Abstract
G protein-coupled receptors (GPCR) comprise a diverse superfamily of over 800 proteins that have gained relevance as biological targets for pharmaceutical drug design. Although these receptors have been investigated for decades, three-dimensional structures of GPCR have only recently become available. In this review, we focus on the technological advancements that have facilitated efforts to gain insights into GPCR structure. Progress in these efforts began with the initial crystal structure determination of rhodopsin (PDB: 1F88) in 2000 and has continued to the most recently published structure of the A1AR (PDB: 5UEN) in 2017. Numerous experimental developments over the past two decades have opened the door for widespread GPCR structural characterization. These efforts have resulted in the determination of three-dimensional structures for over 40 individual GPCR family members. Herein we present a comprehensive list and comparative analysis of over 180 individual GPCR structures. This includes a summary of different GPCR functional states crystallized with agonists, dual agonists, partial agonists, inverse agonists, antagonists, and allosteric modulators.
Keywords: G protein-coupled receptors, fusion proteins, x-ray crystallography, ligand binding, agonists, antagonists, inverse agonists, allosteric modulators, extracellular loops, intracellular loops, 7TM domain
Introduction
G protein-coupled receptors (GPCR) comprise a superfamily of over 800 proteins that are the largest family of cell surface receptors in the human genome [1–3]. These proteins share a characteristic seven transmembrane spanning, alpha-helical structure [4]. The GPCR superfamily is commonly subdivided, based on sequence comparisons, into five distinct families: Rhodopsin (class A), Adhesion (class B), Secretin (class B), Glutamate (class C), and Frizzled/Taste2 (class F) [3, 5]. More details on the sequence-based analyses that led to these phylogenetic divisions are further discussed in the section titled “Phylogenetic classification/structure” in this review. GPCR have been implicated in numerous biological processes such as cognitive responses [6], cardiovascular functions [7], and cancer growth and development [8]. GPCR implication in human disease is reflected by the estimated 50% of pharmaceutical drugs that interact with these receptors [9].
GPCR mediate signal transduction cascades initiated by numerous extracellular molecules through which they produce downstream physiological responses [4, 8, 10]. However, receptor activity is not solely stimulated by binding of extracellular ligands. Constitutively active basal signaling [11] has been demonstrated in over 60 wild-type GPCR, including ADRB2; A2AR; and CB1 [12]. Some GPCR, such as taste receptors [13, 14], adopt active state conformations upon interaction with other receptors [15]. Further details regarding GPCR activation can be found in the “GPCR function” section.
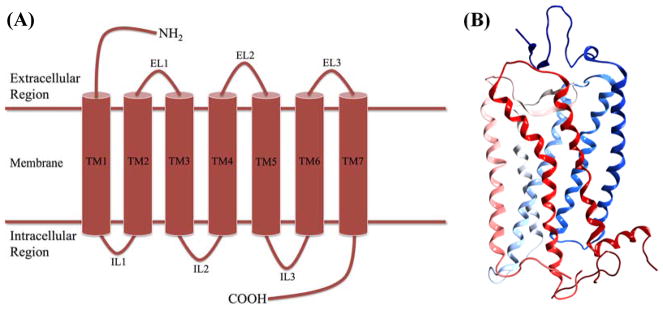
Although these receptors have diverse roles in cellular signaling and in physiology and pathophysiology, they share similar structural topology. This topology includes common structural features shown in figure 1 that include extracellular loops (EL1-3) and intracellular loops (IL1-3) that alternately connect a characteristic seven 251660288251659264 transmembrane (7TM) α-helical bundle (figure 1B) [3, 4]. Crystallographic approaches described in more detail in the section titled “Structural characterization” have revealed structural similarities shared by many GPCR found mainly within the 7TM domain. This structural homology, described in more detail in later sections titled by class, suggests similar means for cell signaling. Although GPCR have a shared topology and 7TM structure, there is diversity within the superfamily in regards to sequence composition and length, producing structural variations that are associated with functional specificity in regards to ligand binding or G protein coupling for different types of GPCR. Select structural differences are also described in more detail in later sections titled by class.
Figure 1.
(A) Topological cartoon representing the N-terminal domain, EL 1–3, 7TM domain in cylinders, IL1-3, and C-terminal domain. (B) Ribbon structure of rhodopsin (PDB: 1F88) demonstrating the arrangement of the 7TM α-helical domains within the characteristic membrane-spanning bundle. Here the helices are colored blue starting at the N-terminus fading to red at the C-terminus.
GPCR function
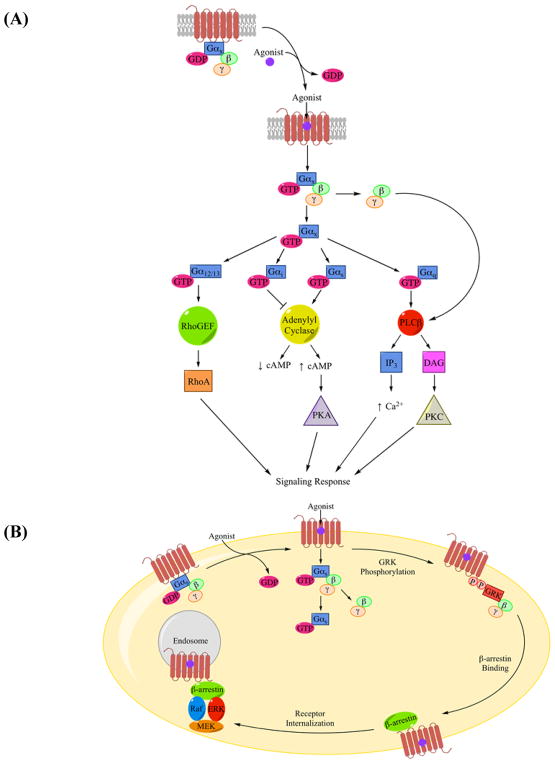
Although this review focuses on GPCR structure and structural characterization, GPCR play a significant role in signal transduction cascades that warrants a generalized overview of their functional and regulatory mechanisms. GPCR are a critical mediator in overall cell signaling, both through ligand-dependent and ligand-independent mechanisms. The majority of these receptors relay signals initiated by binding of extracellular ligands. These ligands are either naturally produced (endogenous) or externally administered (exogenous). Ligands are classified as agonists, inverse agonists, or antagonists. Agonists bind to the receptors and induce a conformational change to an active state, which increases signaling effects. Inverse agonists shift the receptor conformational equilibrium toward inactive conformations, thereby inhibiting basal activity. Antagonists, on the other hand, prevent binding of agonists or inverse agonists without affecting the dynamic conformational equilibrium, which prevents agonist-dependent receptor activation [16]. Changes in conformation within the TM domain, in turn, initiate a conformational change in the intracellular region. IL2 and IL3 have been found to contain critical interaction sites for G proteins or other cytoplasmic effectors [17]. This is illustrated in the crystal structure of ADRB2-Gαs complex (PDB: 3SN6). Residues located in IL2, TM5, and TM6 of ADRB2 demonstrate an interaction interface with the α4 helix, α5-helix, αN-β1 junction, and β3 strand of Gαs [18]. In the classical understanding of GPCR signaling, agonist binding promotes the formation of the activated receptor-G protein complex that modulates functional effects. A detailed analysis of G protein structure, regulation, and their involvement in signaling has been effectively summarized by Gilman [19]. The heterotrimeric G protein complex is made up of α, β, and γ subunits where the α subunit falls into four subfamilies: Gαs, which stimulates adenylyl cyclase activity; Gαi, which inhibits adenylyl cyclase activity; Gαq, which activates phospholipase Cβ; and Gα12/13, which promotes Rho activation. An activated receptor can interact with heterotrimeric G protein complexes. Receptors function as guanine nucleotide exchange factors (GEF) to promote dissociation of GDP from the Gα subunit in exchange for GTP (figure 2A) [4]. In the activated heterotrimeric G protein complex, the Gα-GTP bound subunit dissociates from the βγ subunit resulting in propagation of signaling cascades. However, subsequent studies have shown some receptors are able to reach an activated state through the formation of dimers and oligomers without agonist binding [4, 16]. Alternatively, GPCR activation can occur through interactions with adaptor and scaffolding proteins. Through specific binding sites, adaptor/scaffolding proteins, such as A-kinase anchoring proteins (AKAP) and β-arrestins (figure 2B), mediate interactions between the receptor and downstream second messengers by scaffolding a network of proteins which then operate as a large molecular complex [4]. Furthermore, receptors including the cannabinoid receptor 1 (CB1) exhibit high basal signaling, independent of agonist activation [20]. High basal signaling might be indicative of conformational flexibility [21]; however, a study with a constitutively active ADRB2 mutant has linked conformational flexibility with structural instability [11].
Figure 2.
(A) A classical example of GPCR signaling pathway. Receptors, such as ADRB2, are in an inactive state until an agonist binds to activate the receptor. The activated receptor-G protein complex is formed resulting in GDP to GTP exchange by the G protein complex followed by the dissociation of the Gα subunit from the Gβγ dimer. Both Gα and Gβγ subunits, in turn, activate downstream effectors. Gαx represents the general Gα subunit followed by diagrams for Gα12/13, Gαi, Gαs, and Gαq pathways. (B) β-arrestins can function as adaptor/scaffolding molecules activating the ERK (MAPK) cascade. GRK phosphorylates the activated receptor recruiting β-arrestin to the phosphorylation sites along with Raf, ERK (MAPK), and MEK. This association triggers activation of the ERK (MAPK) cascade leading to cytosolic signaling pathways. Figures 2A and 2B were adapted from Pierce, et al. [4].
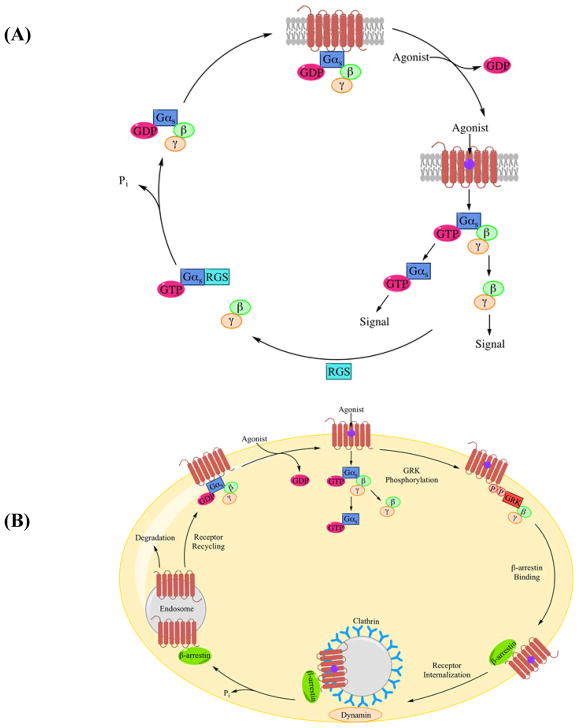
Receptor signaling is modulated by various mechanisms to control cellular functions. Three such processes are highlighted here - deactivation, desensitization, and receptor internalization. At the receptor level, members of the RGS family of proteins serve as GPTase activating proteins (GAPs) and accelerate deactivation by enhancing the rate of Gα-catalyzed hydrolysis of GTP to GDP by factors up to 2000-fold (figure 3A) [22]. In other cases of continuous agonist stimulation, the receptor can be phosphorylated by downstream second-messenger protein kinases or a family of G protein-coupled receptor kinases (GRK). Following phosphorylation, β-arrestin is recruited to the receptor leading to the inhibition of receptor-G protein interaction via steric hindrance. The 251659264 receptor-β-arrestin complex is targeted and removed from the cell surface through endocytosis. The receptor is then either degraded or recycled back to the cell surface in the inactive conformation (figure 3B). Receptor internalization is mediated by β-arrestin, clathrin-coated pits, and caveolae through diverse mechanisms [23]. Further in-depth evaluations of GPCR function can be found in excellent reviews by Pierce, et al. [4] and Syrovatkina, et al. [16].
Figure 3.
(A) Receptor deactivation is mediated by the RGS family of proteins. RGS alters the conformation of Gα-GTP complex making it a better hydrolase, which accelerates the hydrolysis of GTP to GDP. (B) GPCR desensitization through internalization occurs when GRK phosphorylates the activated receptor promoting β-arresting binding, which sterically hinders receptor-G protein interaction. The receptor is either degraded or recycled back to the cell surface. Figure 3A was adapted from Neubig, et al. [191] and 3B was adapted from Pierce, et al. [4].
Structural characterization
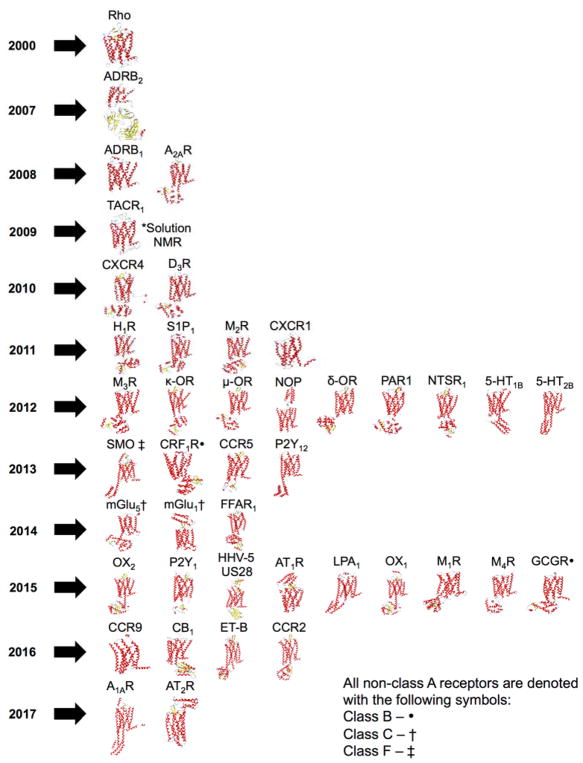
Due to its high endogenous concentration, the GPCR rhodopsin was purified from bovine rod outer segment (ROS) membranes [24] leading to its crystallization in 2000 (figure 4; PDB: 1F88) [25]. To date, rhodopsin is the only GPCR that has been purified from its natural source for structural studies. The next GPCR successfully crystallized was ADRB2 (PDB: 2RH1 [26], 2R4R/2R4S [27]) seven years later. This delay was due to a need for numerous technological advances required to express, purify and crystallize these lipid-soluble targets. The most critical of these advances were improved lipid phases [28–30], lipidic cubic phase (LCP) methods for crystallization [31], and the incorporation of soluble fusion partners [31, 32].
Figure 4.
Timeline of availability for individual representative GPCR crystal structure structures including protein fusion partners. Rho (1F88), ADRB2 (2R4R), ADRB1 (2VT4), A2AR (3EML), TACR1 (2KS9), CXCR4 (3ODU), D3R (3PBL), H1R (3RZE), S1P1 (3V2W), M2R (3UON), CXCR1 (2LNL), M3R (4DAJ), κ-OR (4DJH), μ-OR (4DKL), NOP (4EA3), δ-OR (4EJ4), PAR1 (3VW7), NSTR1 (3ZEV), 5-HT1B (4IAQ), 5-HT2B (4IB4), SMO (4JKV), CRF1R (4K5Y), CCR5 (4MBS) P2Y12 (4NTJ), mGlu5 (4OO9), mGlu1 (4OR2), FFAR1 (4PHU), OX2 (4S0V), P2Y1 (4XNW), HHV-5 US28 (4XT1), AT1R (4YAY), LPA1 (4Z34), OX1 (4ZJ8), M1R (5CXV), M4R (5DSG), GCGR (5EE7), CCR9 (5LWE), ET-B (5GLH), CCR2 (5T1A), CB1 (5TGZ), A1AR (5UEN), AT2R (5UNF).
Purifying hydrophobic membrane proteins requires additional steps compared to water-soluble proteins. Prior to purification, membrane proteins must be removed from their native lipid environment. Although solubilizing detergents are required to enable efficient extraction of GPCR from the membranes, it is essential to select a mild detergent that discourages protein denaturation. It is important to note that detergent protocols used for one receptor may not be appropriate to other receptors, thus making detergent screening a critical step in the obtaining viable protein. Rhodopsin (PDB: 1F88) was effectively solubilized in a mixed micellar solution containing heptane-1,2,3-triol (HPTO), nonyl glucoside, and zwitterionic lauryldimethylamine-N-oxide (LDAO) [24]. Both ADRB2 structures from 2007 (PDB: 2RH1 [26], 2R4R/2R4S [27]) were solubilized in dodecylmaltoside. Often, detergents used for extraction are not sufficient to stabilize protein and a detergent exchange is required [31]. This method was observed in the crystallization trials of the ADRB2-Gs complex (PDB: 3SN6) where the complex was formed in dodecylmaltoside solution and exchanged into maltose neopentylglycerol detergent (MNG-3) [18].
Crystallization of membrane proteins has been accomplished through the use of different lipid phases including micelles, bicelles, and nanodiscs. These different lipid phases allow for protein stabilization and promote crystal lattice formation [28–30]. Detergent micelles surround the hydrophobic GPCR structural regions and allow crystal lattice contacts to form between exposed protein loop structures [28]. Lipid-based bicelles and nanodiscs, on the other hand, act as lipidic mimics of a native bilayer environment. Bicelles are formed via the combination of a detergent or short-chain lipid with a long chain lipid, such as dimyristoyl phosphatidylcholine (DMPC) [29]. A nanodisc is a non-covalently assembled phospholipid bilayer encircled by membrane scaffold proteins, such as plasma lipoproteins [33]. Rhodopsin (PDB: 1F88 [24]) and ADRB2 (PDB: 2R4R/2R4S [27]) were crystallized in micelles and DMPC/CHAPSO bicelles, respectively. Nanodiscs were used in the crystallization of the nanobody-stabilized ADRB2 (PDB: 3P0G).
Following many detergent optimization efforts, rhodopsin was crystallized by vapor diffusion [24]. Vapor diffusion forces protein solutions to reach supersaturation prior to crystallization. In this method, protein solutions are mixed with a crystallization solution and positioned in a sitting-drop or hanging-drop orientation in an airtight chamber that also contains the crystallization solution. The chemical equilibrium between the drop and the chamber results in the evaporation of volatile species, which diffuse from the drop to the well. This diffusion leads to a slow saturation of protein within the drop allowing the protein-detergent complexes to form crystal lattice contacts with neighboring molecules [34]. In lipidic cubic phase (LCP) methods, proteins are placed in a membrane-like environment where they can diffuse and interact with each other to form crystal lattice contacts on both complementary hydrophobic and hydrophilic regions [31]. Crystals of rhodopsin (PDB: 1F88 [25]) and ADRB2 (PDB: 2R4R/2R4S [27]) were grown by hanging drop diffusion, whereas ADRB2 (PRB: 2RH1 [26]) and ADRB2-Gs complex (PDB: 3SN6 [18]) crystallization experiments implemented LCP methods.
In determining crystallographic structures, molecular replacement has been successfully utilized to help in determining the phase of an unknown target by applying the phase of a closely related and previously characterized target. This approach was implemented in the analysis of the ADRB2 structure [26]. Multi-wavelength anomalous scattering (MAD) and single-wavelength anomalous scattering (SAD) methods have impacted atomic-level structure determinations of GPCR when a heavy atom anomalous scatterer is incorporated into the protein structure. Structural determination using MAD phasing was implemented in the first crystal structure of rhodopsin (PDB: 1F88) [25].
Developments in protein engineering and heterologous protein expression have accelerated structural determinations of GPCR. Site-directed mutagenesis has been beneficial for the stabilization of receptor structure, as well as determination of the functional activity of many GPCR [31]. Mutagenesis has also enabled the introduction of fusion partners into protein sequences. Fusion partners are designed to be highly stable, compact, and easily crystallized. These partners maintain essential surface contacts and increase protein solubility, making them suitable replacements for flexible domains [31, 32]. The position of the fusion partner and the number of residues conjoining the fusion partner to the protein may alter expression and/or stability; nevertheless, optimizing the number of linker residues can minimize adverse effects. A Fab5 epitope was engineered into IL3 of ADRB2 (PDB: 2R4R/2R4S) by mutagenesis, which allowed a Fab5 monoclonal antibody to bind to the epitope [27]. Fab5 stabilized the receptor conformation and increased the polar surface area necessary for crystal lattice contacts. Since the initial use of the Fab5 antibody, many GPCR have been successfully engineered with fusion partners accelerating the number solved of crystallographic structures in the last two decades as illustrated in the timeline in figure 4. T4 lysozyme has been the most widely used fusion partner in multiple receptors such as adenosine A2A receptor (A2AR; PDB: 3EML) [37] and metabotropic glutamate receptor 5 (mGlu5; PDB: 4OO9) [38]. Thermostabilized apocytochrome b562RIL (BRIL) is a highly versatile fusion partner that has been appended to the N-terminal domain of nociception receptor 1 (NOP; PDB: 4EA3) [39] and C-terminal domain of smoothened receptor (SMO; PDB: 4JKV) [40]. Pyrococcus abyssi glycogen synthase (PGS) and rubredoxin emerged as fusion partners later in the timeline of GPCR characterization. These tools have been successfully used in analysis of the orexin 1 (OX1; PDB: 4ZJ8) [41] and purinergic receptor 1 (P2Y1; PDB: 4XNW) receptors [42]. Although the addition of fusion protein sequences have been effective in the analysis of many GPCR, it is important to note that engineering designs are often specific for one protein and require optimization for each application to new receptor sequences.
GPCR loops and termini, which are characteristically flexible and difficult to crystallize, can result in additional challenges. Proteins truncated or modified at these areas can reduce flexibility. However, simple truncations can also reduce the polar surface area of the protein, which is a characteristic crucial for forming the crystal lattice contacts required for crystal formation. Rhodopsin has a relatively short IL3 with ~3 amino acids and C-terminal domain with ~25 amino acids, in contrast, ADRB2 has a lengthy IL3 with ~28 amino acids and a C-terminal domain ~70 amino acids [43]. Two early structures of ADRB2 were engineered differently in these regions to achieve crystal lattice formation during crystallization. In the ADRB1-Fab5 complex (PDB: 2R4R/2R4S), truncating the final 48 amino acids in the unstructured C-terminal domain optimized crystal size and uniformity [27]. In another structure of ADRB2 (PDB: 2RH1), IL3 was completely removed and replaced with the soluble fusion partner, T4 lysozyme, which promoted the growth of diffraction quality crystals [26]. Amino acid truncations have also been found to have a variable effect on protein expression. Truncations at the N-terminus reduced expression. However, replacing the N-terminus with BRIL maintained similar expression as constructs with the complete N-terminus [39]. This was the case with NOP (PDB: 4EA3) where an N-terminal replacement with BRIL and a 31 amino acid deletion at the C-terminus significantly increased protein expression [31, 39].
There have been challenges in GPCR structure determination due to their hydrophobic nature and instability outside of the hydrophobic membrane environment [44, 45]. Since the first complete x-ray crystallographic structure of a GPCR (bovine rhodopsin; PDB: 1F88) was solved in 2000 [25], there are now over 180 comprehensive structures of GPCR in the Protein Data Bank (www.rcsb.org [46]) listed in table 1. More than 150 of these structures co-crystallized with ligands are described in more detail in table 2. However, less than 45 distinct family members are represented out of a superfamily of over 800 proteins. This limitation in representation suggests that significantly more work is yet to be done.
Table 1.
A comprehensive list of GPCR crystal structures according to date deposited into the Protein Data Bank as of April 19, 2017.
| GPCR Crystal Structures | |||||||
|---|---|---|---|---|---|---|---|
| Individual Receptors | Family | Class/Subclass | Species | Receptor | PDB [46] ID | Resolution (Å) | Date Deposited |
| 1 | Rhodopsin | Class Aα | Bovine | Rho | 1F88 [25] | 2.80 | 6/29/00 |
| 2 | Rhodopsin | Class Aα | Bovine | Rho | 1HZX [94] | 2.80 | 1/26/01 |
| 3 | Rhodopsin | Class Aα | Bovine | Rho | 1JFP [95] | Solution NMR | 6/21/01 |
| 4 | Rhodopsin | Class Aα | Bovine | Rho | 1L9H [96] | 2.60 | 3/23/02 |
| 5 | Rhodopsin | Class Aα | Bovine | Rho | 1LN6 [97] | Solution NMR | 5/3/02 |
| 6 | Rhodopsin | Class Aα | Bovine | Rho | 1GZM [98] | 2.65 | 5/24/02 |
| 7 | Rhodopsin | Class Aα | Bovine | Rho | 1U19 [99] | 2.20 | 7/15/04 |
| 8 | Rhodopsin | Class Aα | Bovine | Rho | 2G87 [100] | 2.60 | 3/2/06 |
| 9 | Rhodopsin | Class Aα | Bovine | Rho | 2HPY [101] | 2.80 | 7/18/06 |
| 10 | Rhodopsin | Class Aα | Bovine | Rho | 2I35 [102] | 3.80 | 8/17/06 |
| 11 | Rhodopsin | Class Aα | Bovine | Rho | 2I36 [102] | 4.10 | 8/17/06 |
| 12 | Rhodopsin | Class Aα | Bovine | Rho | 2I37 [102] | 4.15 | 8/17/06 |
| 13 | Rhodopsin | Class Aα | Bovine | Rho | 2J4Y [103] | 3.40 | 9/7/06 |
| 14 | Rhodopsin | Class Aα | Bovine | Rho | 2PED [104] | 2.95 | 4/2/07 |
| 15 | Rhodopsin | Class Aα | Bovine | Rho | 3C9L [105] | 2.65 | 2/16/08 |
| 16 | Rhodopsin | Class Aα | Bovine | Rho | 3C9M [105] | 3.40 | 2/16/08 |
| 17 | Rhodopsin | Class Aα | Bovine | Rho | 3CAP [73] | 2.90 | 2/20/08 |
| 18 | Rhodopsin | Class Aα | Bovine | Rho | 3DQB [106] | 3.20 | 7/9/08 |
| 19 | Rhodopsin | Class Aα | Bovine | Rho | 2X72 [107] | 3.00 | 2/22/10 |
| 20 | Rhodopsin | Class Aα | Bovine | Rho | 3OAX[108] | 2.60 | 8/5/10 |
| 21 | Rhodopsin | Class Aα | Bovine | Rho | 3PQR [109] | 2.85 | 11/26/10 |
| 22 | Rhodopsin | Class Aα | Bovine | Rho | 3PXO [109] | 3.00 | 12/10/10 |
| 23 | Rhodopsin | Class Aα | Bovine | Rho | 4A4M [110] | 3.30 | 10/17/11 |
| 24 | Rhodopsin | Class Aα | Bovine | Rho | 4J4Q [111] | 2.65 | 2/7/13 |
| 25 | Rhodopsin | Class Aα | Bovine | Rho | 4BEY [112] | 2.90 | 3/12/13 |
| 26 | Rhodopsin | Class Aα | Bovine | Rho | 4BEZ [112] | 3.30 | 3/12/13 |
| 27 | Rhodopsin | Class Aα | Bovine | Rho | 4PXF [113] | 2.75 | 3/23/14 |
| 28 | Rhodopsin | Class Aα | Bovine | Rho | 4X1H [114] | 2.29 | 11/24/14 |
| 29 | Rhodopsin | Class Aα | Human | Rho | 4ZWJ [115] | 3.30 | 5/19/15 |
| 30 | Rhodopsin | Class Aα | Bovine | Rho | 5DYS [116] | 2.30 | 9/25/15 |
| 31 | Rhodopsin | Class Aα | Bovine | Rho | 5EN0 [116] | 2.81 | 11/8/15 |
| 32 | Rhodopsin | Class Aα | Bovine | Rho | 5TE3 [117] | 2.70 | 9/20/16 |
| 33 | Rhodopsin | Class Aα | Bovine | Rho | 5TE5 [117] | 4.01 | 9/21/16 |
| 34 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2VT4 [72] | 2.70 | 5/9/08 |
| 35 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2Y00 [118] | 2.50 | 11/30/10 |
| 36 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2Y01[118] | 2.60 | 11/30/10 |
| 37 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2Y02 [118] | 2.60 | 11/30/10 |
| 38 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2Y03 [118] | 2.85 | 11/30/10 |
| 39 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2Y04 [118] | 3.05 | 11/30/10 |
| 40 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2YCW [74] | 3.00 | 3/17/11 |
| 41 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2YCX [74] | 3.25 | 3/17/11 |
| 42 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2YCY [74] | 3.15 | 3/17/11 |
| 43 | Rhodopsin | Class Aα | Turkey | ADRB1 | 2YCZ [74] | 3.65 | 3/17/11 |
| 44 | Rhodopsin | Class Aα | Turkey | ADRB1 | 3ZPQ [119] | 2.80 | 3/1/13 |
| 45 | Rhodopsin | Class Aα | Turkey | ADRB1 | 3ZPR [119] | 2.70 | 3/1/13 |
| 46 | Rhodopsin | Class Aα | Turkey | ADRB1 | 4AMI [120] | 3.20 | 3/11/12 |
| 47 | Rhodopsin | Class Aα | Turkey | ADRB1 | 4AMJ [120] | 2.30 | 3/12/12 |
| 48 | Rhodopsin | Class Aα | Turkey | ADRB1 | 4BVN [69] | 2.10 | 6/26/13 |
| 49 | Rhodopsin | Class Aα | Turkey | ADRB1 | 4GPO [121] | 3.50 | 8/21/12 |
| 50 | Rhodopsin | Class Aα | Turkey | ADRB1 | 5A8E [122] | 2.40 | 7/15/15 |
| 51 | Rhodopsin | Class Aα | Turkey | ADRB1 | 5F8U [123] | 3.35 | 12/9/15 |
| 52 | Rhodopsin | Class Aα | Human | ADRB2 | 2R4R [27] | 3.40 | 8/31/07 |
| 53 | Rhodopsin | Class Aα | Human | ADRB2 | 2R4S [27] | 3.40 | 8/31/07 |
| 54 | Rhodopsin | Class Aα | Human | ADRB2 | 2RH1 [26] | 2.40 | 10/5/07 |
| 55 | Rhodopsin | Class Aα | Human | ADRB2 | 3D4S [124] | 2.80 | 5/14/08 |
| 56 | Rhodopsin | Class Aα | Human | ADRB2 | 3KJ6 [125] | 3.40 | 11/2/09 |
| 57 | Rhodopsin | Class Aα | Human | ADRB2 | 3NY8 [126] | 2.84 | 7/14/10 |
| 58 | Rhodopsin | Class Aα | Human | ADRB2 | 3NY9 [126] | 2.84 | 7/14/10 |
| 59 | Rhodopsin | Class Aα | Human | ADRB2 | 3NYA [126] | 3.16 | 7/14/10 |
| 60 | Rhodopsin | Class Aα | Human | ADRB2 | 3P0G [127] | 3.50 | 9/28/10 |
| 61 | Rhodopsin | Class Aα | Human | ADRB2 | 3PDS [128] | 3.50 | 10/24/10 |
| 62 | Rhodopsin | Class Aα | Human | ADRB2 | 3SN6 [18] | 3.20 | 6/28/11 |
| 63 | Rhodopsin | Class Aα | Human | ADRB2 | 4GBR [129] | 3.99 | 7/27/12 |
| 64 | Rhodopsin | Class Aα | Human | ADRB2 | 4LDE [130] | 2.79 | 6/24/13 |
| 65 | Rhodopsin | Class Aα | Human | ADRB2 | 4LDL [130] | 3.10 | 6/24/13 |
| 66 | Rhodopsin | Class Aα | Human | ADRB2 | 4LDO [130] | 3.20 | 6/24/13 |
| 67 | Rhodopsin | Class Aα | Human | ADRB2 | 4QKX [131] | 3.30 | 6/10/14 |
| 68 | Rhodopsin | Class Aα | Human | ADRB2 | 5D5A [132] | 2.48 | 8/10/15 |
| 69 | Rhodopsin | Class Aα | Human | ADRB2 | 5D5B [132] | 3.80 | 8/10/15 |
| 70 | Rhodopsin | Class Aα | Human | ADRB2 | 5D6L [133] | 3.20 | 8/12/15 |
| 71 | Rhodopsin | Class Aα | Human | ADRB2 | 5JQH [134] | 3.20 | 5/5/16 |
| 72 | Rhodopsin | Class Aα | Human | H1R | 3RZE [135] | 3.10 | 5/11/11 |
| 73 | Rhodopsin | Class Aα | Human | D3R | 3PBL [6] | 2.89 | 10/20/10 |
| 74 | Rhodopsin | Class Aα | Human | 5-HT1B | 4IAQ [136] | 2.80 | 12/7/12 |
| 75 | Rhodopsin | Class Aα | Human | 5-HT1B | 4IAR [136] | 2.70 | 12/7/12 |
| 76 | Rhodopsin | Class Aα | Human | 5-HT2B | 4IB4 [137] | 2.70 | 12/7/12 |
| 77 | Rhodopsin | Class Aα | Human | 5-HT2B | 4NC3 [138] | 2.80 | 10/23/13 |
| 78 | Rhodopsin | Class Aα | Human | 5-HT2B | 5TVN [139] | 2.90 | 11/9/16 |
| 79 | Rhodopsin | Class Aα | Human | M1R | 5CXV [140] | 2.70 | 7/29/15 |
| 80 | Rhodopsin | Class Aα | Human | M2R | 3UON [141] | 3.00 | 11/16/11 |
| 81 | Rhodopsin | Class Aα | Human | M2R | 4MQS [142] | 3.50 | 9/16/13 |
| 82 | Rhodopsin | Class Aα | Human | M2R | 4MQT [142] | 3.70 | 9/16/13 |
| 83 | Rhodopsin | Class Aα | Rat | M3R | 4DAJ [143] | 3.40 | 1/12/12 |
| 84 | Rhodopsin | Class Aα | Rat | M3R | 4U14 [144] | 3.57 | 7/15/14 |
| 85 | Rhodopsin | Class Aα | Rat | M3R | 4U15 [144] | 2.80 | 7/15/14 |
| 86 | Rhodopsin | Class Aα | Rat | M3R | 4U16 [144] | 3.70 | 7/15/14 |
| 87 | Rhodopsin | Class Aα | Human | M4R | 5DSG [140] | 2.60 | 9/17/15 |
| 88 | Rhodopsin | Class Aα | Human | A1AR | 5UEN [145] | 3.20 | 1/3/17 |
| 89 | Rhodopsin | Class Aα | Human | A2AR | 3EML [37] | 2.60 | 9/24/08 |
| 90 | Rhodopsin | Class Aα | Human | A2AR | 3PWH [146] | 3.30 | 12/8/10 |
| 91 | Rhodopsin | Class Aα | Human | A2AR | 3QAK [68] | 2.71 | 1/11/11 |
| 92 | Rhodopsin | Class Aα | Human | A2AR | 2YDO [147] | 3.00 | 3/23/11 |
| 93 | Rhodopsin | Class Aα | Human | A2AR | 2YDV [147] | 2.60 | 3/24/11 |
| 94 | Rhodopsin | Class Aα | Human | A2AR | 3REY [146] | 3.31 | 4/5/11 |
| 95 | Rhodopsin | Class Aα | Human | A2AR | 3RFM [146] | 3.60 | 4/6/11 |
| 96 | Rhodopsin | Class Aα | Human | A2AR | 3VG9 [148] | 2.70 | 8/4/11 |
| 97 | Rhodopsin | Class Aα | Human | A2AR | 3VGA [148] | 3.10 | 8/4/11 |
| 98 | Rhodopsin | Class Aα | Human | A2AR | 3UZA [149] | 3.27 | 12/7/11 |
| 99 | Rhodopsin | Class Aα | Human | A2AR | 3UZC [149] | 3.34 | 12/7/11 |
| 100 | Rhodopsin | Class Aα | Human | A2AR | 4EIY [66] | 1.80 | 4/6/12 |
| 101 | Rhodopsin | Class Aα | Human | A2AR | 4UG2 [150] | 2.60 | 3/21/15 |
| 102 | Rhodopsin | Class Aα | Human | A2AR | 4UHR [150] | 2.60 | 3/25/15 |
| 103 | Rhodopsin | Class Aα | Human | A2AR | 5IU4 [151] | 1.72 | 3/17/16 |
| 104 | Rhodopsin | Class Aα | Human | A2AR | 5IU7 [151] | 1.90 | 3/17/16 |
| 105 | Rhodopsin | Class Aα | Human | A2AR | 5IU8 [151] | 2.00 | 3/17/16 |
| 106 | Rhodopsin | Class Aα | Human | A2AR | 5IUA [151] | 2.20 | 3/17/16 |
| 107 | Rhodopsin | Class Aα | Human | A2AR | 5IUB [151] | 2.10 | 3/17/16 |
| 108 | Rhodopsin | Class Aα | Human | A2AR | 5K2A [152] | 2.50 | 5/18/16 |
| 109 | Rhodopsin | Class Aα | Human | A2AR | 5K2B [152] | 2.50 | 5/18/16 |
| 110 | Rhodopsin | Class Aα | Human | A2AR | 5K2C [152] | 1.90 | 5/18/16 |
| 111 | Rhodopsin | Class Aα | Human | A2AR | 5K2D [152] | 1.90 | 5/18/16 |
| 112 | Rhodopsin | Class Aα | Human | A2AR | 5G53 [153] | 3.40 | 5/19/16 |
| 113 | Rhodopsin | Class Aα | Human | A2AR | 5UIG [154] | 3.50 | 1/13/17 |
| 114 | Rhodopsin | Class Aα | Human | S1P1 | 3V2W [155] | 3.35 | 12/12/11 |
| 115 | Rhodopsin | Class Aα | Human | S1P1 | 3V2Y [155] | 2.80 | 12/12/11 |
| 116 | Rhodopsin | Class Aα | Human | LPA1 | 4Z34 [156] | 3.00 | 3/30/15 |
| 117 | Rhodopsin | Class Aα | Human | LPA1 | 4Z35 [156] | 2.90 | 3/30/15 |
| 118 | Rhodopsin | Class Aα | Human | LPA1 | 4Z36 [156] | 2.90 | 3/30/15 |
| 119 | Rhodopsin | Class Aα | Human | CB1 | 5TGZ [157] | 2.80 | 9/28/16 |
| 120 | Rhodopsin | Class Aα | Human | CB1 | 5U09 [158] | 2.60 | 11/23/16 |
| 121 | Rhodopsin | Class Aβ | Rat | NTSR1 | 4GRV [159] | 2.80 | 8/27/12 |
| 122 | Rhodopsin | Class Aβ | Rat | NTSR1 | 3ZEV [160] | 3.00 | 12/7/12 |
| 123 | Rhodopsin | Class Aβ | Rat | NTSR1 | 4BUO [160] | 2.75 | 6/21/13 |
| 124 | Rhodopsin | Class Aβ | Rat | NTSR1 | 4BV0 [160] | 3.10 | 6/24/13 |
| 125 | Rhodopsin | Class Aβ | Rat | NTSR1 | 4BWB [160] | 3.57 | 7/1/13 |
| 126 | Rhodopsin | Class Aβ | Rat | NTSR1 | 4XEE [161] | 2.90 | 12/23/14 |
| 127 | Rhodopsin | Class Aβ | Rat | NTSR1 | 4XES [161] | 2.60 | 12/24/14 |
| 128 | Rhodopsin | Class Aβ | Rat | NTSR1 | 5T04 [162] | 3.30 | 8/16/16 |
| 129 | Rhodopsin | Class Aβ | Human | OX1 | 4ZJ8 [41] | 2.75 | 4/29/15 |
| 130 | Rhodopsin | Class Aβ | Human | OX1 | 4ZJC [41] | 2.83 | 4/29/15 |
| 131 | Rhodopsin | Class Aβ | Human | OX2 | 4S0V [163] | 2.50 | 1/6/15 |
| 132 | Rhodopsin | Class Aβ | Human | TACR1 | 2KS9 [164] | Solution NMR | 12/31/09 |
| 133 | Rhodopsin | Class Aβ | Human | TACR1 | 2KSA [164] | Solution NMR | 12/31/09 |
| 134 | Rhodopsin | Class Aβ | Human | TACR1 | 2KSB [164] | Solution NMR | 12/31/09 |
| 135 | Rhodopsin | Class Aβ | Human | ET-B | 5GLI [165] | 2.5 | 7/11/16 |
| 136 | Rhodopsin | Class Aβ | Human | ET-B | 5GLH [165] | 2.8 | 7/11/16 |
| 137 | Rhodopsin | Class Aγ | Human | CXCR1 | 2LNL [166] | Solid-State NMR | 12/31/11 |
| 138 | Rhodopsin | Class Aγ | Human | CCR2 | 5T1A [167] | 2.81 | 8/18/16 |
| 139 | Rhodopsin | Class Aγ | Human | CXCR4 | 3ODU [168] | 2.50 | 8/11/10 |
| 140 | Rhodopsin | Class Aγ | Human | CXCR4 | 3OE0 [168] | 2.90 | 8/12/10 |
| 141 | Rhodopsin | Class Aγ | Human | CXCR4 | 3OE6 [168] | 3.20 | 8/12/10 |
| 142 | Rhodopsin | Class Aγ | Human | CXCR4 | 3OE8 [168] | 3.10 | 8/12/10 |
| 143 | Rhodopsin | Class Aγ | Human | CXCR4 | 3OE9 [168] | 3.10 | 8/12/10 |
| 144 | Rhodopsin | Class Aγ | Human | CXCR4 | 4RWS [169] | 3.10 | 12/5/14 |
| 145 | Rhodopsin | Class Aγ | Human | CCR5 | 4MBS [170] | 2.71 | 8/19/13 |
| 146 | Rhodopsin | Class Aγ | Human | CCR9 | 5LWE [171] | 2.80 | 9/16/16 |
| 147 | Rhodopsin | Class Aγ | Human | NOP | 4EA3 [39] | 3.01 | 3/22/12 |
| 148 | Rhodopsin | Class Aγ | Human | NOP | 5DHG [172] | 3.00 | 8/30/15 |
| 149 | Rhodopsin | Class Aγ | Human | NOP | 5DHH [172] | 3.00 | 8/31/15 |
| 150 | Rhodopsin | Class Aγ | Human | κ-OR | 4DJH [173] | 2.90 | 2/1/12 |
| 151 | Rhodopsin | Class Aγ | Mouse | μ-OR | 4DKL [174] | 2.80 | 2/3/12 |
| 152 | Rhodopsin | Class Aγ | Mouse | μ-OR | 5C1M[175] | 2.10 | 6/15/15 |
| 153 | Rhodopsin | Class Aγ | Mouse | δ-OR | 4EJ4 [176] | 3.40 | 4/6/12 |
| 154 | Rhodopsin | Class Aγ | Human | δ-OR | 4N6H [70] | 1.80 | 10/12/13 |
| 155 | Rhodopsin | Class Aγ | Human | δ-OR | 4RWA [177] | 3.28 | 12/1/14 |
| 156 | Rhodopsin | Class Aγ | Human | δ-OR | 4RWD [177] | 2.70 | 12/2/14 |
| 157 | Rhodopsin | Class Aγ | Human | AT1R | 4YAY [178] | 2.90 | 2/18/15 |
| 158 | Rhodopsin | Class Aγ | Human | AT1R | 4ZUD [179] | 2.80 | 5/15/15 |
| 159 | Rhodopsin | Class Aγ | Human | AT2R | 5UNF [180] | 2.80 | 1/30/17 |
| 160 | Rhodopsin | Class Aγ | Human | AT2R | 5UNG [180] | 2.80 | 1/30/17 |
| 161 | Rhodopsin | Class Aγ | Human | AT2R | 5UNH [180] | 2.90 | 1/30/17 |
| 162 | Rhodopsin | Class Aγ | HHV-5 | HHV-5 US28 | 4XT1 [181] | 2.89 | 1/22/15 |
| 163 | Rhodopsin | Class Aγ | HHV-5 | HHV-5 US28 | 4XT3 [181] | 3.80 | 1/22/15 |
| 164 | Rhodopsin | Class Aδ | Human | P2Y1 | 4XNW [182] | 2.70 | 1/16/15 |
| 165 | Rhodopsin | Class Aδ | Human | P2Y1 | 4XNV [182] | 2.20 | 1/16/15 |
| 166 | Rhodopsin | Class Aδ | Human | P2Y12 | 4NTJ [42] | 2.62 | 12/2/13 |
| 167 | Rhodopsin | Class Aδ | Human | P2Y12 | 4PXZ [183] | 2.50 | 3/25/14 |
| 168 | Rhodopsin | Class Aδ | Human | P2Y12 | 4PY0 [183] | 3.10 | 3/25/14 |
| 169 | Rhodopsin | Class Aδ | Human | PAR1 | 3VW7 [71] | 2.20 | 8/7/12 |
| 170 | Rhodopsin | Class Aδ | Human | FFAR1 | 4PHU [184] | 2.33 | 5/7/14 |
| 171 | Secretin | Class B | Human | GCGR | 4L6R [185] | 3.30 | 6/12/13 |
| 172 | Secretin | Class B | Human | GCGR | 5EE7 [78] | 2.50 | 10/22/15 |
| 173 | Secretin | Class B | Human | CRF1R | 4K5Y [186] | 2.98 | 4/15/13 |
| 174 | Secretin | Class B | Human | CRF1R | 4Z9G [187] | 3.18 | 4/10/15 |
| 175 | Glutamate | Class C | Human | mGlu1 | 4OR2 [188] | 2.80 | 2/10/14 |
| 176 | Glutamate | Class C | Human | mGlu5 | 4OO9 [38] | 2.60 | 1/31/14 |
| 177 | Glutamate | Class C | Human | mGlu5 | 5CGC [189] | 3.10 | 7/9/15 |
| 178 | Glutamate | Class C | Human | mGlu5 | 5CGD [189] | 2.60 | 7/9/15 |
| 179 | Frizzled/Taste2 | Class F | Human | SMO | 4JKV [40] | 2.45 | 3/11/13 |
| 180 | Frizzled/Taste2 | Class F | Human | SMO | 4N4W [190] | 2.80 | 10/8/13 |
| 181 | Frizzled/Taste2 | Class F | Human | SMO | 4O9R [131] | 3.20 | 1/2/14 |
| 182 | Frizzled/Taste2 | Class F | Human | SMO | 4QIM [190] | 2.61 | 5/31/14 |
| 183 | Frizzled/Taste2 | Class F | Human | SMO | 4QIN [190] | 2.60 | 5/31/14 |
| 184 | Frizzled/Taste2 | Class F | Human | SMO | 5L7D [93] | 3.20 | 6/3/16 |
| 185 | Frizzled/Taste2 | Class F | Human | SMO | 5L7I [93] | 3.30 | 6/3/16 |
Table 2.
A comprehensive list of GPCR crystallized with ligands deposited into the Protein Data Bank as of April 19, 2017.
| PDB [46] ID | Class/Subclass | GPCR | Ligand Name | Ligand Type | |
|---|---|---|---|---|---|
| 1 | 1F88 [25] | Class Aα | Rho | 11-cis-Retinal | |
| 2 | 1GZM [98] | Class Aα | Rho | 11-cis-Retinal | |
| 3 | 1HZX [94] | Class Aα | Rho | 11-cis-Retinal | |
| 4 | 1JFP [95] | Class Aα | Rho | all-trans-Retinal | |
| 5 | 1L9H [96] | Class Aα | Rho | 11-cis-Retinal | |
| 6 | 1LN6 [97] | Class Aα | Rho | all-trans-Retinal | |
| 7 | 1U19 [99] | Class Aα | Rho | 11-cis-Retinal | |
| 8 | 2G87 [100] | Class Aα | Rho | all-trans-Retinal | |
| 9 | 2HPY [101] | Class Aα | Rho | all-trans-Retinal | |
| 10 | 2I35 [102] | Class Aα | Rho | 11-cis-Retinal | |
| 11 | 2J4Y [103] | Class Aα | Rho | 11-cis-Retinal | |
| 12 | 2PED [104] | Class Aα | Rho | 9-cis-Retinal | |
| 13 | 2X72 [107] | Class Aα | Rho | 11-cis-Retinal | |
| 14 | 3C9L [105] | Class Aα | Rho | 11-cis-Retinal | |
| 15 | 3C9M [105] | Class Aα | Rho | 11-cis-Retinal | |
| 16 | 3OAX [108] | Class Aα | Rho | 11-cis-Retinal | |
| 17 | 3PQR [109] | Class Aα | Rho | all-trans-Retinal | |
| 18 | 3PXO [109] | Class Aα | Rho | all-trans-Retinal | |
| 19 | 4A4M [110] | Class Aα | Rho | all-trans-Retinal | |
| 20 | 5DYS [116] | Class Aα | Rho | all-trans-Retinal | |
| 21 | 5EN0 [116] | Class Aα | Rho | all-trans-Retinal | |
| 22 | 5TE5 [117] | Class Aα | Rho | (2E)-{(4E)-4-[(3E)-4-(2,6,6-trimethylcyclohex- 1-en-1-yl)but-3-en-2-ylidene]cyclohex-2-en- 1-ylidene}acetaldehyde | |
| 23 | 4J4Q [111] | Class Aα | Rho | B-Octylglucoside | Agonist-stabilized active receptor conformation |
| 24 | 2VT4 [72] | Class Aα | ADRB1 | Cyanopindolol | Antagonist |
| 25 | 2Y00 [118] | Class Aα | ADRB1 | Dobutamine | Partial Agonist |
| 26 | 2Y01 [118] | Class Aα | ADRB1 | Dobutamine | Partial Agonist |
| 27 | 2Y02 [118] | Class Aα | ADRB1 | Carmoterol | Agonist |
| 28 | 2Y03 [118] | Class Aα | ADRB1 | Isoprenaline | Agonist |
| 29 | 2Y04 [118] | Class Aα | ADRB1 | Salbutamol | Partial Agonist |
| 30 | 2YCW [74] | Class Aα | ADRB1 | Carazolol | Antagonist |
| 31 | 2YCX [74] | Class Aα | ADRB1 | Cyanopindolol | Antagonist |
| 32 | 2YCY [74] | Class Aα | ADRB1 | Cyanopindolol | Antagonist |
| 33 | 2YCZ [74] | Class Aα | ADRB1 | Iodocyanopindolol | Antagonist |
| 34 | 3ZPQ [119] | Class Aα | ADRB1 | 4-(Piperazin-1- yl)-1H-indole | Antagonist |
| 35 | 3ZPR [119] | Class Aα | ADRB1 | 4-Methyl-2-(piperazin-1-yl)quinoline | Antagonist |
| 36 | 4AMI [120] | Class Aα | ADRB1 | Bucindolol | Agonist |
| 37 | 4AMJ [120] | Class Aα | ADRB1 | Carvedilol | Agonist |
| 38 | 4BVN [69] | Class Aα | ADRB1 | Cyanopindolol | Antagonist |
| 39 | 5A8E [122] | Class Aα | ADRB1 | 7-methylcyanopindolol | Agonist |
| 40 | 5F8U [123] | Class Aα | ADRB1 | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile | Antagonist |
| 41 | 2RH1 [26] | Class Aα | ADRB2 | Carazolol | Antagonist |
| 42 | 3D4S [124] | Class Aα | ADRB2 | Timolol maleate | Antagonist |
| 43 | 3NY8 [126] | Class Aα | ADRB2 | ICI 118,551 | Antagonist |
| 44 | 3NY9 [126] | Class Aα | ADRB2 | Ethyl 4-({(2S)-2-hydroxy-3-[(1-methylethyl)amino]propyl}oxy)- 3-methyl-1-benzofuran-2-carboxylate | Antagonist |
| 45 | 3NYA [126] | Class Aα | ADRB2 | Alprenolol | Antagonist |
| 46 | 3P0G [127] | Class Aα | ADRB2 | BI 167107 | Agonist |
| 47 | 3PDS [128] | Class Aα | ADRB2 | FAUC50 | Agonist |
| 48 | 3SN6 [18] | Class Aα | ADRB2 | BI 167107 | Agonist |
| 49 | 4GBR [129] | Class Aα | ADRB2 | BI 167107 | Agonist |
| 50 | 4LDE [130] | Class Aα | ADRB2 | BI 167107 | Agonist |
| 51 | 4LDL [130] | Class Aα | ADRB2 | Hydroxybenzyl isoproterenol | Agonist |
| 52 | 4LDO [130] | Class Aα | ADRB2 | Epinephrine | Agonist |
| 53 | 4QKX [131] | Class Aα | ADRB2 | FAUC37 | Agonst |
| 54 | 5D5A [132] | Class Aα | ADRB2 | Carazolol | Antagonist |
| 55 | 5D5B [132] | Class Aα | ADRB2 | Carazolol | Antagonist |
| 56 | 5D6L [133] | Class Aα | ADRB2 | Carazolol | Antagonist |
| 57 | 5JQH [134] | Class Aα | ADRB2 | Carazolol | Antagonist |
| 58 | 3RZE [135] | Class Aα | H1R | Doxepin (E/Z) | Antagonist |
| 59 | 3PBL [6] | Class Aα | D3R | Eticlopride | Antagonist |
| 60 | 4IAQ [136] | Class Aα | 5-HT1B | Dihydroergotamine | Agonist |
| 61 | 4IAR [136] | Class Aα | 5-HT1B | Ergotamine | Agonist |
| 62 | 4IB4 [137] | Class Aα | 5-HT2B | Ergotamine | Agonist |
| 63 | 4NC3 [138] | Class Aα | 5-HT2B | Ergotamine | Agonist |
| 64 | 5TVN [139] | Class Aα | 5-HT2B | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline- 8-carboxamide | Agonist |
| 65 | 5CXV [140] | Class Aα | M1R | Tiotropium | Antagonist |
| 66 | 3UON [141] | Class Aα | M2R | 3-quinuclidinyl-benzilate | Antagonist |
| 67 | 4MQS [142] | Class Aα | M2R | Iperoxo | Agonist |
| 68 | 4MQT [142] | Class Aα | M2R | Iperoxo/LY2119620 | Agonist/Allosteric modulator |
| 69 | 4DAJ [143] | Class Aα | M3R | Tiotropium | Antagonist |
| 70 | 4U14 [144] | Class Aα | M3R | Tiotropium | Antagonist |
| 71 | 4U15 [144] | Class Aα | M3R | Tiotropium | Antagonist |
| 72 | 4U16 [144] | Class Aα | M3R | N-methyl scopolamine | Antagonist |
| 73 | 5DSG [140] | Class Aα | M4R | Tiotropium | Antagonist |
| 74 | 5UEN [145] | Class Aα | A1AR | DU172 | Antagonist |
| 75 | 2YDO [147] | Class Aα | A2AR | Adensoine | Agonist |
| 76 | 2YDV [147] | Class Aα | A2AR | NECA | Agonist |
| 77 | 3EML [37] | Class Aα | A2AR | ZM241385 | Antagonist |
| 78 | 3PWH [146] | Class Aα | A2AR | ZM241385 | Antagonist |
| 79 | 3QAK [68] | Class Aα | A2AR | UK-432097 | Agonist |
| 80 | 3REY [146] | Class Aα | A2AR | XAC | Antagonist |
| 81 | 3RFM [146] | Class Aα | A2AR | Caffeine | Antagonist |
| 82 | 3UZA [149] | Class Aα | A2AR | 6-(2,6-Dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine | Antagonist |
| 83 | 3UZC [149] | Class Aα | A2AR | 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol | Antagonist |
| 84 | 3VG9 [148] | Class Aα | A2AR | ZM241385 | Antagonist |
| 85 | 3VGA [148] | Class Aα | A2AR | ZM241385 | Antagonist |
| 86 | 4EIY [66] | Class Aα | A2AR | ZM241385 | Antagonist |
| 87 | 4UG2 [150] | Class Aα | A2AR | CGS21680 | Agonist |
| 88 | 4UHR [150] | Class Aα | A2AR | CGS21680 | Agonist |
| 89 | 5G53 [153] | Class Aα | A2AR | DB03719 | Agonist |
| 90 | 5IU4 [151] | Class Aα | A2AR | ZM241385 | Antagonist |
| 91 | 5IU7 [151] | Class Aα | A2AR | Compound 12c | Antagonist |
| 92 | 5IU8 [151] | Class Aα | A2AR | Compound 12f | Antagonist |
| 93 | 5IUA [151] | Class Aα | A2AR | Compound 12b | Antagonist |
| 94 | 5IUB [151] | Class Aα | A2AR | Compound 12x | Antagonist |
| 95 | 5K2A [152] | Class Aα | A2AR | ZM241385 | Antagonist |
| 96 | 5K2B [152] | Class Aα | A2AR | ZM241385 | Antagonist |
| 97 | 5K2C [152] | Class Aα | A2AR | ZM241385 | Antagonist |
| 98 | 5K2D [152] | Class Aα | A2AR | ZM241385 | Antagonist |
| 99 | 5UIG [154] | Class Aα | A2AR | 5-amino-N-[(2-methoxyphenyl)methyl]-2-(3- methylphenyl)-2H-1,2,3-triazole-4-carboximidamide | Angatonist |
| 100 | 3V2W [155] | Class Aα | S1P1 | ML056 | Antagonist |
| 101 | 3V2Y [155] | Class Aα | S1P1 | ML056 | Antagonist |
| 102 | 4Z34 [156] | Class Aα | LPA1 | ONO9780307 | Antagonist |
| 103 | 4Z35 [156] | Class Aα | LPA1 | ONO-9910539 | Antagonist |
| 104 | 4Z36 [156] | Class Aα | LPA1 | ONO-3080573 | Antagonist |
| 105 | 5TGZ [157] | Class Aα | CB1 | AM6538 | Antagonist |
| 106 | 5U09 [158] | Class Aα | CB1 | Taranabant | Antagonist |
| 107 | 3ZEV [160] | Class Aβ | NTSR1 | Neurotensin | Agonist |
| 108 | 4BUO [160] | Class Aβ | NTSR1 | Neurotensin | Agonist |
| 109 | 4BV0 [160] | Class Aβ | NTSR1 | Neurotensin | Agonist |
| 110 | 4BWB [160] | Class Aβ | NTSR1 | Neurotensin | Agonist |
| 111 | 4GRV [159] | Class Aβ | NTSR1 | Neurotensin | Agonist |
| 112 | 4XEE [161] | Class Aβ | NTSR1 | Neurotensin | Agonist |
| 113 | 4XES [161] | Class Aβ | NTSR1 | Neurotensin | Agonist |
| 114 | 5T04 [162] | Class Aβ | NTSR1 | Neurotensin | Agonist |
| 115 | 4ZJ8 [41] | Class Aβ | OX1 | Suvorexant | Dual Antagonist |
| 116 | 4ZJC [41] | Class Aβ | OX1 | SB-674042 | Antagonist |
| 117 | 4S0V [163] | Class Aβ | OX2 | Suvorexant | Dual Antagonist |
| 118 | 5GLH [165] | Class Aβ | ET-B | Endothelin-1 | Agonist |
| 119 | 5T1A [167] | Class Aγ | CCR2 | BMS-681/CCR2-RA-[R] | Orthosteric/allosteric antagonist |
| 120 | 3ODU [168] | Class Aγ | CXCR4 | IT1t | Antagonist |
| 121 | 3OE0 [168] | Class Aγ | CXCR4 | CVX15 | Antagonist |
| 122 | 3OE6 [168] | Class Aγ | CXCR4 | IT1t | Antagonist |
| 123 | 3OE8 [168] | Class Aγ | CXCR4 | IT1t | Antagonist |
| 124 | 3OE9 [168] | Class Aγ | CXCR4 | IT1t | Antagonist |
| 125 | 4MBS [170] | Class Aγ | CCR5 | Maraviroc | Antagonist |
| 126 | 5LWE [171] | Class Aγ | CCR9 | Vercirnon | Antagonist |
| 127 | 4EA3 [39] | Class Aγ | NOP | Banyu Compound-24 (C-24) | Antagonist |
| 128 | 5DHG [172] | Class Aγ | NOP | Compound-35 (C-35) | Antagonist |
| 129 | 5DHH [172] | Class Aγ | NOP | SB-612111 | Antagonist |
| 130 | 4DJH [173] | Class Aγ | κ-OR | JDTic | Antagonist |
| 131 | 4DKL [174] | Class Aγ | μ-OR | Beta-FNA | Antagonist |
| 132 | 5C1M [175] | Class Aγ | μ-OR | BU72 | Agonist |
| 133 | 4EJ4 [176] | Class Aγ | δ-OR | Naltrindole | Antagonist |
| 134 | 4N6H [70] | Class Aγ | δ-OR | Naltrindole | Antagonist |
| 135 | 4RWA [177] | Class Aγ | δ-OR | Bifunctional Peptide H-Dmt(1)-Tic(2)-Phe(3)-Phe(4)-NH2 (DIPP-NH2) | Antagonist |
| 136 | 4RWD [177] | Class Aγ | δ-OR | Bifunctional Peptide H-Dmt(1)-Tic(2)-Phe(3)-Phe(4)-NH2 (DIPP-NH2) | Antagonist |
| 137 | 4YAY [178] | Class Aγ | AT1R | ZD7155 | Antagonist |
| 138 | 4ZUD [179] | Class Aγ | AT1R | Olmesartan | Inverse Agonist |
| 139 | 5UNF [180] | Class Aγ | AT2R | N-benzyl-N-(2-ethyl-4-oxo-3-{[2′-(2H-tetrazol- 5-yl)[1,1′-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin- 6-yl)thiophene-2-carboxamide | |
| 140 | 5UNG [180] | Class Aγ | AT2R | N-benzyl-N-(2-ethyl-4-oxo-3-{[2′-(2H-tetrazol- 5-yl)[1,1′-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin- 6-yl)thiophene-2-carboxamide | |
| 141 | 5UNH [180] | Class Aγ | AT2R | N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl- 3-{[2′-(2H-tetrazol-5-yl)[1,1′-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide | |
| 142 | 4XNW [182] | Class Aδ | P2Y1 | MRS2500 | Antagonist |
| 143 | 4XNV [182] | Class Aδ | P2Y1 | BPTU | Antagonist |
| 144 | 4NTJ [42] | Class Aδ | P2Y12 | AZD1283 | Antagonist |
| 145 | 4PXZ [183] | Class Aδ | P2Y12 | 2MeSADP | Agonist |
| 146 | 4PY0 [183] | Class Aδ | P2Y12 | 2MeSADP | Agonist |
| 147 | 3VW7 [71] | Class Aδ | PAR1 | Vorapaxar | Antagonist |
| 148 | 5EE7 [78] | Class B | GCGR | MK-0893 | Antagonist |
| 149 | 4K5Y [186] | Class B | CRF1R | CP-376395 | Antagonist |
| 150 | 4Z9G [187] | Class B | CRF1R | CP-376395 | Antagonist |
| 151 | 4OR2 [188] | Class C | mGlu1 | FITM | Negative Allosteric Modulator |
| 152 | 4OO9 [38] | Class C | mGlu5 | Mavoglurant | Negative Allosteric Modulator |
| 153 | 5CGC [189] | Class C | mGlu5 | HTL14242 | Antagonist |
| 154 | 5CGD [189] | Class C | mGlu5 | HTL14242 | Antagonist |
| 155 | 4JKV [40] | Class F | SMO | LY2940680 | Antagonist |
| 156 | 4N4W [190] | Class F | SMO | SANT-1 | Antagonist |
| 157 | 4O9R [131] | Class F | SMO | Cyclopamine | Antagonist |
| 158 | 4QIM [190] | Class F | SMO | Anta XV | Antagonist |
| 159 | 4QIN [190] | Class F | SMO | SAG1.5 | Agonist |
| 160 | 5L7D [93] | Class F | SMO | Cholesterol | |
| 161 | 5L7I [93] | Class F | SMO | Vismodegib | Antagonist |
Phylogenetic classification/structure
Prior to the availability of three-dimensional structural information, GPCR classification methods initially utilized primary amino acid sequences to phylogenetically categorize receptors. This approach ultimately laid the foundation for the most-commonly used GPCR classification system. The sequences of seven hydrophobic regions (represented as cylinders in figure 1A) were used to design a fingerprinting method to identify sequences not previously categorized as rhodopsin-like receptors [47]. In the method, an individual conserved hydrophobic region served as a single “feature.” Multiple features grouped together comprised a signature “fingerprint” within a sequence. A feature, when used by a scanning algorithm to search a database, is then referred to as a “discriminator” and fingerprints are referred to as “composite discriminators. The search results using the discriminators generate an output hit list for each hydrophobic region. A hit list correlation was used to differentiate between true members, where all features of the fingerprints matched, and noise, where zero, one or two random matches occur. A second database was built from the sequences resulting from the previous search and scans were repeated with the new discriminators. This process was repeated until the composite discriminator continuously distinguished between true members and noise. As a result, 240 sequences were identified as members of the superfamily. Sequences for pheromone receptors did not match any of the discriminators and cAMP receptors exhibited only two matches, thus falling within the noise [47]. These sequences were previously identified as GPCR [48] suggesting they were members of distantly related families [47]. The hit-list fingerprint correlation was later expanded to distinguish partial matches. Upon this expansion, sequences belonging the GPCR superfamily increased from 240 to 393. In addition, the pheromone, cAMP, and secretin-like families were established as rhodopsin-like receptors [49].
The rhodopsin-like family rapidly became overly complex as more diverse receptors were discovered, leading to the establishment of the GPCR superfamily and formation of the ‘class’ system. This A-F system includes GPCR found in both vertebrates and invertebrates [3]. In 2001, the first draft of the human genome became available [1, 2] and allowed further GPCR to be classified using the most prevalent classification system with most of the human GPCR categorized into five families shown in italics: Glutamate (class C), Rhodopsin (class A), Adhesion (class B), Frizzled/Taste2 (class F), and Secretin (class B); also recognized as GRAFS [3].
Rhodopsin (Class A)
The Rhodopsin (class A) family, the largest of the GPCR classes [3], is divided into α, β, γ, and δ subgroups [5]. The α-subgroup is the largest group in class A and is comprised of the prostaglandin, amine, opsin, melatonin, and MECA receptor clusters [3]. Generally, ligands bind to these receptors within a pocket inside the transmembrane cavity involving residues contained in TM3, TM5, and TM6 [50–53]. An example of this target binding domain can be found in the ADRB2 structure in complex with carazolol (PDB: 2RH1) [26]. Biogenic monoamine receptors, such as the adrenergic; cannabinoid; muscarinic; serotonin; dopamine; and histamine receptors, are important drug targets for cardiovascular drugs, antipsychotics, and anti-histamines [54, 55]. Novel drugs that lack receptor specificity for particular amine receptors pose a risk for cardiovascular side effects due to off-target interactions with adrenergic receptors that show expression in many tissues [56]. Since the α-subgroup receptors outside the amine receptor subset are divergent from the amine receptors, side effects of novel aminergic drugs are most likely to occur only through the amine subset [5].
The β-subgroup of the Rhodopsin (class A) family has no branching subgroups and includes the hypocretin receptor, neuropeptide FF receptor, tachykinin receptor, cholecystokinin receptor, neuropeptide Y receptor, endothelin-related peptide receptor, gastrin-releasing peptide receptor, neuromedin receptor, thyrotropin releasing hormone receptor, ghrelin receptor, arginine vasopressin/oxytocin receptor, gonadotropin-releasing hormone receptor, and orphan receptors. Orphan receptors have been established as GPCR based on DNA sequence but have unidentified endogenous ligands [57]. Members of the β-subgroup mainly bind peptides with a high specificity-binding profile and are pursued as drug targets for treatments including pulmonary arterial hypertension [58] and hormone-related cancer [59]. Agonist drug design for β-subgroup members has been a challenge since it is difficult to design novel ligands that are flexible enough to mimic the magnitude of interaction sites engaged by endogenous peptide agonists [3].
The γ-subgroup of Rhodopsin (class A) contains the SOG, MCH, and chemokine receptor clusters, which bind both peptide and small ligand-like compounds. The SOG receptor cluster members include GPR54, the somatostatin receptors (SSTRs), and the opioid receptors [3]. The opioid receptors are important drug targets for the treatment of pain, cough, and alcoholism [54]. The MCH cluster includes receptors that branch off the SOG cluster. These receptors bind melanin-concentrating hormone (MCH), whereas the chemokine receptor cluster contains the chemokine receptors, the angiotensin/bradykinin-related receptors, and additional orphan receptors. Most ligands that interact with these receptors are peptides such as the chemokines and angiotensin. The chemokine receptors are drug targets due to their roles in acute and chronic inflammation [3] and as co-receptors engaged by some HIV strains [60]. While there are chemokine receptor-targeting drugs in clinical stages, maraviroc, an antagonist for CCR5, is the only currently FDA-approved drug on the market [61] targeting this class.
The δ-subgroup of Rhodopsin (class A) includes four main groups, the MAS-related receptor, glycoprotein receptor, purine receptor, and the olfactory receptor clusters. The MAS-receptor cluster includes the MAS1 oncogene receptor and the MAS-related receptors. The glycoprotein receptor cluster contains the classic glycoprotein hormone receptors and the leucine-rich-repeat-containing G protein-coupled receptors, whereas the purine receptor cluster is the largest in the δ-subgroup made up of the formyl peptide receptors, the nucleotide receptors, and a number of orphan receptors [3].
Extracellular region
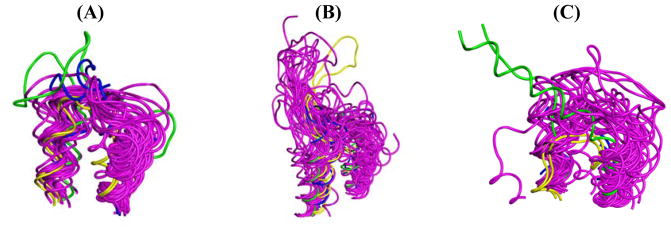
The extracellular region of GPCR contains the N-terminus and three loops (ECL1-3) that shape the opening to the ligand-binding pocket. The ligand-binding region is found within the extracellular region of the 7TM bundle. EL2 (figure 5B) is known to vary in length between GPCR classes resulting in distinct conformations, while EL1 (figure 5A) and EL3 (figure 5C) are short and often have disordered structures [62]. A highly conserved disulfide bond between EL2 and C3.25 (see description of the Ballesteros-Weinstein numbering system in the 7TM region section) has been observed in a majority of crystallized GPCR structures. This covalent modification limits movement and stabilizes the conformation of EL2 [62, 63]. Compared to other classes, class A receptors have a shorter N-terminus [63].
Figure 5.
Superpositions of 31 class A (fuchsia), 2 class B (blue), 2 class C (yellow), and 1 class F (green) extracellular loops that are connected to the first two membrane-embedded helical turns are shown in ribbon structures where structures chosen did not have an N-terminal fusion partner. (A) EL1 and (C) EL3 are short in classes A through C and form mainly disordered structures that are mostly conserved among all classes. (B) EL2 is the longest of the EL, and varies in length and conformation in different GPCR. Class A GPCR: Rho (1F88), ADRB1 (2VT4), ADRB2 (2R4R), H1R (3RZE), D3R (3PBL), 5-HT1B (4IAQ), 5-HT2B (4IB4), M1R (5CXV), M2R (3UON), M3R (4DAJ), M4R (5DSG), A2AR (3EML), S1P1 (3V2W), LPA1 (4Z32), CB1 (5TGZ), NTSR1 (3ZEV), OX1 (4ZJ8), OX2 (4S0V), TACR1 (2KSP), CXCR1 (2LNL), CXCR4 (3ODU), CCR5 (4MBS), CCR9 (5LWE), κ-OR (4DJH), μ-OR (4DKL), δ-OR (4EJ4), HHV-5 US28 (4XT1), P2Y1 (4XNW), P2Y12 (4NTJ), PAR1 (3VW7), FFAR1 (4PHU). Class B GPCR: GCGR (5EE7), CRF1R (4K5Y). Class C GPCR: mGlu1 (4OR2), mGlu5 (4OO9). Class F GPCR: SMO (4QIN).
7TM region
Despite differences in primary structure, a majority of the proteins in class A/Rhodopsin share conserved residues found within the 7TM domain [5]. The Ballesteros-Weinstein numbering system is often used to assign numbers to common residues that are conserved in both sequence and structure-based alignments of different receptors [64]. This indexing system consists of two numbers separated by a period. The first value represents the TM helix in which the residue is found and has values ranging from 1 to 7. The second number denotes the residue position relative to the most conserved residue within that TM segment, which is defined as position 50. The value decreases as you move through the amino acid sequence toward the N-terminus and increases as you move though the amino acid sequence toward the C-terminus. Class A/Rhodopsin receptors have highly conserved residues in each TM segment: N1.50, D2.50, R3.50, W4.50, P5.50, P6.50, and P7.50 [64]. In addition to these residues, class A/Rhodopsin contains two fingerprint regions: the D/ERY motif at positions 3.49–3.51 and the NPXXY motif at positions 7.49–7.53 [3]. In 2000, x-ray crystallography studies on rhodopsin (PDB: 1F88) confirmed that conserved residues formed interhelical networks important for stabilization and activation [25] as had previously been suggested [65]. Seven years later, two structures of ADRB2 (PDB: 2RH1, 2R4R/2R4S) were determined and revealed TM structural similarity to rhodopsin [26, 27]. The basic canonical architecture of this region has now been observed in numerous examples of crystallized GPCR as is shown in figure 4. It is interesting to note that an allosteric sodium ion binding pocket involving two conserved residues D2.50 and S3.39 was identified in the 1.8Å crystal structure of A2A (PDB: 4EIY) [66, 67]. This central Na+ pocket was formed by D2.50, S3.39, and three water molecules. Liu, et al., compared their inactive A2A structure to an active A2A (PDB 3QAK [68]) structure and found that the size of the central pocket in the active form could only support three water molecules without sufficient room for Na+ coordination. This suggests that Na+ may stabilize the inactive conformation of A2A [66]. After the discovery of the Na+ pocket in A2A, the same characteristic was seen in other inactive GPCR crystal structures. These other structures include representatives of the α, γ, and δ subgroups of class A [ADRB1 (PDB: 4BVN [69]), delta-opioid (δ-OR; PDB: 4N6H [70]), and protease-activated receptor 1 (PAR1; PDB: 3VW7 [71])], suggesting the sodium ion binding pocket is a common feature in class A GPCR.
Conformational changes in the 7TM bundle are required for signal transduction across the cell membrane following ligand binding. Experimentally determined crystal structures of ADRB2 reflect a variety of activation states. In these structures, TM5, TM6, and TM7 have a critical function in GPCR activation due to clear differences in helical arrangement. The active state of ADRB2 (PDB: 3P0G) exhibits altered conformations of TM5 and TM7 and a prominent outward shift of TM6 [18] in contrast to the inactive state (PDB: 2RH1) [26].
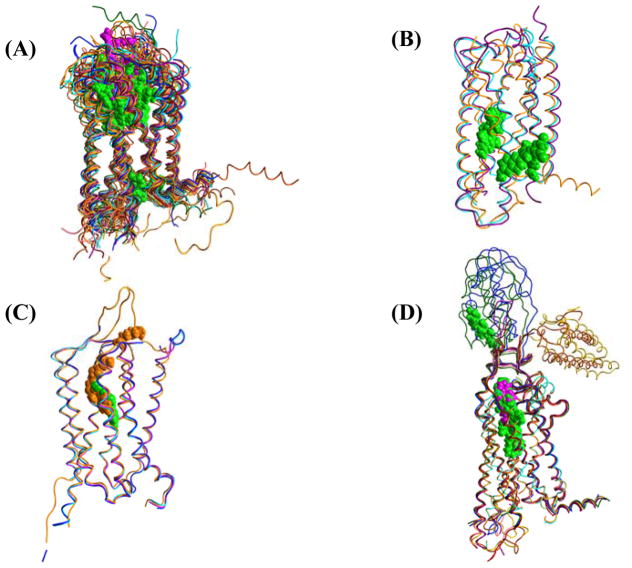
The 7TM region forms distinctive ligand-binding pockets for different receptors where varying size, shape, and electrostatics provide receptor-ligand selectivity. Aminergic and nucleotide receptors have small binding pockets buried inside the 7TM bundle while peptide receptors have large, more accessible binding pockets near the extracellular surface [62]. These diverse ligand binding pocket profiles are demonstrated with 29 class A GPCR in figure 6A.
Figure 6.
Superpositions of GPCR represented as thin ribbon structures with ligands as space-filling atoms colored accordingly by antagonists as green, agonists as fuchsia, and negative allosteric modulators as orange. (A) Structures of 29 individual class A GPCR. (B) Structures of 3 class B GPCR. (C) Structures of 4 class C GPCR. (D) Structures of 7 class F GPCR. Class A GPCR: Rho (1F88), ADRB1 (2VT4), ADRB2 (2RH1), H1R (3RZE), D3R (3PBL), 5-HT1B (4IAQ), 5-HT2B (4IB4), M1R (5CXV), M2R (3UON), M3R (4DAJ), M4R (5DSG), A2AR (2YDO), S1P1 (3V2W), LPA1 (4Z34), CB1 (5TGZ), NTSR1 (3ZEV), OX1 (4ZJ8), OX2 (4S0V), CCR4 (3ODU), CCR5 (4MBS), CCR9 (5LWE), NOP (4EA3), κ-OR (4DJH), μ-OR (4DKL), δ-OR (4EJ4), AT1R (4YAY), P2Y1 (4XNW), P2Y12 (4NTJ), PAR1 (3VW7). Class B GPCR: GCGR (5EE7), CRF1R (4K5Y, 4Z9G). Class C GPCR: mGlu1 (4OR2, 5CGC, 5CGD), mGlu5 (4OO9) Class F GPCR: SMO (4JKV, 4N4W, 4OR9, 4QIM, 4QIN, 5L7D, 5L7I).
Intracellular region
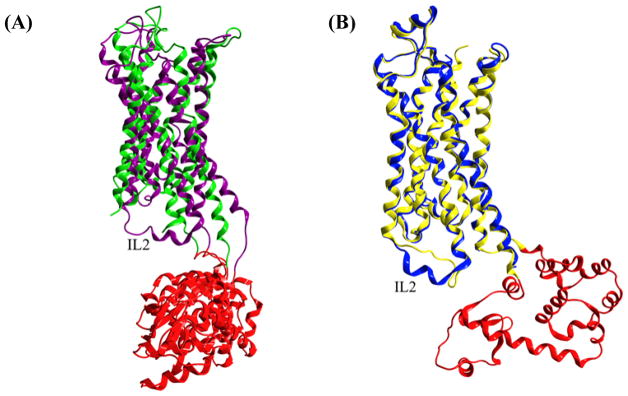
The intracellular region of GPCR includes the C-terminus and three loops (ICL1-3) that interact with G proteins, β-arrestins, and other downstream effectors [4, 10, 62]. GPCR crystal structures have shown structural conservation in the short IL1 chain, though high levels of variability have been observed within IL2 and IL3 suggesting dynamic and/or unstable conformations. Differences have been observed in IL2 in both the D3R and ADRB receptor structures. In a crystal structure of D3R (PDB: 3PBL) [6], where there were two copies of the protein from the same unit cell, IL2 had 2.5 turns of an α-helical conformation for chain A in contrast to a disordered loop lacking electron density for chain B (figure 7A). ADRB1 [72] and ADRB2 [26], which have an overall percent identity of 80% and nearly identical IL2 sequences, have displayed an α-helical conformation in ADRB1 and unstructured conformation in ADRB2 (figure 7B; PDB: ADRB1 – 2VT4, ADRB2 – 2RH1). The three dimensional structure of IL2 may be dependent upon the functional state of the protein, as well as interactions with intracellular partners. IL3 exhibits the greatest length variability amongst IL, ranging from five to hundreds of residues and has been implicated in G protein selectivity. IL3 251664384251663360 has been observed to form a disordered conformation or, more often, has been replaced by a fusion partner for increased conformational homogeneity to enable crystal formation in several solved GPCR structures [10]. There are crystallographic structures of rhodopsin (PDB: 3CAP) [73], ADRB1 (PDB: 2YCW, 2YCX, 2YCY, 2YCZ) [74], and δ-opioid receptor (PDB: 4N6H) [70] where IL3 adopts an α-helical conformation resulting in extended TM5 and TM6 helices [10, 62]. The IL region undergoes considerable conformational changes required for G protein interaction and the consequent initiation of the signal transduction cascade.
Figure 7.
(A) Superposition of D3R crystal structures (PBD: 3PBL) showing a 2.5 helical turn for chain A (purple) and no electron density for chain B (green) for IL2. The T4 lysozyme fusion partner replacing IL3 is shown in red for both chains. (B) Superposition of crystal structures comparing ADRB1 (PDB: 2VT4 chain B; blue) and ADRB2 (PDB: 2RH1 chain A; yellow and T4 lysozyme in red) where IL2 exhibits an ordered structure in ADRB1 and a disordered structure in ADRB2.
Secretin and Adhesion (Class B)
Class B, the second largest class within the Rhodopsin family, is comprised of the Secretin and Adhesion families. Secretin and Adhesion receptors have been classified together due to sequence similarities between their 7TM regions, although they have distinctions elsewhere that establish them as separate families. The Secretin family has an extracellular hormone-binding domain that interacts with peptide hormones [5]. The members of this family include the calcitonin/calcitonin-like receptors, the corticotropin-releasing hormone receptors, the glucagon receptor, the gastric inhibitory polypeptide receptor, the glucagon-like peptide receptors, the growth-hormone-releasing hormone receptor, the adenylyl cyclase activating polypeptide hormone receptor, the parathyroid hormone receptors, the secretin receptor, the vasoactive intestinal peptide receptors, and additional orphan receptors [3, 5]. The Secretin family includes potential targets for drug development due to their involvement in central homeostatic functions. Members of this family have been connected to appetite regulation and type-2 diabetes [5].
The Adhesion family includes the epidermal growth factor receptors and lectomedin receptors, though a majority of the members are currently classified as orphan receptors [5]. This GPCR family exhibits a highly variable number of amino acids at the N-terminal region, ranging from 200–2800 in length, making this family phylogenetically and structurally different from the rest of the class B GPCR. The “Adhesion” family name is related to the N-terminal region that contains sequence motifs, such as the GPCR proteolysis (GPS) motif, which serve as intracellular autocatalytic processing sites that participate in cellular adhesion [3, 5]. Adhesion family receptors bind extracellular matrix molecules rather than peptide hormones [5]. These receptors are believed to be involved in cell proliferation/migration, as well as immune system function via the mediation of leukocyte and neutrophil interactions. Adhesion receptors that contain long N-termini have become targets of monoclonal antibodies used as drugs candidates [5]. Numerous receptors from this family are localized in the central nervous system (CNS) [75] though their functional role in the CNS is not fully understood [5].
Structure
Within class B, sequence alignments show that Adhesion and Secretin families contain structural differences at the N-terminal region. Adhesion receptors contain distinctive O- and N-glycosylation sites, as well as EGF and GPS motifs [76]. Secretin family members have a 60–80 amino acid N-terminal domain [3] that include a hormone-binding (HRM) domain that is believed to have a key role in binding peptide hormones [63]. The Ballesteros-Weinstein numbering system somewhat extends to class B/Adhesion/Secretin where residues E3.50 and W4.50 are conserved [77]. The binding pockets of class B GPCR are broader and deeper inside the 7TM bundle compared to class A [45], as illustrated in figure 6B, in order to accommodate endogenous peptide ligands. On the other hand, the crystal structure for GCGR (PDB: 5EE7) shows an allosteric binding site outside of the 7TM domain between TM6 and TM7 [78].
Glutamate (Class C)
The Glutamate (class C) family of receptors consists of the metabotropic glutamate receptors, GABA receptors, single calcium-sensing receptors, and sweet and umami taste receptors. The known endogenous ligands of this family are known to bind to the N-terminal region, but many allosteric ligands have been discovered to interact with TM3, TM5, TM6, and TM7 [79–81]. In addition, Ca2+ can bind to the extracellular region [82] and enhance the effects of glutamate in some glutamate receptors [83, 84]. Many of the Ca2+ interacting residues are conserved within the “Venus flytrap” region of the Glutamate (class C) receptors [82] and have potential significance in drug design targeting depression learning, and memory [85].
Structure
In class C/Glutamate receptors, the 280–580 amino acid N-terminus [3] forms a cavity surrounded by two lobes which close upon ligand binding through a process known as the “Venus flytrap” mechanism (VFTM) [5, 86]. This mechanism involves a ligand binding-induced conformational change that results in the formation of disulfide bonds between the N-terminus and 7TM domain [63, 87]. Receptors in this class lack the conserved residues defined by the Ballesteros-Weinstein numbering scheme seen extensively in Class A and to a lesser extent in Class B GPCR. Although the conserved ligand binding pocket of most class C receptors is located within the extracellular region, there are allosteric binding sites within the 7TM bundle [5] as seen in figure 6C. These allosteric binding sites may be a way to achieve receptor specificity using allosteric modulators.
Frizzled/Taste2 (Class F)
The Frizzled/Taste2 (class F) group of receptors includes frizzled and smoothened receptors (SMO) [3, 5]. The frizzled receptors are known to bind secreted glycoproteins [88] while the SMO receptor functions in a ligand-independent manner through the SMO and sonic hedgehog (SHH) complex [89]. Frizzled receptors are involved in cell fate, proliferation, and polarity through association with secreted glycoproteins at the cysteine-rich N-terminus [3, 5]. Although the cysteine-rich region is highly conserved in the frizzled receptor, there is evidence that there are additional binding sites located in the extracellular loops of the TM regions [90]. SMO receptors are structurally similar to frizzled receptors [5]. The cysteine-rich region found in the frizzled receptors, as well as the chemical properties of residues that bind secreted glycoproteins, are conserved in SMO [90, 91]. Class F GPCR, though not well understood, have been linked to cancer development and are potential targets for cancer therapy [5].
Structure
Based on sequence analysis, class F/Frizzled/Taste2 is the most highly conserved class within the GPCR superfamily [63]. These proteins have a distinctive N-terminus that spans 200–320 amino acids in length [3, 5], in addition to a variable linker region between the EL and TM domains [5]. While the Ballesteros-Weinstein numbering scheme does not apply to this class, these proteins still share the common structure of the 7TM hydrophobic core [92]. There is limited structural information about this class of receptors since SMO is the only class F GPCR to be crystallized to date. The binding pocket is observed to be narrow and elongated in the currently available crystal structures (figure 6D). In one SMO crystal structure (PDB: 5L7D), cholesterol is bound to the extracellular cysteine-rich domain (CRD) that is highly conserved in vertebrates. An oxysterol was observed to displace cholesterol and bind to the CRD groove leading to SMO activation and Hedgehog (Hh) signaling. Cholesterol has been proposed as an endogenous ligand that stabilizes the inactive SMO conformation [93].
Conclusions/further perspectives
Since the initial crystallization of rhodopsin in 2000, numerous technological advances have significantly impacted GPCR structure determination efforts, resulting in crystal structures of 42 individual receptors (to date). Currently > 180 structures of GPCR have been solved and made available through the Protein Data Bank (table 1). Of these, over 150 protein-ligand complexes are available (table 2), representing the entire spectrum of ligand functions from inverse agonist to agonist. These data sets give insights into structural features and ligand recognition within this diverse protein superfamily. Specifically, the majority of crystallized ligand complexes with GPCR exhibit a ligand binding pocket near the extracellular end of the TM helical bundle, as shown in figure 6. The TM helical bundle structure shows considerable structural similarity across the family (figure 4), while EL2 shows the greatest structural diversity in the vicinity of the ligand binding pocket (figure 5). Thus, differences in ligand selectivity between GPCR family members are driven both by sidechain differences in the TM helical bundle (rather than backbone conformational differences) as well as overall EL2 fold. GPCR have essential biological roles, and many have been confirmed to have value as therapeutic targets. Therefore solved, three-dimensional GPCR structures have tremendous potential to influence drug discovery. Structure-based drug discovery approaches can be applied directly to crystallized GPCR family members as therapeutic targets. Additionally, the crystallized GPCR structures exhibit close sequence identity to additional GPCR family members and can serve as templates for the development of reliable and predictive homology models. Validated models allow structure-based drug discovery approaches to be used against this even broader set of target GPCR members. Although efforts in GPCR crystal structure determination over the past two decades have been fruitful, vast amounts of work remain to characterize unrepresented family members that have lower sequence identities to currently crystallized GPCR family members. GPCR structural characterization will continue to be a rich research area in need of further advances and innovative approaches into the foreseeable future.
Acknowledgments
Research reported in this publication was supported by the National Institute of Mental Health of the National Institutes of Health under Award Number R15MH109034. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- A1AR
Adenosine A1A receptor
- A2AR
adenosine A2A receptor
- ADRB1
adrenergic β1 receptor
- ADRB2
adrenergic β2 receptor
- AT1R
angiotensin II receptor type 1
- AT2R
angiotensin II receptor type 2
- CB1
cannabinoid receptor 1
- CCR2
chemokine receptor 2
- CXCR4
chemokine receptor 4
- CCR5
chemokine receptor 5
- CCR9
chemokine receptor 9
- CRF1R
corticotropin-releasing factor 1
- δ-OR
δ-opioid receptor
- D3R
dopamine D3 Receptor
- ET-B
endothelin receptor type-B
- CXCR1
chemokine receptor 1
- EL
extracellular loop
- FFAR1
free fatty acid receptor 1
- GPCR
g protein-coupled receptors
- GCGR
glucagon receptor
- mGlu1
glutamate receptor 1
- mGlu5
glutamate receptor 5
- H1R
histamine H1 receptor
- κ-OR
κ-opioid receptor
- LPA1
lysophosphatidic acid receptor 1
- μ-OR
μ-opioid receptor
- M1R
muscarinic receptor 1
- M2R
muscarinic receptor 2
- M3R
muscarinic receptor 3
- M4R
muscarinic receptor 4
- NTSR1
neurotensin receptor 1
- NOP
nociceptin receptor
- OX1
orexin receptor 1
- OX2
orexin receptor 2
- PAR1
protease-activated receptor 1
- P2Y1
purinergic P2 receptor 1
- P2Y12
purinergic P2 receptor 12
- Rho
rhodopsin
- 5-HT1B
serotonin 5HT1B
- 5-HT2B
serotonin 5HT2B
- SMO
smoothened receptor
- S1P1
sphingosine 1-phosphate receptor 1
- TACR1
tachykinin receptor 1
- TM
transmembrane
- IL
intracellular loop
- HHV-5 US28
viral GPCR US28
References
- 1.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Fredriksson R, Lagerstrom MC, Lundin LG, et al. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 4.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 5.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 6.Chien EY, Liu W, Zhao Q, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Villar VA, Armando I, et al. G Protein-Coupled Receptor Kinases: Crucial Regulators of Blood Pressure. J Am Heart Assoc. 2016;5:10. doi: 10.1161/JAHA.116.003519. 1161/JAHA.116.003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bar-Shavit R, Maoz M, Kancharla A, et al. G Protein-Coupled Receptors in Cancer. Int J Mol Sci. 2016;17:1320. doi: 10.3390/ijms17081320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flower DR. Modelling G-protein-coupled receptors for drug design. Biochim Biophys Acta. 1999;1422:207–234. doi: 10.1016/s0304-4157(99)00006-4. [DOI] [PubMed] [Google Scholar]
- 10.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gether U, Ballesteros JA, Seifert R, et al. Structural instability of a constitutively active G protein-coupled receptor. Agonist-independent activation due to conformational flexibility. J Biol Chem. 1997;272:2587–2590. doi: 10.1074/jbc.272.5.2587. [DOI] [PubMed] [Google Scholar]
- 12.Seifert R, Wenzel-Seifert K. Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch Pharmacol. 2002;366:381–416. doi: 10.1007/s00210-002-0588-0. [DOI] [PubMed] [Google Scholar]
- 13.Nelson G, Hoon MA, Chandrashekar J, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 14.Nelson G, Chandrashekar J, Hoon MA, et al. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 15.Milligan G. G Protein-Coupled Receptor Dimerization: Function and Ligand Pharmacology. Mol Pharmacol. 2004:66. doi: 10.1124/mol.104.000497.. [DOI] [PubMed] [Google Scholar]
- 16.Syrovatkina V, Alegre KO, Dey R, et al. Regulation, Signaling, and Physiological Functions of G-Proteins. J Mol Biol. 2016;428:3850–3868. doi: 10.1016/j.jmb.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wess J. G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition. FASEB J. 1997;11:346–354. [PubMed] [Google Scholar]
- 18.Rasumussen SG, DeVree BT, Zou Y, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 20.Nie J, Lewis DL. Structural domains of the CB1 cannabinoid receptor that contribute to constitutive activity and G-protein sequestration. J Neurosci. 2001;21:8758–8764. doi: 10.1523/JNEUROSCI.21-22-08758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 23.Claing A, Laporte SA, Caron MG, et al. Endocytosis of G protein-coupled receptors: roles of G protein-coupled receptor kinases and beta-arrestin proteins. Prog Neurobiol. 2002;66:61–79. doi: 10.1016/s0301-0082(01)00023-5. [DOI] [PubMed] [Google Scholar]
- 24.Okada T, Le Trong I, Fox BA, et al. X-Ray diffraction analysis of three-dimensional crystals of bovine rhodopsin obtained from mixed micelles. J Struct Biol. 2000;130:73–80. doi: 10.1006/jsbi.1999.4209. [DOI] [PubMed] [Google Scholar]
- 25.Palczewski K, Kumasaka T, Hori T, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 26.Cherezov V, Rosenbaum DM, Hanson MA, et al. High Resolution Crystal Structure of an Engineered Human Beta2-Adrenergic G protein-Couple Receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen SG, Choi HJ, Rosenbaum DM, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 28.Stroud RM. New tools in membrane protein determination. F1000 Biol Rep. 2011;3:8–8. doi: 10.3410/B3-8. Epub 2011 Apr 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ujwal R, Bowie JU. Crystallizing membrane proteins using lipidic bicelles. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denisov IG, Sligar SG. Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol. 2016;23:481–486. doi: 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang J, Chun E, Liu C, et al. Successful Strategies to Determine High-Resolution Structures of GPCRs. Trends Pharmacol Sci. 2016;37:1055–1069. doi: 10.1016/j.tips.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Rawlings AE. Membrane proteins: always an insoluble problem? Biochem Soc Trans. 2016;44:790–795. doi: 10.1042/BST20160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 34.Delmar JA, Bolla JR, Su CC, et al. Crystallization of membrane proteins by vapor diffusion. Methods Enzymol. 2015;557:363–392. doi: 10.1016/bs.mie.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendrickson WA. Anomalous diffraction in crystallographic phase evaluation. Q Rev Biophys. 2014;47:49–93. doi: 10.1017/S0033583514000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor GL. Introduction to phasing. Acta Crystallogr D Biol Crystallogr. 2010;66:325–338. doi: 10.1107/S0907444910006694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaakola VP, Griffith MT, Hanson MA, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dore AS, Okrasa K, Patel JC, et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014;511:557–562. doi: 10.1038/nature13396. [DOI] [PubMed] [Google Scholar]
- 39.Thompson AA, Liu W, Chun E, et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485:395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Wu H, Katritch V, et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin J, Babaoglu K, Brautigam CA, et al. Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat Struct Mol Biol. 2016;23:293–299. doi: 10.1038/nsmb.3183. [DOI] [PubMed] [Google Scholar]
- 42.Zhang K, Zhang J, Gao Z, et al. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature. 2014:509. doi: 10.1038/nature13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isberg V, Mordalski S, Munk C, et al. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkv1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lundstrom K. Latest development in drug discovery on G protein-coupled receptors. Curr Protein Pept Sci. 2006;7:465–470. doi: 10.2174/138920306778559403. [DOI] [PubMed] [Google Scholar]
- 45.Shonberg J, Kling RC, Gmeiner P, et al. GPCR crystal structures: Medicinal chemistry in the pocket. Bioorg Med Chem. 2015;23:3880–3906. doi: 10.1016/j.bmc.2014.12.034. [DOI] [PubMed] [Google Scholar]
- 46.Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucl Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Attwood TK, Findlay JB. Design of a discriminating fingerprint for G-protein-coupled receptors. Protein Eng. 1993;6:167–176. doi: 10.1093/protein/6.2.167. [DOI] [PubMed] [Google Scholar]
- 48.Chee MS, Satchwell SC, Preddie E, et al. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature. 1990;344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- 49.Attwood TK, Findlay JB. Fingerprinting G-protein-coupled receptors. Protein Eng. 1994;7:195–203. doi: 10.1093/protein/7.2.195. [DOI] [PubMed] [Google Scholar]
- 50.Strader CD, Sigal IS, Dixon RA. Structural basis of beta-adrenergic receptor function. FASEB J. 1989;3:1825–1832. doi: 10.1096/fasebj.3.7.2541037. [DOI] [PubMed] [Google Scholar]
- 51.Liapakis G, Ballesteros JA, Papachristou S, et al. The forgotten serine. A critical role for Ser-2035.42 in ligand binding to and activation of the beta 2-adrenergic receptor. J Biol Chem. 2000;275:37779–37788. doi: 10.1074/jbc.M002092200. [DOI] [PubMed] [Google Scholar]
- 52.Swaminath G, Xiang Y, Lee TW, et al. Sequential binding of agonists to the beta2 adrenoceptor. Kinetic evidence for intermediate conformational states. J Biol Chem. 2004;279:686–691. doi: 10.1074/jbc.M310888200. [DOI] [PubMed] [Google Scholar]
- 53.Baldwin JM. Structure and function of receptors coupled to G proteins. Curr Opin Cell Biol. 1994;6:180–190. doi: 10.1016/0955-0674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 54.Tyndall JD, Sandilya R. GPCR agonists and antagonists in the clinic. Med Chem. 2005;1:405–421. doi: 10.2174/1573406054368675. [DOI] [PubMed] [Google Scholar]
- 55.Jacoby E, Bouhelal R, Gerspacher M, et al. The 7 TM G-protein-coupled receptor target family. ChemMedChem. 2006;1:761–782. doi: 10.1002/cmdc.200600134. [DOI] [PubMed] [Google Scholar]
- 56.Spiss CK, Maze M. Adrenoreceptors. Anaesthesist. 1985;34:1–10. [PubMed] [Google Scholar]
- 57.Civelli O, Reinscheid RK, Zhang Y, et al. G protein-coupled receptor deorphanizations. Annu Rev Pharmacol Toxicol. 2013;53:127–146. doi: 10.1146/annurev-pharmtox-010611-134548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barst RJ, Langleben D, Frost A, et al. Sitaxsentan therapy for pulmonary arterial hypertension. Am J Respir Crit Care Med. 2004;169:441–447. doi: 10.1164/rccm.200307-957OC. [DOI] [PubMed] [Google Scholar]
- 59.Kotake T, Usami M, Akaza H, et al. Goserelin acetate with or without antiandrogen or estrogen in the treatment of patients with advanced prostate cancer: a multicenter, randomized, controlled trial in Japan. Zoladex Study Group. Jpn J Clin Oncol. 1999;29:562–570. doi: 10.1093/jjco/29.11.562. [DOI] [PubMed] [Google Scholar]
- 60.Onuffer JJ, Horuk R. Chemokines, chemokine receptors and small-molecule antagonists: recent developments. Trends Pharmacol Sci. 2002;23:459–467. doi: 10.1016/s0165-6147(02)02064-3. [DOI] [PubMed] [Google Scholar]
- 61.Fatkenheuer G, Pozniak AL, Johnson MA, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 62.Lu M, Wu B. Structural studies of G protein-coupled receptors. IUBMB Life. 2016;68:894–903. doi: 10.1002/iub.1578. [DOI] [PubMed] [Google Scholar]
- 63.Strotmann R, Schrock K, Boselt I, et al. Evolution of GPCR: change and continuity. Mol Cell Endocrinol. 2011;331:170–178. doi: 10.1016/j.mce.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 64.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in Neurosciences. 1995;25:366–428. [Google Scholar]
- 65.Zhang D, Weinstein H. Polarity conserved positions in transmembrane domains of G-protein coupled receptors and bacteriorhodopsin. FEBS Lett. 1994;337:207–212. doi: 10.1016/0014-5793(94)80274-2. [DOI] [PubMed] [Google Scholar]
- 66.Liu W, Chun E, Thompson AA, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katritch V, Fenalti G, Abola EE, et al. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci. 2014;39:233–244. doi: 10.1016/j.tibs.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu F, Wu H, Katritch V, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller-Gallacher JL, Nehme R, Warne T, et al. The 2.1 A resolution structure of cyanopindolol-bound beta1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor. PLoS One. 2014;9:e92727. doi: 10.1371/journal.pone.0092727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fenalti G, Giguere PM, Katritch V, et al. Molecular Control of Delta-Opioid Receptor Signaling. Nature. 2014;506:191–196. doi: 10.1038/nature12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang C, Srinivasan Y, Arlow DH, et al. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012;492:387–392. doi: 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warne T, Serrano-Vega MJ, Baker JG, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park JH, Scheerer P, Hofmann KP, et al. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 74.Moukhametzianov R, Warne T, Edwards PC, et al. Two distinct conformations of helix 6 observed in antagonist-bound structures of a beta-1 adrenergic receptor. Proc Natl Acad Sci U S A. 2011;108:8228–8232. doi: 10.1073/pnas.1100185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bjarnadottir TK, Geirardsdottir K, Ingemansson M, et al. Identification of novel splice variants of Adhesion G protein-coupled receptors. Gene. 2007;387:38–48. doi: 10.1016/j.gene.2006.07.039. [DOI] [PubMed] [Google Scholar]
- 76.Lin HH, Chang GW, Davies JQ, et al. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 77.Isberg V, Vroling B, van der Kant R, et al. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 2014;42:D422–5. doi: 10.1093/nar/gkt1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jazayeri A, Dore AS, Lamb D, et al. Extra-helical binding site of a glucagon receptor antagonist. Nature. 2016;533:274–277. doi: 10.1038/nature17414. [DOI] [PubMed] [Google Scholar]
- 79.Gasparini F, Kuhn R, Pin JP. Allosteric modulators of group I metabotropic glutamate receptors: novel subtype-selective ligands and therapeutic perspectives. Curr Opin Pharmacol. 2002;2:43–49. doi: 10.1016/s1471-4892(01)00119-9. [DOI] [PubMed] [Google Scholar]
- 80.Malherbe P, Kratochwil N, Zenner MT, et al. Mutational analysis and molecular modeling of the binding pocket of the metabotropic glutamate 5 receptor negative modulator 2-methyl-6-(phenylethynyl)-pyridine. Mol Pharmacol. 2003;64:823–832. doi: 10.1124/mol.64.4.823. [DOI] [PubMed] [Google Scholar]
- 81.Litschig S, Gasparini F, Rueegg D, et al. CPCCOEt, a noncompetitive metabotropic glutamate receptor 1 antagonist, inhibits receptor signaling without affecting glutamate binding. Mol Pharmacol. 1999;55:453–461. [PubMed] [Google Scholar]
- 82.Silve C, Petrel C, Leroy C, et al. Delineating a Ca2+ binding pocket within the venus flytrap module of the human calcium-sensing receptor. J Biol Chem. 2005;280:37917–37923. doi: 10.1074/jbc.M506263200. [DOI] [PubMed] [Google Scholar]
- 83.Hermans E, Challiss RA. Structural, signalling and regulatory properties of the group I metabotropic glutamate receptors: prototypic family C G-protein-coupled receptors. Biochem J. 2001;359:465–484. doi: 10.1042/0264-6021:3590465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- 85.Riedel G, Platt B, Micheau J. Glutamate receptor function in learning and memory. Behav Brain Res. 2003;140:1–47. doi: 10.1016/s0166-4328(02)00272-3. [DOI] [PubMed] [Google Scholar]
- 86.Kunishima N, Shimada Y, Tsuji Y, et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 87.Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bhanot P, Brink M, Samos CH, et al. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 89.Murone M, Rosenthal A, de Sauvage FJ. Sonic hedgehog signaling by the patched-smoothened receptor complex. Curr Biol. 1999;9:76–84. doi: 10.1016/s0960-9822(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 90.Chen CM, Strapps W, Tomlinson A, et al. Evidence that the cysteine-rich domain of Drosophila Frizzled family receptors is dispensable for transducing Wingless. Proc Natl Acad Sci U S A. 2004;101:15961–15966. doi: 10.1073/pnas.0407103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nakano Y, Nystedt S, Shivdasani AA, et al. Functional domains and sub-cellular distribution of the Hedgehog transducing protein Smoothened in Drosophila. Mech Dev. 2004;121:507–518. doi: 10.1016/j.mod.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 92.Isberg V, de Graaf C, Bortolato A, et al. Generic GPCR Residue Numbers - Aligning Topology Maps Minding The Gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Byrne EF, Sircar R, Miller PS, et al. Structural basis of Smoothened regulation by its extracellular domains. Nature. 2016;535:517–522. doi: 10.1038/nature18934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Teller DC, Okada T, Behnke CA, et al. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yeagle PL, Choi G, Albert AD. Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms. Biochemistry. 2001;40:11932–11937. doi: 10.1021/bi015543f. [DOI] [PubMed] [Google Scholar]
- 96.Okada T, Fujiyoshi Y, Silow M, et al. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choi G, Landin J, Galan JF, et al. Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin. Biochemistry. 2002;41:7318–7324. doi: 10.1021/bi025507w. [DOI] [PubMed] [Google Scholar]
- 98.Li J, Edwards PC, Burghammer M, et al. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 99.Okada T, Sugihara M, Bondar AN, et al. The Retinal Conformation and its Environment in Rhodopsin in Light of a New 22 A Crystal Structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 100.Nakamichi H, Okada T. Crystallographic analysis of primary visual photochemistry. Angew Chem Int Ed Engl. 2006;45:4270–4273. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- 101.Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:12729–12734. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Salom D, Lodowski DT, Stenkamp RE, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Standfuss J, Xie G, Edwards PC, et al. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372:1179–1188. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakamichi H, Buss V, Okada T. Photoisomerization mechanism of rhodopsin and 9-cis-rhodopsin revealed by x-ray crystallography. Biophys J. 2007;92:L106–8. doi: 10.1529/biophysj.107.108225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stenkamp RE. Alternative models for two crystal structures of bovine rhodopsin. Acta Crystallogr D Biol Crystallogr. 2008;D64:902–904. doi: 10.1107/S0907444908017162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scheerer P, Park JH, Hildebrand PW, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 107.Standfuss J, Edwards PC, D’Antona A, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Makino CL, Riley CK, Looney J, et al. Binding of more than one retinoid to visual opsins. Biophys J. 2010;99:2366–2373. doi: 10.1016/j.bpj.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choe HW, Kim YJ, Park JH, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 110.Deupi X, Edwards P, Singhal A, et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci U S A. 2012;109:119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park JH, Morizumi T, Li Y, et al. Opsin, a structural model for olfactory receptors? Angew Chem Int Ed Engl. 2013;52:11021–11024. doi: 10.1002/anie.201302374. [DOI] [PubMed] [Google Scholar]
- 112.Singhal A, Ostermaier MK, Vishnivetskiy SA, et al. Insights into congenital stationary night blindness based on the structure of G90D rhodopsin. EMBO Rep. 2013;14:520–526. doi: 10.1038/embor.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Szczepek M, Beyriere F, Hofmann KP, et al. Crystal structure of a common GPCR-binding interface for G protein and arrestin. Nat Commun. 2014;5:4801. doi: 10.1038/ncomms5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Blankenship E, Vahedi-Faridi A, Lodowski DT. The High-Resolution Structure of Activated Opsin Reveals a Conserved Solvent Network in the Transmembrane Region Essential for Activation. Structure. 2015;23:2358–2364. doi: 10.1016/j.str.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kang Y, Zhou XE, Gao X, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Singhal A, Guo Y, Matkovic M, et al. Structural role of the T94I rhodopsin mutation in congenital stationary night blindness. EMBO Rep. 2016;17:1431–1440. doi: 10.15252/embr.201642671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gulati S, Jastrzebska B, Banerjee S, et al. Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism. Proc Natl Acad Sci U S A. 2017;114:E2608–E2615. doi: 10.1073/pnas.1617446114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Warne T, Moukhametzianov R, Baker JG, et al. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Christopher JA, Brown J, Dore AS, et al. Biophysical fragment screening of the beta1-adrenergic receptor: identification of high affinity arylpiperazine leads using structure-based drug design. J Med Chem. 2013;56:3446–3455. doi: 10.1021/jm400140q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Warne T, Edwards PC, Leslie AG, et al. Crystal structures of a stabilized beta1-adrenoceptor bound to the biased agonists bucindolol and carvedilol. Structure. 2012;20:841–849. doi: 10.1016/j.str.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang J, Chen S, Zhang JJ, et al. Crystal structure of oligomeric beta1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat Struct Mol Biol. 2013;20:419–425. doi: 10.1038/nsmb.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sato T, Baker J, Warne T, et al. Pharmacological Analysis and Structure Determination of 7-Methylcyanopindolol-Bound beta1-Adrenergic Receptor. Mol Pharmacol. 2015;88:1024–1034. doi: 10.1124/mol.115.101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leslie AG, Warne T, Tate CG. Ligand occupancy in crystal structure of beta1-adrenergic G protein-coupled receptor. Nat Struct Mol Biol. 2015;22:941–942. doi: 10.1038/nsmb.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hanson MA, Cherezov V, Griffith MT, et al. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;6:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bokoch MP, Zou Y, Rasmussen SG, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463:108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wacker D, Fenalti G, Brown MA, et al. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rasmussen SG, Choi HJ, Fung JJ, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rosenbaum DM, Zhang C, Lyons JA, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zou Y, Weis WI, Kobilka BK. N-Terminal T4 Lysozyme Fusion Facilitates Crystallization of a G Protein Coupled Receptor. PLos ONE. 2012:7. doi: 10.1371/journal.pone.0046039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ring AM, Manglik A, Kruse AC, et al. Adrenaline-activated structure of beta2-adrenoreceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weichert D, Kruse AC, Manglik A, et al. Covalent agonists for studying G protein-coupled receptor activation. Proc Natl Acad Sci U S A. 2014;111:10744–10748. doi: 10.1073/pnas.1410415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang CY, Olieric V, Ma P, et al. In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures. Acta Crystallogr D Struct Biol. 2016;72:93–112. doi: 10.1107/S2059798315021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ma P, Caffrey M. beta2AR-T4L - CIM. TO BE PUBLISHED. [Google Scholar]
- 134.Staus DP, Strachan RT, Manglik A, et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016;535:448–452. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shimamura T, Shiroishi M, Weyand S, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang C, Jiang Y, Ma J, et al. Structural basis for molecular recognition at serotonin receptors. Science. 2013;340:610–614. doi: 10.1126/science.1232807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wacker D, Wang C, Katritch V, et al. Structural features for functional selectivity at serotonin receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu W, Wacker D, Gati C, et al. Serial femtosecond crystallography of G protein-coupled receptors. Science. 2013;342:1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wacker D, Wang S, McCorvy JD, et al. Crystal Structure of an LSD-Bound Human Serotonin Receptor. Cell. 2017;168:377–389 e12. doi: 10.1016/j.cell.2016.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thal DM, Sun B, Feng D, et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016;531:335–340. doi: 10.1038/nature17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Haga K, Kruse AC, Asada H, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kruse AC, Ring AM, Manglik A, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kruse AC, Hu J, Pan AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Thorsen TS, Matt R, Weis WI, et al. Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis. Structure. 2014;22:1657–1664. doi: 10.1016/j.str.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Glukhova A, Thal DM, Nguyen AT, et al. Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity. Cell. 2017;168:867–877 e13. doi: 10.1016/j.cell.2017.01.042. [DOI] [PubMed] [Google Scholar]
- 146.Dore AS, Robertson N, Errey JC, et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–1293. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lebon G, Warne T, Edwards PC, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hino T, Arakawa T, Iwanari H, et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482:237–240. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Congreve M, Andrews SP, Dore AS, et al. Discovery of 1,2,4-triazine derivatives as adenosine A(2A) antagonists using structure based drug design. J Med Chem. 2012;55:1898–1903. doi: 10.1021/jm201376w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lebon G, Edwards PC, Leslie AG, et al. Molecular Determinants of CGS21680 Binding to the Human Adenosine A2A Receptor. Mol Pharmacol. 2015;87:907–915. doi: 10.1124/mol.114.097360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Segala E, Guo D, Cheng RK, et al. Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength. J Med Chem. 2016;59:6470–6479. doi: 10.1021/acs.jmedchem.6b00653. [DOI] [PubMed] [Google Scholar]
- 152.Batyuk A, Galli L, Ishchenko A, et al. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci Adv. 2016;2:e1600292. doi: 10.1126/sciadv.1600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Carpenter B, Nehme R, Warne T, et al. Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature. 2016;536:104–107. doi: 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun B, Bachhawat P, Chu ML, et al. Crystal structure of the adenosine A2A receptor bound to an antagonist reveals a potential allosteric pocket. Proc Natl Acad Sci U S A. 2017;114:2066–2071. doi: 10.1073/pnas.1621423114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hanson MA, Roth CB, Jo E, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chrencik JE, Roth CB, Terakado M, et al. Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell. 2015;161:1633–1643. doi: 10.1016/j.cell.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hua T, Vemuri K, Pu M, et al. Crystal Structure of the Human Cannabinoid Receptor CB1. Cell. 2016;167:750–762 e14. doi: 10.1016/j.cell.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shao Z, Yin J, Chapman K, et al. High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature. 2016 doi: 10.1038/nature20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.White JF, Noinaj N, Shibata Y, et al. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Egloff P, Hillenbrand M, Klenk C, et al. Structure of signaling-competent neurotensin receptor 1 obtained by directed evolution in Escherichia coli. Proc Natl Acad Sci U S A. 2014;111:E655–62. doi: 10.1073/pnas.1317903111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Krumm BE, White JF, Shah P, et al. Structural prerequisites for G-protein activation by the neurotensin receptor. Nat Commun. 2015;6:7895. doi: 10.1038/ncomms8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Krumm BE, Lee S, Bhattacharya S, et al. Structure and dynamics of a constitutively active neurotensin receptor. Sci Rep. 2016;6:38564. doi: 10.1038/srep38564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yin J, Mobarec JC, Kolb P, et al. Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature. 2015;519:247–250. doi: 10.1038/nature14035. [DOI] [PubMed] [Google Scholar]
- 164.Gayen A, Goswami SK, Mukhopadhyay C. NMR evidence of GM1-induced conformational change of Substance P using isotropic bicelles. Biochim Biophys Acta. 2011;1808:127–139. doi: 10.1016/j.bbamem.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 165.Shihoya W, Nishizawa T, Okuta A, et al. Activation mechanism of endothelin ETB receptor by endothelin-1. Nature. 2016;537:363–368. doi: 10.1038/nature19319. [DOI] [PubMed] [Google Scholar]
- 166.Park SH, Das BB, Casagrande F, et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491:779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zheng Y, Qin L, Zacarias NV, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016;540:458–461. doi: 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu B, Chien EY, Mol CD, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Qin L, Kufareva I, Holden LG, et al. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347:1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tan Q, Zhu Y, Li J, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Oswald C, Rappas M, Kean J, et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 2016;540:462–465. doi: 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- 172.Miller RL, Thompson AA, Trapella C, et al. The Importance of Ligand-Receptor Conformational Pairs in Stabilization: Spotlight on the N/OFQ G Protein-Coupled Receptor. Structure. 2015;23:2291–2299. doi: 10.1016/j.str.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wu H, Wacker D, Mileni M, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Manglik A, Kruse AC, Kobilka TS, et al. Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Huang W, Manglik A, Venkatakrishnan AJ, et al. Structural insights into micro-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Granier S, Manglik A, Kruse AC, et al. Structure of the delta-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fenalti G, Zatsepin NA, Betti C, et al. Structural basis for bifunctional peptide recognition at human delta-opioid receptor. Nat Struct Mol Biol. 2015;22:265–268. doi: 10.1038/nsmb.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang H, Unal H, Gati C, et al. Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell. 2015;161:833–844. doi: 10.1016/j.cell.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang H, Unal H, Desnoyer R, et al. Structural Basis for Ligand Recognition and Functional Selectivity at Angiotensin Receptor. J Biol Chem. 2015;290:29127–29139. doi: 10.1074/jbc.M115.689000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang H, Han GW, Batyuk A, et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017 doi: 10.1038/nature22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Burg JS, Ingram JR, Venkatakrishnan AJ, et al. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 2015;347:1113–1117. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang D, Gao ZG, Zhang K, et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature. 2015;520:317–321. doi: 10.1038/nature14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhang J, Zhang K, Gao ZG, et al. Agonist-bound structure of the human P2Y12 receptor. Nature. 2014;509:119–122. doi: 10.1038/nature13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Srivastava A, Yano J, Hirozane Y, et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature. 2014;513:124–127. doi: 10.1038/nature13494. [DOI] [PubMed] [Google Scholar]
- 185.Siu FY, He M, de Graaf C, et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013;499:444–449. doi: 10.1038/nature12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hollenstein K, Kean J, Bortolato A, et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013;499:438–443. doi: 10.1038/nature12357. [DOI] [PubMed] [Google Scholar]
- 187.Dore AS, Bortolato A, Hollenstein K, et al. Decoding Corticotropin-Releasing Factor Receptor Type 1 Crystal Structures. doi: 10.2174/1874467210666170110114727. TO BE PUBLISHED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wu H, Wang C, Gregory KJ, et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Christopher JA, Aves SJ, Bennett KA, et al. Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile) J Med Chem. 2015;58:6653–6664. doi: 10.1021/acs.jmedchem.5b00892. [DOI] [PubMed] [Google Scholar]
- 190.Wang C, Wu H, Evron T, et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat Commun. 2014;5:4355. doi: 10.1038/ncomms5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Neubig RR, Siderovski DP. Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discov. 2002;1:187–197. doi: 10.1038/nrd747. [DOI] [PubMed] [Google Scholar]