Abstract
Eukaryotic tRNAs are transcribed as precursors. A 5′-end leader and 3′-end trailer are endonucleolytically removed by RNase P and 3′-tRNase before 3′-end CCA addition, aminoacylation, nuclear export and translation. 3′-End -CC can be a 3′-tRNase anti-determinant with the ability to prevent mature tRNA from recycling through 3′-tRNase. Twenty-two tRNAs punctuate the two rRNAs and 13 mRNAs in long, bidirectional mitochondrial transcripts. Accurate mitochondrial gene expression thus depends on endonucleolytic excision of tRNAs. Various mitochondrial diseases and syndromes could arise from defective tRNA end processing. The U7445C substitution in the human mitochondrial L-strand transcript (U74C directly following the discriminator base of tRNASer(UCN)) causes non-syndromic deafness. The sequence of the precursor (G↓UCU) becomes G↓CCU, resembling a 3′-tRNase anti-determinant. We demonstrate that a tRNASer(UCN) precursor with the U7445C substitution cannot be processed in vitro by 3′-tRNase from human mitochondria. A 3′-end processing defect in this tRNA precursor could thus be responsible for mitochondrial disease.
INTRODUCTION
tRNA end processing and a 3′-tRNase anti-determinant
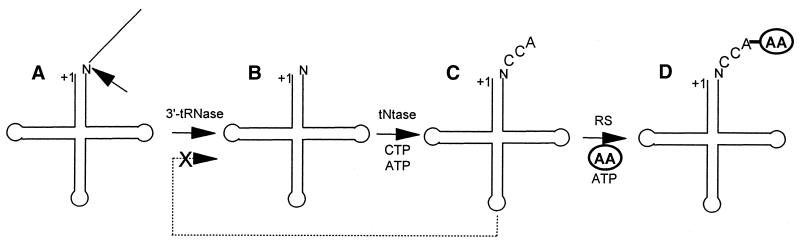
Removal of a 5′-end leader and 3′-end trailer from tRNA precursors are early, required steps in tRNA maturation. RNase P is typically a ribonucleoprotein enzyme which exists as a ribozyme in bacteria (1); its RNA component is also essential in yeast (2). 3′-tRNase, a eukaryotic 3′-end processing endonuclease (3–7), can precisely remove the 3′-end trailer from tRNA precursors (Fig. 1A and B). In eukaryotes, CCA addition to the 3′-end by tRNA nucleotidyltransferase (tNtase; Fig. 1B and C) is required before aminoacylation (Fig. 1C and D) because this sequence is not genomically encoded. The 3′-end -CC of -CCA in mature tRNA has been shown to be an anti-determinant for 3′-tRNase from mouse, pig and fruit fly (7,8). This anti-determinant could prevent mature tRNA from recycling through 3′-tRNase (dashed arrow from C to A, B with an X through it in Fig. 1), thus ensuring smooth progress of tRNA precursors through end maturation to aminoacylation. A tRNA precursor with -CCA- following the discriminator base accumulates when microinjected into Xenopus oocytes (E. Lund and J. Dahlberg, personal communication), consistent with this interpretation.
Figure 1.
Eukaryotic tRNA end processing from the 3′-tRNase reaction through aminoacylation. The tRNA has been processed by RNase P to +1. (A) The RNase P product is a substrate for 3′-tRNase (←). N, discriminator base. (B) The 3′-tRNase product is a substrate for CCA addition by tRNA nucleotidyltransferase (tNtase). (C) The tNtase product (mature tRNA) is a substrate for aminoacylation by aminoacyl tRNA synthetase (RS). (D) Charged tRNA. The dashed arrow from (C) to (A, B) with an X through it signifies that mature tRNA with CCA at its 3′-end is a 3′-tRNase anti-determinant (8); it is neither a substrate nor a good inhibitor.
Mitochondrial gene expression and tRNA end processing
Maternally transmitted diseases and syndromes correlate with numerous mutations in human mitochondrial tRNA genes (for a review see 9; mitomap web site at www.infinity.gen.emory.edu/mitomap). Twenty-two tRNAs punctuate the 13 mRNAs and two rRNAs (10). There is only one mitochondrial tRNA for each of 18 amino acids and two each for Leu (UUR and CUN) and Ser (AGY and UCN). No cytoplasmic tRNAs are imported into human mitochondria. A tRNA processing defect could thus reduce mitochondrial gene expression and cause disease.
Ten mitochondrial tRNAs require precise cleavage at their 5′-ends because the neighboring nucleotide upstream forms the 3′-end of another mature RNA species (11). Three of the mRNAs upstream of a tRNA (ND1, ND3 and ND6) require addition of one or two A residues to complete the termination codon. Similarly, tRNATyr must have an A (the discriminator base) added to its 3′-end after endonucleolytic cleavage of tRNACys at +1 (12). Ten tRNAs (not always the same ones) require precise cleavage at their 3′-ends because the next nucleotide downstream is at the mature 5′-end of another RNA (13).
Defective tRNA processing models for mitochondrial diseases have been investigated. King et al. (14) found RNA 19, a large precursor, associated with the G3243A (MELAS) mutation (G14A in tRNALeu(UUR)). Bindoff et al. (15) also observed RNA 19 in mitochondria with a A3302G mutation (A71G in the same tRNA). Reid et al. (16) and Guan et al. (17) noted a reduced steady-state level of tRNASer(UCN) (25–30% of normal) in cell lines homoplasmic for the T7445C substitution, which causes non-syndromic deafness. The T7445C substitution (U7445C in L-strand RNA and U74C in the tRNASer(UCN) precursor; see Fig. 2 and below) was suggested to cause a 3′-end tRNA processing defect (16,17). A7445G in the H-strand transcript is predicted to be a silent mutation in the termination codon of the COX1 gene. Interestingly, several kilobases of L-strand RNA separate tRNASer(UCN) from the neighboring ND6 mRNA upstream and from the cluster of four tRNAs starting with tRNATyr downstream. These long spacers could explain why discrete precursors of U7445C tRNASer(UCN) were not observed by northern blotting (17).
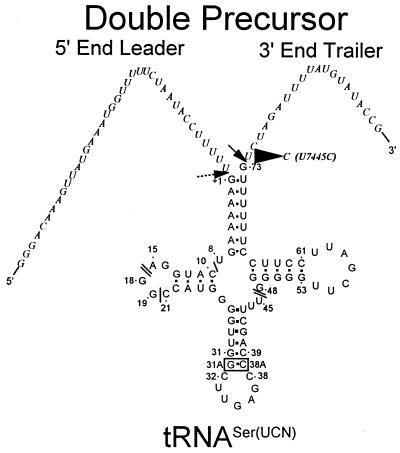
Figure 2.
Human mitochondrial tRNASer(UCN) secondary structure and sequence of the double precursor (DP). Horizontal and diagonal arrows signify RNase P and 3′-tRNase cleavage sites, respectively. Thirty-eight nucleotides of 5′-end leader and 20 nt of 3′-end trailer are shown, including the U7445C substitution which changes the sequence GØUCU to GØCCU. /s indicate the positions of nucleotides in canonical tRNA (27,28) which are missing from tRNASer(UCN) and the box encloses an extra base pair (31A, 38A) in the anticodon stem.
In vitro studies of mammalian mitochondrial tRNA end processing
Rat mitochondrial tRNA end processing was investigated by Manam and Van Tuyle (18) and human mitochondrial tRNA end processing by Rossmanith et al. (19). tRNA modification is not generally required for end processing, but human mitochondrial tRNALys requires modification to prevent incorrect folding (20). Efforts to characterize human mitochondrial RNase P have been controversial (21; compare with 18,19,22); a recent report (23) suggests that human mitochondrial and nuclear RNase P have the same RNA component. Some of the disease-causing substitutions in tRNAs reduce the ability of the precursors to be processed by RNase P (24). While mitochondrial tRNA 3′-end processing has been less thoroughly investigated, the production of mature tRNA using extracts from a precursor that has both a 5′-end leader and a 3′-end trailer has been observed and, in some cases, the RNA 3′-end trailer was detected (see for example 18,19). Although the order of tRNA end processing reactions in human mitochondria is uncertain, several reports favor initial cleavage of cytoplasmic (4,5,25) and mitochondrial (18,19) tRNA precursors by RNase P.
A substitution in the tRNASer(UCN) precursor proximal to the in vitro 3′-tRNase cleavage site, which causes non-syndromic deafness, resembles a 3′-tRNase anti-determinant
The U7445C mutation in tRNASer(UCN), located 1 nt downstream from the discriminator base (Fig. 2), would be removed by 3′-end processing (16,17) and thus could not affect the structure or function of mature tRNA. This substitution changes the precursor sequence G↓UCU (↓ indicates the in vitro 3′-tRNase processing site) to G↓CCU (Fig. 2), which resembles a 3′-tRNase anti-determinant (8).
We investigated 3′-end processing of wild-type and U7445C tRNASer(UCN) precursors in vitro, which required precursor design and fractionation of human mitochondrial 3′-tRNase. The wild-type precursor with a mature 5′-end and an 18 nt 3′-end trailer could be efficiently processed by 3′-tRNase, but variant processing was undetectable.
MATERIALS AND METHODS
Preparation of tRNASer(UCN) precursors
A double precursor (Fig. 2) with a 38 nt 5′-end leader and a 20 nt 3′-end trailer was efficiently transcribed using a template amplified from total human (including mitochondrial) DNA. The T7 promoter and three G residues were introduced at the 5′-end of the 5′-end leader and an MspI run-off site was introduced at the 3′-end of the 3′-end trailer. EcoRI and BamHI sites were used for subcloning. Accuracy was confirmed by DNA sequencing.
Templates for T7 initiation at +1 of mature tRNASer(UCN) and/or run-off at +73 gave no RNAs of the expected sizes, perhaps due to the unusual homopolymeric runs (A5 at +2–6, G6 at 48–53 and U6 at 67–72; see Fig. 2). Attempts at T7 transcription with four different labeled NTPs, use of a pGpA primer and unlabeled transcription with a higher concentration of NTPs were all unsuccessful.
To obtain the needed additional RNAs for this project, we constructed tRNA precursors from synthesized -tRNAs by ligation (26). Oligonucleotides +1–38 (5′ -tRNA, oligo 1) and +38A–89 (with an extra paired nucleotide in the anticodon stem; 27,28) (3′ -tRNA, oligo 2) were synthesized by Dharmacon on the 0.2 µmol scale and gel purified by us. A phosphate was attached to the 5′-end of oligo 2 by polynucleotide kinase; oligo 1 and kinased oligo 2 were mixed, annealed and ligated with T4 RNA ligase.
We also used cis-acting ribozymes (29; S. Searles, personal communication) to efficiently obtain well-defined 5′- and 3′-ends of the tRNA precursor. The hammerhead ribozyme was linked to the 5′-end of a tRNASer(UCN) gene by construction using overlapping oligonucleotides and the hepatitis delta virus ribozyme template (the PstI–HindIII segment from pSS419, a kind gift of S. Searles, MRC, Cambridge, UK) was added to the 3′-end to obtain a cleavage site at +94. After T7 transcription, ribozymes were activated by increasing the concentration of MgCl2 to 25 mM and heating to 90°C for 5 min, followed by incubation at 55°C for 30 min. The resulting tRNAs were gel purified twice.
RNA labeling and processing reactions
The human mitochondrial tRNASer(UCN) double precursor (Fig. 2, DP in Fig. 3A and C) was internally labeled by incorporation of [α-32P]UTP during T7 transcription as previously described (6). 5′-End-labeling of precursors using T4 polynucleotide kinase and [γ-32P]ATP was also performed as described. 3′-End-labeling of tRNA produced by -tRNA ligation was performed using T4 RNA ligase and [32P]Cp (30).
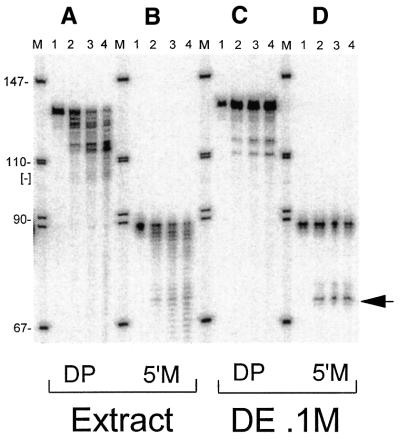
Figure 3.
A template with a mature 5′-end and fractionation of mitochondrial extract are required to observe 3′-tRNase activity. (A) The 127 nt double precursor (DP) with a 38 nt 5′-end leader and a 20 nt 3′-end trailer was incubated at 37°C with human mitochondrial extract for 0, 10, 20 and 30 min in lanes 1–4, respectively. [–] at left indicates the expected position of the 107 nt intermediate that would be produced by the activity of 3′-tRNase alone. An 89 nt intermediate would be produced by the activity of RNase P alone. Mature tRNA produced by the activity of both RNase P and 3′-tRNase would be 69 nt. (B) The precursor with a mature 5′-end and an 18 nt 3′-end trailer (produced by ligation of -tRNAs) was incubated with extract as in (A). (C) The double precursor [same substrate as in (A)] was incubated with the DEAE–Sepharose 0.1 M KCl fraction. (D) Precursor with a mature 5′-end [same substrate as in (B)] was incubated with the DEAE–Sepharose 0.1 M KCl fraction. ← to the right of (D) indicates the predicted tRNA product of reaction with 3′-tRNase.
Preparation and fractionation of human culture cell mitoplast extract
A seed culture of HeLa cells (a kind gift of J. Manley, Columbia) was grown in minimal essential medium supplemented with 5% calf serum in spinner flasks and up to 8 l of cells were harvested weekly at a density of 5 × 105/ml. Mitochondria were isolated from fresh cells using sucrose density step gradients (31). The outer membrane was stripped from the mitochondria by treatment with digitonin, producing mitoplasts. The extract was prepared by treatment of mitoplasts with 0.5% Triton X-100 and 0.15 M KCl followed by centrifugation (19).
We used the activity of lactate dehydrogenase (LDH), a marker enzyme for cytosol, to estimate the cytosolic contamination of mitochondria (not shown). LDH activity in the mitoplast extract (8 ml) was 2.4% of the concentration in cytosol (400 ml). We used PCR of a 305 bp mitochondrial DNA segment (5587–5891) spanning the L-strand tRNA gene cluster from tRNATyr to tRNAAla to estimate the enrichment of mitochondrial DNA in mitoplast extract relative to cytosol. Amplified band intensities from mitoplast extract and cytosol matched with an ∼150 times lower volume of mitoplast template.
Mitochondrial DNA could contaminate the cytosol because the centrifugation used to prepare the cytosolic post-mitochondrial supernatant is insufficient to remove all mitochondria and/or because some mitochondria may be damaged during cell lysis. Moreover, mitochondrial DNA may not be quantitatively extracted from mitoplasts in the above preparation. Nonetheless, the low level of LDH (cytoplasmic) contamination of the mitoplast extract suggests that the observed 3′-tRNase activity comes from mitochondria.
Mitoplast extract was adjusted to 30 mM K–HEPES, pH 8, 1 mM MgCl2, 1 mM DTT, 0.1% Tween 20, 0.1 mM PMSF and 10% glycerol (CB) containing 50 mM KCl (50 mM KCl–CB) by passing it over a Sephadex G25 column equilibrated with the same buffer. Adjusted extract was loaded on a DEAE–Sepharose column equilibrated with 50 mM KCl–CB and step eluted with 0.1, 0.2, 0.5 and 1 M KCl–CB. The peak of 3′-tRNase activity elutes in 0.1 M KCl–CB, as previously reported for fruit fly nuclear 3′-tRNase (6).
Processing reactions
Labeled tRNA precursors were incubated with mitoplast extract or with the DEAE–Sepharose 0.1 M KCl fraction at 37°C in 50 mM KCl–CB containing 40 U RNasin/ml. Reactions with 3′-tRNase at a volume ratio of 1:20 were terminated after 0, 10, 20 and 30 min and recovered for electrophoresis on 6% denaturing polyacrylamide gels. Dried gels were exposed to a storage phosphor screen and scanned with a PhosphorImager. In a Michaelis–Menten experiment, 5′-end-labeled wild-type tRNASer(UCN) precursor was mixed with unlabeled tRNA covering a concentration range from 1 to 50 × 10–8 M substrate. Km was determined by Eadie–Hofstee analysis as previously described (30).
Structure probing
End-labeled wild-type and U7445C precursor tRNAs were mixed with 1.25 µg unlabeled yeast tRNA, denatured at 90°C in water and refolded by adding buffer to make 10 µl at a final concentration of 15 mM Tris–HCl, pH 8, 175 mM KCl, 3 mM MgCl2 and cooling to room temperature. Labeled tRNAs were added to an equal volume of RNase T1 (to a final concentration of 0.5, 1 or 2 U/ml) or RNase A (to a final concentration of 2, 4 or 8 × 10–3 U/ml) in the same buffer. After incubation for 5 min at room temperature, RNAs were deproteinized, recovered and electrophoresed on a 6% denaturing gel. Images were obtained by exposure to a storage phosphor screen.
RESULTS
Substrate with a mature 5′-end and fractionated extract are both required for in vitro cleavage by 3′-tRNase
We prepared wild-type and U7445C tRNASer(UCN) precursors for in vitro reactions using a human cell culture mitoplast extract (Fig. 3). Various bands were observed when the internally labeled 127 nt double precursor (Fig. 2) with a 38 nt 5′-end leader and a 20 nt 3′-end trailer (see Materials and Methods) was incubated with extract (Fig. 3A), but not in the position predicted for a 107 nt 3′-tRNase intermediate ([–] at left of Fig. 3A). The same pattern was observed using DP with the U7445C substitution (data not shown).
Incubation with extract of a substrate which has a 32P-labeled mature 5′-end and an 18 nt 3′-end trailer (constructed using -tRNA oligonucleotides; see Materials and Methods) produced a less complex pattern than that obtained with the double precursor (Fig. 3B, compare with Fig. 3A), consisting of the 69 nt 3′-tRNase product (← at right of Fig. 3) superimposed on a background of bands produced by 3′-exonuclease activity. The 0.1 M KCl DEAE–Sepharose fraction (see Materials and Methods) displays less contaminating nuclease activity (Fig. 3C and D), but products obtained with the double precursor are still uninterpretable (Fig. 3C), suggesting that the presence of a 38 nt 5′-end leader interferes with recognition of the 3′-tRNase cleavage site. Only the combination of fractionated extract with simplified substrate gives a prominent cleavage at the expected position (Fig. 3D, bands designated by ← at right).
Faint bands surround the in vitro 3′-tRNase product. The main 3′-tRNase product, N (←), has 72% of the intensity in the region: (N–1), 12%; (N–2), 2%; (N+1), 6%; (N+2), 8%. These products could arise from cleavage site heterogeneity or residual 3′-exonuclease. Heterogeneity of the 3′-tRNase cleavage site is more likely a characteristic of the tRNA substrate than of the human mitochondrial 3′-tRNase. The pattern obtained using a fruit fly cytoplasmic tRNAHis precursor with human mitochondrial and fruit fly nuclear 3′-tRNase is identical (32; data not shown); human 3′-tRNase also produces the predicted bands from mitochondrial tRNAIle and tRNALeu(UUR) precursors in vitro (data not shown). The Km for human mitochondrial 3′-tRNase is 75 nM (data not shown), comparable to that of fruit fly and pig nuclear 3′-tRNase under similar reaction conditions (8,30).
The U7445C substitution abolishes the ability of the tRNASer(UCN) precursor to be processed by 3′-tRNase
A 5′-end-labeled tRNA precursor with G↓CCU substituted for G↓UCU was incubated with the mitoplast DEAE–Sepharose 0.1 M KCl 3′-tRNase fraction (Fig. 4B) alongside the wild-type precursor (Fig. 4A). No processing products were obtained from the U7445C precursor (Fig. 4B; note the absence of a band at 69 nt, identified by → at left), while the wild-type precursor could be efficiently processed in vitro (Fig. 4A, compare Fig. 3D).
Figure 4.
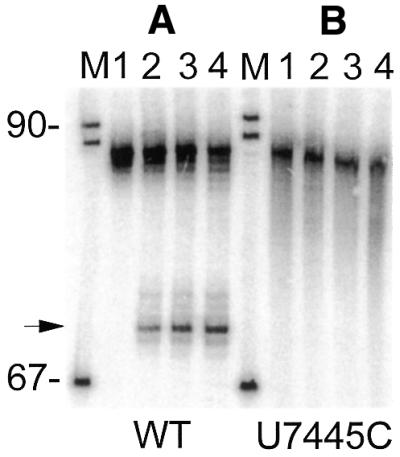
tRNA precursor with the U7445C substitution is not a 3′-tRNase substrate. (A and B) 5′-End-labeled wild-type and U7445C precursors were incubated with 3′-tRNase (DEAE–Sepharose 0.1 M KCl fraction) as in Figure 3D and reactions were sampled after 0, 10, 20 and 30 min (lanes 1–4, respectively). An arrow to the left indicates the 3′-tRNase product.
Eukaryotic 3′-tRNase has been demonstrated to be an endonuclease in numerous cases (3–7,33), but prokaryotic tRNA precursors are sometimes cleaved by endonucleases followed by trimming of their 3′-ends by exonucleases (34), and 3′-exonucleases could also be involved with eukaryotic tRNA processing (see Discussion). Incubation of 3′-end-labeled wild-type and U7445C variant precursors with 3′-tRNase (Fig. 5A and B) reveals the predicted 18 nt 3′-end trailer (→ at left of Fig. 5A) with wild-type (Fig. 5A) but not with U7445C (Fig. 5B) tRNA. Faint bands are also seen below the wild-type 3′-end trailer (–s at left of Fig. 5A). A band in the unincubated wild-type control (dot at left of Fig. 5A) may arise from background RNase A-like cleavage at U76 (see Fig. 6).
Figure 5.
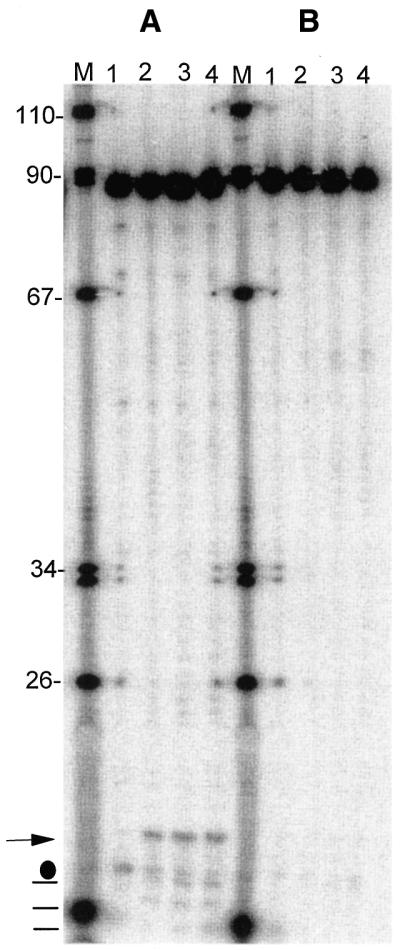
Human mitochondrial 3′-tRNase makes an endonucleolytic cut at the expected processing site. (A and B) As in Figure 4A and B except that tRNAs were labeled at the 3′-end and the gel was run so as to retain the 3′-end trailer fragment [horizontal arrow at left of (A)]. Dot designates a degradation product in the prepared substrate (unincubated lane 1). –s indicate bands that arise from cleavages downstream from the 3′-tRNase site.
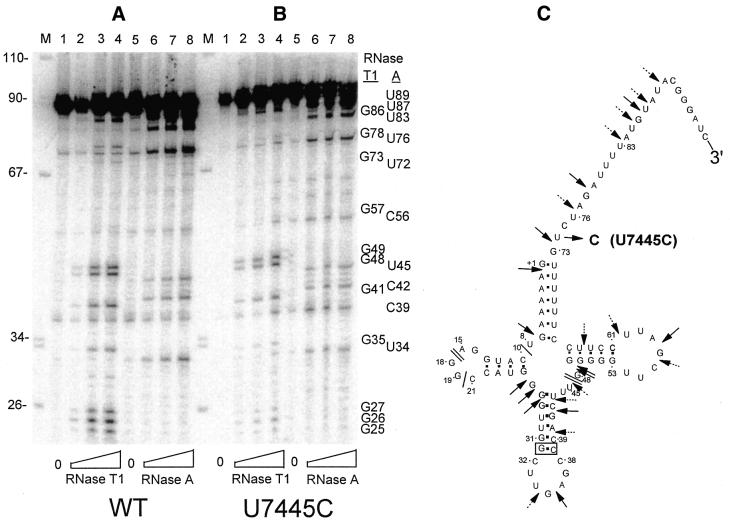
Figure 6.
Structure probing of 5′-end-labeled wild-type and U7445C tRNASer(UCN) precursors. Precursor tRNASer(UCN) prepared using the double ribozyme method was labeled at its 5′-end and incubated at room temperature for 5 min with increasing concentrations of RNase T1 (0.5, 1 and 2 U/ml) or RNase A (2, 4 and 8 × 10–3 U/ml) as indicated. RNase T1 and RNase A bands are identified by nt number at right. (A) Wild-type; (B) U7445C. (C) Susceptibility of the wild-type and U7445C tRNA precursors to structure probing nucleases. Solid and dashed arrows designate sites susceptible to RNase T1 and A, respectively.
RNase susceptibility of tRNASer(UCN) is consistent with canonical tRNA structure
Wild-type and U7445C tRNA precursors with a mature 5′-end and a 3′-end trailer prepared using cis-acting ribozymes (see Materials and Methods) were 5′-end-labeled and incubated with RNase T1 or RNase A (Fig. 6). Susceptible G residues and pyrimidines (mainly U residues) are identified at the right. Results with 3′-end-labeled tRNAs produced by -tRNA ligation and with 5′-end-labeled double precursors (which allowed cleavages to be observed as far upstream as +1) were substantially the same (data not shown). G86 displayed the greatest RNase T1 susceptibility and U89, U87, U83 and U76 displayed the greatest RNase A susceptibility, showing the 3′-end trailer to be relatively unstructured. Background cleavage of control tRNAs (incubated without added RNase; lanes 1 and 5 of Fig. 6A and B) are observed at U76 and elsewhere. The 3′-tRNase site (G73) is barely susceptible to T1. The distribution of susceptible and inaccessible sites is consistent with canonical tRNA structure. For example, G13, G14, G18 and G19 in the D loop (data not shown) and nt 57 in the T loop are cleaved relatively weakly, presumably due to tRNA tertiary structure. The most accessible sites are G49, G48 and G41 (lanes 2–4) and U45, C42 and U34 (lanes 6–8) in the anticodon and V loops. RNase susceptibilities in the U7445C precursor are similar to those in wild-type tRNASer(UCN) (Fig. 6B, compare with Fig. 6A).
DISCUSSION
Potential importance of 3′-tRNase in tRNA end processing
The existence of H-strand RNAs whose 5′-ends are contiguous with the discriminator base of tRNAs provides a strong argument for the importance of a 3′-end tRNA processing endonuclease in mitochondrial RNA maturation (11). Our finding that a U7445C tRNASer(UCN) precursor cannot be processed by mitochondrial 3′-tRNase in vitro (Figs 4 and 5) is also consistent with the interpretation that endonucleolytic cleavage by this enzyme could be central to mitochondrial RNA processing. U7445C changes the sequence G↓UCU to G↓CCU, which has -CC of -CCA (a demonstrated anti-determinant for mouse, pig and fruit fly 3′-tRNase; 7,8) following the discriminator base. The 3′-tRNase processing defect we observed in vitro could be functionally important in vivo. If so, this 3′-end processing defect could cause non-syndromic deafness by reducing the steady-state concentration of functional tRNASer(UCN) below that required to support mitochondrial translation in tissues which require the greatest respiratory activity, including cochlear cells of the inner ear, as previously suggested (16,17).
A possible alternative pathway for 3′-end processing of U7445C tRNASer(UCN)
If the 5′-end of tRNATyr was processed by RNase P or if an endonuclease cleaved elsewhere in the ∼2 kb spacer between tRNASer(UCN) and tRNATyr, an exonuclease could trim the 3′-end of the tRNASer(UCN) U7445C precursor to nucleotides 73, 74 or 75 for repair by tNtase, much as in Escherichia coli (34). Nuclear and cytoplasmic 3′-exonucleases have important functions (35,36), but less is known about mitochondrial 3′-exonucleases. 3′-Exonuclease activity is mixed with 3′-tRNase in the unfractionated mitoplast extract (Fig. 3B) and endonucleolytic cleavages may occur downstream from the 3′-tRNase site in the presence of a 5′-end leader in the wild-type or U7445C precursor (Fig. 3A and data not shown). If endonucleolytic 3′-end processing of U7445C tRNASer(UCN) at G73 by 3′-tRNase is prevented in vivo as it is in vitro (Figs 4 and 5), mature tRNASer(UCN) could still be produced by an endo/exonucleolytic end processing mechanism. Exonucleases would not be expected to salvage most other defective mitochondrial tRNA 3′-end processing, however, because of consequences for functional RNAs downstream.
RNase accessibility and tRNASer(UCN) structure
Cleavage with sequence- and structure-sensitive RNases can be used to confirm the sequence of a constructed RNA, to suggest whether a tRNA is correctly folded and to reveal changes in structure arising from a substitution or other sequence change. Guanines and pyrimidines were assigned in wild-type and variant tRNASer(UCN) (Fig. 6); susceptible positions are generally consistent with the canonical tRNA structure. Interestingly, the 3′-tRNase cleavage site is a weak T1 site in wild-type tRNASer(UCN). The position-specific RNase susceptibilities of wild-type and U7445C tRNASer(UCN) (Fig. 6A and B) do not suggest a major difference in folding or an obvious structural basis for failure of the mutant tRNA precursor as an in vitro substrate for 3′-tRNase (Figs 4 and 5).
The structure of 3′-tRNase is unknown, but human mitochondrial 3′-tRNase is a good candidate for further characterization because of the anti-determinant phenomenon, due to its theorized importance for maturation of mitochondrial RNAs and because human nuclear 3′-tRNase is also practically uncharacterized.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to D. Bogenhagen (SUNY at Stony Brook), D. Engelke (University of Michigan), C. Florentz (IBMC, Strasbourg), S. Kelley (Boston College), A. Mohan, M. Nashimoto (Tsukuba), E. Schon (Columbia) and O. Uhlenbeck (Boulder) for helpful discussions, to E. Lund and J. Dahlberg (University of Wisconsin) for communicating results prior to publication, to C. Florentz for critical reading of the manuscript, to J. Manley (Columbia) for the gift of the HeLa cell seed culture, to S. Searles (MRC, Cambridge) for the gift of pSS419, to V. Greene, A. Hopper (Penn State) and E. Schon for advice about estimating mitochondrial purity and to H. Hawkins, T. Koyejo and M. Ousman for technical assistance. This work was supported by NIH grants SO6GM08153, R15GM57617 and T34GM08498 and NSF grant MCB9904589.
References
- 1.Guerrier-Takada C., Gardiner,K., Marsh,T., Pace,N. and Altman,S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 35, 849–857. [DOI] [PubMed] [Google Scholar]
- 2.Lee, J.Y., Rohlman,C.E., Molony,L.A. and Engelke,D.R. (1991) Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell. Biol., 11, 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solari A. and Deutscher,M.P. (1983) Identification of multiple RNases in Xenopus laevis oocytes and their possible role in tRNA processing. Mol. Cell. Biol., 3, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castaño J.G., Tobian,J.A. and Zasloff,M. (1985) Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNAi–Met (3′Pre-tRNase). J. Biol. Chem., 260, 9002–9008. [PubMed] [Google Scholar]
- 5.Frendewey D., Dingermann,T., Cooley,L. and Söll,D. (1985) Processing of precursor tRNAs in Drosophila. Processing of the 3′ end involves an endonucleolytic cleavage and occurs after 5′ end maturation. J. Biol. Chem., 260, 449–454. [PubMed] [Google Scholar]
- 6.Levinger L., Vasisht,V., Greene,V., Bourne,R., Birk,A. and Kolla,S. (1995) Sequence and structure requirements for Drosophila tRNA 5′ and 3′-end formation. J. Biol. Chem., 270, 18903–18909. [DOI] [PubMed] [Google Scholar]
- 7.Nashimoto M. (1997) Distribution of both length and 5′ terminal nucleotides of mammalian pre-tRNA 3′ trailers reflect properties of 3′ processing endoribonuclease. Nucleic Acids Res., 25, 1148–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohan A., Whyte,S., Wang,X., Nashimoto,M. and Levinger,L. (1999) The 3′ end CCA of mature tRNA is an anti-determinant for eukaryotic 3′-tRNase. RNA, 5, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- 10.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F., Schreier,P.H., Smith,A.J., Staden,R. and Young,I.G. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 11.Montoya J., Ojala,D. and Attardi,G. (1981) Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature, 290, 465–470. [DOI] [PubMed] [Google Scholar]
- 12.Reichert A., Rothbauer,U. and Mör,M. (1998) Processing and editing of overlapping tRNAs in human mitochondria. J. Biol. Chem., 273, 31977–31984. [DOI] [PubMed] [Google Scholar]
- 13.Ojala D., Montoya,J. and Attardi,G. (1981) Transfer RNA punctuation model of RNA processing in human mitochondria. Nature, 290, 470–474. [DOI] [PubMed] [Google Scholar]
- 14.King M.P., Koga,Y., Davidson,M. and Schon,E.A. (1992) Defects in mitochondrial protein synthesis and respiratory chain activity segregate with tRNALeu(UUR) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and strokelike episodes. Mol. Cell. Biol., 12, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindoff L.A., Howell,N., Poulton,J., McCullough,D.A., Morten,K.J., Lightowlers,R.N., Turnbull,D.M. and Weber,K. (1993) Abnormal RNA processing associated with a novel tRNA mutation in mitochondrial DNA. J. Biol. Chem., 268, 19559–19564. [PubMed] [Google Scholar]
- 16.Reid F.M., Rovio,A., Holt,I.J. and Jacobs,H.T. (1997) Molecular phenotype of a human lymphoblastoid cell-line homoplasmic for the np-7445 deafness-associated mitochondrial mutation. Hum. Mol. Genet., 6, 443–449. [DOI] [PubMed] [Google Scholar]
- 17.Guan M.-X., Enriquez,J.A., Fischel-Ghodsian,N., Puranam,R.S., Lin,C.P., Law,M.A. and Attardi,G. (1998) The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol. Cell. Biol., 18, 5868–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manam S. and Van Tuyle,G.C. (1987) Separation and characterization of 5′ and 3′-tRNA processing nucleases from rat liver mitochondria. J. Biol. Chem., 262, 10272–10279. [PubMed] [Google Scholar]
- 19.Rossmanith W., Tullo,A., Potushak,T., Karwan,R. and Sbisa,E. (1995) Human mitochondrial tRNA processing. J. Biol. Chem., 270, 12885–12891. [DOI] [PubMed] [Google Scholar]
- 20.Helm M., Giegé,R. and Florentz,C. (1999) A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry, 38, 13338–13346. [DOI] [PubMed] [Google Scholar]
- 21.Doersen C.J., Guerrier-Takada,C., Altman,S. and Attardi,G. (1985) Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J. Biol. Chem., 260, 5942–5949. [PubMed] [Google Scholar]
- 22.Rossmanith W. and Karwan,R.M. (1998) Characterization of human mitochondrial RNase P: novel aspects in tRNA processing. Biochem. Biophys. Res. Commun., 247, 234–241. [DOI] [PubMed] [Google Scholar]
- 23.Puranam R.S. and Attardi,G. (2001) The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol. Cell. Biol., 21, 548–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossmanith W. and Karwan,R.M. (1998) Impairment of tRNA processing by point mutations in mitochondrial tRNALeu(UUR) associated with mitochondrial diseases. FEBS Lett., 433, 269–274. [DOI] [PubMed] [Google Scholar]
- 25.Nashimoto M., Wesemann,D.R., Geary,S., Tamura,M. and Kaspar,R.L. (1999) Long 5′ leaders inhibit removal of a 3′ trailer from a precursor tRNA by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res., 27, 2770–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romaniuk P.J. and Uhlenbeck,O.C. (1983) Joining of RNA molecules with RNA ligase. Methods Enzymol., 100, 52–59. [DOI] [PubMed] [Google Scholar]
- 27.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helm M., Brulé,H., Friede,D., Giegé,R., Pütz,D. and Florentz,C. (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA, 6, 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fechter P., Rudinger,J., Giege,R. and Theobald-Dietrich,A. (1998) Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett., 436, 99–103. [DOI] [PubMed] [Google Scholar]
- 30.Levinger L., Bourne,R., Kolla,S., Cylin,E., Russell,K., Wang,X. and Mohan,A. (1998) Matrices of paired substitutions show the effects of tRNA D/T loop sequence on Drosophila RNase P and 3′-tRNase processing. J. Biol. Chem., 273, 1015–1025. [DOI] [PubMed] [Google Scholar]
- 31.Tapper D.P., Van Etten,R.A. and Clayton,D.A. (1983) Isolation of mammalian mitochondrial DNA and RNA and cloning of the mitochondrial genome. Methods Enzymol., 97, 426–434. [DOI] [PubMed] [Google Scholar]
- 32.Levinger L., Mohan,A., Jacobs,O. and Ousman,M. (1999) A test of the 3′-tRNase anti-determinant hypothesis using the human mitochondrial tRNASer(UCN) A7445G mutant which causes non-syndromic deafness. Nucleic Acids Symp. Ser., 41, 110–112. [Google Scholar]
- 33.Mayer M., Schiffer,S. and Marchfelder,A. (2000) tRNA 3′ processing in plants: nuclear and mitochondrial activities differ. Biochemistry, 39, 2096–2105. [DOI] [PubMed] [Google Scholar]
- 34.Li Z. and Deutscher,M.P. (1996) Maturation pathway for E. coli tRNA precursors: a random multienzyme process in vivo. Cell, 86, 503–512. [DOI] [PubMed] [Google Scholar]
- 35.Van Hoof A. and Parker,R. (1999) The exosome: a proteasome for RNA? Cell, 99, 347–350. [DOI] [PubMed] [Google Scholar]
- 36.Allmang C., Petfabski,E., Podtelejinikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999) The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]