Abstract
Members of the basic helix–loop–helix (bHLH) family of transcription factors regulate a wide array of developmental processes in many cell types, including cell fate specification, differentiation and morphogenesis. Our studies describe the cloning of a gene from the nematode Caenorhabditis elegans that is closely related to the vertebrate-activated B-cell factor (ABF) gene. The nematode gene product CeABF-1 was detected by northern blot analysis from RNA isolated from pooled nematodes representing different developmental stages. The developmental expression profile of CeABF-1 was shown by RT–PCR analysis to be predominantly expressed in the larval stages L3 and L4, with lower levels observed in the L2 larval stage and adult. We also show that CeABF-1 is capable of forming heterodimers with E2A proteins and binding E-box target sites. Mammalian cells transfected with CeABF-1 expression plasmids were capable of blocking E2A-mediated gene transcription, but full repression activity required the presence of two conserved amino acid residues found within the first helix of the CeABF-1 bHLH domain. These results suggest a conserved mechanism of gene repression between certain class II bHLH and class I bHLH proteins found in vertebrates and invertebrates.
INTRODUCTION
The helix–loop–helix (HLH) family of transcription regulatory proteins are key regulators in a wide number of developmental processes. To date, over 240 HLH proteins have been identified in organisms ranging from yeast Saccharomyces cerevisiae to humans (1). Analyses in Xenopus laevis, Drosophila melanogaster and Mus musculis have provided insight into the participation of HLH proteins in developmental events such as cellular differentiation, lineage commitment and sex determination. In particular, in multicellular organisms, HLH factors are required for a variety of critical developmental processes, including myogenesis, neurogenesis, hematopoiesis and pancreatic, heart and spleen development (2–5).
HLH proteins are classified upon tissue distribution, dimerization properties and DNA-binding specificities (6). Class I HLH proteins, also referred to as E proteins, include E12, E47, HEB, E2-2 and Daughterless. These genes are expressed in many tissues and their products are capable of forming homo- or heterodimers. Class II HLH proteins, which include family members MyoD, myogenin, NeuroD/BETA2 and SCL/TAL1 display more tissue-restricted patterns of expression. In general, they are unable to form homodimers and preferentially heterodimerize with the E proteins. Recently, a novel class II HLH protein dubbed activated B-cell factor 1 (ABF-1) was identified in human B-lymphocyte cell lines transformed with Epstein–Barr virus (EBV) (7). ABF-1 has been shown to form heterodimers with E47, a product of the human E2A gene. E2A gene products are important regulators of B-cell formation and T-cell development (2,8). In vivo ABF-1–E2A heterodimers have been detected in nuclear lysates prepared from EBV-infected B lymphocytes and their role as important regulators of B-cell proliferation following antigenic stimulation has been proposed. Recently, the mouse homolog of ABF-1, musculin/MyoR, has been cloned and displays a slightly different expression profile, being detected in developing skeletal muscle as well as the spleen (9). A role for MyoR as a lineage-restricted basic (b)HLH transcriptional repressor of the muscle differentiation program has been reported (10). The amino acid sequence of human ABF-1 also shares homology to the bHLH protein capsulin and the Drosophila bHLH factor bHLH54F. Capsulin is expressed in smooth muscle cell precursors during mouse embryogenesis whereas the Drosophila bHLH factor bHLH54F is detected in visceral and skeletal muscle cell precursors (11).
In order to search for the ABF-1 homolog in Caenorhabditis elegans, we used the predicted human ABF-1 polypeptide sequence to search the C.elegans genome database for a related gene product. Recent analysis of the open reading frames present in the C.elegans genome database has revealed 24 potential HLH proteins (12). A number of these genes have been identified previously in genetic screens. The gene lin-32 encodes a bHLH protein that is required for proper formation of the male tail rays; these are important peripheral sense organs critical for mating (13). Also, the bHLH protein encoded by lin-22 is important for proper nervous system patterning (14). Despite the characterization of a few bHLH genes in C.elegans, the majority of the computer-predicted bHLH genes remain to be verified as bona fide gene products. Following our computer search of the C.elegans database, our results yielded one potential candidate gene with high homology to the human ABF-1 gene.
In this study we report the cloning of a C.elegans gene that encodes a bHLH protein related to human ABF-1. We have named this protein C.elegans ABF-1 or CeABF-1. Relative to human ABF-1, CeABF-1 shares 72% amino acid similarities within the bHLH domain and is capable of forming heterodimers with E47, the mammalian homolog of the C.elegans protein CeE. The CeABF-1 transcript was detected by northern blot analysis from RNA collected from pooled nematodes representative of different developmental stages, including embryo, larval and adult. Developmentally, CeABF-1 expression was detected in the larval stages L2 through L4 as well as in the adult, with the highest levels observed in the L3 and L4 stages. Similarly to the transcriptional properties of human ABF-1, we demonstrate that CeABF-1 can function as a strong transcription repressor capable of blocking E47-mediated gene transcription at an E-box binding site in HeLa cells. We also provide evidence for the importance of two conserved amino acid residues, serine-lysine (SK), found within the first helix of the bHLH domain of CeABF-1 that participate in the inhibition of E2A gene activation. These findings suggest that CeABF-1 may have a conserved function as a negative regulator of gene transcription for developmental programs ranging from nematodes to humans.
MATERIALS AND METHODS
RT–PCR analysis and DNA sequencing
To isolate the CeABF-1 cDNA, total cytoplasmic RNA was isolated from the nematodes according to the manufacturer’s instructions (Qiagen Inc., Valencia, CA). Approximately 1 µg of total RNA was heated with an oligo(dT) primer to 65°C for 10 min in a total volume of 10 µl and then incubated with 1× first-strand buffer, 10 mM dithiothreitol, 0.5 mM each deoxynucleotide triphosphate, 20 U of RNasin (Promega, Madison, WI) and 20 U of MMLV reverse transcriptase (New England Biolabs, Beverly, MA) in a total volume of 20 µl at 42°C for 1 h. After reverse transcription (RT), one-quarter of the RT reaction was then mixed with 0.5 µM of each primer, 0.5 mM dNTPs, 5% dimethyl sulfoxide (DMSO) and 2 U Pfu DNA polymerase in a 50 µl reaction volume and amplified by a conventional three-step PCR protocol for 30 cycles. The CeABF-1 forward and reverse primers contain the nucleotide sequence 5′-ccg aat tca tgc aag tga tgg aat cat gtg g-3′ and 5′-ccg tcg act tac ttt cgt aac gtt ttt gca cat tcc-3′, respectively. The parameters for the three-step PCR protocol were as follows: heat denaturation at 92°C for 45 s, annealing at 65°C for 45 s and extension at 72°C for 90 s. Following PCR, the cDNA was gel purified and inserted into the pCRTopoII cloning vector (Invitrogen, Carlsbad, CA). The CeABF-1 cDNA was sequenced using an oligonucleotide primer corresponding to the polylinker of the pCRTopoII-cloning vector essentially as described previously (15). The DNA sequence for CeABF-1 has been deposited in GenBank.
For the developmental studies, total RNA was isolated from the different larval stages as well as the adult according to the manufacturer’s instructions (Qiagen Inc.). First-strand cDNA was generated essentially as described above except 300 ng of total RNA was reverse transcribed using Sensiscript™ reverse transcriptase (Qiagen Inc.), followed by PCR using the HotStarTaq DNA polymerase (Qiagen Inc.). Southern blot analysis and probe labeling was performed as described previously (15). To create the CeE probe, two primers, designated CeE-F1 and CeE-R1, were synthesized and used for conventional RT–PCR analysis as described above. The amplified CeE cDNA was gel purified and subcloned into pBluescript KS (Stratagene, La Jolla, CA) and the gene sequence verified by DNA sequencing. The CeE-F1 and CeE-R1 primers had the nucleotide sequence 5′-cca agc tta tgg cgg atc caa ata gcc aac tta-3′ and 5′-cca gat ctt taa aac cgt gga tgt cca aac tgc-3′, respectively.
Collection of staged animals
Animals of genotype him-5(e1490) were grown on Nematode Growth Media plates under standard conditions (16). Animals were rinsed off the plates using M9 buffer, precipitated in a clinical centrifuge, and placed on a 10 µ nitex filter for 10–15 min. At the end of this time, the animals that had passed through the filter were examined in a dissecting scope and confirmed to be L1 larvae. These larvae were placed onto plates and allowed to develop until the majority of animals had reached a given stage, at which time they were once again collected from the plates in M9 buffer and quick frozen until needed for RNA isolation for RT–PCR analysis.
Northern blot analysis
Total cellular RNA was prepared using the RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen Inc.). Ten micrograms of total cellular RNA were resolved by electrophoresis on a 1.5% agarose gel supplemented with formaldehyde and the RNA fragments transferred to a GeneScreen nylon membrane according to the manufacturer’s protocol (New England Nuclear Research Products, Boston, MA). Following the transfer of RNA to the membrane, the RNA fragments were covalently cross-linked to the membrane using the UV Stratalinker 1800 (autolink-setting). Then, the RNA was hybridized with 5.0 × 106 c.p.m. 32P-radiolabeled probe. The blotting, hybridization and washing were carried out as recommended by the manufacturer’s directions. RNA transcripts of known size were used as markers to confirm the size of the hybridized species.
Cell culture
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% (v/v) fetal bovine serum supplemented with 100 µg/ml penicillin and 100 µg/ml streptomycin. For transient transfections, ∼4.0 × 106 cells were seeded onto 6-cm diameter plates 1 day before transfection. All cells were transfected with purified plasmid DNA mixed with Superfect (Qiagen Inc.). Forty-eight hours after transfection, cells were harvested and equivalent volumes assayed for luciferase activity as previously described (15). The amount of luciferase activity was normalized to the β-galactosidase activity for each sample tested.
Plasmid constructions
The CeABF-1 cDNA was amplified from total RNA using RT–PCR. Following RT–PCR, the cDNA was blunt-end ligated into the cloning vector pCRTopoII (Invitrogen). After the insertion of the cDNA into pCRTopoII, the CeABF-1 cDNA was excised from the plasmid by cutting the DNA with the restriction enzymes EcoRI and SalI. The full-length CeABF-1 cDNA was inserted into the eukaryotic expression vector pCMV-TAG2B (Stratagene) to create pCMV-CeABF-FL. For the construction of the N- and C-terminal CeABF-1 proteins, the following strategy was used. To create the N-terminal portion of CeABF-1, pCMV-CeABF-FL was linearized with XmnI and the DNA ends filled using the Klenow fragment. A 219 bp insert, which encodes the N-terminal 73 amino acids, was released by an EcoRI digestion, gel purified and inserted into the eukaryotic expression vector pCMV-TAG4B (Stratagene) and dubbed pCMV-CeABF-N73. For the creation of the C-terminal portion of CeABF-1, the pCMV-CeABF-FL vector was digested with EcoRV followed by SalI digestion. The 291 bp cDNA fragment was inserted into pCMV-TAG2B to create pCMV-CeABF-C97. This piece of DNA encodes the C-terminal 97 amino acids; this region contains the bHLH domain. For the construction of the CeABF-1 SK to asparagine-glutamic acid (NE) mutant, site-directed mutagenesis was performed using overlapping PCR as previously described (17). The two internal primers used for the introduction of the mutation were designated CeABF-F2 SK-NE and CeABF-R2 SK-NE and contained the nucleic acid sequences 5′-ctt aac gaa atg ttt gat caa ttg aga gtc tgc-3′ and 5′-tct caa ttg atc aaa cat ttc gtt aag ttg ttg aac tct att-3′, respectively. Following the introduction of the mutation, the cDNA was inserted into pCMV-TAG2B to create pCMV-CeABF SK-NE.
Gel-shift assays
For gel-shift assays, 2 µl of rabbit reticulocyte lysate programmed with RNA was incubated on ice for 15 min in binding buffer consisting of 10 mM Tris pH 7.5, 75 mM NaCl, 1–5 µg poly[d(I–C)], 6% (v/v) glycerol and 10 µg bovine serum albumen (fraction V) before addition of the radiolabeled probe (100 000 c.p.m./reaction; ∼2 ng). Binding reactions were carried out in a final volume of 15 µl for 15 min and electrophoresed at room temperature on 6% non-denaturing polyacrylamide gels containing 0.25× Tris–borate–EDTA (TBE) pH 8.5, at 13 V/cm for 1.5 h. For antibody-binding experiments, 1 µl of an anti-E2A polyclonal antiserum or 1 µl of anti-FLAG monoclonal antibody (0.36 mg/ml; Sigma, St Louis, MO) was used. Sense and antisense oligonucleotides were annealed and used as blunt-ended, double-stranded DNA molecules. The sequence of the probe used in electrophoretic mobility shift assay (EMSA) is listed as follows: µE5 element, 5′-GGCCAGAACACCTGCAGACG-3′.
In vitro transcription and translation
pCMV-CeABF-FL, pAR5/2 (E47), pCMV-ABF-FL and pCMV-CeABF SK-NE were added separately or in different combinations to the coupled TNT rabbit reticulocyte lysate system and transcribed with either T3 or T7 RNA polymerase followed by translation (Promega).
RESULTS
Cloning the CeABF-1 gene
In order to identify gene products showing similarities to vertebrate ABF-1, we searched the C.elegans genome database. After computer analysis, one candidate showed strong homology to vertebrate ABF-1 within the bHLH domain as well as a portion of the N-terminus. To determine whether the identified DNA segment represented a bona fide gene in C.elegans, we performed RT–PCR analysis. Total RNA was isolated from C.elegans and used to generate first-strand cDNA. Using the putative gene sequence found in the C.elegans genome database, we designed two primers corresponding to pieces of the hypothetical gene and performed RT–PCR analysis. Following RT–PCR analysis, a single amplified DNA product corresponding to ∼500 bp was observed by agarose gel electrophoresis. The PCR product was subsequently cloned and sequenced, revealing substantial homology within the bHLH domain of human ABF-1 and its mouse homolog musculin (Fig. 1B). Based upon the structural similarities, we have named the protein product CeABF-1. Across the bHLH domain, CeABF-1 has a stretch of 23 out of 44 matches, or 52% identities, to human ABF-1. Outside the bHLH domain, there is also a second region sharing homology with the vertebrate ABF-1. In the N-terminus, there is a stretch of 6 out of 14 identities, or 43% similarity (Fig. 2). The functional significance of this region remains to be elucidated. Conceptual translation of the open reading frame of the apparent full-length cDNA results in a 170 amino acid protein of Mr 19 584 and pI = 5.62. There are a few other notable protein sequence domains within CeABF-1. One is a glutamic- and lysine-rich region in the C-terminus, whereas a second is a glutamine-rich region in the N-terminus. Analysis of the protein sequence using the computer program ScanProsite revealed potential phosphorylation sites for serine/threonine protein kinases cAMP, PKC and CK2.
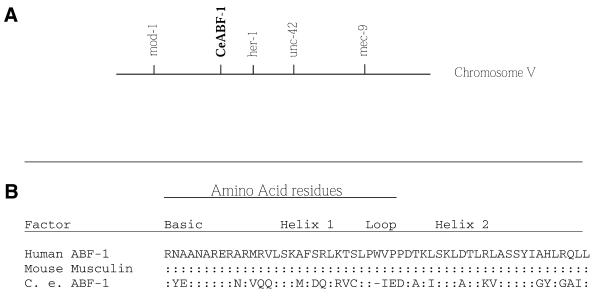
Figure 1.
The CeABF-1 gene. (A) Genomic map position of the CeABF-1 gene. (B) A comparison of the bHLH domains of the ABF-1 regulatory genes shows conservation within the dimerization motif. The bHLH domain of C.elegans ABF-1 is compared with the human and mouse homologs. The human ABF-1 has been used as the canonical sequence. A colon indicates amino acid identities whereas the dash indicates the introduction of a gap into the sequence to maximize amino acid alignment; differences are indicated. Sources for protein sequences are as follows: for human ABF-1 (7) and for mouse musculin/MyoR (9,10).
Figure 2.
For comparison, the human ABF-1 protein has been aligned with the C.elegans homolog, CeABF-1. The asterisk indicates identical amino acids; the colon symbol represents amino acids with similar side-chain properties; the single dot represents weaker side-chain similiarities relative to amino acids with a colour symbol. The dashes indicate the introduction of gaps into the sequence to maximize amino acid alignment.
CeABF-1 gene structure and transcript size
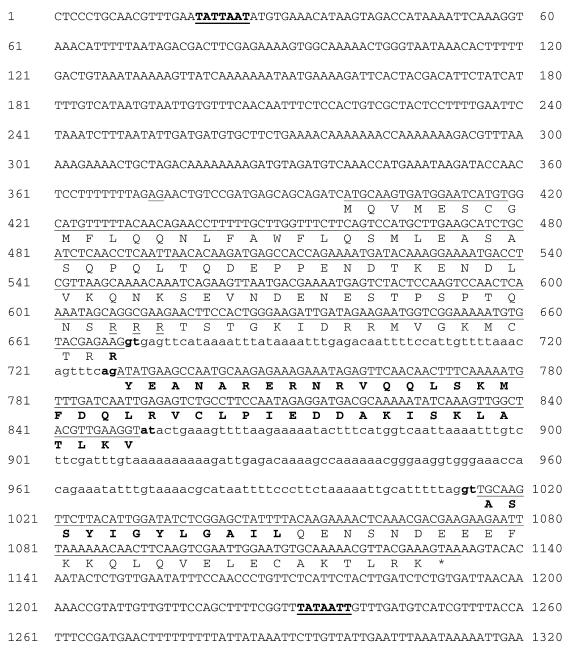
The CeABF-1 gene maps to the cosmid ZK682, which is located between the cloned genes mod-1 and her-1 on chromosome 5. A number of genes have been mapped to this region (Fig. 1A). A comparison of the C.elegans genome database with the CeABF-1 cDNA reveals an ∼1250 bp transcription unit composed of three conventional exons and two introns (Fig. 3). The intron splice donor and acceptor sites conformed to the typical C.elegans consensus sequences (18). In addition, there is a potential trans-splicing acceptor site located at position 373 (–25 relative to the initiator methionine) (19). The intron/exon structure of CeABF-1 shows little similarity to the human ABF-1 gene sequenced previously (15). Upstream of the putative initiator methionine of CeABF-1 there is a potential TATA box beginning at nucleotide position 19 (–372 relative to the initiator methionine). At the 3′-end of the gene there is a potential polyadenylation signal sequence (TATAAA) located downstream of the translational termination codon (Fig. 3).
Figure 3.
The DNA sequence and flanking regions of the CeABF-1 gene. The genomic sequence is shown with the amino acid sequence of the encoded protein printed below the line using the one-letter code notation. The column of numbers to the left and right of each line sequence indicates the nucleotide positions. The putative initiator methionine begins at nucleotide position 399 and the termination codon begins at nucleotide position 1130. Lower case lettering and boldface type indicate the 3′ acceptor sites as well as the 5′ donor sites. A potential TATA box is underlined at the 5′-end of the gene (bold line) with boldface type, as well as the potential polyadenylation signal sequence at the 3′-end of the gene. The longest open reading frame encodes a protein 170 amino acids in length. The conceptual translation product predicts a 19.6 kDa protein with an estimated pI = 5.62. In addition to the bHLH motif (boldface single letter notation) there is a putative nuclear localization signal (dashed underline) and potential trans-splicing acceptor site (underlined AG at position –25 relative to translational initiation site).
We have used the CeABF-1 cDNA to make a probe to examine the expression by northern blot analysis. Total RNA was isolated from nematodes representing all stages of development, including embryos, larvae and adults. In northern analysis the probe hybridized weakly to an ∼1200 base, poly(A)+ message (Fig. 4). This size is consistent with the cDNA length plus a poly(A) tail, assuming the putative promoter and potential polyadenylation signal sequence are utilized and no trans-splicing occurs (Fig. 3).
Figure 4.
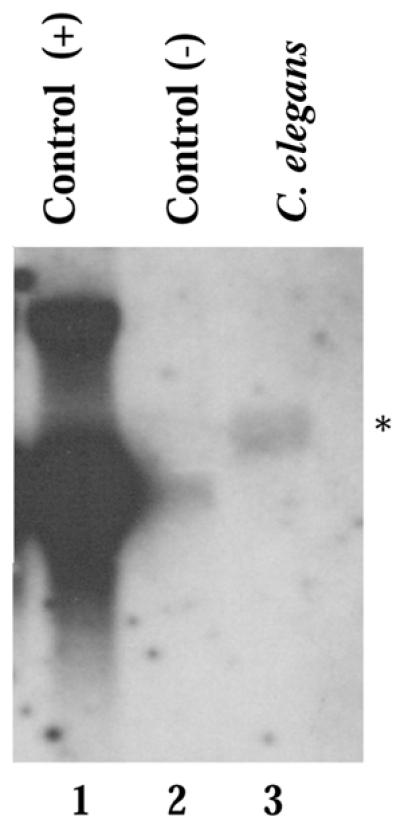
Northern blot analysis detects CeABF-1 expression. The expression pattern of CeABF-1 in nematodes was analyzed from pooled organisms found at the embryo, larval and adult stages. Ten micrograms of total RNA was isolated and analyzed by northern blot analysis. As a control for size markers, HeLa cells were transfected with an inducible cloning vector carrying full-length human ABF-1. HeLa cells were either induced with tetracycline (+) or not induced (–). Total RNA was prepared from the HeLa cells and examined by northern blot analysis on the same gel. Following hybridization with either the human ABF-1 probe or CeABF-1 probe, the membranes were aligned and the human transcript used as size markers. The asterisk designates the detected CeABF-1 transcript.
Developmental expression profile of CeABF-1
To determine the expression pattern of CeABF-1 in C.elegans during development, we performed RT–PCR analysis. Total RNA was isolated from the four stages (commonly referred to as larval stages) L1–L4 as well as from the adult nematodes (includes hermaphrodites and males). Equivalent amounts of total RNA were reverse transcribed from each stage to generate first-strand cDNA followed by the amplification of the cDNA encoding CeABF-1 by PCR. Following RT–PCR, the amplified products were analyzed and confirmed by Southern blot analysis using a CeABF-1-specific probe. The CeABF-1 expression profile was found to be the strongest in the larval stages L3 and L4 (Fig. 5). In addition, relative to the L3 and L4 stages, the L2 larval stage and adult showed moderately lower levels of CeABF-1 expression. Essentially no CeABF-1 was detected in the L1 larval stage (Fig. 5). To control for the quality of the RT reaction, the CeE gene, which represents the C.elegans homolog of the mammalian gene E2A, was amplified with CeE gene-specific primers using the same RT reaction as CeABF-1. Following RT–PCR analysis, the CeE cDNA products were verified by Southern blot analysis using a CeE radiolabeled probe. All developmental stages showed similar levels of amplified CeE (Fig. 5). The detection of CeE at all developmental stages has been reported previously (20). Taken together, the data suggest CeABF-1 is most highly expressed in the L3 and L4 larval stages.
Figure 5.
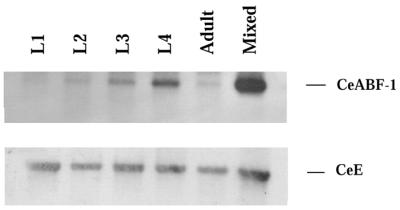
CeABF-1 is expressed during different stages of C.elegans development. The expression pattern of CeABF-1 in nematodes was analyzed from larval stages L1–L4, adult and a mixed population (includes all stages) of nematodes. 300 ng of total RNA was reversed transcribed using Sensiscript™ reverse transcriptase to generate first-strand cDNA. Following first-strand cDNA production, gene-specific primers corresponding to either CeABF-1 or CeE were used to amplify expressed gene products. After PCR, the amplified products were verified using Southern blot analysis with radiolabeled probes specific for the CeABF-1 or CeE cDNAs.
CeABF-1 has the ability to bind as a heterodimer to DNA
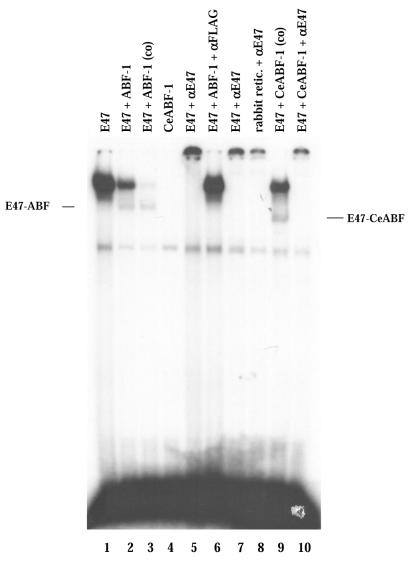
To investigate the capability for heterodimerization by CeABF-1 and E47, we assayed protein interactions by gel-shift assays using a canonical E-box binding site (CAGGTG). The cDNAs encoding CeABF-1, human ABF-1 and E47 were transcribed and translated in vitro and analyzed by SDS–polyacrylamide gel electrophoresis (PAGE). Each of the gene products was translated efficiently (data not shown). Subsequently, the in vitro-translated proteins were incubated with a 32P-labeled µE5-containing oligonucleotide probe derived from the IgH intronic enhancer and analyzed by EMSA. The µE5 site is closely related to the NdE boxes found within the egl-15 promoter, a gene product that encodes a member of the Fibroblast Growth Factor receptor family that is required for the proper migration of the sex myoblasts in the nematode (21). As expected, E47 was capable of binding to the µE5 site as a homodimer (Fig. 6). In addition, when E47 and human ABF-1 were cotranslated, E47 was capable of forming heterodimers with human ABF-1 as reported previously (7). This complex was eliminated by the addition of both anti-FLAG or anti-E47 antibodies, confirming the shifted complex contained both human ABF-1 and E47 molecules (Fig. 6). To examine whether CeABF-1 could bind DNA as a homodimer, in vitro produced CeABF-1 was combined with the probe. The CeABF-1 homodimer failed to bind the target site, whereas when CeABF-1 and E47 were cotranslated heterodimer formation was observed (Fig. 6). The inability of CeABF-1 to bind the E-box sequence as a homodimer is consistent with the binding properties of human ABF-1 to the µE5 box (7). Interestingly, relative to the human ABF-1–E47 heterodimer, the CeABF-1–E47 heterodimer displayed a different migration profile, perhaps reflecting the slight difference in size between human ABF-1 and CeABF-1 or a different heterodimeric protein-complex conformation. These results suggest that the nematode bHLH protein CeABF-1 retains similar dimerization properties as human ABF-1 and is capable of binding target sites containing the consensus E-box sequence.
Figure 6.
CeABF-1 binds as a heterodimer to an E-box in vitro. EMSA analysis of human ABF-1 and CeABF-1 heterodimeric complexes formed on the µE5 probe. Lines indicate the position of E47–ABF-1 and E47–CeABF-1 heterodimers. The symbol (co) indicates the proteins E2A and ABF-1 or CeABF-1 were cotranslated. Addition of an antibody specific to E47 or the FLAG-tagged ABF-1 is indicated by the labeling αE47 and αFLAG, respectively.
CeABF-1 is a transcriptional repressor
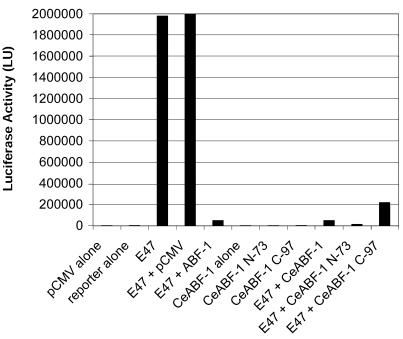
Previous experiments have demonstrated that human ABF-1 and E47 form heterodimers in vivo. This heterodimer functions as a transcriptional repressor, presumably by blocking E2A homodimer formation or blocking the binding of the E2A homodimer to target sites (7). To determine whether CeABF-1 could also function as a transcriptional repressor, we transiently cotransfected HeLa cells with expression vectors capable of producing CeABF-1 and E47, the equivalent of the nematode homolog CeE, along with a reporter vector carrying the firefly luciferase gene driven by a multimerized E-box binding site with a minimal promoter. As a control, HeLa cells were transfected with the eukaryotic expression vector encoding E47 along with the reporter vector. Expression of E47 readily activated the reporter gene (Fig. 7). Consistent with previous reports, when human ABF-1 was expressed in the presence of E47, it repressed E2A-mediated gene activation of the E-box-driven promoter (7,22). In addition, when CeABF-1 was expressed in the presence of E47, similar to human ABF-1, E2A-mediated gene activation was reduced, demonstrating that the nematode homolog also functions as a transcriptional repressor (Fig. 7). In all experiments performed, CeABF-1 was not capable of activating the reporter gene alone, suggesting that alone CeABF-1 lacks transcriptional activation potential. Because the gel-shift data failed to detect either human ABF-1 or CeABF-1 bound to the E-box site as a homodimer, these data are consistent with conventional class II bHLH proteins and their general inability to homodimerize and active gene transcription.
Figure 7.
CeABF-1 functions as a transcriptional repressor in mammalian cells similar to human ABF-1. HeLa S3 cells were transiently transfected with 1 µg of (µE5-µE3-µE2)4-Luc reporter plasmid and different combinations of either the E47 expression vector (0.5 µg) with expression vector FLAG-human ABF-1, FLAG-CeABF-1, CeABF-1 N-73 or CeABF-1 C-97 (0.25 µg). Twenty-five microliters of Superfect reagent was used for each transfection. Equal amounts of protein extract were assayed for firefly luciferase activity. Experiments were performed in triplicates and run at least three independent times. Luciferase activity was normalized to β-galactosidase levels as a control for transfection efficiency.
To address the issue of how CeABF-1 might inhibit E47-mediated transcriptional activation, we created two different truncations of CeABF-1. The C-terminal deletion of CeABF-1 (CeABF-1 N-73), which lacks the bHLH domain, surprisingly repressed the activity of the reporter gene by blocking the activity of E47. This result is consistent with a weak repression domain mapping to the N-terminus of human ABF-1 (22). In addition, the N-terminal deletion of CeABF-1 (CeABF-1 C-97), which contained the bHLH domain, also repressed the activity of the reporter gene by attenuating E47 transactivation. These repression activities were specific and required the presence of E47 proteins (data not shown). In the absence of E47, the CeABF-1 deletion mutants failed to activate the reporter gene alone (Fig. 7). Previous studies with the human ABF-1 protein have shown that the C-terminus, which contains the bHLH domain, is capable of repressing E47 activity. The ability of the C-terminus of CeABF-1 to inhibit E2A transcription is consistent with the human ABF-1 protein and its reported transcriptional properties.
Conserved amino acid pair within the HLH domain of CeABF-1 contributes to the repression of E47-mediated gene transcription
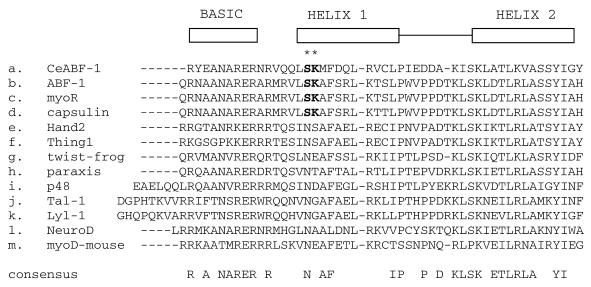
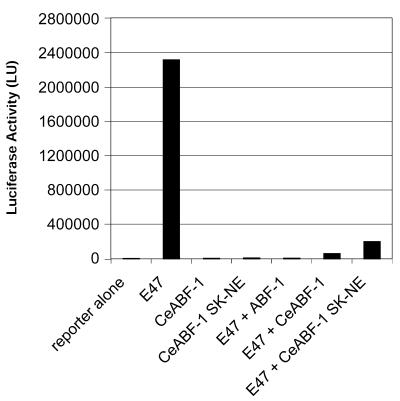
Because a large portion of the repressing activity of the CeABF-1 polypeptide maps to the region corresponding to the C-terminal bHLH domain, and a similar repressing activity has been observed in the bHLH domain of mouse MyoR/musculin and human homolog ABF-1 (7,10,22), we analyzed the bHLH domain of the human ABF-1, mouse MyoR and nematode CeABF-1 polypeptides for sequence similarities responsible for the repressing activity. Interestingly, following the comparison of the amino acid sequences, we identified two conserved amino acids, SK, within helix one of the bHLH domain (Fig. 8). Because these two conserved amino acids are absent in nearly all other bHLH family members, we investigated the biological relevance of the SK residues and their participation in repressing E2A-mediated gene activation. To address the biological function of the SK residues, we performed site-directed mutagenesis to change the residues from SK to NE. The mutation of the SK residues to NE was selected because many other class II bHLH family members often contain NE at this position and are not transcriptional repressors, e.g. MyoD. Following the introduction of the mutation within the CeABF-1 cDNA by overlapping PCR, the gene segment was inserted into the mammalian expression vector pCMV-TAG2B. After confirmation of the mutation by DNA sequencing, transient transfection assays were performed. The expression vector encoding the mutant form of CeABF-1, designated CeABF-1 SK-NE, was transfected into HeLa cells along with the reporter vector carrying the firefly luciferase gene driven by four copies of the µE5-µE3-µE2 E-box elements derived from the IgH gene enhancer. The expression vector encoding the mutant CeABF-1 SK-NE, in the absence of the plasmid encoding E47, failed to transactivate the LUC reporter gene (Fig. 9). Similar results were obtained when the expression vector encoding the wild-type CeABF-1 was introduced (Fig. 9). As expected, when the E47 expression vector was transfected into HeLa cells along with the reporter gene construct, strong transactivation of the reporter vector resulted (Fig. 9). When the wild-type CeABF-1 and E47 expression plasmids were cotransfected along with the LUC reporter plasmid, a dramatic inhibition of E47-mediated transactivation was observed, reaffirming the function of CeABF-1 as a transcriptional repressor (Fig. 9). To address the functional significance of the SK amino acids within helix 1 of CeABF-1, the expression plasmids encoding CeABF-1 SK-NE and E47 were cotransfected along with the LUC reporter vector, and the transcriptional activity measured. The mutant CeABF-1 SK-NE was still capable of repressing E47-mediated gene activation; however, when compared with the wild-type CeABF-1, the mutant repressed E2A somewhat less efficiently (Fig. 9). These results indicate the SK residues found within helix 1 of the wild-type CeABF-1 may contribute to the inhibition of E47 transactivation, although other residues contribute significantly to repression of E2A activity as well.
Figure 8.
Sequence alignment of bHLH domain of CeABF-1 with the class II family of bHLH proteins. Multiple sequence alignment of the bHLH region of nematode ABF-1 (a.) with the most related class II bHLH proteins as determined by the Clustal algorithm. Interestingly, CeABF-1, human ABF-1 (7), MyoR (10) and capsulin (11), all contain the two conserved amino acids, SK, near the more N-terminal side (see asterisks).
Figure 9.
SK residues found within the first helix of the bHLH domain of CeABF-1 help mediate repression of E47 activity. HeLa S3 cells were transiently transfected with 1 µg of (µE5-µE3-µE2)4-Luc reporter vector and different combinations of the E47 expression vector (0.5 µg), FLAG-CeABF-1, and FLAG-CeABF-1 SK-NE expression plasmids (0.25 µg). Twenty-five microliters of Superfect reagent was used for each transfection and equal amounts of protein extract were assayed for firefly luciferase activity. Experiments were performed in triplicate and represent the means of two independent assays. Luciferase activity was normalized to β-galactosidase levels as a control for transfection efficiency. The mean of the firefly luciferase value for E47 + CeABF-1 was 60 248 light units and E47 + CeABF-1 SK-NE was 198 818 light units.
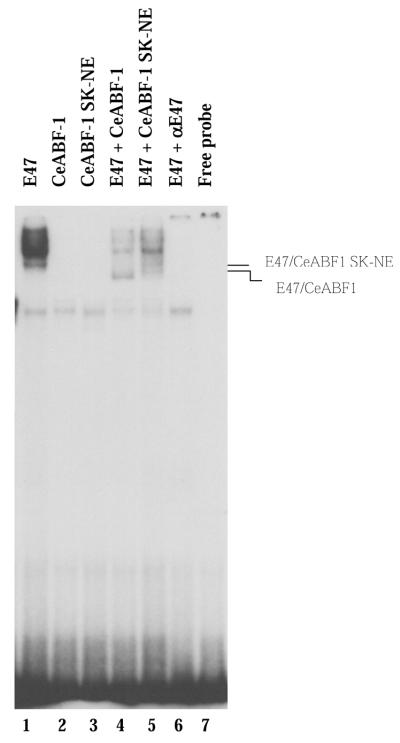
To assess whether the mutant CeABF-1 SK-NE could still dimerize with E47 and bind DNA, we performed an EMSA using the µE5-box site as a probe. Each of the gene products was translated efficiently (data not shown). Subsequently, the in vitro-translated proteins were incubated with a 32P-labeled µE5-containing an oligonucleotide probe from the IgH intronic enhancer and analyzed by EMSA. As expected, E47 was capable of binding to the µE5 site as a homodimer (Fig. 10, lane 1). When CeABF-1 and the mutant CeABF-1 SK-NE were incubated individually with the E-box-containing binding site, no DNA binding was observed (Fig. 10, lanes 2 and 3). When CeABF-1 was cotranslated with E47, a complex of intermediate mobility was formed (Fig. 10, lane 4). This complex represents the heterodimer composed of CeABF-1 and E47. To test whether the introduction of the SK to NE mutation affected DNA-binding activity, CeABF-1 SK-NE was cotranslated with E47 and incubated with the radiolabeled µE5 probe. CeABF-1 SK-NE dimerized with E47 and bound DNA as a heterodimer (Fig. 10, lane 5). Interestingly, a subtle difference in the migration profile of the CeABF-1 SK-NE–E47 heterodimer was observed relative to the wild-type CeABF-1–E47 heterodimer. This may represent a different conformation of the CeABF-1 SK-NE–E47 dimer as compared with the CeABF-1–E47 heterodimer. Taken together, the data suggest the mutant CeABF-1 SK-NE can still dimerize with E47 and bind DNA.
Figure 10.
CeABF-1 SK-NE binds DNA as a heterodimer with E47. EMSA analysis of E47–CeABF-1 and E47–CeABF-1 SK-NE heterodimeric complexes formed on the µE5 radiolabeled probe. Lines indicate the position of E47–CeABF1 and E47–CeABF1 SK-NE heterodimers. Addition of an antibody specific to E47 is indicated by the labeling αE47.
DISCUSSION
Recent analysis of open reading frames present in the C.elegans genome has revealed 24 putative HLH proteins. Although 24 putative HLH proteins have been predicted, only about five different bHLH gene products have been investigated thoroughly, including the homologs for MyoD (HLH-1/CeMyoD), NeuroD/BETA-2 (END-1), E2A (HLH-2/CeE protein), Myc and Twist (HLH-8/CeTwist). Many of the 24 putative HLH proteins have yet to be confirmed as bona fide gene products. To determine whether C.elegans contained a gene closely related to the human ABF-1 gene, we searched the C.elegans genome database for potential candidates containing similarities to human ABF-1. The search identified several bHLH gene products, including LIN-32, achaete-scute (HLH-3) and CeTwist (HLH-8) that showed homology to human ABF-1 within the bHLH domain. In particular, the highest degree of homology was observed for a predicted gene encoding a polypeptide containing 52% identities within the bHLH domain. Closer analysis of the entire predicted product revealed 51% (87/170) similarity over the entire length of the protein relative to human ABF-1.
In these studies, we have cloned a novel gene from the nematode C.elegans. Because this gene product is closely related to the vertebrate ABF-1 gene product and has similar transcriptional properties and dimerization partners, we have dubbed the gene CeABF-1. The CeABF-1 gene contains three exons and two introns. The splice junctions follow normal GT/AG 5′ donor and 3′ acceptor rules (Fig. 3). In vitro transcription and translation of CeABF-1 produced two different protein products similar in size, presumably reflecting alternative translational initiation (unpublished results). The predicted size of the CeABF-1 polypeptide correlates closely to the reported sizes of human and mouse ABF-1, which contain 218 and 201 amino acids, respectively (7,9). The conservation between vertebrate and invertebrate ABF-1 is observed within the C-terminal bHLH domain and a second region residing in the N-terminus. The bHLH domain contains 23 out of 44 identities, or 52% identities whereas the N-terminal region shows six out of 14 identities, or 43% conservation between the nematode and human (Fig. 2). Compared with all other vertebrate gene products, CeABF-1 shows the highest number of identities to human ABF-1. Interestingly, one other vertebrate gene product that shows relatively strong homology to CeABF-1 is capsulin/POD-1/epicardin; however, this homology is exclusively restricted to the bHLH domain and has no obvious similarities outside this region (23,24). Mice homozygous for the capsulin null mutation fail to form a spleen, demonstrating the importance of this bHLH factor as a key regulator of spleen organogenesis (25). Functionally, capsulin appears to have different transcriptional properties relative to CeABF-1. capsulin has been reported to bind E-box binding sites and activate a reporter gene driven by minimal promoter (24). These results seem to indicate that capsulin can function as a transcriptional activator.
Studies of nematode or fly bHLH proteins in vertebrates have provided significant functional data. The evolutionary conservation of the bHLH domain in CeMyoD has been reported to be sufficient to drive myogenesis in mouse embryonic cells transfected with viral constructs capable of producing invertebrate MyoD, whereas NAUTILUS, the Drosophila homolog of MyoD, is unable to initiate myogenesis in mouse cells (26). These studies have been important for revealing conserved pathways in biological systems used by nematodes and humans. Aside from these studies, limited information has been reported regarding regulatory domains of invertebrate HLH proteins and their relationship to vertebrate homologs. In these studies, we have shown that CeABF-1 can function as a transcriptional repressor, similar to human ABF-1 and the mouse ortholog musculin/MyoR. Our gel-shift studies suggest at least two possible mechanisms whereby CeABF-1 can block E-box-mediated gene expression which include (i) the formation of CeABF-1/E protein heterodimers that bind target sites but fail to activate gene transcription, and (ii) the formation of non-DNA-binding heterodimeric complexes between CeABF-1 and E proteins.
The relationship between the CeABF-1–E47 heterodimeric species detected by gel-shift assays and the strong repression of E2A activity observed in the transient transfection assays is intriguing. If DNA binding of the CeABF-1–E47 heterodimer is solely responsible for decreasing E2A-mediated gene activation, one might expect to detect more shifted CeABF-1–E47 heterodimer on EMSA. However, our gel-retardation studies show only a minimal amount of shifted heterodimeric species bound to the E-box. Perhaps one explanation for the weak binding could be that our gel-shift conditions are not optimal for binding the CeABF-1–E47 heterodimer and/or the proteins require some type of post-translational modification for optimal binding. Alternatively, perhaps CeABF-1 functions as a potent transcriptional repressor of E47 via the formation non-DNA-binding heterodimers. Only a few class II bHLH proteins have been reported to function as negative regulators of E47 activity. These class II bHLH factors include human ABF-1, mouse musculin, and now, the nematode protein CeABF-1. Interestingly, all these factors require the presence of the bHLH domain for optimal repressing activity. Following the inspection of the bHLH region of these factors, we discovered the presence of two conserved amino acids, SK, within helix one of the bHLH domain (Fig. 8). In order to investigate the role of these residues on repressing E2A activity, we used site-directed mutagenesis to alter the SK residues to NE and tested their function by transient transfections. Our experiments suggest that these two residues within helix 1 of CeABF-1 contribute, in part, to CeABF-1’s ability to repress the activity of E47. However, the significant repressing activity that remains in the presence of the SK to NE mutation suggests that other regions within the bHLH domain are also critical for transcriptional repression. As assayed by gel-shift analysis, the CeABF-1 mutant could still efficiently dimerize with E47, resulting in a diminished E47 homodimer formation relative to the DNA-binding activity observed with E47 alone (Fig. 10, compare lanes 1 and 5). In addition, the CeABF-1 SK-NE mutant was still observed to bind DNA as a heterodimer with E47 (Fig. 10, lane 5). The slight difference in the migration profile between the CeABF-1–E47 and CeABF-1 SK-NE–E47 heterodimers as observed by EMSA most likely reflects a different conformational state of the heterodimers bound to DNA—as gel-shift assays are capable of detecting subtle differences in protein conformation (Fig. 10, compare lanes 4 and 5). The difference in the ability of the mutant CeABF-1 to function as efficiently as a repressor of E47 activity relative to the wild-type CeABF-1 could possibly be explained by one or both of the following: (i) the SK residues function to allow optimal dimerization of CeABF-1 with E47 or (ii) the SK residues help contribute to the neutralization of the transactivation domain of E47 following dimerization. Our gel-shift assay data are consistent with both of these models as possibilities. The question arises whether there are other examples of critical amino acid residues within the bHLH domain that are functionally important in regulating gene transcription. Interestingly, for class II bHLH proteins that serve as transcriptional activators, the importance of two conserved amino acids within the bHLH domain, alanine-threonine, has already been clearly demonstrated for the myogenic bHLH proteins and muscle-specific gene activation (27).
The reported expression patterns for human and mouse musculin have provided some insight to gene function. Human ABF-1 has been detected in tumor B-cell lines, primary lymphoid tissue and the aorta (7). In mice, mouse musculin has been detected in embryonic skeletal muscle and the spleen (9). To date, disruption of the musculin gene in mice has not been reported. The lack of a characterized immune system in the nematode would seem to suggest human ABF-1 has evolved, perhaps, multiple biological roles in vertebrates. The most likely interacting partner in C.elegans for CeABF-1 is CeE (HLH-2), the worm ortholog of the human E2A gene products, E12 and E47. Although no null mutations have been identified in hlh-2, the expression pattern has been localized to neurons and non-striated muscle (20). Presumably, CeE proteins heterodimerize with a variety of different bHLH proteins during development, which include the C.elegans achaete-scute-like factors HLH-3, -4, -6 and -8 (12,20,28). Our data supports a developmental expression profile of CeABF-1 in the L3 and L4 larval stages. The expression profile of CeABF-1 in the late larval stages L3 and L4 may be suggestive of a role in sexual maturation, a process that occurs late in development.
Future studies involving the formation of transgenic animals for expression studies should provide important clues regarding the function of the CeABF-1 gene in C.elegans and better define its functional relationship to human ABF-1.
Acknowledgments
ACKNOWLEDGEMENTS
We are thankful for the critical reading of the manuscript by Dr Lisa Wrischnik and Dr Paul Richmond for preparation of the figures. Also, we would like to thank Andrew Fire (Carnegie Institution of Washington) for the cloning vector carrying the unc-22 gene for the RNA-I assays. Many of the strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources of the NIH. This work was supported by a RUI-National Science Foundation Research grant MCB (MCB 0111069) and AREA-National Institute of Health grant NIAID (R15 AI45767-01A1).
References
- 1.Massari M.E. and Murre,C. (2000) Helix–loop–helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol., 20, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain G., Maandag,E.C., Izon,D.J., Amsen,D., Kruisbeek,A.M., Weintraub,B.C., Krop,I., Schlissel,M.S., Feeney,A.J., van Roon,M. et al. (1994) E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell, 79, 885–892. [DOI] [PubMed] [Google Scholar]
- 3.Porcher C., Swat,W., Rockwell,K., Fujiwara,Y., Alt,F.W. and Orkin,S.H. (1996) The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell, 86, 47–57. [DOI] [PubMed] [Google Scholar]
- 4.Naya F.J., Huang,H.P., Qiu,Y., Mutoh,H., DeMayo,F.J., Leiter,A.B. and Tsai,M.J. (1997) Diabetes, defective pancreatic morphogenesis and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev., 11, 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudnicki M.A., Schnegelsberg,P.N., Stead,R.H., Braun,T., Arnold,H.H. and Jaenisch,R. (1993) MyoD or Myf-5 is required for the formation of skeletal muscle. Cell, 75, 1351–1359. [DOI] [PubMed] [Google Scholar]
- 6.Murre C., McCaw,P.S., Vaessin,H., Caudy,M., Jan,L.Y., Jan,Y.N., Cabrera,C.V., Buskin,J.N., Hauschka,S.D., Lassar,A.B. et al. (1989) Interactions between heterologous helix–loop–helix proteins generate complexes that bind specifically to a common DNA sequence. Cell, 58, 537–544. [DOI] [PubMed] [Google Scholar]
- 7.Massari M.E., Rivera,R.R., Voland,J.R., Quong,M.W., Breit,T.M., van Dongen,J.J., de Smit,O. and Murre,C. (1998) Characterization of ABF-1, a novel basic helix–loop–helix transcription factor expressed in activated B lymphocytes. Mol. Cell. Biol., 18, 3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bain G., Engel,I., Robanus Maandag,E.C., te Riele,H.P., Voland,J.R., Sharp,L.L., Chun,J., Huey,B., Pinkel,D. and Murre,C. (1997) E2A deficiency leads to abnormalities in alpha beta T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol., 17, 4782–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robb L., Hartley,L., Wang,C.C., Harvey,R.P. and Begley,C.G. (1998) Musculin: a murine basic helix–loop–helix transcription factor gene expressed in embryonic skeletal muscle. Mech. Dev., 76, 197–201. [DOI] [PubMed] [Google Scholar]
- 10.Lu J., Webb,R., Richardson,J.A. and Olson,E.N. (1999) MyoR: a muscle-restricted basic helix–loop–helix transcription factor that antagonizes the actions of MyoD. Proc. Natl Acad. Sci. USA, 96, 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J., Richardson,J.A. and Olson,E.N. (1998) Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech. Dev., 73, 23–32. [DOI] [PubMed] [Google Scholar]
- 12.Ruvkun G. and Hobert,O. (1998) The taxonomy of developmental control in Caenorhabditis elegans. Science, 282, 2033–2041. [DOI] [PubMed] [Google Scholar]
- 13.Zhao C. and Emmons,S.W. (1995) A transcription factor controlling development of peripheral sense organs in C. elegans. Nature, 373, 74–78. [DOI] [PubMed] [Google Scholar]
- 14.Wrischnik L.A. and Kenyon,C.J. (1997) The role of lin-22, a hairy/enhancer of split homolog, in patterning the peripheral nervous system of C. elegans. Development, 124, 2875–2888. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell B., Mugiya,M., Youngblom,J., Funes-Duran,M., Miller,R., Ezpeleta,J., Rigby,N. and Vierra,C. (2000) The genomic structure and promoter analysis of the human ABF-1 gene. Biochim. Biophys. Acta, 1492, 320–329. [DOI] [PubMed] [Google Scholar]
- 16.Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watkins B.A., Davis,A.E., Cocchi,F. and Reitz,M.S.,Jr (1993) A rapid method for site-specific mutagenesis using larger plasmids as templates. Biotechniques, 15, 700–704. [PubMed] [Google Scholar]
- 18.Fields C. (1990) Information content of Caenorhabditis elegans splice site sequences varies with intron length. Nucleic Acids Res., 18, 1509–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bektesh S., Van Doren,K. and Hirsh,D. (1988) Presence of the Caenorhabditis elegans spliced leader on different mRNAs and in different genera of nematodes, Genes Dev., 2, 1277–1283. [DOI] [PubMed] [Google Scholar]
- 20.Krause M., Park,M., Zhang,J.M., Yuan,J., Harfe,B., Xu,S.Q., Greenwald,I., Cole,M., Paterson,B. and Fire,A. (1997) A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development, 124, 2179–2189. [DOI] [PubMed] [Google Scholar]
- 21.Stern M.J. and Horvitz,H.R. (1991) A normally attractive cell interaction is repulsive in two C. elegans mesodermal cell migration mutants. Development, 113, 797–803. [DOI] [PubMed] [Google Scholar]
- 22.Wong J.M.F.D., Ahlberg,J., Round,J., O’Connell,R., Miller,R., Chen,E., Richmond,P.A. and Vierra,C.A. (2001) Characterization of a basic helix–loop–helix protein ABF-1: nuclear localization, transcriptional properties and interaction with Id-2. DNA Cell. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 23.Quaggin S.E., Schwartz,L., Cui,S., Igarashi,P., Deimling,J., Post,M. and Rossant,J. (1999) The basic-helix–loop–helix protein pod1 is critically important for kidney and lung organogenesis. Development, 126, 5771–5783. [DOI] [PubMed] [Google Scholar]
- 24.Hidai H., Bardales,R., Goodwin,R., Quertermous,T. and Quertermous,E.E. (1998) Cloning of capsulin, a basic helix–loop–helix factor expressed in progenitor cells of the pericardium and the coronary arteries. Mech. Dev., 73, 33–43. [DOI] [PubMed] [Google Scholar]
- 25.Lu J., Chang,P., Richardson,J.A., Gan,L., Weiler,H. and Olson,E.N. (2000) The basic helix–loop–helix transcription factor capsulin controls spleen organogenesis. Proc. Natl Acad. Sci. USA, 97, 9525–9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J.M., Chen,L., Krause,M., Fire,A. and Paterson,B.M. (1999) Evolutionary conservation of MyoD function and differential utilization of E proteins. Dev. Biol., 208, 465–472. [DOI] [PubMed] [Google Scholar]
- 27.Davis R.L., Cheng,P.F., Lassar,A.B. and Weintraub,H. (1990) The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell, 60, 733–746. [DOI] [PubMed] [Google Scholar]
- 28.Corsi A.K., Kostas,S.A., Fire,A. and Krause,M. (2000) Caenorhabditis elegans twist plays an essential role in non-striated muscle development. Development, 127, 2041–2051. [DOI] [PubMed] [Google Scholar]
- 29.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]