Abstract
G-quadruplex (G4) is a DNA/RNA conformation that consists of two or more G-tetrads resulting from four-guanine bases connected by Hoogsteen-type hydrogen bonds, which is often found in the telomeres of chromatin, as well as in the promoter regions of genes. The function of G4 in the genomic DNA is being elucidated and some G4-protein interactions have been reported; these are believed to play a role in vital cellular functions. In this study, we focused on CpG methylation, a well-known epigenetic modification of the genomic DNA, especially found in the promoter regions. Although many G4-forming sequences within the genomic DNA harbor CpG sites, the relationship between CpG methylation and the binding properties of associated proteins remains unclear. We demonstrated that the binding ability of vascular endothelial growth factor (VEGF) G4 DNA to VEGF165 protein was significantly decreased by CpG methylation. We identified the binding activity of G4 DNA oligonucleotides derived from gene promoter regions to SP1, a transcription factor that interacts with a G4-forming DNA and is also altered by CpG methylation. The effect of methylation on binding affinity was accompanied by changes in G4 structure and/or topology. Therefore, this study suggested that CpG methylation might be involved in protein binding to G4-forming DNA segments for purposes of transcriptional regulation.
Keywords: G-quadruplex, CpG methylation, vascular endothelial growth factor, SP1, protein-DNA binding
1. Introduction
G-quadruplex (G4) is a non-canonical nucleic acid structure formed in G-rich sequences [1]. G4 is formed by stacking of G-tetrads to form a planar array of four guanine bases connected by Hoogsteen bonding. It is well-known that the thermostability of G4 structures depends on the concentration and type of monovalent cations [2]. Telomeric DNA folds into a G4 structure to maintain telomere length [3]. The topology of human telomeric DNA depends on the type of cations: G4 folds into a basket-type mixed antiparallel/parallel -stranded G4 structure or hybrid-type mixed antiparallel/parallel-stranded G4 structure in the presence of Na+ or K+, respectively [4,5]. These results suggest that the topology and stability are related to the biological function of G4. G4-forming DNAs have also been identified in several promoter regions [6,7,8] and are involved in transcriptionally activating [9] or silencing gene expression [10,11,12]. Moreover, genome-wide G4-forming DNA identification analysis revealed that G4-forming DNAs are enriched in gene regulatory elements, including gene promoters and CpG islands in genomic DNA [13,14,15,16,17,18,19,20].
Several G4-binding proteins have been identified such as CNBP and nucleolin [21]. We previously reported that the vascular endothelial growth factor 165 (VEGF165) protein binds to G4 DNA formed in the VEGF promoter [22] and G4 RNA formed in VEGF pre-mRNA [23]. Moreover, it has been reported that specificity protein 1 (SP1), a C2H2-type zinc finger transcription factor bound to the c-KIT G4 structure with an affinity similar to that of the duplex structure [24]. The consensus sequence of SP1 is [5′-(G/T)GGGCGG(G/A)(G/A)(G/T)-3′] [25], and SP1 is ubiquitously expressed to not only maintain basal transcription of housekeeping genes, but also regulate tissue-specific gene expression [26]. In several cancers, SP1 is often overexpressed and the target genes are mainly involved in oncogenesis [27]. Genome-wide analysis revealed that SP1 binding sites were enriched in the promoter and CpG island [28]. In silico analysis revealed a correlation between G4-forming sequences upstream of genes and the occurrence of the SP1-binding element [29,30]. These results suggest that G4 is involved in controlling gene expression by SP1.
DNA methylation is an epigenetic modification recognized by methylated CpG binding proteins to recruit the enzymatic machinery needed to establish silent chromatin [31,32,33]. DNA methylation affects not only the thermodynamic properties of DNA duplex structures [34,35] but also G4 structures. For example, the thermostability of quadruplex structures formed by d(CGCG3GCG) oligonucleotides [36], FMR1 repeats [37], C9orf72 repeats [38], and BCL-2 G4 [39] is increased by DNA methylation, whereas the thermostability of MEST G4 is decreased by DNA methylation [40]. We previously reported that the initial elongation efficiency of PCR decreased when template DNA containing a VEGF G4-forming sequence was methylated, suggesting that the thermostability of VEGF G4 was increased by CpG methylation [41]. These results suggest that the thermostability and topology of G4 structures are affected by CpG methylation, resulting in altered binding ability of G4-binding proteins to G4 structures. In this study, we investigated the binding ability of VEGF165 to methylated VEGF G4 DNA [42,43] and the binding ability of SP1 to methylated BCL-2, VEGF, c-KIT, HRAS1, and HRAS2 G4 structures [44,45,46].
2. Results
2.1. Effect of CpG Methylation of VEGF G4 DNA on Binding Ability for VEGF165
In this study, unmethylated and methylated 20-mer oligonucleotides of VEGF DNA containing three CpG sites were analyzed (Table 1). We previously reported that VEGF165 protein bound to the VEGF G4 DNA in vitro [22]. Binding domain analysis revealed that VEGF G4 bound to the heparin-binding domain of VEGF165. The G4 structure has twice as much negative charge density as the double-stranded DNA structure [47] and the heparin-binding motif contains several basic amino acid residues that interact with the negatively charged sulfo groups on heparin. These results suggest that G4 structures preferentially bind to the heparin-binding domain via electrostatic interactions.
Table 1.
DNA sequences used in this study.
| Name | Sequence (5′–3′) |
|---|---|
| VEGF G4 | GGGGCGGGCCGGGGGCGGGG |
| c-KIT G4 | GGCGAGGAGGGGCGTGGCCGGC |
| BCL-2 G4 | CGGGCGCGGGAGGAAGGGGGCGGGAGC |
| HRAS1 G4 | TCGGGTTGCGGGCGCAGGGCACGGGCG |
| HRAS2 G4 | CGGGGCGGGGCGGGGGCGGGGGCG |
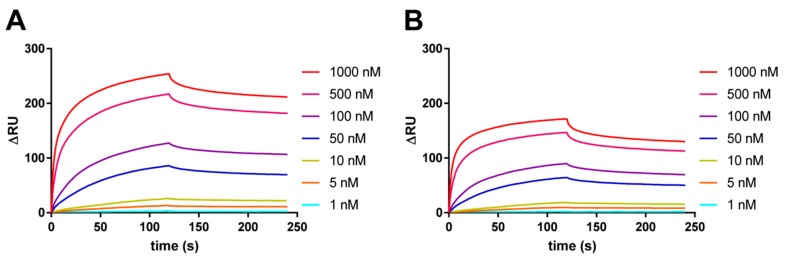
To investigate whether the interaction is affected by CpG methylation of the G4 structure, surface plasmon resonance (SPR) binding analysis was performed and binding constant (KD) was elucidated. The VEGF165 protein was immobilized on a CM5 sensor chip and then unmethylated or methylated VEGF G4 DNA was injected onto the VEGF165-immobilized chip. As previously reported, binding activity of unmethylated VEGF G4 to VEGF165 protein was confirmed, with a KD of 13 ± 0.43 nM (Figure 1). In contrast, the KD of the methylated VEGF G4 to VEGF165 protein increased to 25 ± 5.7 nM, indicating that binding activity was decreased by DNA methylation.
Figure 1.
Binding analysis of VEGF165 to VEGF G4 DNA by SPR. Representative SPR binding signal of unmethylated (A) or methylated VEGF G4 DNA (B) to VEGF165 immobilized on a CM5 chip.
Methylated CpG sites are underlined. For the plate assay, G4 DNAs were biotinylated at the 5′ end for immobilization to streptavidin-coated wells. For Native-PAGE, G4 DNAs were modified with TAMRA at the 3′ end for detection of electrophoretogram.
2.2. Effect of CpG Methylation of Promoter-Derived G4-Forming DNA on DNA-Binding Ability of GST-SP1
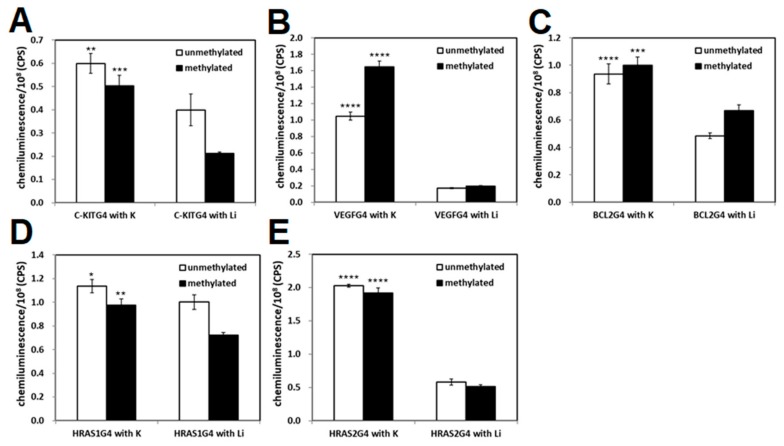
We tested the binding ability of methylated and ummethylated 22-mer c-KIT, 20-mer VEGF, 27-mer BCL-2, 27-mer HRAS1, and 24-mer HRAS2 G4-forming DNAs, which contain 3, 4, 4, and 5 CpG sites, respectively, to the transcription factor GST-SP1. In the c-KIT promoter region, three G4-forming sequences were identified and the middle G4-forming sequence contains an SP1 binding motif; therefore, the middle c-KIT G4-forming DNA was used. SP1 protein was expressed as a GST fusion protein in E. coli as previously reported [48]. The purified GST-SP1 was confirmed by SDS-PAGE (Figure 2). Enzyme-linked oligonucleotide assay (ELONA) revealed that a stronger chemiluminescent signal from HRP was observed to come from the wells with all G4 DNAs in comparison with the negative control wells not harboring G4 DNAs (Figure 3A–E). It was also confirmed that no binding occurred between the GST tag and G4 DNA (data not shown). To investigate the effect of G4 folding on the binding ability of SP1, the potassium ions in the folding buffer were substituted with lithium ions, which do not promote the formation of G4. The binding of SP1 to all G4 DNAs in the presence of Li+ was significantly decreased, indicating that SP1 recognized G4 folding of single-stranded c-KIT, BCL-2, VEGF, HRAS1, and HRAS2 DNAs regardless of the presence of methylated CpG sites (Figure 3A–E). In the presence of K+, methylated VEGF G4 DNA exhibited a 1.5 times stronger chemiluminescent signal from SP1 binding activity than unmethylated VEGF G4 DNA did, whereas the binding signal in the presence of Li+ was not increased by DNA methylation (Figure 3B). Based on these results, we can conclude that SP1 interacted with the five G4-forming DNAs derived from the promoter regions of their genes—specifically c-KIT, BCL-2, VEGF, HRAS1, and HRAS2. Furthermore, it can be concluded that these binding activities between these G4-forming DNAs and SP1 could be affected by CpG methylation of them.
Figure 2.
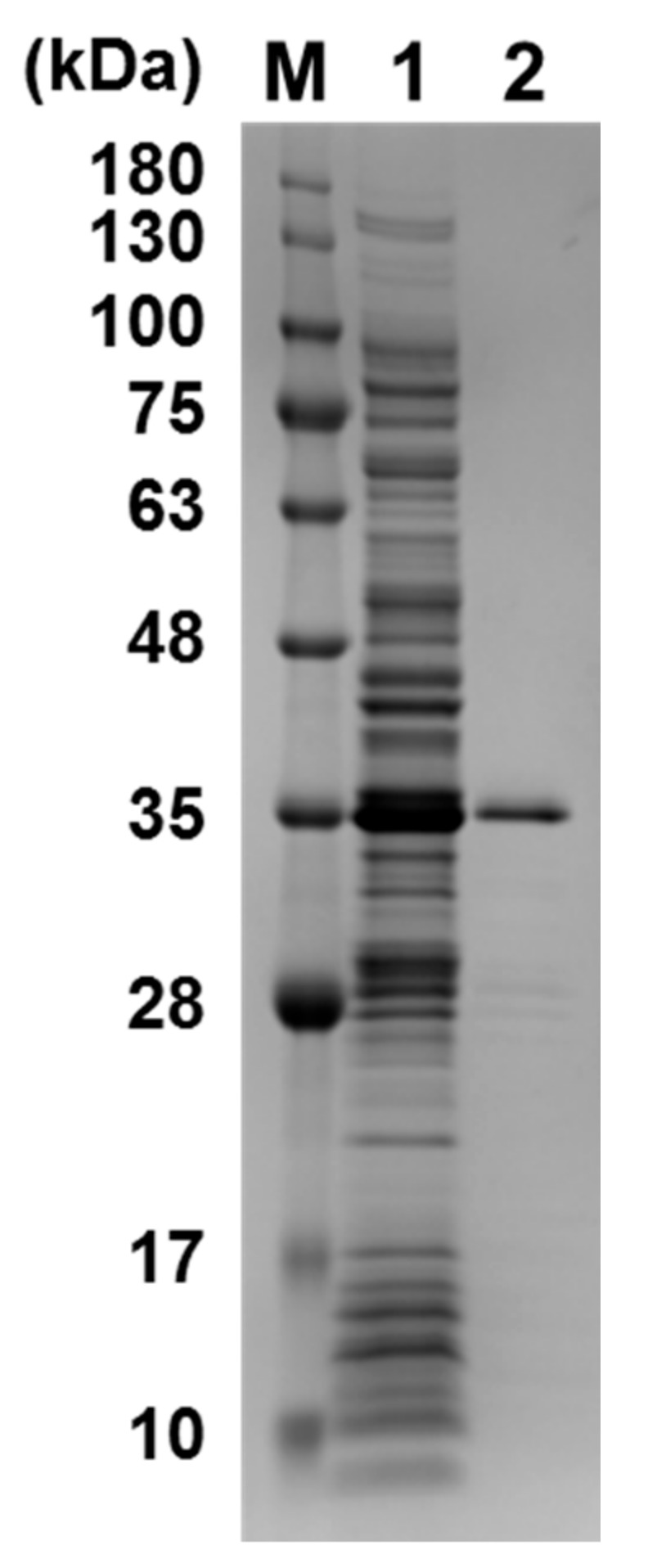
SDS-PAGE of purified GST-SP1 (37.5 kDa). M: protein marker; 1: crude fraction; 2: purified GST-SP1.
Figure 3.
Binding ability of SP1 to c-KIT (A); VEGF (B); BCL-2 (C); HRAS1 (D); and HRAS2 (E) G4 DNAs in the presence of K+ or Li+. Binding of GST-SP1 to G4 DNAs immobilized on the well was detected with an HRP-conjugated anti-GST antibody. As a negative control, a well without immobilized DNA was utilized. Bar plots mean values ± SD (N = 3). Statistical analyses were performed using two-way ANOVA with Tukey’s multiple comparisons post-hoc tests. The significance of differences of observed values from the Li+-treated group are indicated as follows: * p < 0.05; ** p < 0.001; *** p < 0.0005; **** p < 0.0001.
An SPR experiment was performed to elucidate the KD of G4-forming oligonucleotides to SP1 (Figure S1). The binding ability (i.e., KD) of all the G4-forming DNAs to GST-SP1 was confirmed (Table 2). We compared the KD of G4 DNA containing methylation at CpG sites to that of ummethylated G4 DNA. Lower KD values were obtained for c-KIT and VEGF G4 DNA that had undergone CpG methylation compared with their unmethylated forms. The CpG methylation of BCL-2, HRAS1, and HRAS2 G4 DNAs had the opposite effect, and resulted in higher KD values of those G4-forming DNAs when methylated than unmethylated. The binding ability of SP1 to G4-forming DNA increased or decreased by CpG methylation depending on the gene investigated, implying that CpG methylation may function as both a positive or negative regulator of protein-G4 structure binding. This result was in agreement with experimental observations from our study of VEGF G4 DNA binding to VEGF165 protein (Figure 1).
Table 2.
KD of unmethylated or methylated VEGF, c-KIT, BCL-2, HRAS1, and HRAS2 G4 DNAs for GST-SP1.
| KD (nM) | |||||
|---|---|---|---|---|---|
| VEGF G4 | c-KIT G4 | BCL-2 G4 | HRAS1 G4 | HRAS2 G4 | |
| Unmethylated | 14 | 16 | 12 | 10 | 10 |
| Methylated | 11 | 11 | 28 | 36 | 27 |
2.3. Analysis of G4 Folding in the Presence or Absence of CpG Methylation
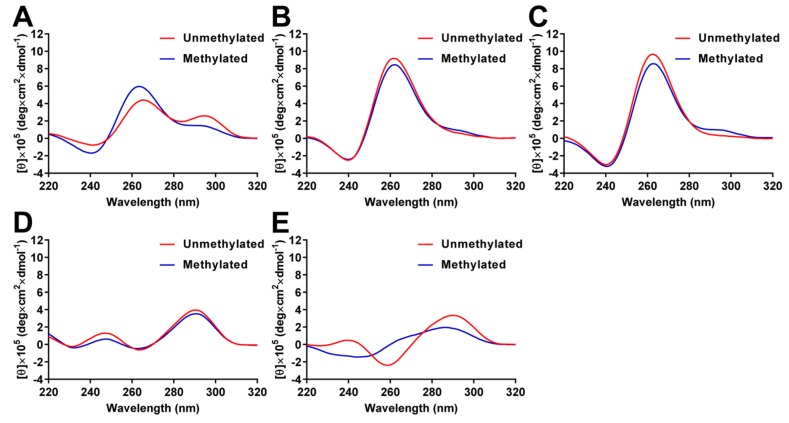
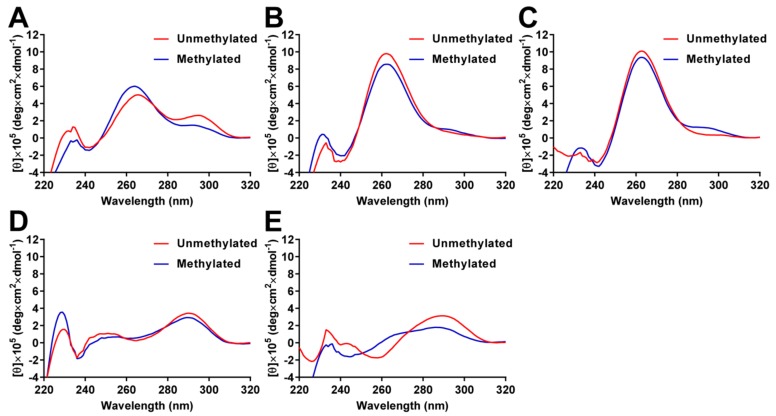
To analyze the effects of CpG methylation on the G4 structures, the circular dichroism (CD) spectra were measured in 10 mM Tris-HCl (pH 7.4) containing 100 mM KCl. CD spectra of unmethylated BCL-2 G4 showed two positive peaks at 265 and 295 nm; the peak at 295 nm was decreased by CpG methylation (Figure 4A). These results indicate that unmethylated BCL-2 G4 folds into a mixed-type G4 structure, while methylated BCL-2 G4 mainly folds into a parallel G4 structure, as previously reported [39]. It has been reported that VEGF and HRAS2 G4 DNAs fold into parallel G4 structures and c-KIT and HRAS1 G4 DNAs fold into anti-parallel G4 structures in 100 mM KCl. The topology of unmethylated VEGF and HRAS2 G4 DNAs were confirmed by CD spectral analysis, as VEGF and HRAS2 G4 DNA showed a positive peak at approximately 262 nm, which is typical for a parallel G4 structure (Figure 4B,C). In contrast, the positive peaks decreased and a shoulder between 280 and 300 nm was detected by CpG methylation, indicating that methylated G4 DNAs were present. Unmethylated c-KIT and HRAS1 G4 DNAs showed a positive peak at approximately 290 nm, which indicates a typical fold into a parallel G4 structure, but antiparallel topology spectra for an anti-parallel G4 structure (Figure 4D,E). By methylating c-KIT G4 DNA, the positive peak at approximately 290 nm decreased, indicating that methylated c-KIT G4 DNA folds into an anti-parallel G4 structure that differs from the unmethylated structure. For HRAS1 G4 DNA, the positive peak at 290 nm was shifted to 286 nm and the negative peak at 259 nm and positive peak at 239 nm were not detected after DNA methylation, suggesting that methylated HRAS1 G4 does not fold into a G4 structure. We measured the CD spectra of G4 DNAs in the presence of GST-SP1 to investigate whether SP1 recognizes the G4 structures or single-stranded DNA structures. CD spectra of both unmethylated and methylated G4 DNAs were not affected by addition of GST-SP1 (Figure 5), indicating that G4 DNAs folded into the G4 structure in the presence of GST-SP1 and SP1 recognized the G4 structures of c-KIT, BCL-2, VEGF, HRAS1, and HRAS2. These results indicated that methylation of cytosine in G4 structures gave rise to different G4 topology, whereby the binding abilities of VEGF165 and SP1 to the G4 DNAs were altered by methylation.
Figure 4.
CD spectra of unmethylated and methylated BCL-2 (A); VEGF (B); HRAS2 (C); c-KIT (D); and HRAS1 G4 DNAs (E) in 10 mM Tris-HCl, 100 mM KCl, pH 7.4 at 25 °C.
Figure 5.
CD spectra of the unmethylated and methylated BCL-2 (A); VEGF (B); HRAS2 (C); c-KIT (D); and HRAS1 G4 DNAs (E) in the presence of the GST-SP1 in 10 mM Tris-HCl, 100 mM KCl, pH 7.4 at 25 °C.
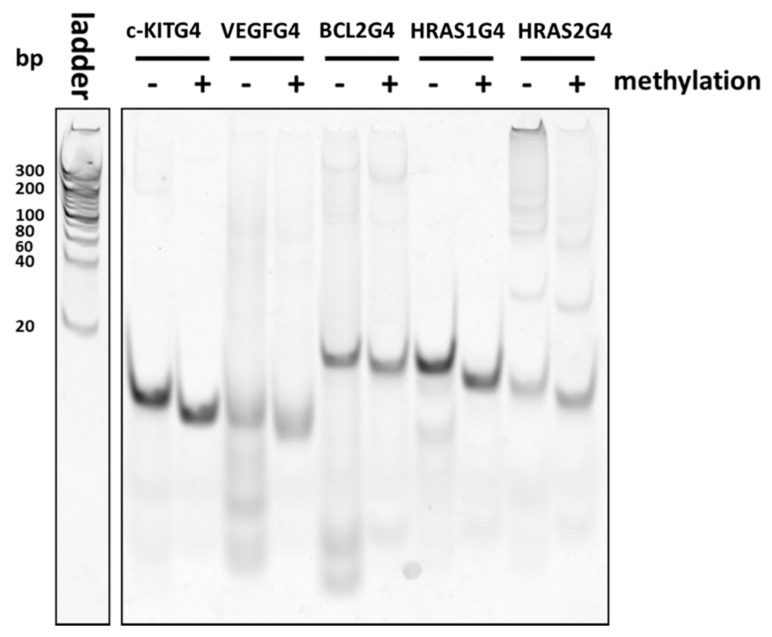
Native-PAGE analysis showed that CpG methylation of DNA within G4 structures caused distinct mobility in the gel (Figure 6). This was especially apparent when methylated VEGF, BCL-2, and HRAS2 G4 DNAs separated into multiple bands, which were distinct from those of the unmethylated DNAs. Thus, G4-associated DNAs can fold into many different types of G4 types, as observed among the G4 DNAs in this study. G4-forming sequences can fold into several types of structures with differing topology, multimer components, or structures folded by different guanine bases. Interestingly, even in the presence of the complementary strand of each G4-forming DNA, we observed multiple bands in Native-PAGE (Figure S2). Some SP1-binding sites that are generally thought to form dsDNA therefore seem to form different, distinct structures apart as ssDNA from those formed while in the dsDNA state.
Figure 6.
Native-PAGE of the unmethylated and methylated c-KIT, BCL-2, VEGF, HRAS1, and HRAS2 G4 DNAs. Oligonucleotides fluorescently labelled with TAMRA were detected. Ladder: DNA marker.
3. Discussion
In this study, we demonstrated that CpG methylation in G4 DNA structures was involved in mediating the binding ability of VEGF165 and SP1 to G4 DNAs. The affinity of VEGF165 to VEGF G4 DNA was decreased by CpG methylation. The binding of GST-SP1 to c-KIT, BCL-2, VEGF, HRAS1, and HRAS2 G4 DNAs were confirmed by ELONA and SPR to have KD values of 10–36 nM, which are altered by the presence of methylated cytosine. CD spectra analysis indicated that the topology of different G4 showed different spectra, with some change to these by CpG methylation in all G4 DNAs. Additionally, the electrophoretogram showed that the CpG methylation altered the distribution of several states of G4. These results suggested that the effect of CpG methylation on binding activity of the proteins studied to G4 DNA led to changes in the mixed states of G4 structures. We have previously reported on DNA aptamers, which fold into some G4 structures different from typical G4 topology, were able to increase the affinity for target molecules (e.g., proteins) by changing G4 topology in such a way to make it more suitable for binding by them [49]. Thus, among the several different states of a given G4 DNA, there should be a structure that has high binding ability for GST-SP1. If the mixed states were altered by DNA methylation, the structure binding to GST-SP1 was increased/decreased by the cytosine methylation. This might result in the enhancement/inhibition of binding ability of G4 DNA against GST-SP1. Methylation at the C-5 position of cytosine increases the molecular polarizability of pyrimidine to enhance stacking interactions [36]. Moreover, hydrophobic methyl groups on cytosine affect the electrostatic potential of the major groove [50] and methylation may add CH-π interactions, which have important roles in the G4 structure [51]. It was suggested that the CH-π interactions by methylation can stabilize structure or topology of G4. These properties would affect not only the G4 structure, but also the protein binding activity.
It has been reported that the binding activity of SP1 to double-stranded DNA containing the consensus sequence was not affected by DNA methylation [52,53]. We determined SP1 recognized the G4 structures of c-KIT, BCL-2, VEGF, HRAS1, and HRAS2 and its bindings may be caused by the change of mixed G4 states by DNA methylation, suggesting that G4 structure containing CpG methylation site is involved in transcriptional regulation. DNA methylation on promoters is generally recognized as an epigenetic modification for transcriptional repression, as CpG binding proteins bind to methylated regions to recruit the enzymatic machinery needed to establish silent chromatin. In contrast, DNA methylation profiling analyses have identified hypermethylated low-CpG content promoters with transcriptional activity [54,55], suggesting that the regions are in an open chromatin state. CpG methylation profiling analyses also indicated that 31% of experimentally verified SP1 binding sites were hypermethylated, suggesting that SP1 binding occurs independently of methylation [54]. These results suggest that G4 structures are formed in methylated promoter regions when the regions are in an open chromatin state and that transcriptional activity is regulated via SP1 binding to methylated G4 structures. We also expect that CpG methylation in VEGF G4 DNA could have some roles in cell function, which are controlled by its binding to the VEGF protein.
4. Materials and Methods
4.1. Expression and Purification of GST Fused SP1
Escherichia coli BL21 (DE3) were transformed with pGEX2T-SP1 [48]. GST-SP1 fusion protein was expressed using the Overnight Express™ autoinduction system at 20 °C for 30 h. The cell pellets were collected by centrifugation at 3000× g for 10 min and resuspended in cell lysis buffer (10 mM Na2PO4, 1.8 mM KH2PO4, 140 mM NaCl, 2.7 mM KCl, 90 µM ZnCl2, 5 mM DTT, 4 mM Pefabloc, 1% Triton X-100, pH 7.3). The sample was homogenized using a French press (Ohtake, Tokyo, Japan) and centrifuged at 8000× g for 30 min at 4 °C. The supernatant was filtered through a 0.2-µm nitrocellulose filter (Advantec, Dublin, CA, USA). The filtered sample was loaded onto two tandemly connected GSTrap HP 1 mL columns (GE Healthcare, Little Chalfont, UK) to purify GST-SP1. The column was washed with 10 mL of wash buffer (10 mM Na2PO4, 1.8 mM KH2PO4, 140 mM NaCl, 2.7 mM KCl, 90 µM ZnCl2, 5 mM DTT, pH 7.3). GST-SP1 was eluted using 8 mL of elution buffer (50 mM Tris-HCl, 10 mM reduced glutathione, 90 µM ZnCl2, 5 mM DTT, 4 mM Pefabloc, pH 8.0). The purified sample was analyzed on a 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gradient gel (WAKO, Osaka Japan) and visualized with Coomassie Brilliant Blue (Kanto Chemical Co., Inc., Tokyo, Japan). The purified protein was dialyzed against TBS buffer (10 mM Tris-HCl, 150 mM NaCl, 5 mM KCl, 90 µM ZnCl2). Protein concentration was measured by using Bradford reagent (Sigma, St. Louis, MO, USA).
4.2. Binding Analysis of VEGF165 to VEGF G4 DNAs by SPR
All oligonucleotides employed in this study (Table 1) were synthesized elsewhere (Eurofins Genomics K.K., Tokyo, Japan; TAKARA BIO INC., Shiga, Japan). Approximately 1500 RU of VEGF165 was immobilized on a sensor chip CM5 (GE Healthcare, Little Chalfont, UK) using an amine coupling procedure in 10 mM acetate buffer (pH 6.0). The oligonucleotides were diluted to 10 µM in TK buffer (10 mM Tris-HCl, 100 mM KCl, pH 7.4). These DNA samples were denatured at 95 °C for 10 min and then allowed to cool to 25 °C for 30 min. Various concentrations of oligonucleotides were injected onto the VEGF165-immobilized chip at a flow rate of 30 μL/min at 25 °C. The VEGF165-immobilized chip was regenerated by injection of a mixture of 1 M NaCl and 1 mM NaOH. Dissociation constant (Kd) values were calculated by fitting the association and dissociation rates using BIA evaluation software (GE Healthcare). SPR binding analysis was performed four times to evaluate the Kd value (mean ± SD, N = 4).
4.3. Binding Analysis of SP1 to Methylated G4 DNAs by ELONA and SPR
The oligonucleotides were modified with biotin at the 5′ end. The biotinylated oligonucleotides (1 µM) were folded by heat treatment in TK buffer or Tris-LiCl buffer (10 mM Tris-HCl, 100 mM LiCl, pH 7.4) as described above. The oligonucleotides were diluted in rinse buffer (TBS (10 mM Tris-HCl, 150 mM NaCl, 5 mM KCl), 90 µM ZnCl2, pH 7.3) to 250 nM and then 100 µL of the diluted oligonucleotide samples were added to a streptavidin-coated 96 well plate (Nalge Nunc International, Rochester, NY, USA). After washing the wells with 100 µL of washing buffer (TBS, 0.05% Tween 20, 90 µM ZnCl2, pH 7.3), 100 µL of GST-SP1 (250 nM) was added and then incubated for 1 h at RT. After washing the wells with 100 µL of washing buffer four times followed by 100 µL of rinse buffer twice, 100 µL of 10,000-fold diluted horseradish peroxidase (HRP)-conjugated anti-GST antibody (Abcam, Cambridge, UK) was added. After 1 h incubation at RT, the wells were washed with 100 µL of the washing buffer six times followed by 100 µL of rinse buffer twice, and then 100 µL of SuperSignal ELISA Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Waltham, MA, USA) was added. The chemiluminescence intensities were measured at RT with a microplate reader (Perkin Elmer, Waltham, MA, USA).
For binding analysis of SP1 to double-stranded DNAs, the oligonucleotide that could form G4 was modified with biotin at the 5′ end, while the complementary strand of G4-forming oligonucleotide was not biotinylated. Both oligonucleotides were mixed and folded by heat treatment in TK buffer as described above. Final concentrations of both oligonucleotides were 1 µM. We confirmed the formation of double-stranded structures by electrophoresis (data not shown).
In SPR analysis, an anti-GST antibody was immobilized on a CM5 sensor chip, and then GST tag or GST-SP1 were captured on a reference cell or a flow cell, respectively. VEGF, c-KIT, BCL-2, HRAS1, and HRAS2 G4 DNAs were injected onto the sensor chip. KD of G4 DNAs against GST-SP1 was determined using a single-cycle kinetics approach. A dilution series of G4 DNAs was prepared at concentrations of 10, 20, 40, 80, and 160 nM.
4.4. Circular Dichroism Spectroscopy
The oligonucleotides (1 µM) were folded by heat treatment in TK buffer as described above. GST-SP1 (final concentration (f.c.) 0 or 1.5 µM) was added to the DNA samples (f.c. 1.5 µM) and incubated for 30 min at room temperature. Circular dichroism (CD) spectra was measured using a J-820 spectropolarimeter (JASCO, Tokyo, Japan) and a quartz cell of 10 mm optical path length (JASCO) at 25 °C.
4.5. Native-PAGE Analysis
The G4 DNAs were modified with addition of a TAMRA label at the 3′ end. The folded oligonucleotides (f.c. 500 nM) in TK buffer were separated in a 20% polyacrylamide gel and visualized with fluorescence of TAMRA. To analyze dsDNA forms of the G4 DNA, the complementary strand of each G4-forming DNA was mixed in just before heat treatment.
Supplementary Materials
The following are available online, Figure S1: Binding analysis of GST-SP1 to c-KIT, BCL-2, VEGF, HRAS1, and HRAS2 G4 DNA by SPR, Figure S2: Native-PAGE of double-stranded, unmethylated and methylated c-KIT, BCL-2, VEGF, HRAS1, and HRAS2 G4 DNAs.
Author Contributions
K.T., S.S., and K.I. conceived and designed the experiments. S.S. performed the experiments. All authors analyzed the data. K.T., S.S., W.Y., and K.I. wrote the manuscript. All authors reviewed the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds (pGEX2T-SP1) are available from the authors.
References
- 1.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: Sequence, topology and structure. Nucl. Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharyya D., Mirihana Arachchilage G., Basu S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016;4:38. doi: 10.3389/fchem.2016.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson E., Hardin C.C., Walk S.K., Tinoco I., Jr., Blackburn E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 5.Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucl. Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eddy J., Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucl. Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucl. Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catasti P., Chen X., Moyzis R.K., Bradbury E.M., Gupta G. Structure-function correlations of the insulin-linked polymorphic region. J. Mol. Biol. 1996;264:534–545. doi: 10.1006/jmbi.1996.0659. [DOI] [PubMed] [Google Scholar]
- 10.Brooks T.A., Kendrick S., Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onel B., Carver M., Wu G., Timonina D., Kalarn S., Larriva M., Yang D. A New G-Quadruplex with Hairpin Loop Immediately Upstream of the Human BCL2 P1 Promoter Modulates Transcription. J. Am. Chem. Soc. 2016;138:2563–2570. doi: 10.1021/jacs.5b08596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bay D.H., Busch A., Lisdat F., Iida K., Ikebukuro K., Nagasawa K., Karube I., Yoshida W. Identification of G-quadruplex structures that possess transcriptional regulating functions in the Dele and Cdc6 CpG islands. BMC Mol. Biol. 2017;18:17. doi: 10.1186/s12867-017-0094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucl. Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iida K., Nakamura T., Yoshida W., Tera M., Nakabayashi K., Hata K., Ikebukuro K., Nagasawa K. Fluorescent-ligand-mediated screening of G-quadruplex structures using a DNA microarray. Angew. Chem. 2013;52:12052–12055. doi: 10.1002/anie.201305366. [DOI] [PubMed] [Google Scholar]
- 17.Lam E.Y., Beraldi D., Tannahill D., Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 19.Rigo R., Palumbo M., Sissi C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta. 2017;1861:1399–1413. doi: 10.1016/j.bbagen.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida W., Saikyo H., Nakabayashi K., Yoshioka H., Bay D.H., Iida K., Kawai T., Hata K., Ikebukuro K., Nagasawa K., et al. Identification of G-quadruplex clusters by high-throughput sequencing of whole-genome amplified products with a G-quadruplex ligand. Sci. Rep. 2018;8:3116. doi: 10.1038/s41598-018-21514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazda V., Haronikova L., Liao J.C., Fojta M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014;15:17493–17517. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida W., Saito T., Yokoyama T., Ferri S., Ikebukuro K. Aptamer selection based on G4-forming promoter region. PLoS ONE. 2013;8:e65497. doi: 10.1371/journal.pone.0065497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito T., Yoshida W., Yokoyama T., Abe K., Ikebukuro K. Identification of RNA Oligonucleotides Binding to Several Proteins from Potential G-Quadruplex Forming Regions in Transcribed Pre-mRNA. Molecules. 2015;20:20832–20840. doi: 10.3390/molecules201119733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raiber E.A., Kranaster R., Lam E., Nikan M., Balasubramanian S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucl. Acids Res. 2012;40:1499–1508. doi: 10.1093/nar/gkr882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beishline K., Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282:224–258. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 26.Wierstra I. Sp1: Emerging roles—Beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 27.Vizcaino C., Mansilla S., Portugal J. Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol. Ther. 2015;152:111–124. doi: 10.1016/j.pharmthera.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Gilmour J., Assi S.A., Jaegle U., Kulu D., van de Werken H., Clarke D., Westhead D.R., Philipsen S., Bonifer C. A crucial role for the ubiquitously expressed transcription factor Sp1 at early stages of hematopoietic specification. Development. 2014;141:2391–2401. doi: 10.1242/dev.106054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todd A.K., Neidle S. The relationship of potential G-quadruplex sequences in cis-upstream regions of the human genome to SP1-binding elements. Nucleic Acids Res. 2008;36:2700–2704. doi: 10.1093/nar/gkn078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar P., Yadav V.K., Baral A., Kumar P., Saha D., Chowdhury S. Zinc-finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide. Nucl. Acids Res. 2011;39:8005–8016. doi: 10.1093/nar/gkr536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 32.Chen R.Z., Pettersson U., Beard C., Jackson-Grusby L., Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 33.Bogdanovic O., Veenstra G.J. DNA methylation and methyl-CpG binding proteins: Developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sowers L.C., Shaw B.R., Sedwick W.D. Base stacking and molecular polarizability: Effect of a methyl group in the 5-position of pyrimidines. Biochem. Biophys. Res. Commun. 1987;148:790–794. doi: 10.1016/0006-291X(87)90945-4. [DOI] [PubMed] [Google Scholar]
- 35.Perez A., Castellazzi C.L., Battistini F., Collinet K., Flores O., Deniz O., Ruiz M.L., Torrents D., Eritja R., Soler-Lopez M., et al. Impact of methylation on the physical properties of DNA. Biophys. J. 2012;102:2140–2148. doi: 10.1016/j.bpj.2012.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardin C.C., Corregan M., Brown B.A., 2nd, Frederick L.N. Cytosine-cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry. 1993;32:5870–5880. doi: 10.1021/bi00073a021. [DOI] [PubMed] [Google Scholar]
- 37.Fry M., Loeb L.A. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamiri B., Mirceta M., Bomsztyk K., Macgregor R.B., Jr., Pearson C.E. Quadruplex formation by both G-rich and C-rich DNA strands of the C9orf72 (GGGGCC)8*(GGCCCC)8 repeat: Effect of CpG methylation. Nucl. Acids Res. 2015;43:10055–10064. doi: 10.1093/nar/gkv1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin J., Hou J.Q., Xiang H.D., Yan Y.Y., Gu Y.C., Tan J.H., Li D., Gu L.Q., Ou T.M., Huang Z.S. Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: Implications for epigenetic regulation. Biochem. Biophys. Res. Commun. 2013;433:368–373. doi: 10.1016/j.bbrc.2012.12.040. [DOI] [PubMed] [Google Scholar]
- 40.Stevens A.J., Stuffrein-Roberts S., Cree S.L., Gibb A., Miller A.L., Doudney K., Aitchison A., Eccles M.R., Joyce P.R., Filichev V.V., et al. G-quadruplex structures and CpG methylation cause drop-out of the maternal allele in polymerase chain reaction amplification of the imprinted MEST gene promoter. PLoS ONE. 2014;9:e113955. doi: 10.1371/journal.pone.0113955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida W., Yoshioka H., Bay D.H., Iida K., Ikebukuro K., Nagasawa K., Karube I. Detection of DNA Methylation of G-Quadruplex and i-Motif-Forming Sequences by Measuring the Initial Elongation Efficiency of Polymerase Chain Reaction. Anal. Chem. 2016;88:7101–7107. doi: 10.1021/acs.analchem.6b00982. [DOI] [PubMed] [Google Scholar]
- 42.Sun D., Guo K., Rusche J.J., Hurley L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucl. Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo K., Gokhale V., Hurley L.H., Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucl. Acids Res. 2008;36:4598–4608. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Membrino A., Cogoi S., Pedersen E.B., Xodo L.E. G4-DNA formation in the HRAS promoter and rational design of decoy oligonucleotides for cancer therapy. PLoS ONE. 2011;6:e24421. doi: 10.1371/journal.pone.0024421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cogoi S., Shchekotikhin A.E., Xodo L.E. HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 2014;42:8379–8388. doi: 10.1093/nar/gku574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platella C., Riccardi C., Montesarchio D., Roviello G.N., Musumeci D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta. 2017;1861:1429–1447. doi: 10.1016/j.bbagen.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gatto B., Palumbo M., Sissi C. Nucleic acid aptamers based on the G-quadruplex structure: Therapeutic and diagnostic potential. Curr. Med. Chem. 2009;16:1248–1265. doi: 10.2174/092986709787846640. [DOI] [PubMed] [Google Scholar]
- 48.Osawa Y., Ikebukuro K., Motoki H., Matsuo T., Horiuchi M., Sode K. The simple and rapid detection of specific PCR products from bacterial genomes using Zn finger proteins. Nucl. Acids Res. 2008;36:e68. doi: 10.1093/nar/gkn274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsukakoshi K., Ikuta Y., Abe K., Yoshida W., Iida K., Ma Y., Nagasawa K., Sode K., Ikebukuro K. Structural regulation by a G-quadruplex ligand increases binding abilities of G-quadruplex-forming aptamers. Chem. Commun. 2016;52:12646–12649. doi: 10.1039/C6CC07552E. [DOI] [PubMed] [Google Scholar]
- 50.Dantas Machado A.C., Zhou T., Rao S., Goel P., Rastogi C., Lazarovici A., Bussemaker H.J., Rohs R. Evolving insights on how cytosine methylation affects protein-DNA binding. Brief. Funct. Genom. 2015;14:61–73. doi: 10.1093/bfgp/elu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tateishi-Karimata H., Ohyama T., Muraoka T., Podbevsek P., Wawro A.M., Tanaka S., Nakano S.I., Kinbara K., Plavec J., Sugimoto N. Newly characterized interaction stabilizes DNA structure: Oligoethylene glycols stabilize G-quadruplexes CH-pi interactions. Nucl. Acids Res. 2017;45:7021–7030. doi: 10.1093/nar/gkx299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrington M.A., Jones P.A., Imagawa M., Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc. Natl. Acad. Sci. USA. 1988;85:2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holler M., Westin G., Jiricny J., Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- 54.Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.