Abstract
Laser-capture microdissection was coupled with PCR to define the mitochondrial genotype of aged muscle fibers exhibiting mitochondrial enzymatic abnormalities. These electron transport system (ETS) abnormalities accumulate with age, are localized segmentally along muscle fibers, are associated with fiber atrophy and may contribute to age-related fiber loss. DNA extracted from single, 10 µm thick, ETS abnormal muscle fibers, as well as sections from normal fibers, served as templates for PCR-based deletion analysis. Large mitochondrial (mt) DNA deletion mutations (4.4–9.7 kb) were detected in all 29 ETS abnormal fibers analyzed. Deleted mtDNA genomes were detected only in the regions of the fibers with ETS abnormalities; adjacent phenotypically normal portions of the same fiber contained wild-type mtDNA. In addition, identical mtDNA deletion mutations were found within different sections of the same abnormal region. These findings demonstrate that large deletion mutations are associated with ETS abnormalities in aged rat muscle and that, within a fiber, deletion mutations are clonal. The displacement of wild-type mtDNAs with mutant mtDNAs results in concomitant mitochondrial enzymatic abnormalities, fiber atrophy and fiber breakage.
INTRODUCTION
Defects in the mitochondrial genome accumulate with age and are thought to have a causal role in aging processes (1–4). The mitochondrial genome is a 16.3 kb closed circular DNA molecule and it encodes 22 tRNAs, two rRNAs and 13 polypeptides of the electron transport system (ETS). Two to 10 copies of the genome are found in each mitochondrion (5). The mitochondrial genome is susceptible to mutation damage because the circular DNA is located near the source of reactive oxygen species production, it lacks histone protection (6) and DNA repair systems are limited in mitochondria (5,7–9). Mitochondrial (mt) DNA rearrangements have been implicated in ETS abnormalities observed in mitochondrial myopathies (reviewed in 10). These mutations include duplications of regions of the mitochondrial genome (11), point mutations (12) and deletions of large segments of the genome. mtDNA deletion mutations accumulate with age and are commonly detected in post-mitotic tissues, such as brain, heart and skeletal muscle, which rely heavily on oxidative metabolism (13–15).
Age-associated defects of mitochondrial respiratory function accumulate in skeletal muscle of humans, rhesus monkeys and rodents (3,15–18). These defects accrue in a subset of muscle fibers necessitating histological approaches for detection. Commonly used markers for mitochondrial ETS abnormalities include the histochemical activities of cytochrome c oxidase (COX, complex IV) and succinate dehydrogenase (SDH, complex II). Two abnormalities in these activities have been observed with age, a loss of COX activity (COX–) and concomitant increase in SDH activity (SDH hyperreactive regions, also known as a ragged red phenotype). This phenotype (COX– and SDH++) suggests a lesion in the mtDNA as three of the COX subunits are encoded by the mitochondrial genome whereas SDH is entirely nuclear encoded. The distribution of COX–/SDH++ regions is not homogeneous in all muscle fibers but rather the abnormalities accumulate focally in a subset of muscle fibers. The abundance of these abnormalities ranges from 15% of rectus femoris muscle fibers in old rats (15) to an estimated 60% of vastus lateralis fibers in very old rhesus monkeys (18). The physiological consequence of ETS abnormalities is suggested in studies that examine individual fibers along their length. Fiber atrophy leading to fiber breakage appears to be a result of mtDNA deletion mutations and associated ETS abnormalities (15,18).
The majority of mtDNA mutation studies in aging tissues have been performed on tissue homogenates involving thousands of cells. The calculated abundance of specific mtDNA deletion products compared with total DNA present in the homogenates is exceedingly low (<0.1%) (3,13,19). Inherent to these tissue homogenate studies is the assumption that there is a homogeneous distribution of aberrant mitochondria. Mitochondrial enzymatic abnormalities, however, accumulate focally in muscle fibers as evidenced by distinct, localized and segmental phenotypic changes (15,18). mtDNA deletion mutations are also distributed in the same mosaic manner. This has been demonstrated in defined fiber number studies (20), in situ hybridization analyses (3,18,21) and by characterization of individual cardiomyocytes (22,23). In situ hybridization analyses indicate an association of age-related mitochondrial ETS abnormalities with mitochondrial deletion mutations (3,18,21); however, such hybridization analyses cannot precisely define the deletion event.
Manual techniques for the isolation of individual muscle fibers prior to PCR analysis have been employed in studies examining mitochondrial myopathy patients (24–28). The development of laser-capture microdissection (LCM) allows for the precise identification, isolation and subsequent molecular characterization of a specific cell or portion of a cell from a histological section. In the work presented here, histological analyses and LCM were combined with PCR-based mtDNA analyses to determine the mitochondrial genotype in ragged red regions of aged skeletal muscle. mtDNA deletion mutations were detected, using whole genome PCR amplification in the ETS abnormal region, in all 29 ragged red fibers analyzed. Wild-type mtDNA genomes were present in ETS normal fibers and in the ETS normal segments of ragged red fibers, only genomes with deletion mutations were detected in the ragged red regions. Our results demonstrate a strict correlation between the mtDNA deletion mutations and the ragged red phenotype in aged rat skeletal muscle. The presence of a specific deleted genome in an individual ragged red fiber suggests a clonal expansion of deleted mtDNA genomes in ETS abnormal regions of muscle fibers.
MATERIALS AND METHODS
Animals and sample preparation
Male Fischer 344 × Brown Norway F1 hybrid rats were purchased from the National Institute on Aging colony maintained by Harlan Sprague Dawley (Indianapolis, IN). Five 38-month-old rats were anesthetized with sodium pentobarbital and killed by exsanguination. The rectus femoris muscles were dissected, transected at the mid-belly, embedded in optimal cutting temperature (OCT) medium (Miles Inc., Elkhart, IN) and flash frozen in liquid nitrogen-cooled isopentane. Samples were stored at –80°C until analyzed.
Histochemistry
Rectus femoris muscle samples in OCT were raised to the temperature of cryostat (–20°C). Two hundred consecutive, 10 µm thick serial sections were cut and placed on Probe-on Plus Slides (Fisher Scientific, Pittsburgh, PA). Sections were stored at –80°C until analyzed. Histochemical staining for COX and SDH activities were performed according to Seligman et al. (29) and Dubowitz (30). Ragged red fibers exhibiting COX– and SDH++ phenotypes were identified and characterized along the length of muscle fiber at 60 µm intervals.
Laser-capture microdissection
Frozen sections, immediately adjacent to those used for the detection of ragged red fibers, were stained for SDH activity as described above. Sections were then dehydrated through an ethanol and xylene series. Slides were air dried and immediately used for LCM. Individual skeletal muscle fiber sections were microdissected using a PixCell II laser-capture microscope (Arcturus). Settings for LCM were a laser spot size of 15–30 µm in diameter, a pulse power of 30 mV and a pulse width of 50 ms. A single-fiber section was captured on LCM transfer film on a CapSure cap. Any extraneous non-captured material was removed from the cap using the CapSure pad (Arcturus).
DNA isolation and PCR-based deletion analysis
Digestion solution (1 µl) containing 2.0 mg/ml proteinase K, 0.5% SDS and 10 mM EDTA was added to each sample to release DNA from individual fiber sections. The samples were incubated at 37°C for 30 min in a humidified chamber. The released DNA was recovered with 10 µl water. One microliter of the DNA solution from an individual fiber section was analyzed by PCR. Short extension PCR (93°C for 15 s, 60°C for 30 s, 72°C for 2 min, 35 cycles) was employed to amplify small mtDNA fragments (<2 kb) using the Taq polymerase PCR system (Promega). Long extension PCR (93°C for 15 s, 62°C for 30 s, 68°C for 15 min, 25 cycles) was performed to amplify the whole mtDNA genome using Expand Long Template PCR System (Roche). Primers used for PCR analyses are summarized in Table 1. For whole mitochondrial genome amplification, 25 cycles of primary PCR were followed by 25 cycles of nested PCR. The outer primer pairs for primary PCR, F15671 and R15377, amplify a 16 007-bp fragment of the rat mitochondrial genome. The inner primer pairs for nested PCR, F15826 and R15233, amplify a 15 708-bp fragment. The amplification products from long extension PCR were ligated into the pGEM®-T Easy vector (Promega) and breakpoints were defined following sequence analysis at the University of Wisconsin-Madison DNA Sequencing Center.
Table 1. Primers used for PCR analyses.
| Primers |
Sequences |
Nucleotide locationa |
|
F15671 |
5′-CCATCCTCCGTGAAATCAACAACCCG-3′ |
15671–15696 |
|
R15377 |
5′-CTTTGGGTGTTGATGGTGGGGAGGTAG-3′ |
15377–15350 |
|
F15826 |
5′-AAGACATCTCSATGGTAACGGGTC-3′ |
15826–15849 |
|
R15233 |
5′-CCAGAGATTGGTATGAGAATGAGG-3′ |
15233–15209 |
| F5881 |
5′-AGCCGTCCTACTACTTCTCTCAC-3′ |
5881–5903 |
|
F14165 |
5′-CAACCACTCCTTTATCGACCTACC-3′ |
14165–14188 |
| R15144 | 5′-GGCCTCCGATTCATGTTAAGACTA-3′ | 15144–15121 |
aThe numbering system for each primer is based on rat mitochondrial genome sequences (1–16 300 bp, GenBank accession no. X14848). F represents forward primer, while R represents reverse primer.
RESULTS
Age-associated ragged red fibers in rat skeletal muscle
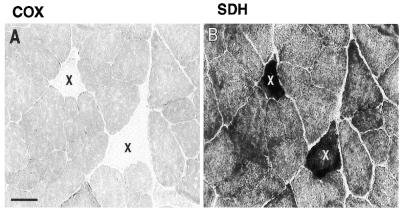
Serial sections of 38-month-old rat rectus femoris muscle samples were examined at 60-µm intervals for COX and SDH activities to identify and analyze the distribution of ETS normal and abnormal regions of individual fibers. Twenty-nine fibers containing regions exhibiting concomitant COX– and SDH++ phenotypes were identified. All were mosaically and segmentally distributed among 7000 fibers over a 2000-µm thick region of 38-month-old rectus femoris (Fig. 1).
Figure 1.
Identification of ETS abnormal fibers. Enzymatic staining for COX and SDH activities in the rectus femoris muscle from a 38-month-old rat. Adjacent 10 µm sections showing examples of two fibers (X) exhibiting the COX– (A) and SDH++ (B) phenotypes. Black bar represents 50 µm.
Detection of mtDNA deletion products in ragged red skeletal muscle fibers using whole mtDNA genome PCR amplification
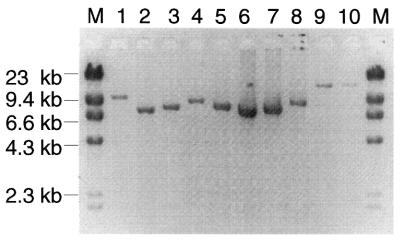
LCM was used to isolate individual ragged red skeletal muscle fibers from the 10-µm thick muscle sections. Total DNA from 29 ragged red fiber sections and 10 randomly selected ETS normal control fiber sections were analyzed by PCR amplification using long-extension PCR. With 25 cycles of primary amplification followed by 25 cycles of nested PCR amplification, the wild-type mitochondrial genome was successfully amplified from the ETS normal fibers (Fig. 2, lanes 9 and 10). In all 29 ragged red regions, smaller than wild-type mtDNA genomes were detected. Single amplification products were detected in all but one of the ragged red fibers, which contained two deletion products (Table 2). The deletion products ranged in size from 4.4 to 9.7 kb, corresponding to mtDNA genomes of 11.9–6.6 kb (Fig. 2, lanes 1–8). Deletion mutations were not detected in the ETS normal fibers; conversely, wild-type mtDNA genomes were not detected in the ragged red regions. Confirmation of the size of the mitochondrial genomes associated with the ETS abnormal fiber regions was performed in a second amplification of the LCM template using major arc primers (F5881 and R15144, Table 1) in a 35 cycle short-extension PCR. Figure 3A presents five amplification products from ETS abnormal regions detected using nested long-extension PCR (lanes 1–5). When the same fiber sections were amplified by short-extension PCR, a single, appropriately sized deletion product, based on the long-extension reactions, was amplified in four fibers (Fig. 3B). A deletion product was not observed in the other fiber (Fig. 3B, lane 3). Subsequent sequence analysis demonstrated that the deletion in this sample included the F5881 primer site (Table 2).
Figure 2.
Long-extension PCR analysis of mtDNA deletions. Total DNA from laser-captured ragged red fiber sections (lanes 1–8) and ETS normal fiber sections (lanes 9 and 10) was subjected to whole mitochondrial genome PCR. M denotes the DNA (size) marker.
Table 2. mtDNA deletion breakpoints of 30 deleted mitochondrial genomes from 29 rat skeletal muscle ragged red fibers.
| Fiber number |
Breakpointa |
Sequence at breakpoint |
Deletion (kb) |
Genes removed |
| 1 |
7100–14392 |
AGCTCCCT–TTTTCATC |
7.3 |
COX II–CYT b |
| 2 |
6375–13084 |
ACGAGAAG–AACGCCTA |
6.7 |
COX I–NADH 5 |
| 3 |
6883–14608 |
ACAACTGG–AATGAATC |
7.7 |
COX II–CYT b |
| 4 |
10221–14991 |
CAAAAAAA–GGGGTCGT |
4.7 |
NADH 4–CYT b |
| 5 |
7166–14228 |
GACGCCCA–TCTACTAG |
7.0 |
COX II–CYT b |
| 6b |
9332–9441 |
GTTTGACT—CCTACTTA |
5.6 |
NADH 3–CYT b |
| |
9567–9333 |
ACCCAACA–ATTCCT |
|
|
| |
9338–9343 |
ATTCCT–GTTTCTAT |
|
|
| |
9353–9366 |
TCTATCTA–TACTCCCT |
|
|
| |
9420–15112 |
CTGAAAAA–CAACCTCC |
|
|
| 7 |
5530–14616 |
CTATAATA–TGAGGAGG |
9.1 |
COX I–CYT b |
| 8 |
6872–13867 |
ATCGACCC–ACCCAAAT |
7.0 |
COX II–CYT b |
| 9 |
7268–14151 |
AATAACCC–CTATTCAA |
6.9 |
COX II–CYT b |
| 10 |
6844–15225 |
ATGTAAAA–ATCTCTGG |
8.4 |
COX I–CYT b |
| 11 |
5935–14612 |
CTATACTC–ATCTGAGG |
8.7 |
COX I–CYT b |
| 12 |
7247–13095 |
CTATACTA–TTAGGAAG |
5.8 |
COX II–NADH 5 |
| 13 |
6336–13075 |
CTCATCCC–ATCCAATC |
6.7 |
COX I–NADH 5 |
| 14 |
6549–13914 |
GCCCACTT–ACCCTAAA |
7.4 |
COX I–NADH 6 |
| 15 |
6033–14885 |
CACCCAGA-CAATTATA |
8.8 |
COX I–CYT b |
| 16 |
5251–14984 |
GTAAAAAG–ACTAGGAG |
9.7 |
COX I–CYT b |
| 17 |
8149–14616 |
ATATTTAT–TGAGGAGG |
6.5 |
ATPase 6–CYT b |
| 18 |
6423–14545 |
CTTCCACT–GAGGAGCT |
8.1 |
COX I–CYT b |
| 19 |
7398–15101 |
GTCTATTA–TCCTTTAC |
7.8 |
COX II–CYT b |
| 20 |
5827–12360 |
TAAAACCC–ACAGGACT |
6.5 |
COX I–NADH 5 |
| 21 |
7170–14614 |
CCCAAGAA–TCTGAGGA |
7.4 |
COX II–CYT b |
| 22 |
6250–14399 |
ATCGCAAT–CTGCCTAT |
8.1 |
COX I–CYT b |
| 29 |
7810–15378 |
TTTATTTT–TGATATTC |
7.5 |
ATPase 8–CYT b |
| 30 |
7489–13122 |
CATCCCTT–ATCTCACT |
5.7 |
COX II–NADH 5 |
| 31 |
8201–14613 |
GCTATCTA–ATCTGAGG |
6.4 |
ATPase 6–CYT b |
| 32 |
6349–14219 |
TCTTCTTA–CTTCGGTT |
7.9 |
COX I–CYT b |
| 33 |
8313–15127 |
ATCTCACT–TAACATGA |
6.8 |
ATPase 6–CYT b |
| 34 |
7650–14466 |
ATTTCAGG–TTGGGATC |
6.8 |
COX II–CYT b |
| 35a |
7501–11908 |
AGGGTTAA–TTACTAAC |
4.4 |
COX II–NADH 5 |
| 35b | 6826–13410 | CATTCGAA–AAATCAAC | 6.6 | COX I–CYT b |
aThe numbering system is based upon the rat mtDNA sequence.
bMultiple breakpoints exist in fiber number 6.
Figure 3.
Detection of deleted mtDNA genome with long-extension PCR (LX-PCR) and short extension PCR (SX-PCR). Total DNA from ragged red sections of fiber numbers 18–22 (lanes 1–5) was analyzed with LX-PCR and SX-PCR. (A) Nested PCR (25 + 25 cycles) with whole mtDNA genome LX-PCR amplification. (B) SX-PCR amplification (35 cycles) with F5881 and R15144 primers.
Identification of deletion breakpoints
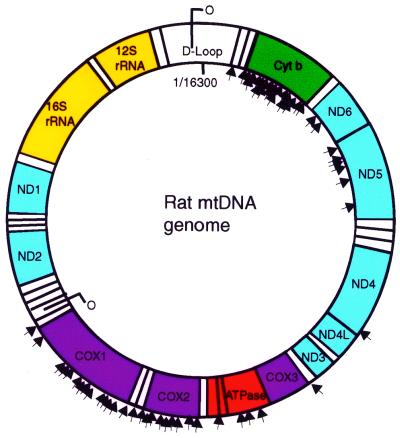
DNA sequence analysis defined the breakpoints of the deletion events. All deletions occurred in the major arc of the mtDNA genome with both the heavy and light strands of origin of replication being conserved (Fig. 4). Large direct repeat and inverted repeat sequences were not present at the deletion breakpoints (Table 2). Each ragged red fiber contained a specific deleted genome with unique breakpoints, confirming the amplification reactions, which indicated that ‘common’ deletions do not appear to be associated with the age-related ragged red phenotype in rat skeletal muscle. A single deletion event with specific breakpoints was identified in 28 mtDNA genomes. One exception was fiber number 6 in which several breakpoints were present and some of the sequences between breakpoints were disordered (Table 2).
Figure 4.
Distribution of mtDNA deletion breakpoints. A representation of the rat mitochondrial genome. Arrowheads on the inner edge of the circle denote the 5′ deletion breakpoints. Arrowheads on the outer edge of the circle represent the 3′ deletion breakpoint.
Longitudinal analysis of mitochondrial genotype and ragged red phenotype
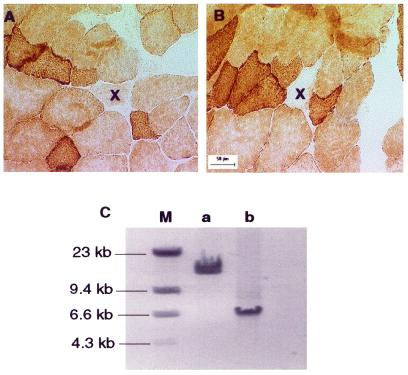
The segmental nature of the RRF phenotype suggested a focal accumulation of mutated mitochondrial genomes. Longitudinal studies were performed to define further the relationship between the mitochondrial genotype and the ragged red phenotype. We used laser capture to sample various regions along the length of a single fiber including both the ragged red region and flanking phenotypically normal regions (Fig. 5A and B). Long-extension PCR analysis showed that wild-type mtDNA was present in ETS normal regions whereas deleted mtDNA genomes were detected only within ragged red regions (Fig. 5C). The same deleted genomes were present at different points within the same ragged red region, demonstrating the clonal nature of the mutated mitochondrial genome in individual ragged red fibers.
Figure 5.
Co-localization of mtDNA deletion with segmental ragged red phenotype. COX and SDH enzymatic staining was performed on muscle fibers from a 38-month-old rat. ‘X’ (A and B) denotes the same fiber that shows normal COX activity in section (A) and negative COX phenotype in section (B). The distance between sections (A) and (B) is 550 µm. (C) The whole mtDNA genome PCR from ETS normal and ragged red sections of the same fiber: a, amplification of ETS normal section; b, amplification of ragged red section; M, DNA marker.
Predominance of deleted mitochondrial genomes in ragged red regions
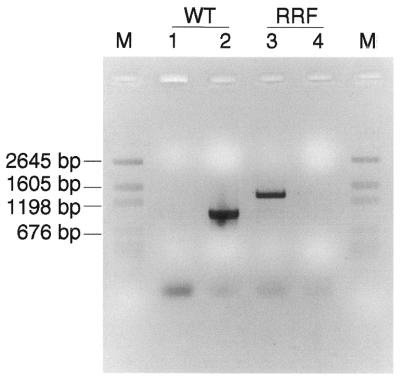
In situ hybridization studies using mtDNA and mtRNA probes to ETS abnormal regions of muscle fibers suggest that mtDNA deletion mutations predominate within the affected portion of the fiber whereas phenotypically normal regions contain wild-type mtDNAs (3,21). The PCR analysis of the ETS abnormal regions also identified the presence of mtDNA deletion mutations whereas wild-type mitochondrial genomes were not easily detected (Fig. 2). As PCR favors the amplification of smaller products, it was possible that the wild-type genomes were not detected by long-extension PCR in the presence of deleted genomes in the ETS abnormal region. To address this question, ETS normal and ragged red regions from the same fiber were laser captured and both the wild-type and deleted genomes were amplified using short-extension PCR. The primer pair F14165 and R15144 amplified a 1-kb fragment from wild-type genomes but not the deleted genome as the F14165 primer site was deleted (breakpoint 6429–14219, 7.8 kb deletion, Table 2). The primer pair F5881 and R15144 amplified a 1.3-kb fragment of the deleted genome but not the wild-type genome as the wild-type PCR products will be 9 kb and are not efficiently amplified in the short-extension PCR reaction. The wild-type mitochondrial genome PCR product was present in the ETS normal fiber section, whereas the deleted genome PCR product was detected in the ragged red region. Interestingly, the 1 kb wild-type amplification product was not detected in ragged red sections after 35 cycles of PCR amplification, indicating a high ratio of mtDNA deletions to wild-type mtDNA (Fig. 6).
Figure 6.
Wild-type mtDNA genome is not detectable in ragged red region. Deletion-specific primer set [5881 and 15144, with deletion products ∼1.3 kb (breakpoint 6349/14219)] and wild-type-specific primer set (14165 and 15144, with wild-type products ∼1.0 kb) were used to amplify deleted genomes and wild-type genomes, respectively, from ragged red and ETS normal sections of fiber number 32. Lanes 1 and 3, amplification with deletion-specific primer set; lanes 2 and 4, amplification with wild-type-specific primer set. WT, ETS normal section; RRF, ragged red section; M, DNA marker.
DISCUSSION
We found a direct correlation between individual age-related ETS abnormal muscle fiber regions and unique mtDNA deletion mutations. mtDNA deletion mutations were detected in all 29 ETS abnormal fibers analyzed. The deleted mitochondrial genomes were concomitant with COX– and SDH++ regions of the affected muscle fibers. Only mtDNA deletion mutations were detected in the ragged red regions of the muscle fiber, whereas only wild-type mtDNA was detected in the ETS normal regions. DNA sequence analysis studies have typically been carried out on tissue homogenates. Numerous deletion products were detected, of which many were defined by DNA sequence analysis (3,31–33); however, as these were homogenate analysis studies, deletion products could not be associated with a known phenotype. LCM and PCR analysis clearly demonstrate that deletion mutations are localized to specific cells identified by histochemical analysis.
The deletion mutations in all 29 ETS abnormal fiber regions from rat rectus femoris were located exclusively in the major arc regions of the mitochondrial genome with both the heavy and light DNA replication origins being conserved. Although the retention of both replication origins would appear to be a prerequisite for mtDNA replication, studies in primates indicate otherwise. In situ hybridization of aged rhesus monkey skeletal muscle, for example, identified a minority of mtDNA deletion mutations that involved the light strand origin of mtDNA replication (3,18). Recently, Bodyak et al. (23) identified deletion mutations in aged human cardiomyocytes exhibiting a similar genotype. In human mitochondrial diseases, large deletion mutations involving the light strand origin have also been characterized (34). These findings suggest the presence of a secondary site in the primate mitochondrial genome that can function as the light strand mtDNA replication of origin. Alternatively, the large size of the deletion mutation combined with the longer lifespan of primates may also facilitate the accumulation of deletion mutations involving the light strand origin.
Based upon the observation that direct repeat sequences are present at the deletion breakpoint, a slip replication mechanism was hypothesized as the means by which deletions are produced (35). In human mitochondria, the 4977 bp ‘common’ deletion, mtDNA4977, was flanked by a 13 bp direct repeat. The mtDNA4977 deletion accumulated with age in several tissues, reaching the highest level in muscle tissues (0.1% of wild-type genome; 13,36). Direct repeat sequences do not appear to be present at the deletion breakpoints in rodents. Interestingly, this is not due to a lack of direct repeat sequences in the rodent mitochondrial genome. Furthermore, large direct repeats are not detected in the deletion breakpoint. Only one breakpoint contained a 7 bp direct repeat and a few of the deleted genomes had 2–4 bp direct repeats present in the breakpoint. In one ragged red fiber, we found several breakpoints present as well as disordered sequences between breakpoints (Table 2). This event is not easily explained by a slip replication mechanism.
Accumulation of the same deleted genomes in individual ragged red fibers and in individual cardiomyocytes from aged humans (22) suggests the clonal expansion of deleted mitochondrial genomes. Two hypotheses have been proposed to explain this clonal phenomenon. De Grey (37) proposed a novel model suggesting that damaged mitochondria are degraded more slowly than intact mitochondria and that abnormal mitochondria accumulate in the cell by ‘survival of slowest’. The low proton gradient in defective mitochondria may reduce the production of oxygen free radicals and, therefore, decrease the damage to the mitochondrial membrane, leading to the accumulation of defective mitochondria in individual cells (37,38). This hypothesis suggests that a variety of mitochondrial genome mutations affecting mitochondrial function would lead to the accumulation of defective mitochondria. In contrast to De Grey’s hypothesis, our studies demonstrated a significant correlation between the ETS abnormal regions and large deletion mutations.
A second hypothesis, based upon the size of the mtDNA deletion mutation, presumes a replicative advantage inherent to smaller genomes (39,40). Given that cells contain hundreds to thousands of mitochondria, it is difficult to explain how a defective mitochondrion, which suffers from chronic energy shortage and a disrupted proton gradient, could display a replicative advantage over wild-type mitochondria. Clearly, however, large mtDNA deletion mutations accumulate and are associated with the RRF phenotype. Homogenate studies (33) identified smaller deletion mutations in rodents, mutations that do not appear to accumulate to levels sufficient to produce the RRF phenotype. Therefore, it would appear that there is a selection mechanism for large deletion mutations.
In conclusion, the mtDNA genotype was examined along the length of muscle fibers exhibiting a ragged red phenotype from aged rat rectus femoris. Large mtDNA deletion mutations, each unique to the specific ETS abnormal fiber examined, were found in all 29 ragged red fibers analyzed. These deleted mtDNA genomes were predominant in the ETS abnormal fiber segment but not detectable in the ETS normal region of the same fibers. The unequivocal relationship between the RRF phenotype and mitochondrial genotype further strengthens the likelihood that mtDNA deletion mutations are a direct cause of the age-associated ETS abnormalities and subsequent fiber atrophy observed in aged mammalian skeletal muscle.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Alan Herbst and Damian Lee for technical assistance and Dr Debbie McKenzie for critical reading of this manuscript. This work is supported by grants RO1 AG11604, P01 AG11915 and T32 AG00213 from the National Institutes of Health.
References
- 1.Linnane A.W., Marzuki,S., Ozawa,T. and Tanaka,M. (1989) Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet, i, 642–645. [DOI] [PubMed] [Google Scholar]
- 2.Cortopassi G.A. and Arnheim,N. (1990) Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res., 18, 6927–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C.M., Lopez,M.E., Weindruch,R. and Aiken,J.M. (1998) Association of age-related mitochondrial abnormalities with skeletal muscle fiber atrophy. Free Radical Biol. Med., 25, 964–972. [DOI] [PubMed] [Google Scholar]
- 4.Cortopassi G.A. and Wang,A. (1999) Mitochondria in organismal aging and degeneration. Biochim. Biophys. Acta, 1410, 183–193. [DOI] [PubMed] [Google Scholar]
- 5.Clayton D.A. (1982) Replication of animal mitochondrial DNA. Cell, 28, 693–705. [DOI] [PubMed] [Google Scholar]
- 6.Richter C. (1988) Do mitochondrial DNA fragments promote cancer and aging? FEBS Lett., 241, 1–5. [DOI] [PubMed] [Google Scholar]
- 7.Clayton D.A., Doda,J.N. and Friedberg,E.C. (1974) The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl Acad. Sci. USA, 71, 2777–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bittles A.H. (1992) Evidence for and against the causal involvement of mitochondrial DNA mutation in mammalian ageing. Mutat. Res., 275, 217–225. [DOI] [PubMed] [Google Scholar]
- 9.Croteau D.L., Stierum,R.H. and Bohr,V.A. (1999) Mitochondrial DNA repair pathways. Mutat. Res., 434, 137–148. [DOI] [PubMed] [Google Scholar]
- 10.Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- 11.Moore C.A., Gudikote,J. and Van Tuyle,G.C. (1998) Mitochondrial DNA rearrangements, including partial duplications, occur in young and old rat tissues. Mutat. Res., 421, 205–217. [DOI] [PubMed] [Google Scholar]
- 12.Liu V.W., Zhang,C., Linnane,A.W. and Nagley,P. (1997) Quantitative allele-specific PCR: demonstration of age-associated accumulation in human tissues of the A→G mutation at nucleotide 3243 in mitochondrial DNA. Hum. Mutat., 9, 265–271. [DOI] [PubMed] [Google Scholar]
- 13.Cortopassi G.A., Shibata,D., Soong,N.W. and Arnheim,N. (1992) A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc. Natl Acad. Sci. USA, 89, 7370–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C., Baumer,A., Maxwell,R.J., Linnane,A.W. and Nagley,P. (1992) Multiple mitochondrial DNA deletions in an elderly human individual. FEBS Lett., 297, 34–38. [DOI] [PubMed] [Google Scholar]
- 15.Wanagat J., Cao,Z., Pathare,P. and Aiken,J.M. (2001) Mitochondrial DNA deletion mutations colocalized with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting and oxidative damage in sarcopenia. FASEB J., 15, 323–332. [DOI] [PubMed] [Google Scholar]
- 16.Muller-Hocker J. (1989) Cytochrome-c-oxidase deficient cardiomyocytes in the human heart—an age-related phenomenon. A histochemical ultracytochemical study. Am. J. Pathol., 134, 1167–1173. [PMC free article] [PubMed] [Google Scholar]
- 17.Muller-Hocker J. (1990) Cytochrome c oxidase deficient fibres in the limb muscle and diaphragm of man without muscular disease: an age-related alteration. J. Neurol. Sci., 100, 14–21. [DOI] [PubMed] [Google Scholar]
- 18.Lopez M.E., Van Zeeland,N.L., Dahl,D.B., Weindruch,R. and Aiken,J.M. (2000) Cellular phenotypes of age-associated skeletal muscle mitochondrial abnormalities in rehesus monkeys. Mutat. Res., 452, 123–138. [DOI] [PubMed] [Google Scholar]
- 19.Aspnes L.E., Lee,C.M., Weindruch,R., Chung,S.S., Roecker,E.B. and Aiken,J.M. (1997) Caloric restriction reduces fiber loss and mitochondrial abnormalities in aged rat muscle. FASEB J., 11, 573–581. [DOI] [PubMed] [Google Scholar]
- 20.Schwarze S.R., Lee,C.M., Chung,S.S., Roecker,E.B., Weindruch,R. and Aiken,J.M. (1995) High levels of mitochondrial DNA deletions in skeletal muscle of old rhesus monkeys. Mech. Ageing Dev., 83, 91–101. [DOI] [PubMed] [Google Scholar]
- 21.Muller-Hocker J., Seibel,P., Schneiderbanger,K. and Kadenbach,B. (1993) Different in situ hybridization patterns of mitochondrial DNA in cytochrome c oxidase-deficient extraocular muscle fibres in the elderly. Virchows Arch. A Pathol. Anat. Histopathol., 422, 7–15. [DOI] [PubMed] [Google Scholar]
- 22.Khrapko K., Bodyak,N., Thilly,W.G., van Orsouw,N.J., Zhang,X., Coller,H.A., Perls,T.T., Upton,M., Vijg,J. and Wei,J.Y. (1999) Cell-by-cell scanning of whole mitochondrial genomes in aged human heart reveals a significant fraction of myocytes with clonally expanded deletions. Nucleic Acids Res., 27, 2434–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodyak N.D., Nekhaeva,E., Wei,J.Y. and Khrapko,K. (2001) Quantification and sequencing of somatic deleted mtDNA in single cells: evidence for partially duplicated mtDNA in aged human tissues. Hum. Mol. Genet., 10, 17–24. [DOI] [PubMed] [Google Scholar]
- 24.Moraes C.T. and Schon,E.A. (1996) Detection and analysis of mitochondrial DNA and RNA in muscle by in situ hybridization and single-fiber PCR. Methods Enzymol., 264, 522–540. [DOI] [PubMed] [Google Scholar]
- 25.Mita S., Tokunaga,M., Uyama,E., Kumamoto,T., Uikawa,K. and Uchino,M. (1998) Single muscle fiber analysis of myoclonus epilepsy with ragged-red fibers. Muscle Nerve, 21, 490–497. [DOI] [PubMed] [Google Scholar]
- 26.Sciacco M., Gasparo-Rippa,P., Vu,T.H., Tanji,K., Shanske,S., Mendell,J.R., Schon,E.A., DiMauro,S. and Bonilla,E. (1998) Study of mitochondrial DNA depletion in muscle by single-fiber polymerase chain reaction. Muscle Nerve, 21, 1374–1381. [DOI] [PubMed] [Google Scholar]
- 27.Silverstri G., Rana,M., Odoardi,F., Modoni,A., Paris,E., Papacci,M., Tonali,P. and Servidei,S. (2000) Single-fiber PCR in MELAS3243 patients: correlation between intratissue distribution and phenotypic expression of the mtDNAA3243G genotype. Am. J. Med. Genet., 94, 201–206. [DOI] [PubMed] [Google Scholar]
- 28.Brierley E.J., Johnson,P.A., Lighttowlers,R.N., James,O.F. and Turnbull,D.M. (1998) Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann. Neurol., 43, 217–223. [DOI] [PubMed] [Google Scholar]
- 29.Seligman A.M., Karnovsky,M.J., Wasserkrug,H.L. and Hanker,J.S. (1968) Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB). J. Cell Biol., 38, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubowitz V. (1985) Muscle Biopsy: A Practical Approach, 2nd Edition. Bailliere Tindall, London, UK.
- 31.Chung S.S., Weindruch,R., Schwarze,S.R., McKenzie,D.I. and Aiken,J.M. (1994) Multiple age-associated mitochondrial DNA deletions in skeletal muscle of mice. Aging, 6, 193–200. [DOI] [PubMed] [Google Scholar]
- 32.Van Tuyle G.C., Gudikote,J.P., Hurt,V.R., Miller,B.B. and Moore,C.A. (1996) Multiple, large deletions in rat mitochondrial DNA: evidence for a major hot spot. Mutat. Res., 349, 95–107. [DOI] [PubMed] [Google Scholar]
- 33.Eimon P.M., Chung,S.S., Lee,C.M., Weindruch,R. and Aiken,J.M. (1996) Age-associated mitochondrial DNA deletions in mouse skeletal muscle: comparison of different regions of the mitochondrial genome. Dev. Genet., 18, 107–113. [DOI] [PubMed] [Google Scholar]
- 34.Ballinger S.W., Shoffner,J.M., Hedaya,E.V., Trounce,I., Polak,M.A., Koontz,D.A. and Wallace,D.C. (1992) Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nature Genet., 1, 11–15. [DOI] [PubMed] [Google Scholar]
- 35.Shoffner J.M., Lott,M.T., Voljavec,A.S., Soueidan,S.A., Costigan,D.A. and Wallace,D.C. (1989) Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc. Natl Acad. Sci. USA, 86, 7952–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corral-Debrinski M., Shoffner,J.M., Lott,M.T. and Wallace,D.C. (1992) Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat. Res., 275, 169–180. [DOI] [PubMed] [Google Scholar]
- 37.De Grey A.D. (1997) A proposed refinement of the mitochondrial free radical theory of aging. BioEssays, 19, 161–166. [DOI] [PubMed] [Google Scholar]
- 38.Kowald A. (1999) The mitochondrial theory of aging: do damaged mitochondria accumulate by delayed degradation? Exp. Gerontol., 34, 605–612. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi J., Ohta,S., Kikuchi,A., Takemitsu,M., Goto,Y. and Nonaka,I. (1991) Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl Acad. Sci. USA, 88, 10614–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takai D., Isobe,K. and Hayashi,J. (1999) Transcomplementation between different types of respiration-deficient mitochondria with different pathogenic mutant mitochondrial DNAs. J. Biol. Chem., 274, 11199–11202. [DOI] [PubMed] [Google Scholar]