Abstract
Recently a number of novel adenoviruses have been isolated from diverse bat species and from diverse geographical locations. We describe the isolation of a novel adenovirus (Family Adenoviridae, genus Mastadenovirus) from a pool of liver and spleen tissue of an apparently healthy wild-caught Egyptian fruit bat (Rousettus aegyptiacus) in South Africa. Genetically the virus is most closely related to four mastadenoviruses recently isolated in China, from Miniopterus schreibersi and Rousettus leschenaultii bats, which are highly divergent from previously identified bat adenoviruses. The length of the Rousettus aegyptiacus adenovirus-3085 (RaegAdV-3085) genome, at 29,342 bp is similar to its closest relatives, and contains 27 open reading frames. The RaegAdV-3085 genome has a low G + C content (36.4%) relative to other viruses in the genus (between 43.6 and 63.9%) but similar to its closest relatives. The inverted terminal repeat (ITR) of RaegAdV-3085 is only 40 bp compared to between 61 and 178 bp of its closest relatives. The discovery of RaegAdV-3085 expands the diversity of known adenoviruses in bats and might represent a member of a new mastadenovirus species in bats.
Introduction
Adenoviruses (family Adenoviridae) are non-enveloped double strand DNA viruses, 70–90 nm in diameter and containing a single linear genome ranging in size from 26 to 48 kb. The genome contains an inverted terminal repetition (ITR) which ranges in size from 36 to 371 bp between different viruses. There are currently five recognized genera: Atadenovirus, Aviadenovirus, Ichtadenovirus, Mastadenovirus and Siadenovirus1. Adenoviruses infect a wide range of hosts. The Mastadenovirus genus is the largest, containing 22 recognized species, isolated exclusively from mammals including human, bovine, bat, canine and equine hosts. Other adenoviruses from mammalian hosts are from species within the Atadenovirus genus which also includes viruses from avian and serpentine hosts. The Aviadenovirus genus comprises exclusively avian viruses; the Ichtadenovirus genus contains one species isolated from sturgeon fish, while Siadenovirus genus includes viruses isolated from frogs and birds. The Mastadenovirus genus includes a number of unclassified viruses isolated from bats2–9. It has been recently proposed and accepted to classify a number of these viruses into two new species, Bat mastadenovirus A and B10.
Adenoviruses have been isolated from bats from various geographical locations and different bat species. The first adenovirus from a bat was isolated in Japan from a fruit-eating flying fox in 2006 (Pteropus dasymallus yayeyamae)9. In China, adenoviruses have been isolated from the insectivorous common bent-wing (Miniopterus schreibersii) and Rickett’s big-footed bats (Myotis ricketti), as well as the fruit-eating Leschnault’s rousette bat (Rousettus leschenaulti)5–7. In Germany these viruses have been isolated from the insectivorous common pipistrelle bat (Pipistrellus pipistrellus)3,4. In the USA a novel adenovirus was isolated from a Rafinesque’s big-eared bat (Corynorhinus rafinesquii)2. Metagenomic or PCR-based detection studies have revealed the presence of adenoviruses in the guano of greater mouse-eared bat (Myotis myoits) in Germany11, common noctule (Nyctalus noctula) and greater horseshoe bats (Rhinolophus ferrumequinum) in Hungary12, and the pallid bat (Antrozous pallidus) in the USA13. One bat associated adenovirus has been isolated from the straw coloured fruit bat (Eidolon helvum) in Ghana, Africa8.
Adenovirus infections usually result in mild disease in different hosts with a few exceptions. In dogs, canine adenovirus 1 (CAdV-1) and CAdV-2 cause infectious hepatitis and respiratory disease respectively14,15. Human diseases caused by adenoviruses include respiratory illness, conjunctivitis, hepatitis and gastroenteritis6. Fatal human pneumonia has also been noted due to adenovirus infection16. A severe adenovirus outbreak in a colony of captive monkeys in a Californian research facility, followed by primary transmission to one of the researchers and secondary transmission to one of his family members highlights the potential of zoonotic spillover of non-human adenoviruses and consequent human-to-human transmission17.
Here we describe the discovery, genomic and electron-microscopic characterization of a novel mastadenovirus, named Rousettus aegyptiacus adenovirus-3085 (RaegAdV-3085), isolated from a pool of liver and spleen tissues of an apparently healthy Egyptian fruit bat (Rousettus aegyptiacus) in South Africa.
Results
Virus isolation
A virus, causing cytopathic effect (CPE) was isolated from the liver/spleen homogenate of bat 3085, after three passages in Vero cells. The Egyptian fruit bat from which the virus was isolated, was collected in December 2013 as part of routine surveillance for bat borne viruses in South African bat populations. The virus infection resulted in disruption of the Vero cell monolayer by day 12 post infection, causing infected cells to shrink, become round and more refractory to light, and finally detach from the tissue culture plastic surface (inset A and B, Fig. 1). The virus replicates relatively slow in VeroE6 cells, reaching a peak in virus titre 15 days after inoculation (Fig. 1). The virus did not replicate in HEK293 cells (results not shown).
Figure 1.
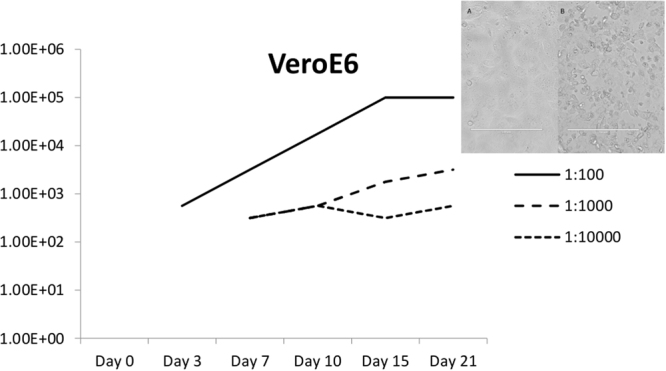
RaegAdV-3085 growth curve in VeroE6 cell culture. Different line types indicate different dilutions of stock virus. Y-axis represents the TCID50/mL of the virus in culture supernatant. The inset shows uninfected (A) and infected (B) VeroE6 cells with CPE at 12 days post inoculation.
Identification and characterization by electron microscopy
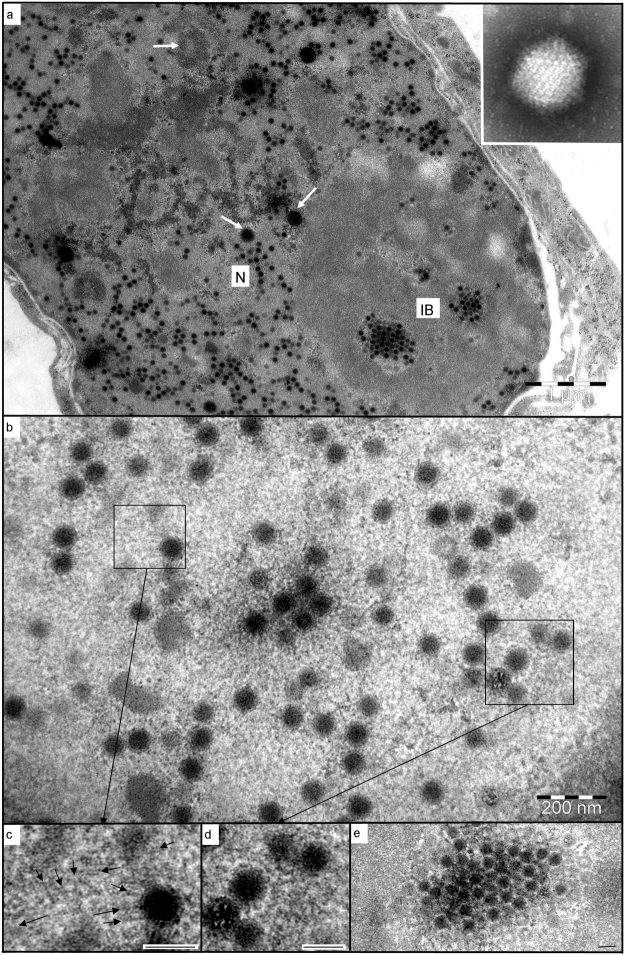
Negatively-stained virions were identifiable as adenovirus, being 70 nm in diameter, non-enveloped, and with spherical capsomers forming distinct facets of the icosahedra (Fig. 2a inset). Resin sections through infected Vero cells confirmed this finding, with numerous virus particles being formed within the nuclei (Fig. 2a), prior to lytic release from the cytoplasm. As the cells were in the late phase of infection, disruption and dispersal of the promyelocytic leukaemia bodies and Cajal bodies had occurred18, with only smaller, scattered remnants of these nuclear bodies evident (Fig. 2a).The similarities between adenovirus replication centres and nucleoli19 were evident in infected nuclei, with the underlying granular component forming a matrix in which virus particles developed in association with fibrillar components of differing osmiophilic densities (Fig. 2b–d). Nuclear actin filaments were seen in association with newly-forming capsids (Fig. 2b,c), similar to those described for alpha-herpesvirus neuronal infections20. Delicate, branched filaments were also visible around developing virus particles (Fig. 2d), consistent with the description and occurrence of viral E4-ORF3 protein21. Developing virus particles were typically filled with viral genomic material resembling host heterochromatin22, but empty capsids were frequently seen around the peripheries of the virus factories (Fig. 2e) – as in both Polyomavirus-infected cells23 and a novel strain of porcine adenovirus24.
Figure 2.
Electron microscopy of RaegAdV-3085 infected Vero cells. (a) Numerous virions forming within the nucleus (N), with crystalline arrays evident within the granular matrix of an inclusion body (IB). Remnants of probable nuclear bodies are scattered throughout the nucleus (arrows). INSET: a negatively-stained virion; (b) developing virus particles within an inclusion body; (c) actin-like filaments (arrows) associated with developing virion; (d) fine, proteinaceous filaments extending between developing virions; (e) one of a series of serial sections through a crystalline array, showing the predominance of empty capsids around the periphery. Scale bars (d,e,f) = 80 nm.
Preliminary identification by non-biased next generation sequencing
Initial attempts to identify the virus, concurrently with electron microscopy, were done by sequence independent single primer amplification (SISPA) as described recently for two novel RNA viruses we identified25,26. NGS following this approach yielded 14 contigs matching adenovirus sequences available on Genbank by dc-megablast. The number of reads making up the 14 contigs ranged from two to 1132 and contig lengths from 103 to 4405 nucleotides (Table 1).
Table 1.
Contigs from Illumina sequencing of RaegAdV-3085, Vero passage 3, using SISPA approach.
| Contig ID | Matching sequence accession number | Matching sequence description on Genbank | Matching sequence start position | Matching sequence end position | Number of reads in contig | Contig length | Percentage sequence identity |
|---|---|---|---|---|---|---|---|
| c3 | KF633445.1 | Human_adenovirus_B_/DEU/HEIM_00086 | 20889 | 20999 | 14 | 161 | 72.97 |
| c16 | AB686663.1 | Human_adenovirus_31_R187 | 162 | 62 | 3 | 104 | 75.25 |
| c24 | DQ630756.1 | Ovine_adenovirus_3_PX611 | 1827 | 1982 | 5 | 157 | 80.38 |
| c25 | AF153447.1 | Ovine_adenovirus_A | 355 | 428 | 7 | 127 | 82.43 |
| c43 | DQ630755.1 | Ovine_adenovirus_2_PX515 | 1847 | 1720 | 6 | 171 | 85.94 |
| c49 | KC692426.1 | Unidentified_adenovirus_PgAdV-10 | 185 | 278 | 2 | 107 | 80.85 |
| c64 | AF258784.1 | Tree_shrew_adenovirus_1 | 18638 | 18537 | 11 | 103 | 74.51 |
| c67 | HQ241818.1 | Simian_adenovirus_48_AJ75 | 23547 | 23621 | 15 | 230 | 82.67 |
| c70 | HQ605912.1 | Simian_adenovirus_20_VR-541 | 23847 | 23728 | 9 | 184 | 74.17 |
| c71 | Y07760.1 | Canine_adenovirus_type_1 | 22885 | 22763 | 7 | 156 | 73.98 |
| c75 | U40839.3 | Ovine_adenovirus_7 | 12098 | 11983 | 2 | 129 | 74.14 |
| c82 | KF268310.1 | Human_adenovirus_C/USA/Pitts_00109/1992/2 | 26840 | 26756 | 7 | 107 | 75.29 |
| c66 | JX885602.1 | Eidolon_helvum_adenovirus_1 | 832 | 1662 | 1132 | 4405 | 73.54 |
| c78 | JX885602.1 | Eidolon_helvum_adenovirus_1 | 2124 | 1948 | 16 | 186 | 77.97 |
Complete genome sequencing, annotation and phylogenetic analysis
The approach of direct Nextera DNA library preparation for MiSeq sequencing from DNA extracted from concentrated adenovirus (ultracentrifuged cell culture virus particles) yielded 716 contigs after de novo assembly, including one contig of 29,301 nucleotides. The 716 contigs were obtained from 912,422 cleaned paired reads, of which the 29,301 nt contig consisted of 232,419 paired reads (25% of total clean reads). The terminal ends of the genome, including a 40 bp ITR, were better resolved after bowtie buildout yielding a sequence of 29,342 nucleotides, encompassing likely the entire genome of RaegAdV-3085 (MG551742). BLAST analysis of this genome revealed highest similarity to Bat mastadenovirus WIV13 complete genome (KT698852)7 and Eidolon helvum adenovirus 1 hexon gene (JX88562)8. The RaegAdV-3085 genome shares a nucleotide identity of 70–74% with complete genomes of Bat mastadenoviruses WIV-12, 13, 17 and −18 (KT698856, KT698852, KX961095, KX961096), and 74% with a partial genome sequence (Hexon gene) of Eidolon helvum adenovirus 1 hexon gene (JX88562).
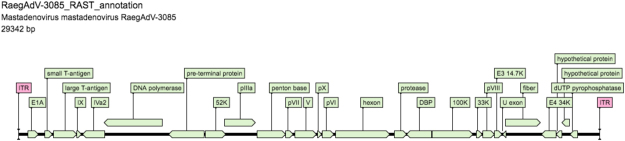
The length of the RaegAdV-3085 genome is smaller than that of most other mastadenoviruses, specifically bat associated adenoviruses, but similar to its closest related viruses from China7. The known ITR length range for most mastadenoviruses is 93 to 371 nt, with lengths of between 50 and 73 noted in bat mastadenoviruses, suggesting that the terminal ends of RaegAdV-3085 might not have been fully resolved2. The RaegAdV-3085 genome has a G + C content of 36.4%, significantly lower than other recognized mastadenoviruses (43.6 to 63.9%)1, but similar to recently isolated bat adenoviruses from China7. The RaegAdV-3085 genome contains 27 open reading frames (ORFs) with recognizable similarity to known adenovirus proteins (Table 2; Fig. 3).
Table 2.
Open reading frames and ITRs in the 29,342 bp genome of RaegAdV-3085.
| ORF number | Gene/protein designation or feature | Predicted description or function of protein | Structural or non-structural | Gene (nt) | Protein (aa) | Genomic position |
|---|---|---|---|---|---|---|
| ITR | Forms panhandles for replication | N/A | 40 | N/A | 1–40 | |
| 1 | E1A | Transccriptional activator | Non-structural | 516 | 171 | 456–971 |
| 2 | E1B small | Small T-antigen | Non-structural | 372 | 123 | 1,324–1,695 |
| 3 | E1B large | Large T-antigen | Non-structural | 1,173 | 390 | 1,755–2,927 |
| 4 | IX | Minor capsid protein | Structural | 249 | 82 | 2,942–3,190 |
| 5 | IVa2 | DNA packaging ATPase | Structural | 1,110 | 369 | 3,214–4,323 (c-strand) |
| 6 | pol | DNA polymerase | Non-structural | 2,922 | 973 | 4,311–7,232 (c-strand) |
| 7 | pTP | Terminal protein | Structural | 1,794 | 597 | 7,568–9,361 (c-strand) |
| 8 | 52 K | DNA packaging protein | Non-structural | 1,014 | 337 | 9,415–10,428 |
| 9 | pIIIa | Minor capsid protein | Structural | 1,566 | 521 | 10,382–11,947 |
| 10 | Penton | Major capsid protein | Structural | 1,425 | 474 | 12,029–13,453 |
| 11 | pVII | Major core protein | Structural | 390 | 129 | 13,459–13,848 |
| 12 | V | Minor core protein | Structural | 1,101 | 366 | 13,929–15,029 |
| 13 | pX (µ) | Minor core protein | Structural | 198 | 65 | 15,084–15,281 |
| 14 | pVI | Minor capsid protein | Structural | 600 | 199 | 15,324–15,923 |
| 15 | Hexon | Major capsid protein | Structural | 2,727 | 908 | 15,988–18,714 |
| 16 | AVP | Protease | Structural | 603 | 200 | 18,943–19,545 |
| 17 | DBP | DNA binding protein | Non-structural | 1,260 | 419 | 19,583–20,842 (c-strand) |
| 18 | 100 K | Hexon scaffold protein | Non-structural | 2,049 | 682 | 20,853–22,901 |
| 19 | 33 K | DNA packaging/assembly protein | Non-structural | 291 | 96 | 23,087–23,377 |
| 20 | pVIII | Minor capsid protein | Structural | 585 | 194 | 23,428–24,012 |
| 21 | E3 14.7 K | Immuno-modulatory protein | Non-structural | 372 | 123 | 24,016–24,387 |
| 22 | U exon | Replication centre protein | Non-structural | 168 | 55 | 24,399–24,566 (c-strand) |
| 23 | Fiber protein | Major capsid protein | Structural | 1,773 | 590 | 24,565–26,337 |
| 24 | E4 ORF34K | p53 and p73 inhibitor | Non-structural | 738 | 245 | 26,368–27,105 (c-strand) |
| 25 | Hypothetical protein | Unknown | Unknown | 309 | 102 | 27,118–27,426 (c-strand) |
| 26 | Hypothetical protein | Unknown | Unknown | 396 | 131 | 27,420–27,815 (c-strand) |
| 27 | dUTP pyrophosphatase | pyrophosphatase | Non-structural | 393 | 130 | 27,809–28,201 (c-strand) |
| ITR | Form panhandles for replication | N/A | 40 | N/A | 29,303–29,342 |
Figure 3.
Genomic organization of RaegAdV-3085 genome (29,342 bp). Open reading frames and predicted gene names are shown.
The putative penton protein of RaegAdV-3085 contains an Arg-Gly-Asp (RGD) motif at position 271–271 (amino acid sequence), which is a common feature of many human adenoviruses involved in binding cell surface integrins27. Similar to BtAdV-WIV12, 13, 17 and 18, RaegAdV-3085 encodes only one putative E3 protein of 14.7 K.
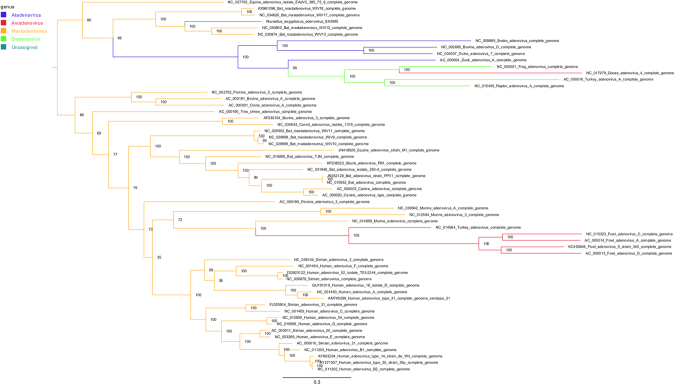
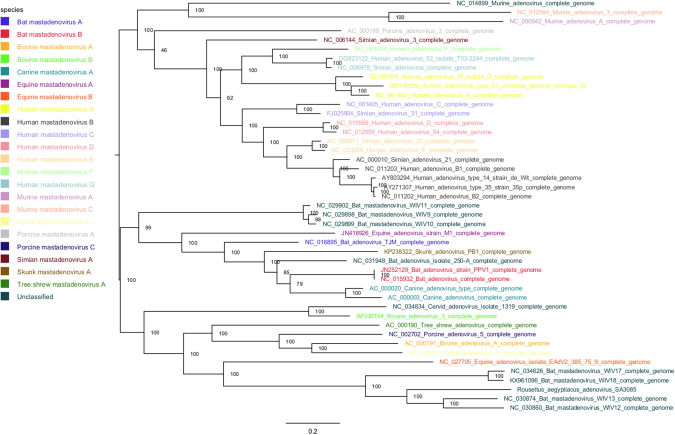
Phylogenetic analysis confirms that the RaegAdV-3085 virus belongs to the Mastadenovirus genus in the Adenoviridae family (Fig. 4). Within the genus, the virus forms a clade with the four viruses isolated from bats in China7. Within the clade, RaegAdV-3085 seems to group more closely with WIV-12 and WIV-13, both of which were isolated from Miniopterus schreibersii bats, as opposed to WIV-17 and WIV-18 that were both isolated from Rousettus leschenaulti bats (Fig. 5).
Figure 4.
Maximum likelihood analysis (RAxML) of the RaegAdV-3085 genome with genomes of representative viruses from all genera within the Adenoviridae family. Branches are coloured according to the genus to which the respective viruses belong, as indicated in the legend. The tree is rooted at the midpoint and nodes ordered in decreasing order.
Figure 5.
Maximum likelihood analysis (RAxML) of the RaegAdV-3085 genome with genomes of representative viruses from different species within the Mastadenovirus genus. Sequence names are coloured according to the species to which they belong (legend). The tree is rooted at the midpoint and nodes ordered in decreasing order.
Discussion
We describe the isolation, identification, electron-microscopic and sequence characterization of a novel adenovirus isolated from an Egyptian fruit bat in South Africa. The source animal was apparently healthy and was sampled as part of surveillance for bat-borne zoonotic pathogens, in a cave situated in Limpopo Province. The virus, provisionally named Rousettus aegyptiacus adenovirus-3085, abbreviated to RaegAdV-3085, groups phylogenetically with the Mastadenovirus genus and shares genome organization characteristics.
Bat-associated adenoviruses are diverse in their DNA sequence as well as genome size and G + C content6,7. The new virus described here further demonstrates this since it is only 74% related to its closest relative based on full genome DNA sequence, and has a relatively short genome with low G + C content. Based on phylogenetic analysis the virus falls within a clade formed by four viruses isolated recently in China. Interestingly, two of these viruses were isolated from the Rousettus leschenaulti fruit bat, a species related to the Egyptian fruit bat, yet RaegAdV-3085 is more closely related to the other two viruses isolated from insectivorous common bent-wing bats (Miniopterus schreibersii). This is surprising considering that adenoviruses are thought to have co-evolved with their hosts7. This observation might be explained by the fact that relatively few bat-associated adenoviruses have been isolated and characterized to date, thus the phylogeny might only be resolved better once more representative adenoviruses from more bat species have been sequenced. In fact, the first adenovirus from bats was only discovered ten years ago9. Another possible explanation might reside in the knowledge of the ecological niche of the particular Egyptian fruit bat population studied here. The cave hosting these bats is co-inhabited by up to three different insectivorous bat species, including a large seasonal colony of the Natal long-fingered bat (Miniopterus natalensis), a close relative of the common bent-wing bat. It is therefore not irrational to hypothesize that the Egyptian fruit bat from which the virus was isolated might have been only coincidently infected through close contact with co-roosting Natal long-fingered bats. This can, however, only be concluded when more sequences become available from these potential hosts. Both of these bat species are widely distributed with overlapping ranges; the Natal long fingered bat is distributed across Southern- and East Africa, into the Arabian Peninsula, while the Egyptian fruit bat is additionally found in West Africa, North Africa and south-west Asia.
The reservoir status will have to be addressed in future studies aimed at surveillance for RaegAdV-3085 infected bats in the cave (and other) through molecular and/or serological testing. Also useful would be experimental infection studies of suspected host reservoir species to evaluate pathological, virological and serological responses. Such studies might also yield data on transmission routes between bats and possible maintenance mechanisms. A wide number of viruses have been detected in or isolated from bats in the last decade due to an exponential increase in the interest in bats and their role in harbouring dangerous zoonotic pathogens. The transmission routes of a number of these viruses between bats, or from bats to other species, are known or can be inferred from the tissues in which they were detected. However, there are still viruses, for which bats have been determined or are hypothesized to be reservoir hosts, such as the filoviruses where the exact transmission routes remain enigmatic28. Elucidating such mechanisms for more bat-associated viruses might shed more light in general on how bats can maintain and transmit pathogens of human and veterinary health importance.
The fact that RaegAdV-3085 contains the Arg-Gly-Asp (RGD) motif in its putative penton protein sequence, usually a common feature of human adenoviruses that plays a role in binding cell surface integrins, and the ability of the virus to replicate in non-human primate cells in vitro (Vero), suggests that the virus might not be restricted only to replication in bats. Although adenoviruses are relatively host-specific, it was shown recently that another bat adenovirus (BtAdV 250-A) replicates efficiently in a range of host cells in vitro, from fox to monkey cells2. Thus it seems like bat associated adenoviruses in general are able to infect a wider range of host cells, including human cells in vitro5.
An interesting aspect to consider in research into a better understanding of maintenance and transmission of dangerous zoonotic pathogens in bat populations is the possibility of co-infections and how these affect the immune status of bats, subsequent shedding patterns, and even recrudescence of possible latent infections due to an overburdened immune system. With the high number of pathogens recently identified in bats, it is possible to imagine the likelihood of a bat being co-infected or infected with different pathogens in short succession, leading to a weakened immune system and possibly increased shedding.
The isolation of RaegAdV-3085 expands the repertoire of adenoviruses isolated from bats as well as their geographical distribution. Additionally, it highlights the likely wealth of undiscovered bat associated (but also other mammalian) adenoviruses and the need to intensify efforts to better resolve our knowledge on the evolution of these viruses as well as cross-species transmission events.
Methods
Virus isolation and source animals
Egyptian fruit bats (Rousettus aegyptiacus) were sampled between March 2013 and March 2014 at Matlapitsi cave in the Matlapitsi Valley, Limpopo Province, South Africa, as described before25,26. Tissues were collected from a total of 102 of these bats. Liver and spleen pools (combined ± 0.1 g) were homogenized (30 Hz for 8 minutes using a Tissuelyzer II and 5 mm stainless steel beads, Qiagen) in Eagle’s minimum essential medium (10% w/v) and clarified supernatants used to inoculate Vero cell cultures, and monitored as described before25,26. A growth curve experiment in VeroE6 cells was performed as follows. Stock RaegAdV-3085 (1 × 105.7 TCID50/mL) was diluted 10−2; 10−3 and 10−4 in culture medium, and 5 mL of each dilution used to inoculate 75 cm2 culture flasks containing 95% confluent VeroE6 monolayers. The inoculum was removed after 1 hour and fresh culture medium added, after which the flasks were incubated at 37 °C in 5% CO2 for 21 days. Aliquots of culture supernatant was collected on the day of inoculation, and thereafter on days 3, 7, 10, 15 and 21 from all three flasks. Supernatant collections were subjected to standard TCID50 titration as described previously26.
Sequencing
Initially, sequencing was performed as described previously using a sequence-independent single primer amplification (SISPA) method, biased towards RNA amplification25. When it became apparent that the unknown virus was a DNA virus, an alternative method was followed. Virus was propagated and concentrated as described previously with some modification29. Infected cells were harvested at 75% CPE, freeze-thawed three times and centrifuged at 3000 × g at 4 °C for 15 minutes to remove cell debris. The virus was pelleted by ultracentrifugation and resuspended in TE buffer (pH 8.0). A volume of 20 ml virus suspension was added to the 30 ml centrifuge tube, and 10 ml ice-cold sucrose solution (30% w/w) layered beneath it. The tube was centrifuged at 141 000 × g in a Beckman SW-28 rotor for 90 minutes at 4 °C. The supernatant was removed and the pellet resuspended in 0.5 ml of TE buffer. Total DNA was then extracted from the concentrated virus stock using a Qiagen DNA blood mini-kit. Libraries were prepared using the Nextera DNA library preparation kit and sequencing performed on an Illumina MiSeq instrument.
Transmission electron microscopy
Processing of infected cell cultures for transmission electron microscopy was performed as described previously25,26. Briefly, culture supernatant was concentrated, adsorbed onto Formvar-coated grids, negatively stained with saturated, aqueous uranyl acetate and viewed. Infected monolayers were routinely processed for ultramicrotomy (primary fixation in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 6.9, post-fixation in 1% osmium tetroxide, embedding in a low-viscosity resin, double-staining of 70 nm sections).
Phylogenetic and sequence analysis
Illumina sequence data was processed as described previously25: quality filtering was conducted with Prinseq-lite and reads assembled into contigs using Ray Meta with kmer length = 25. The complete sequence of RaegAdV-3085 was obtained in one contig (29,342 bp). Alignment with sequences available on the NCBI-Nucleotide database (Genbank) was performed using MAFFT v7.22230. Maximum likelihood analysis was performed using RAxML v8.231. The genome was annotated using Prokka32 and WebDSV (http://www.molbiotools.com/WebDSV/).
Ethical Statement and regulatory requirements
This study was carried out according to the recommendations of the South African National Standards for the Care and Use of Animals for Scientific Purposes (SANS 10386: 2008). The field sampling protocols and transport of Rousettus aegyptiacus and samples collected from this species are approved by the National Health Laboratory Service Animal Ethics Committee (AEC 137/12), University of Pretoria Animal Ethics Committee (EC054–14), Department of Economic Development, Environment and Tourism: Limpopo Province Directorate: Wildlife Trade and Regulation Permit (CPM 006806) and the South African Department of Agriculture, Forestry and Fisheries (Section 20 approval 12/11/1/1/8).
Genbank accession numbers
Acknowledgements
The authors would like to thank the following individuals for their contributions towards fieldwork and technical assistance: Busi Mogodi, Justice Kgatitsoe, Alan Kemp, Stewart McCulloch, Terence Scott, Joe Kgaladi, Marinda Mortlock, Marike Geldenhuys, Jessica Coertse and Andre Coetzer. The project is jointly funded by the following grants awarded to: Janusz T. Paweska (CDC Global Disease Detection program, GDD 5U19 GH000571–05/96667) and Petrus Jansen van Vuren (South African National Research Foundation, Incentive Funding for Rated Researchers, Grant UID 85544). This work was financially supported in part by the National Research Foundation (NRF) of South Africa: the South African Research Chair held by WM, grant no. 98339, as well as grant numbers 92524, 85756, and 91496. The grant holders acknowledge that opinions, findings and conclusions or recommendations expressed in any publication generated by GDD and NRF supported research are those of the authors and that the GDD and NRF accept no liability whatsoever in this regard.
Author Contributions
J.T.P. and P.Jv.V. designed the experiments. P.Jv.V. wrote the manuscript. P.Jv.V., M.A., M.R.W., G.P., A.I., N.S., M.B., W.M. and J.T.P. contributed to field work or laboratory experimental work. P.Jv.V., M.A., M.R.W., G.P., A.I., N.S., M.B., W.M. and J.T.P. edited and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Harrach, B. et al. Family Adenoviridae. In Virus Taxonomy Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses, pp. 125–141. Edited by A. M. Q. King, M. J. Adams, E. B. Carstens & E. J. Lefkowitz. San Diego: Elsevier (2011).
- 2.Hackenbrack N, et al. Evolution and cryo-electron microscopy capsid structure of a North-American bat adenovirus and its relationship to other mastadenoviruses. J Virol. 2017;91(2):e01504–16. doi: 10.1128/JVI.01504-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohl C, et al. Genome analysis of bat adenovirus 2: Indications of interspecies transmission. J Virol. 2012;86(3):1888–1892. doi: 10.1128/JVI.05974-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonntag M, Mühldorfer K, Speck S, Wibbelt G, Kurth A. New adenovirus in bats, Germany. Emerg Infect Dis. 2009;15(12):2052–2055. doi: 10.3201/eid1512.090646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, et al. Host range, prevalence, and genetic diversity of adenoviruses in bats. J Virol. 2010;84(8):3889–3897. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan B, et al. Novel bat adenoviruses with an extremely large E3 gene. J Gen Virol. 2016;97:1625–1635. doi: 10.1099/jgv.0.000470. [DOI] [PubMed] [Google Scholar]
- 7.Tan B, et al. Novel bat adenoviruses with low G + C content shed new light on the evolution of adenoviruses. J Gen Virol. 2017;98(4):739–748. doi: 10.1099/jgv.0.000739. [DOI] [PubMed] [Google Scholar]
- 8.Baker KS, et al. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virol. 2013;441:95–106. doi: 10.1016/j.virol.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda K, et al. Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae) Emerg Infect Dis. 2008;14(2):347–349. doi: 10.3201/eid1402.070932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams MJ, Lefkowitz EJ, King AMQ, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol. 2016;159:2831–2841. doi: 10.1007/s00705-014-2114-3. [DOI] [PubMed] [Google Scholar]
- 11.Drexler JF, et al. Amplification of emerging viruses in a bat colony. Emerg Infect Dis. 2011;17(3):449–456. doi: 10.3201/eid1703.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jánoska M, et al. Novel adenoviruses and herpesviruses detected in bats. Vet J. 2011;189(1):118–121. doi: 10.1016/j.tvjl.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Li L, et al. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. 2010;84(14):6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Appel M, Bistner SI, Menegus M, Albert DA, Carmichael LE. Pathogenicity of low-virulence strains of two canine adenovirus types. Am J Vet Res. 1973;34:543–550. [PubMed] [Google Scholar]
- 15.Swango LJ, Wooding WL, Jr., Binn LN. A comparison of the pathogenesis and antigenicity of infectious canine hepatitis virus and the A26–61 virus strain (Toronto) J Am Vet Med Assoc. 1970;156:1687–1696. [PubMed] [Google Scholar]
- 16.Yatsyshins SB, et al. Complete genome sequence of human adenovirus 7 associated with fatal adult pneumonia. Genome Announc. 2016;4(5):e01204–16. doi: 10.1128/genomeA.01204-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen EC, et al. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 2011;7:e1002155. doi: 10.1371/journal.ppat.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid M, Speiseder T, Dobner T, Gonzalez RA. DNA virus replication compartments. J Virol. 2014;88(3):1404–1420. doi: 10.1128/JVI.02046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hidalgo P, et al. Morphological, biochemical and functional study of viral replication compartments isolated from Adenovirus-infected cells. J Virol. 2016;90(7):3411–3427. doi: 10.1128/JVI.00033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchsova B, Serebryannyy LA, de Lanerolle P. Nuclear actin and myosins in Adenovirus infection. Exp Cell Res. 2015;338(2):170–182. doi: 10.1016/j.yexcr.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou HD, et al. A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors. Cell. 2012;151:304–319. doi: 10.1016/j.cell.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu T, Nagata K, Wodrich H. The role of nuclear antiviral factors against invading DNA viruses: the immediate fate of incoming viral genomes. Viruses. 2016;8:290. doi: 10.3390/v8100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erickson KD, et al. Virion assembly factories in the nucleus of Polyomavirus-infected cells. PLoS Pathog. 2012;8(4):e1002630. doi: 10.1371/journal.ppat.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerman UD, et al. A novel strain of porcine adenovirus detected in urinary bladder urothelial cell culture. Viruses. 2014;6:2505–2518. doi: 10.3390/v6062505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen van Vuren P, et al. Isolation of a novel fusogenic Orthoreovirus from Eucampsipoda africana bat flies in South Africa. Viruses. 2016;8:65. doi: 10.3390/v8030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen van Vuren P, et al. Isolation of a novel orthobunyavirus from bat flies, (Eucampsipoda africana) J Gen Virol. 2017;98(5):935–945. doi: 10.1099/jgv.0.000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berk, A. J. Adenoviruses, p 1704–1731. In Knipe, D. M., Howley & P. M. (eds), Field’s virology, 6th ed. Lippincott/Williams & Wilkins, Philadelphia, PA.
- 28. Paweska, J. T. et al. Lack of Marburg virus transmission from experimentally infected to susceptible in-contact Egyptian fruit bats. J Infect Dis Supplement, S1–10 (2015). [DOI] [PubMed]
- 29.Ojkic D, Nagy E. The long repeat region is dispensable for fowl adenovirus replicationin vitro. Virol. 2001;283:197–206. doi: 10.1006/viro.2000.0890. [DOI] [PubMed] [Google Scholar]
- 30.Katoh K, Stanley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]