.Guida R.A. Candidiasis of the oropharynx and esophagus. Ear Nose Throat J. 1988;67:832–840. [PubMed] [Google Scholar]
- 9.Warrilow A.G., Parker J.E., Kelly D.E., Kelly S.L. Azole Affinity of Sterol 14α-Demethylase (CYP51) Enzymes from Candida albicans and Homo sapiens. Antimicrob. Agents Chemother. 2013;57:1352–1360. doi: 10.1128/AAC.02067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupetti A., Danesi R., Campa M., Del Tacca M., Kelly S. Molecular basis of resistance to azole antifungals. Mol. Med. 2002;8:77–81. doi: 10.1016/S1471-4914(02)02280-3. [DOI] [PubMed] [Google Scholar]
- 11.Moraca F., De Vita D., Pandolfi F., Di Santo R., Costi R., Cirilli R., D’Auria F.D., Panella S., Palamara A.T., Simonetti G., et al. Synthesis, biological evaluation and structureeactivity correlation study of a series of imidazol-based compounds as Candida albicans inhibitors. Eur. J. Med. Chem. 2014;83:665–673. doi: 10.1016/j.ejmech.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Downer-Riley N.K., Jackson Y.A. Recent Advances in the Synthesis of 1,3-Azoles. Curr. Top. Med. Chem. 2016;16:3617–3626. doi: 10.2174/1568026616666160414122349. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H.Z., Gan L.L., Wang H., Zhou C.H. New Progress in Azole Compounds as Antimicrobial Agents. Mini Rev. Med. Chem. 2017;17:122–166. doi: 10.2174/1389557516666160630120725. [DOI] [PubMed] [Google Scholar]
- 14.Kaplancikli Z.A., Turan-Zitouni G., Özdemir A., Revial G. Synthesis and anticandidal activity of some imidazopyridine derivatives. J. Enzym. Inhib. Med. Chem. 2008;23:866–870. doi: 10.1080/14756360701811114. [DOI] [PubMed] [Google Scholar]
- 15.Reis D., Recio Despaigne A.A., Da Silva J.G., Silva N.F., Vilela C.F., Mendes I.C., Takahashi J.A., Beraldo H. Structural Studies and Investigation on the Activity of Imidazole-Derived Thiosemicarbazones and Hydrazones against Crop-Related Fungi. Molecules. 2013;18:12645–12662. doi: 10.3390/molecules181012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niimi M., Firth N.A., Cannon R.D. Antifungal drug resistance of oral fungi. Odontology. 2010;98:15–25. doi: 10.1007/s10266-009-0118-3. [DOI] [PubMed] [Google Scholar]
- 17.Boucher H.W., Groll A.H., Chiou C., Walsh T.J. Newer systemic antifungal agents. Drugs. 2004;64:1997–2020. doi: 10.2165/00003495-200464180-00001. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W., Ramamoorthy Y., Kılıcarslan T., Nolte H., Tyndale R.F., Sellers E.M. Inhıbıtıon of Cytochromes P450 by Antıfungal Imıdazole Derıvatıves Drug Metabolısm And Dısposıtıon. DMD. 2002;30:314–318. doi: 10.1124/dmd.30.3.314. [DOI] [PubMed] [Google Scholar]
- 19.Lopez S.N., Castelli M.V., Zacchino S.A., Domınguez J.N., Lobo G., Charris-Charris J., Corte J.G.C., Ribas J.C., Devia C., Rodrı´guezd A.M., et al. In Vitro Antifungal Evaluation and Structure-Activity Relationships of a New Series of Chalcone Derivatives and Synthetic Analogues, with Inhibitory Properties against Polymers of the Fungal Cell Wall. Bioorg. Med. Chem. 2001;9:1999–2013. doi: 10.1016/S0968-0896(01)00116-X. [DOI] [PubMed] [Google Scholar]
- 20.Batovska D., Parushev S., Slavova A., Bankova V., Tsvetkova I., Ninova M., Najdenski H. Study on the substituents’ effects of a series of synthetic chalcones against the yeast Candida albicans. Eur. J. Med. Chem. 2007;42:87–92. doi: 10.1016/j.ejmech.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Nowakowska Z., Kedzia B., Schroeder G. Synthesis, physicochemical properties and antimicrobial evaluation of new (E)-chalcones. Eur. J. Med. Chem. 2008;43:707–713. doi: 10.1016/j.ejmech.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Boecka P., Leala P.C., Yunesa R.A., Filhob V.C., Lopez S., Sortinoc M., Escalantec A., Furlanc R.L.E., Zacchinoc S. Antifungal Activity and Studies on Mode of Action of Novel Xanthoxyline-Derived Chalcones. Arch. Pharm. Chem. Life Sci. 2005;338:87–95. doi: 10.1002/ardp.200400929. [DOI] [PubMed] [Google Scholar]
- 23.Lahtchev K.L., Batovska D.I., Parushev S.P., Ubiyvovk V.M., Sibirny A.A. Antifungal activity of chalcones: A mechanistic study using various yeast strains. Eur. J. Med. Chem. 2008;43:2220–2228. doi: 10.1016/j.ejmech.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 24.Borra R.C., Lotufo M.A., Gagioti S.M., Barros Fde M., Andrade P.M. A simple method to measure cell viability in proliferation and cytotoxicity assays. Braz. Oral Res. 2009;23:255–262. doi: 10.1590/S1806-83242009000300006. [DOI] [PubMed] [Google Scholar]
- 25.Palomino J.C., Martin A., Camacho M., Guerra H., Swings J., Portaels F. Resazurin microtiter assay plate: Simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2002;46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozkay Y., Tunalı Y., Karaca H., Işıkdağ I. Antimicrobial activity of a new series of benzimidazole derivatives. Arch. Pharm. Res. 2011;34:1427–1435. doi: 10.1007/s12272-011-0903-8. [DOI] [PubMed] [Google Scholar]
- 27.Bard M., Lees N.D., Turi T., Craft D., Cofrin L., Barbuch R., Koegel C., Loper J.C. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids. 1993;28:963–967. doi: 10.1007/BF02537115. [DOI] [PubMed] [Google Scholar]
- 28.Gooday G.W. The Growing Fungus. Chapman & Hall; London, UK: 1995. Cell membrane; pp. 62–64. [Google Scholar]
- 29.Gollapudy R., Ajmani S., Kulkarni S.A. Modeling and interactions of Aspergillus fumigatus lanosterol 14-α demethylase ‘A’ with azole antifungals. Bioorg. Med. Chem. 2004;12:2937–2950. doi: 10.1016/j.bmc.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 30.Kramer J.A., Sagartz J.E., Morris D.L. The application of discovery toxicology and pathology towards the design of safer pharmaceutical lead candidates. Nat. Rev. Drug Discov. 2007;6:636–649. doi: 10.1038/nrd2378. [DOI] [PubMed] [Google Scholar]
- 31.International Organization for Standardization . Biological Evaluation of Medical Devices-Part 5: Tests for In Vitro Cytotoxicity ISO-10993-5. 3rd ed. International Organization for Standardization; Geneva, Switzerland: 2009. [Google Scholar]
- 32.González-Chávez R., Martínez R., Torre-Bouscoulet M.E., Gallo M., González-Chávez M.M. De novo design of non-coordinating indolones as potential inhibitors for lanosterol 14-α-demethylase (CYP51) Chem. Pharm. Bull. 2014;62:16–24. doi: 10.1248/cpb.c13-00003. [DOI] [PubMed] [Google Scholar]
- 33.Podust L.M., Poulos T.L., Waterman M.R. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. USA. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.EUCAST Definitive Document EDef 7.1: Method for the Determination of Broth Dilution MICs of Antifungal Agents for Fermentative Yeasts. Clin. Microbiol. Infect. 2008;14:398. doi: 10.1111/j.1469-0691.2007.01935.x. [DOI] [PubMed] [Google Scholar]
- 35.Breivik O.N., Owades J.L. Spectrophotometric semi-microdetermination of ergosterol in yeast. Agric. Food Chem. 1957;5:360–363. doi: 10.1021/jf60075a005. [DOI] [Google Scholar]
- 36.Karaca Gençer H., Acar Çevik U., Levent S., Sağlık B.N., Korkut B., Özkay Y., Ilgın S., Öztürk Y. New Benzimidazole-1,2,4-Triazole Hybrid Compounds: Synthesis, Anticandidal Activity and Cytotoxicity Evaluation. Molecules. 2017;22:507. doi: 10.3390/molecules22040507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sağlık B.N., Ilgın S., Özkay Y. Synthesis of new donepezil analogues and investigation of their effects on cholinesterase enzymes. Eur. J. Med. Chem. 2016;124:1026–1040. doi: 10.1016/j.ejmech.2016.10.042. [DOI] [PubMed] [Google Scholar]
- 38.Demir Özkay Ü., Can Ö.D., Sağlık B.N., Acar Çevik U., Levent S., Özkay Y., Ilgın S., Atlı Ö. Design, synthesis, and AChE inhibitory activity of new benzothiazole-piperazines. Bioorg. Med. Chem. Lett. 2016;26:5387–5394. doi: 10.1016/j.bmcl.2016.10.041. [DOI] [PubMed] [Google Scholar]
- 39.Patel S., Gheewala N., Suthar A., Shah A. In-vitro cytotoxicity activity of Solanum nigrum extract against Hela cell line and Vero cell line. Int. J. Pharm. Pharm. Sci. 2009;1:38–46. [Google Scholar]
- 40.Maestro. Schrödinger, LLC; New York, NY, USA: 2016. version 10.6. [Google Scholar]
- 41.Schrödinger. LLC; New York, NY, USA: 2016. version 2016-2. [Google Scholar]
- 42.LigPrep. Schrödinger, LLC; New York, NY, USA: 2016. version 3.8. [Google Scholar]
- 43.Glide. Schrödinger, LLC; New York, NY, USA: 2016. version 7.1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.