Abstract
Osteoblasts are bone marrow endosteum-lining niche cells playing important roles in the regulation of hematopoietic stem cells by secreting factors and cell adhesion molecules. Characterization of primary osteoblasts has been achieved through culture of outgrowth of collagenase treated bone. Immunophenotyping and flow-based analysis of long bone osteoblasts offer a simplified and rapid approach to characterize osteoblasts. We describe a modified procedure of isolating mouse bone marrow osteoblastic cells based on cell surface immunophenotyping. The chemokine CXCL12 (also known as stromal-derived factor, SDF-1) together with its receptor CXCR4 are expressed by osteoblasts and bone marrow stroma cells. The CXCL12-CXCR4 axis is important for hematopoietic stem cell retention to their niches ( Sugiyama et al., 2006 ) and for supporting leukemia initiating cell activity ( Pitt et al., 2015 ). Here we describe the procedure of intracellular staining of CXCL12.
Keywords: Bone marrow niche, HSC, Osteoblast, CXCL12, Intracellular staining
Background
The bone marrow niche is a highly organized microenvironment with stroma cells that engage in direct cell-cell interaction with hematopoietic stem cells (HSC) that regulate HSC quiescence, differentiation, and mobilization (Anthony and Link, 2014; Mendelson and Frenette, 2014; Morrison and Scadden, 2014). Multiple cell types in HSC niche may contribute to niche functional support in distinct but maybe overlapping ways. These cells include but are not limited to osteoblasts, osteoclasts, CXCL12-abundant reticular (CAR) cells, Nestin+ stroma cells, leptin receptor+ (LepR+) stroma cells, endothelial cells, macrophages, megakaryocytes, neuronal, and the non-myelinating Schwann cells. Most HSCs are found in the trabecular region of bone marrow, suggesting an important HSC supporting role of the endosteum as well as factors made by osteoblasts and other cells in the endosteum ( Kiel et al., 2005 ; Lo Celso et al., 2009 ). The majority of long-term HSCs are located in close vicinity of the sinusoid in close contact with LepR+ and CXCL12high niche cells, indicating a perivascular niche composed by endothelial or perivascular cells ( Kiel et al., 2005 ; Sugiyama et al., 2006 ; Acar et al., 2015 ). In addition to the perivascular niche associated with sinusoid, mesenchymal cells that surround arterioles in the bone marrow are also important for the maintenance of quiescent HSCs ( Kunisaki et al., 2013 ). Osteoblasts are specialized endosteum-lining cells that are terminally differentiated products of mesenchymal stem cells. Characterization of murine primary osteoblasts has been achieved through culture of outgrowth of collagenase treated bone and retrospective functional analysis (Bakker and Klein-Nulend, 2011). However, culture-based analysis has been complicated by the heterogeneity of the tissue. Immunophenotyping of murine osteoblasts based on defined CD markers is a rapid and prospective approach of phenotypic analysis of osteoblasts in various disease processes. Isolated osteoblasts through flow-based sorting are especially suitable for downstream applications such as gene expression analysis.
The chemokine CXCL12 (also known as stromal-derived factor, SDF-1) together with its receptor CXCR4 are highly expressed by CAR cells but also by osteoblasts and endothelial cells. The CXCL12-CXCR4 axis is important for hematopoietic stem cell retention to their niches ( Sugiyama et al., 2006 ). CXCL12 in the vascular niche has also been shown play a critical role in supporting leukemia-initiating cell (LIC) activity ( Pitt et al., 2015 ). Using a mouse T-ALL model, we reported that leukemia development was accompanied by the drastic suppression of the endosteum-lining osteoblast population. We further showed that aberrant Notch activation negatively regulates the expression of CXCL12 and osteoblastic progenitor differentiation. Here we describe the procedure of sorting mouse bone marrow osteoblastic cells and the procedure of staining intracellular CXCL12 ( Wang et al., 2016 ).
Materials and Reagents
Gauze sponges (Fisher Scientific, FisherbrandTM, catalog number: 22-415-468)
21 G needles
3 ml syringes
70 μm strainer (Fisher Scientific, FisherbrandTM, catalog number: 22-363-548)
1.5 ml microcentrifuge tube (NEST Biotechnology, catalog number: 615601), 15 ml tubes (BioExpress, GeneMateTM, catalog number: C-3394-1), and 50 ml conical tubes (BioExpress, GeneMateTM, catalog number: C-3394-4)
-
Other antibodies:
APC-anti-CD31 (Thermo Fisher Scientific, eBioScienceTM, catalog number: 17-0311-82)
PE-anti-CD51 (BD, BD PharmingenTM, catalog number: 551187)
PE-Cy7-anti-CD45 (BD, BD PharmingenTM, catalog number: 552848)
Biotin-Sca1 (BD, BD PharmingenTM, catalog number: 553334)
Streptavidin APC-Cy7 (BD, BD PharmingenTM, catalog number: 554063)
CXCL12 detection antibodies: H/M CXCL12/SDF-1 Fluorescein (FITC) MAb (Clone 79018) (R&D Systems, catalog number: IC350F)
70% (v/v) ethanol
Hank’s Balanced Salt Solution (HBSS; 1x) (GE Healthcare, HycloneTM, catalog number: SH30588.02)
BSA (Sigma-Aldrich, catalog number: A7906)
Type I collagenase (Worthington, Lakewood, NJ)
DMEM (ATCC®, catalog number: 30-2002TM)
Fixation/Permeabilization Solution Kit with BD GolgiStopTM (BD, BD Cytofix/CytopermTM Plus, catalog number: 554715)
HBSS staining buffer (see Recipes)
Equipment
Pipettes
Scissors
Tweezers
Curved forceps
Tabletop centrifuge (SORVALL Legend RT)
Hemocytometer
FACSAria I
Procedure
-
Preparing single cell suspension from long bones and osteoblast sorting
Euthanize the mouse by CO2 asphyxiation and confirmed death by cervical dislocation.
Sterilize the body with 70% (v/v) ethanol.
Make incisions around the connection between hind limbs and trunk using scissors and tweezers. The whole skin is then removed from the hind limbs by pulling toward the cutting site of the claw (Figure 1A). Carefully remove all the muscles from the long bones (femur and tibia) using dissecting scissors and curved forceps. Excise the long bone and transfer onto sterile gauze.
Clean soft tissue as thoroughly as possible around the bone (Figure 1B).
Cut off the epiphyses.
Fill a 21-gauge needle and 3 ml syringe with 1 ml cold HBSS supplemented with 0.5% BSA and use a syringe to flush out bone marrow (BM) three times (Figure 2A).
Cut the bone into small pieces of approximately 1-2 mm2 using scissors (Figure 2B).
Digest bone fragments obtained from 2 sets of tibias and femurs per mouse in 2 ml HBSS buffer supplemented with collagenase I (3 mg/ml in DMEM) in a 50 ml conical tube for 1 h by shaking (110 rpm) at 37 °C.
Add 3 volume of DMEM (6 ml) supplemented with 10% FBS to terminate collagenase I activity.
Strain the cell solution through a 70 μm strainer in a 50 ml conical tube.
Spin down at 250 × g for 5 min at 4 °C.
Remove supernatant
Wash the cells with 2 ml of HBSS buffer and spin down at 250-400 × g for 5 min at 4 °C.
Discard supernatant and resuspend the cells in 1 ml HBSS buffer.
Determine cell number. Count cells using a hemocytometer.
Spin down at 250-400 × g for 5 min at 4 °C.
The cells are ready for preparing for osteoblast sorting and CXCL12 staining.
Prepare antibody cocktails in HBSS buffer using the following antibodies: FITC- lineage markers (T cells: CD4, CD8; B cells: B220; erythrocytes: Ter119; granulocytes and monocytes: CD11b, and Gr-1), APC-CD31 (endothelial cell), PE-CD51, PE-Cy7-Sca1. Use 100 µl of HBSS buffer containing 1 µl of each antibody for 1 x 106 cells.
Resuspend cells from Step A17 in antibody mixture on ice for 20 min.
Wash cells twice each with 200-500 µl cold HBSS buffer, spinning down at 250-400 × g for 5 min at 4 °C in between washes.
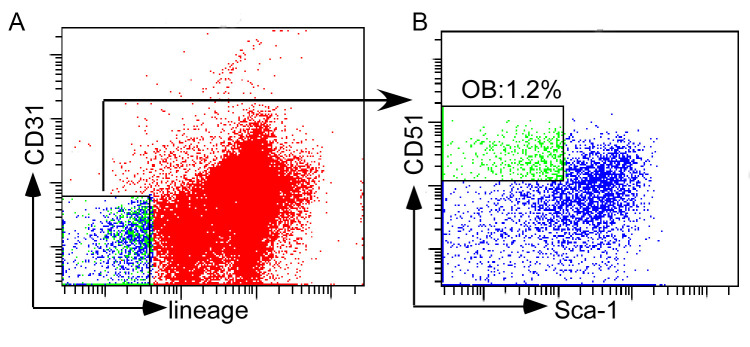
Sort cells with FACSAria I. Osteoblasts are sorted by gating on lin- (CD4, CD8, B220, Ter119, CD11b, Gr-1) CD31-CD51+Sca1- population (Figures 3A and 3B).
-
CXCL12/SDF-1 Intracellular Staining
Resuspend cells from Step A17 from procedure A in HBSS buffer in a 15 ml Falcon tube. Add biotin-Sca1 antibody and leave cells on ice for 20 min. Use 100 µl of HBSS buffer containing 0.5 µl of antibody for 1 x 106 cells.
Wash cells twice each with 200-500 µl HBSS buffer.
Spin down at 250 × g for 5 min at 4 °C. Resuspend cells in HBSS buffer and add APC-CD31, PE-CD51, PE-Cy7-CD45, and Streptavidin APC-Cy7 antibodies, and then incubate on ice for 20 min.
Wash twice each with 1 ml HBSS buffer. Spin down cells at 250 × g for 5 min.
-
Fix and permeabilize cells by resuspending cells in 250 µl of Fixation/Permeabilization solution (included in the BD Cytofix/CytopermTM Plus) for 15-20 min at 4 °C.
Note: Cell aggregation can be avoided by gently vortexing prior to the addition of the Fixation/Permeabilization solution.
-
Wash cells twice each with 1 ml of 1x Perm/WashTM solution (included in the BD Cytofix/CytopermTM Plus), pellet cells by centrifugation at 250 × g for 5 min at 4 °C, and remove supernatant.
Note: Perm/WashTM solution is required in washing steps to maintain cells in a permeabilized state.
Stain intracellular CXCL12 by adding 10 μl of FITC-CXCL12 in 90 μl 1x Perm/WashTM solution and vortex gently.
Incubate cells for 30 min at room temperature in a dark room.
Wash cells twice each with 1 ml 1x Perm/WashTM Buffer. Spin down cells at 250 × g for 5 min at 4 °C.
-
Resuspend cells in 200 μl HBSS buffer for flow cytometric analysis (see Figure 4 below).
Note: For a negative control, a separate set of cells should be stained with an isotype control antibody.
Figure 1. Preparation of mouse long bones.
A. Exposure of hind legs. B. Cleaned long bones (tibia and femur) after removing attached muscles.
Figure 2. Preparation of long bone fragments.
A. Cleaned long bones after flushing out bone marrow; B. Prepared bone fragments.
Figure 3. Sorting of osteoblasts.
A. Gating strategy of lin- (CD4, CD8, B220, Ter119, CD11b, Gr-1) CD31- population; B. Gated osteoblasts (lin- CD31-CD51+Sca1-).
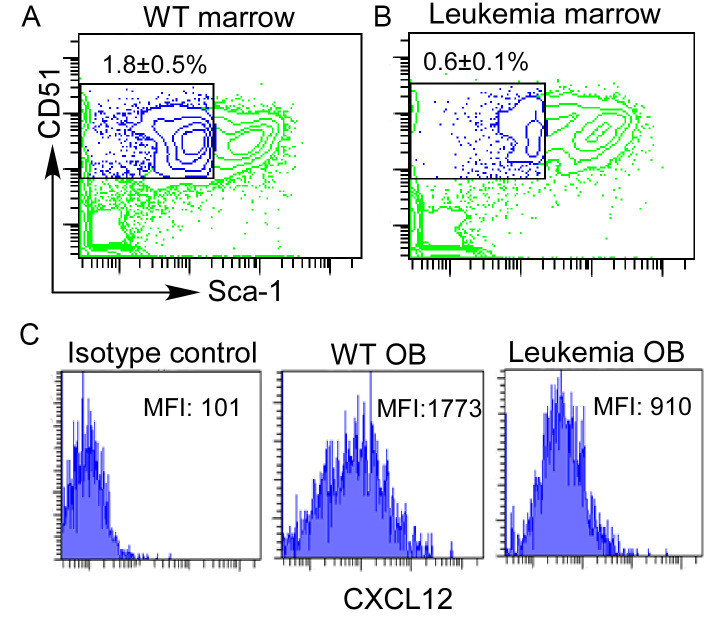
Figure 4. Sorting and CXCL12 staining of bone marrow osteoblasts.
Bone marrow stroma cells were prepared from WT or leukemia-developing mice. Staining of marrow osteoblast (OB) (lin-CD31- CD51+Sca-1-) (A and B) by gating on the lineage-CD31-popualtion. Intracellular expression of CXCL12 was displayed by mean fluorescence intensity (MFI) using isotype control or anti-CXCL12 antibody (C).
Data analysis
From wild type (WT) mice long bones, osteoblasts are readily isolated from the bone marrow stroma after brief digestion with collagenase I (Figure 4A). In comparison, osteoblasts are decreased in the marrow stroma from mice developing T-ALL driven by activated Notch1 (Figure 4B). CXCL12 expression by intracellular staining and flow analysis shows 52% decrease in leukemia mice osteoblasts compared to normal controls (Figure 4C).
Recipes
-
HBSS staining buffer
Hank’s Balanced Salt Solution (HBSS; 1x) supplemented with 0.5% BSA
Acknowledgments
This work was supported by grants from American Cancer Society LIB-125064 (L. Zhou) and NIH HL103827 (L. Zhou). No potential conflicts of interest were disclosed.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Anthony B. A. and Link D. C.(2014). Regulation of hematopoietic stem cells by bone marrow stromal cells. Trends Immunol 35(1): 32-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acar M., Kocherlakota K. S., Murphy M. M., Peyer J. G., Oguro H., Inra C. N., Jaiyeola C., Zhao Z., Luby-Phelps K. and Morrison S. J.(2015). Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526(7571): 126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bakker A. D. and Klein-Nulend J.(2011). Osteoblast isolation from murine calvaria and long bones. Methods Mol Biol 816: 19-29. [DOI] [PubMed] [Google Scholar]
- 4. Kiel M. J., Yilmaz O. H., Iwashita T., Yilmaz O. H., Terhorst C. and Morrison S. J.(2005). SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121(7): 1109-1121. [DOI] [PubMed] [Google Scholar]
- 5. Kunisaki Y., Bruns I., Scheiermann C., Ahmed J., Pinho S., Zhang D., Mizoguchi T., Wei Q., Lucas D., Ito K., Mar J. C., Bergman A. and Frenette P. S.(2013). Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 502(7473): 637-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lo Celso C., Fleming H. E., Wu J. W., Zhao C. X., Miake-Lye S., Fujisaki J., Cote D., Rowe D. W., Lin C. P. and Scadden D. T.(2009). Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457(7225): 92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mendelson A. and Frenette P. S.(2014). Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med 20(8): 833-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morrison S. J. and Scadden D. T.(2014). The bone marrow niche for haematopoietic stem cells. Nature 505(7483): 327-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pitt L. A., Tikhonova A. N., Hu H., Trimarchi T., King B., Gong Y., Sanchez-Martin M., Tsirigos A., Littman D. R., Ferrando A. A., Morrison S. J., Fooksman D. R., Aifantis I. and Schwab S. R.(2015). CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance. Cancer Cell 27(6): 755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sugiyama T., Kohara H., Noda M. and Nagasawa T.(2006). Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25(6): 977-988. [DOI] [PubMed] [Google Scholar]
- 11. Wang W., Zimmerman G., Huang X., Yu S., Myers J., Wang Y., Moreton S., Nthale J., Awadallah A., Beck R., Xin W., Wald D., Huang A. Y. and Zhou L.(2016). Aberrant notch signaling in the bone marrow microenvironment of acute lymphoid leukemia suppresses osteoblast-mediated support of hematopoietic niche function. Cancer Res 76(6): 1641-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]