ABSTRACT
Data on the epidemiology of invasive candidiasis (IC) and the antifungal susceptibility of Candida isolates in China are still limited. Here we report on surveillance for IC from the China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study. Sixty-five tertiary hospitals collected 8,829 Candida isolates from 1 August 2009 to 31 July 2014. Matrix-assisted laser desorption ionization–time of flight mass spectrometry supplemented by ribosomal DNA sequencing was used to define the species, and the fluconazole and voriconazole susceptibilities were determined by the Clinical and Laboratory Standards Institute disk diffusion method. A total of 32 Candida species were identified. Candida albicans was the most common species (44.9%), followed by the C. parapsilosis complex (20.0%), C. tropicalis (17.2%), and the C. glabrata complex (10.8%), with other species comprising <3% of isolates. However, in candidemia, the proportion of cases caused by C. albicans was only 32.3%. C. albicans and C. parapsilosis complex isolates were susceptible to fluconazole and voriconazole (<6% resistance), while fluconazole and azole cross-resistance rates were high in C. tropicalis (13.3% and 12.9%, respectively), C. glabrata complex (18.7% and 14%, respectively), and uncommon Candida species (44.1% and 10.3%, respectively) isolates. Moreover, from years 1 to 5 of the study, there was a significant increase in the rates of resistance to fluconazole among C. glabrata complex isolates (12.2% to 24.0%) and to both fluconazole (5.7% to 21.0%) and voriconazole (5.7% to 21.4%) among C. tropicalis isolates (P < 0.01 for all comparisons). Geographic variations in the causative species and susceptibilities were noted. Our findings indicate that antifungal resistance has become noteworthy in China, and enhanced surveillance is warranted.
KEYWORDS: invasive candidiasis, epidemiology, antifungal susceptibility, fluconazole, voriconazole, China
INTRODUCTION
Invasive candidiasis (IC) is a life-threatening disease with high rates of morbidity and mortality, especially among immunocompromised and critically ill patients (1, 2). Worldwide, Candida albicans remains the predominant pathogen causing IC, but the prevalence of infection due to non-albicans Candida species is on the rise, with non-albicans Candida species accounting for over 50% of cases of IC in many geographic regions (1, 3). Of note, non-albicans Candida species are often more resistant to antifungal drugs than C. albicans (2, 4), which is concerning with respect to clinical outcomes.
Early and appropriate therapy in IC is essential to improve the overall outcomes (5–7). However, initiation of such targeted antifungal therapy is contingent on the timely diagnosis of IC. Because rapid IC diagnostic assays, such as molecular-based tests, have not yet reached the bedside in many hospitals, most clinicians still rely on insensitive culture-based methods to direct patient management (1, 7). Robust local epidemiological data, including knowledge of antifungal susceptibility profiles and trends, are therefore essential for the selection of initial antifungal therapy (1–4). This hinges upon effective surveillance networks.
The China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study was the first and has continued to be the largest national surveillance program for invasive fungal infections, including IC, in mainland China. Initiated in 2009 (8), over 8,000 isolates were collected during the first to fifth surveillance years. We have previously reported results from limited surveillance or on the epidemiology relevant to selected species (9–14) but not the overall results from the entire CHIF-NET study. Here we provide a perspective on the overall comparative species distribution of Candida pathogens and azole antifungal susceptibility data for Candida isolates collected during the first 5 years of the study.
MATERIALS AND METHODS
The CHIF-NET study.
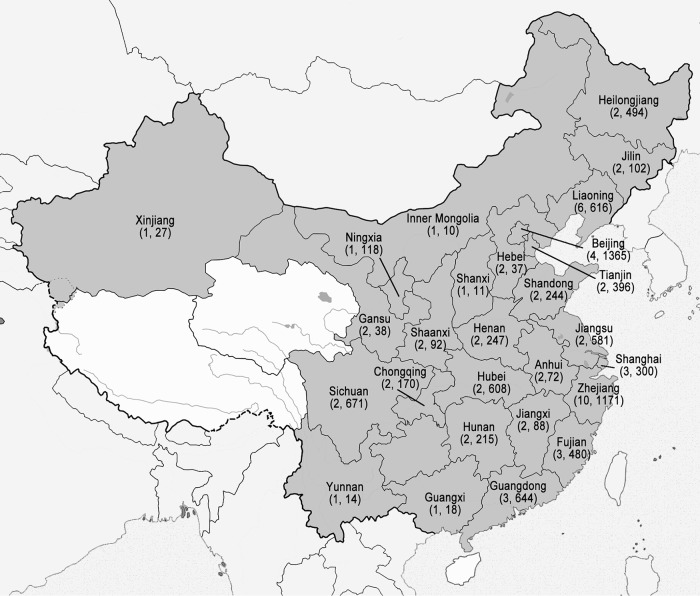
The CHIF-NET study is a prospective, laboratory-based, multicenter study of invasive yeast infections, including IC, initiated in 2009 (as described above). Each surveillance year began on 1 August of the year and continued to 31 July of the following year. Sixty-five tertiary general hospitals from 27 provinces in China participated in the first 5 years (Fig. 1). The number of participating hospitals increased from 12 in the first year to 22, 22, 48, and 61 in the second to fifth years, respectively.
FIG 1.
Geographic regions of the CHIF-NET study covered (27 provinces, in dark gray). The first number in parentheses under the province name indicates the number of hospitals that participated in the CHIF-NET study in each province, and the second number indicates the number of isolates collected.
The study inclusion criteria were as previously described (8). In each surveillance year, all Candida isolates from eligible patients with IC were forwarded to the central laboratory, Department of Clinical Laboratory, Peking Union Medical College Hospital, for species confirmatory identification and antifungal susceptibility testing. The study was approved by the Human Research Ethics Committee of the Peking Union Medical College Hospital (S-263).
Species identification.
To ensure the accuracy of identification, all invasive Candida isolates were identified to the species level in the central laboratory. In year 1 of the study, isolates were identified by DNA sequencing of the fungal ribosomal DNA internal transcribed spacer (ITS) regions (8), and isolates from years 2 to 5 were identified by a combination of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), using a Vitek MS system (bioMérieux, Marcy l'Étoile, France), supplemented by ITS sequencing (15).
Antifungal susceptibility testing.
Susceptibility to fluconazole and voriconazole was determined using the Clinical and Laboratory Standards Institute (CLSI) M44-A2 disk diffusion method (16). For all isolates from all years, species-specific MIC clinical breakpoint (CBP) interpretive criteria were applied to C. albicans, Candida tropicalis, the Candida parapsilosis complex, the Candida glabrata complex, and Candida krusei according to the guidelines in the reference CLSI M60 document (17), while the susceptibilities of the other Candida species were interpreted in accordance with the CLSI M44-S3 document guidelines (18). The quality control strains were C. albicans ATCC 90028, Candida parapsilosis ATCC 22019, and Candida krusei ATCC 6258.
Statistical analysis.
All comparisons were performed using SPSS software (version 12.0; SPSS Inc., Chicago, IL, USA). Comparisons of continuous variables were performed by using the Mann-Whitney test, and comparisons of categorical variables were performed by using a chi-square test or Fisher's exact test, as appropriate. A P value of 0.05 was significant.
RESULTS
Demographics.
A total of 8,829 nonrepetitive (i.e., nonduplicate) Candida isolates from separate patients were collected; 37.8% of the isolates were cultured from male patients, and 62.2% of the isolates were cultured from female patients. The patients' ages ranged from 0 to 103 years (median, 50 years; interquartile range, 45 to 72 years).
Candida species.
Thirty-two species of Candida were identified among the 8,829 isolates. C. albicans was the most common (3,965 isolates, 44.9%), with no significant trend in frequency being observed over the 5 years (P > 0.05) (Table 1). Non-albicans Candida species accounted for 4,864 (55.1%) isolates (Table 1). Of these, C. parapsilosis complex isolates were the most frequent 1,762 (20.0%) and consisted of C. parapsilosis sensu stricto (1,526/8,829 isolates, 17.3%), Candida metapsilosis (1.9%), and Candida orthopsilosis and Lodderomyces elongisporus (0.4% each; Table 1). C. tropicalis was the third most common species (1,515 isolates, 17.2%), followed by the C. glabrata complex (955 isolates, 10.8%); of the latter, 98.5% were C. glabrata sensu stricto isolates (n = 941), while Candida nivariensis and Candida bracarensis were rare (0.1% and <0.1%, respectively; Table 1). Other species were rare (<2.1%; Table 1). No significant trends in frequency were seen for non-albicans Candida species (P > 0.05 for all comparisons).
TABLE 1.
Species distribution of Candida isolates over 5 years
| Candida species | Overall |
Yr 1 |
Yr 2 |
Yr 3 |
Yr 4 |
Yr 5 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| Candida albicans | 3,965 | 44.9 | 284 | 38.6 | 556 | 48.9 | 704 | 47.4 | 1,051 | 43 | 1,370 | 45.3 |
| C. parapsilosis complex | 1,762 | 20 | 172 | 23.4 | 184 | 16.2 | 241 | 16.2 | 538 | 22 | 627 | 20.7 |
| C. parapsilosis sensu stricto | 1,526 | 17.3 | 142 | 19.3 | 161 | 14.2 | 202 | 13.6 | 460 | 18.8 | 561 | 18.5 |
| C. metapsilosis | 167 | 1.9 | 23 | 3.1 | 14 | 1.2 | 25 | 1.7 | 54 | 2.2 | 51 | 1.7 |
| C. orthopsilosis | 35 | 0.4 | 4 | 0.5 | 7 | 0.6 | 6 | 0.4 | 12 | 0.5 | 6 | 0.2 |
| Lodderomyces elongisporus | 34 | 0.4 | 3 | 0.4 | 2 | 0.2 | 8 | 0.5 | 12 | 0.5 | 9 | 0.3 |
| C. tropicalis | 1,515 | 17.2 | 122 | 16.6 | 218 | 19.2 | 267 | 18 | 413 | 16.9 | 495 | 16.4 |
| C. glabrata complex | 955 | 10.8 | 90 | 12.2 | 115 | 10.1 | 178 | 12 | 260 | 10.6 | 312 | 10.3 |
| C. glabrata sensu stricto | 941 | 10.7 | 88 | 12 | 115 | 10.1 | 176 | 11.8 | 254 | 10.4 | 308 | 10.2 |
| C. nivariensis | 13 | 0.1 | 2 | 0.3 | 1 | <0.1 | 6 | 0.2 | 4 | 0.1 | ||
| C. bracarensis | 1 | <0.1 | 1 | <0.1 | ||||||||
| C. guilliermondii | 186 | 2.1 | 12 | 1.6 | 16 | 1.4 | 20 | 1.3 | 53 | 2.2 | 85 | 2.8 |
| C. krusei | 125 | 1.4 | 18 | 2.4 | 16 | 1.4 | 24 | 1.6 | 25 | 1 | 42 | 1.4 |
| C. pelliculosa | 123 | 1.4 | 13 | 1.8 | 10 | 0.9 | 12 | 0.8 | 39 | 1.6 | 47 | 1.6 |
| C. lusitaniae | 50 | 0.6 | 6 | 0.8 | 4 | 0.4 | 18 | 1.2 | 12 | 0.5 | 10 | 0.3 |
| C. lipolytica | 36 | 0.4 | 9 | 1.2 | 3 | 0.3 | 5 | 0.3 | 10 | 0.4 | 9 | 0.3 |
| C. haemulonii | 24 | 0.3 | 1 | 0.1 | 3 | 0.3 | 6 | 0.4 | 10 | 0.4 | 4 | 0.1 |
| C. intermedia | 20 | 0.2 | 3 | 0.3 | 3 | 0.2 | 12 | 0.5 | 2 | <0.1 | ||
| C. norvegensis | 13 | 0.1 | 1 | 0.1 | 3 | 0.2 | 6 | 0.2 | 3 | 0.1 | ||
| C. fabianii | 11 | 0.1 | 1 | 0.1 | 3 | 0.2 | 7 | 0.3 | ||||
| C. inconspicua | 8 | <0.1 | 2 | 0.2 | 3 | 0.1 | 3 | 0.1 | ||||
| C. rugosa | 7 | <0.1 | 1 | <0.1 | 1 | <0.1 | 1 | <0.1 | 4 | 0.1 | ||
| C. fermentati | 6 | <0.1 | 2 | 0.2 | 2 | <0.1 | 2 | <0.1 | ||||
| C. quercitrusa | 4 | <0.1 | 3 | 0.4 | 1 | <0.1 | ||||||
| C. catenulata | 4 | <0.1 | 2 | 0.3 | 1 | <0.1 | 1 | <0.1 | ||||
| C. aaseri | 3 | <0.1 | 3 | 0.1 | ||||||||
| C. famata | 3 | <0.1 | 1 | 0.1 | 1 | <0.1 | 1 | <0.1 | ||||
| C. kefyr | 3 | <0.1 | 1 | 0.1 | 2 | <0.1 | ||||||
| C. opuntiae | 1 | <0.1 | 1 | <0.1 | ||||||||
| C. freyschussii | 1 | <0.1 | 1 | <0.1 | ||||||||
| C. magnoliae | 1 | <0.1 | 1 | <0.1 | ||||||||
| C. palmioleophila | 1 | <0.1 | 1 | <0.1 | ||||||||
| C. utilis | 1 | <0.1 | 1 | <0.1 | ||||||||
| C. diddensiae | 1 | <0.1 | 1 | <0.1 | ||||||||
| Total | 8,829 | 100 | 736 | 100 | 1,137 | 100 | 1,486 | 100 | 2,444 | 100 | 3,026 | 100 |
Species distribution according to specimen type.
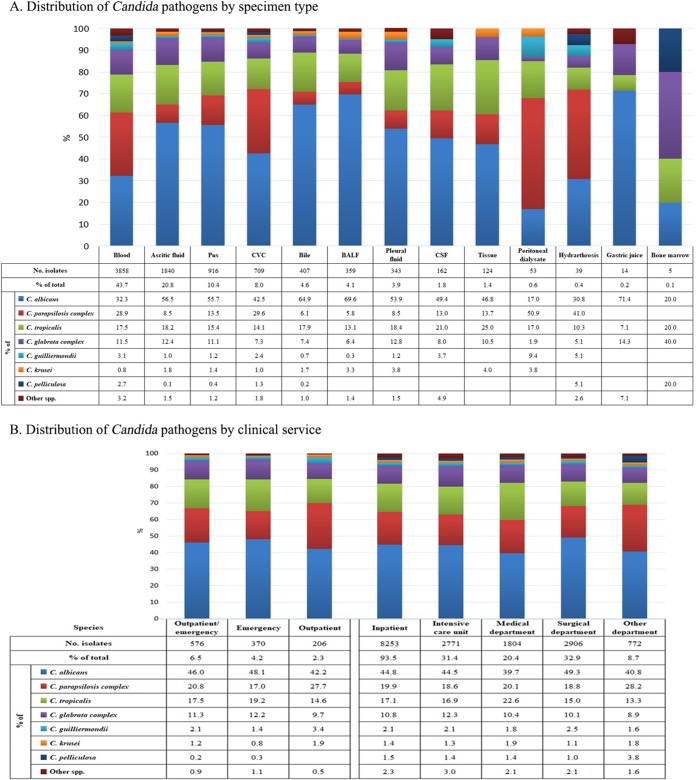
Of the various specimen types, over 40% of invasive Candida isolates (3,858/8,829 isolates, 43.7%) were recovered from blood, followed by ascitic fluid (20.8%), pus (10.4%), central venous catheter tips (CVC; 8.0%), bile (4.6%), bronchoalveolar lavage fluid (4.1%), pleural fluid (3.9%), cerebrospinal fluid (1.8%), and tissue (1.4%) (Fig. 2).
FIG 2.
Distribution of Candida pathogens by specimen type (A) and clinical service (B). Abbreviations: BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; CVC, central venous catheter.
The proportion of non-albicans Candida isolates recovered from blood cultures (2,612/3,858 isolates, 67.7%) was significantly higher than that recovered from other specimen types (2,252/2,719 isolates, 45.3%) (P < 0.01) (Fig. 2). More specifically, the difference in the relative proportion was the largest for C. parapsilosis complex isolates (25.2% versus 13.0%, P < 0.01) (Fig. 2). The frequency between blood source and non-blood source isolates was similar for C. tropicalis (28.9% versus 16.9%, P > 0.05) and the C. glabrata complex (11.5% versus 10.3%, P > 0.05). In addition, over the 5 years of the study, the C. parapsilosis complex was recovered at a higher frequency than C. albicans and became the most predominant species among isolates causing candidemia in years 1 and 4. Significantly higher proportions of C. parapsilosis complex isolates were also observed in CVC (29.6%) and peritoneal dialysate (50.9%) specimens compared with this species' average frequency (20%) (P < 0.01 for both comparisons) (Fig. 2). Of the uncommon Candida species, some were found in higher proportions in blood culture specimens than in non-blood culture specimens, e.g., Candida guilliermondii (3.1% versus 1.3%, P < 0.01), Candida pelliculosa (2.7% versus 0.4%, P < 0.01), and Candida lipolytica (0.8% versus 0.1%, P < 0.01).
Species distribution by patient location and geographic regions.
Of the Candida isolates recovered, 93.5% were from hospital inpatients (including those in intensive care units [ICUs] [31.4%], medical wards [20.4%], and surgical wards [32.9%]) and 6.5% were from patients in outpatient/emergency settings (Fig. 2). In all cases, C. albicans was the predominant species (39.7% to 49.3%). The second and third most common species in different clinical settings were either the C. parapsilosis complex or C. tropicalis (13.3% to 28.2%). The C. glabrata complex was the fourth most common species in all clinical settings (prevalence, 8.9% to 12.3%). Other Candida species were rare (<4%; Fig. 2).
Geographic variation in the species distribution was observed. For instance, C. albicans was most common in 57 of 65 (87.7%) hospitals, but its frequency varied widely from 12.5% to 100% in different hospitals. In the eight hospitals where C. albicans was not the dominant species, the most common species were the C. parapsilosis complex, C. tropicalis, and C. pelliculosa in four hospitals, three hospitals and one hospital, respectively.
In vitro susceptibilities.
Of 8,829 Candida isolates, 80.0%, 11.2%, and 8.8% of the isolates were susceptible, susceptible dose-dependent (SDD), and resistant to fluconazole, respectively (Table 2). In comparison, 94.0% of isolates were susceptible to voriconazole or of the wild-type (WT) phenotype, 0.6% were SDD or intermediate, and 5.4% were resistant or non-wild-type (NWT) (Table 2). Cross-resistance occurred in 5.2% (457/8,829) of the isolates (Table 2), and 59.0% (457/774) of fluconazole-resistant isolates were azole cross-resistant. Across the different hospitals, the fluconazole and voriconazole susceptibility rates ranged from 50% to 100% and 43% to 100%, respectively.
TABLE 2.
In vitro susceptibilities of Candida spp. to fluconazole and voriconazole as determined by CLSI disk diffusion method
| Species | Fluconazolea |
Voriconazolea |
Cross-resistant |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S |
R |
S/WT |
R/NWT |
|||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | |
| C. albicans | 3,927 | 99.0 | 20 | 0.5 | 3,928 | 99.1 | 30 | 0.8 | 19 | 0.5 |
| C. parapsilosis complex | 1,600 | 90.8 | 94 | 5.3 | 1,710 | 97.0 | 44 | 2.5 | 44 | 2.5 |
| C. parapsilosis sensu stricto | 1,425 | 93.4 | 61 | 4.0 | 1,485 | 97.3 | 34 | 2.2 | 34 | 2.2 |
| C. metapsilosis | 117 | 70.1 | 23 | 13.8 | 166 | 99.4 | 1 | 0.6 | 1 | 0.6 |
| C. orthopsilosis | 24 | 68.6 | 10 | 28.6 | 25 | 71.4 | 9 | 25.7 | 9 | 25.7 |
| L. elongisporus | 34 | 100 | 34 | 100 | ||||||
| C. tropicalis | 1,285 | 84.8 | 202 | 13.3 | 1,295 | 85.5 | 200 | 13.2 | 195 | 12.9 |
| C. glabrata complex | 179 | 18.7 | 814 | 85.2 | 141 | 14.8 | 134 | 14.0 | ||
| C. glabrata sensu stricto | 179 | 19.0 | 800 | 85.0 | 141 | 15.0 | 134 | 14.2 | ||
| C. nivariensis | 13 | 100 | ||||||||
| C. bracarensis | 1 | 100 | ||||||||
| C. guilliermondii | 71 | 38.2 | 54 | 29.0 | 157 | 84.4 | 23 | 12.4 | 23 | 12.4 |
| C. krusei | 125 | 100 | 118 | 94.4 | 4 | 3.2 | 4 | 0.3 | ||
| C. pelliculosa | 68 | 55.3 | 24 | 19.5 | 102 | 82.9 | 12 | 9.8 | 12 | 9.8 |
| C. lusitaniae | 48 | 96.0 | 50 | 100 | ||||||
| C. lipolytica | 6 | 16.7 | 25 | 69.4 | 24 | 66.7 | 12 | 33.3 | 12 | 33.3 |
| C. haemulonii | 5 | 20.8 | 18 | 75.0 | 15 | 62.5 | 9 | 37.5 | 9 | 37.5 |
| C. intermedia | 18 | 90.0 | 2 | 10.0 | 20 | 100 | ||||
| C. norvegensis | 3 | 23.1 | 7 | 53.8 | 13 | 100 | ||||
| C. fabianii | 11 | 100 | 11 | 100 | ||||||
| C. inconspicua | 1 | 12.5 | 7 | 87.5 | 8 | 100 | ||||
| C. rugosa | 3 | 42.9 | 4 | 57.1 | 7 | 100 | ||||
| C. fermentati | 4 | 66.7 | 2 | 33.3 | 6 | 100 | ||||
| C. catenulata | 3 | 75.0 | 1 | 25.0 | 3 | 75.0 | 3 | 75.0 | ||
| C. quercitrusa | 3 | 75.0 | 4 | 100 | ||||||
| C. aaseri | 2 | 66.7 | 3 | 100 | ||||||
| C. kefyr | 3 | 100 | 3 | 100 | ||||||
| C. famata | 2 | 66.7 | 1 | 33.3 | 3 | 100 | ||||
| C. magnoliae | 1 | 100 | 1 | 100 | ||||||
| C. utilis | 1 | 100 | 1 | 100 | ||||||
| C. diddensiae | 1 | 100 | 1 | 100 | 1 | 100 | ||||
| C. palmioleophila | 1 | 100 | 1 | 100 | 1 | 100 | ||||
| C. opuntiae | 1 | 100 | 1 | 100 | ||||||
| C. freyschussii | 1 | 100 | 1 | 100 | ||||||
| Total | 7,059 | 80.0 | 774 | 8.8 | 8,296 | 94.0 | 480 | 5.4 | 457 | 5.2 |
Species-specific MIC clinical breakpoint (CBP) interpretive criteria were applied to C. albicans, Candida tropicalis, the C. parapsilosis complex, the Candida glabrata complex, and Candida krusei according to the guidelines in the reference CLSI M60 document (17), and the susceptibilities of the other Candida species were interpreted in accordance with CLSI M44-S3 document guidelines (18). S, susceptible; WT, wild type; R, resistant; NWT, non-wild type.
Among the common Candida species, >99% of C. albicans isolates were susceptible to both fluconazole and voriconazole (Table 2), as were C. parapsilosis complex isolates. However, within the C. parapsilosis complex, 13.8% of the C. metapsilosis isolates were fluconazole resistant, whereas the rate was 4.0% among C. parapsilosis sensu stricto isolates (P < 0.01); C. orthopsilosis isolates also had a high frequency of resistance to both fluconazole (28.6% versus 4.0% for C. parapsilosis sensu stricto isolates, P < 0.01) and voriconazole (25.7% versus 2.2% for C. parapsilosis sensu stricto isolates, P < 0.01). Other species that accounted for >2% of the collection, including C. tropicalis, the C. glabrata complex, and C. guilliermondii, had higher rates of fluconazole resistance (>13%), voriconazole resistance (>12%), and azole cross-resistance (>12%) than C. albicans and the C. parapsilosis complex (Table 2). For C. krusei, 94.4% of isolates were susceptible to voriconazole (Table 2). The overall fluconazole resistance rate for other rare Candida species reached up to 31.2% (100/321), and the voriconazole resistance rate was 11.8% (38/321).
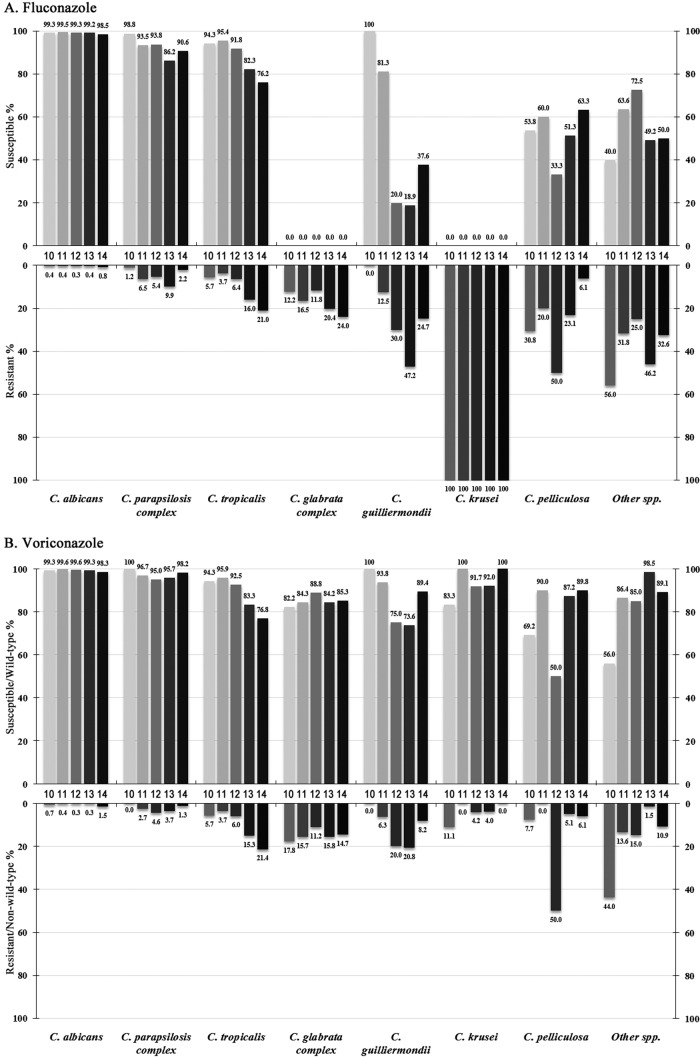
Trends of fluconazole and voriconazole resistance over 5 years.
Over the 5-year study period, the overall fluconazole susceptibility rates were 81.0%, 85.0%, 81.2%, 78.1%, and 78.7%, respectively, and the voriconazole susceptibility/WT rates were 94.2%, 96.5%, 95.0%, 93.4%, and 92.9% respectively. C. albicans remained highly susceptible to both azoles over 5 years (susceptibility rate > 98%) (Fig. 3). The fluconazole susceptibility rate of C. parapsilosis complex isolates significantly decreased from 98.8% in the first year to 86.2% in the fourth year (P < 0.01), although it climbed back to 90.6% in the fifth year (Fig. 3A), while in contrast, the rates of susceptibility to voriconazole remained similar (>95.0% of isolates were susceptible; Fig. 3B). Similar trends were found for the C. glabrata complex (the fluconazole resistance rate increased from 12.2% to 24.0%, P < 0.01) (Fig. 3A), but the proportion of isolates WT for susceptibility to voriconazole remained at 82.2% to 88.8% (Fig. 3B).
FIG 3.
Trends of fluconazole (A) and voriconazole (B) susceptibility and resistance rates of Candida species determined through the CHIF-NET study (2010 to 2014).
However, the susceptibility of C. tropicalis to both fluconazole and voriconazole decreased continuously, from 94.3% (for both azoles) in year 1 to 76.2% and 76.8%, respectively, in year 5 (P < 0.01 for both comparisons) (Fig. 2A and B). In addition, the fluconazole susceptibility rate of C. guilliermondii dropped from 100% to 37.6% (P < 0.01) (Fig. 3A), and its voriconazole susceptibility rate decreased from 100% to 89.4% (P < 0.01) (Fig. 3B). C. pelliculosa and other rare Candida species exhibited generally high rates of resistance to fluconazole and voriconazole, but no significant trends were observed (Fig. 3).
DISCUSSION
This large study has provided valuable data to inform the management of IC in Chinese hospitals. As expected, the four major Candia pathogens, C. albicans, the C. parapsilosis complex, C. tropicalis, and the C. glabrata complex, accounted for 92.9% of isolates and predominated in all hospitals except one. Generally, C. albicans remained the most common species, and no trends toward a decrease in frequency were observed over the 5 years. In addition, the species was susceptible to both fluconazole and voriconazole, which was comparable to global data obtained during the same time period (>99% susceptibility rates) (3, 19–21). C. albicans accounted for 44.9% of the isolates collected in this study, which is similar to the prevalence in North America, Latin America, and other regions in the Asia-Pacific region (40% to 45%) but lower than that in Europe (>50%) (20–22). Of note, the prevalence of C. albicans in the Asia-Pacific region has decreased by about 20% compared with that determined from data obtained from 2001 to 2007 (23). The proportion of C. albicans isolates as the causative agent among candidemia cases was even lower (32.3%).
On the other hand, the overall frequency of non-albicans Candida species as a cause of IC was high in China, with the members of the C. parapsilosis complex being the second most predominant species in this study (20%). Of note, this frequency was even higher than that of C. albicans among candidemia patients in year 1 (8) and year 4. C. parapsilosis complex isolates are notable for their ability to adhere to catheters and other medical devices, to develop biofilms, and to colonize human skin, all of which may facilitate nosocomial outbreaks (1, 12, 24). A reassuring finding was that azole resistance (<6%) was uncommon in the present study, similar to global surveillance data (0 to 5.4%) (3, 19, 21). However, differences in azole resistance rates were noted among the different species within the C. parapsilosis complex, with C. metapsilosis and C. orthopsilosis showing the highest rates of resistance, as has been reported in previous studies (25, 26).
C. tropicalis was the third most common species in the study (17.2%). Of note, this species has become one of the more common non-albicans Candida species worldwide, and its prevalence in Latin America (13% to 18%) and the Asia-Pacific region (about 12%) is generally higher than that in North America (7% to 9%) and Europe (4% to 9%) (20, 23). There is a general consensus that C. tropicalis strains may exhibit a moderate level of azole resistance. Resistance rates have remained stable in North America and Europe (generally, <10%) (20, 21, 27). However, in the present study, a notable trend of increasing rates of azole resistance among C. tropicalis isolates was observed in China (<6% in year 1 to >20% in year 5). This was also observed using data obtained by the broth microdilution method from 10 hospitals which consistently participated in the CHIF-NET study over the 5-year period (13). In addition, a higher azole cross-resistance rate was observed in C. tropicalis isolates (96.5%) than in C. parapsilosis complex (46.8%) and C. glabrata complex (74.9%) isolates. Worldwide, azole resistance in C. tropicalis has been mainly noted in the Asia-Pacific region (3, 13, 28), whereas azole resistance rates among C. tropicalis isolates in North America or European countries remain low (<10%) (3, 23, 29). As C. tropicalis infection is associated with higher rates of mortality and more adverse outcomes (30), consideration may be given to the use of echinocandins as first-line agents in treating C. tropicalis infections in China.
C. glabrata complex isolates accounted for 10.8% of the collection in the present study, similar to the situation in Europe (10% to 16%) but less than that in the United States, where C. glabrata was the most common non-albicans Candida species (20% to 26%) (20, 21, 23). This species is well-known for its high rates of azole resistance, mainly due to the upregulation of drug transporters and the overexpression or alteration of the drug target (2, 4). In this study, the rate of fluconazole resistance among the isolates was 18.7%, which is a rate higher than the global average (8% to 16%) (3, 20, 21, 23). Moreover, 14.0% of isolates were cross-resistant to both fluconazole and voriconazole. A significant increase in the rate of fluconazole resistance in C. glabrata complex isolates was observed over the 5 years of this study, and this has also been noted using broth microdilution methods in hospitals which consistently participated in the CHIF-NET study (14).
Other Candida species, although uncommon, exhibited high fluconazole (44.1%) and voriconazole (10.3%) resistance rates. In addition, many of the less common species that were highly resistant to fluconazole, e.g., C. pelliculosa and C. lipolytica, were more commonly isolated from blood samples than non-blood sources. However, less common Candida species were more likely to be misidentified by phenotypic and biochemical-based identification methods (8, 15, 31). Although MALDI-TOF MS has good accuracy, its capacity to identify an ever expanding range of pathogens largely relies on its protein mass spectral database (15, 32, 33). The CHIF-NET study has also provided a valuable isolate repository including more novel or uncommon species that may be used to expand and build local MALDI-TOF MS identification databases (11, 31, 34) for future surveillance.
There were several limitations in our study. First, the study employed the CLSI disk diffusion assay for antifungal susceptibility testing. The methodology was developed and verified in the 10.5-year ARTEMIS global surveillance program, and the results of that methodology showed a good correlation with those of the “gold standard” broth microdilution method (9, 23). However, to date, azole species-specific CBPs of the disk diffusion method are available only for the most common Candida species, i.e., C. albicans, C. parapsilosis, C. tropicalis, C. glabrata, and C. krusei (17), and the use of old non-species-specific CBPs (18) for other Candida species may introduce incorrect interpretations of the isolates' susceptibilities. Building up epidemiological cutoff values for less common Candida species in China is the next-step goal of the program. In addition, the antifungal agents tested in the present study were limited to only two azoles. However, with the increasing prevalence of azole-resistant Candida isolates, echinocandins have become the first-line treatment of IC (4, 14, 24). In mainland China, echinocandin-nonsusceptible C. glabrata cases have also been identified (9, 14). To address these limitations, we envisage performing broth microdilution to examine the susceptibilities to a broader range of antifungal agents for the next 5 years of the CHIF-NET surveillance study (year 6 to year 10). Moreover, further investigations on antifungal resistance mechanisms, e.g., mutations in the ERG11 and FKS genes and overexpression of drug efflux pumps (2, 4, 29), would enhance the value of the in vitro susceptibility results. Another potential limitation is that there were disparities between the numbers of isolates collected from different provinces, which may influence the accurate geographic picture of the species distribution or antifungal resistance. In order to obtain more representative regional IC data with less bias, the CHIF-NET Study Group has now established subsidiary surveillance programs in each province of China (35).
In conclusion, this study has provided useful data on the epidemiology of IC in mainland China. Although C. albicans remained the most common species, non-albicans Candida species were responsible for about 55% of cases of IC and over 67% of candidemia cases. Fluconazole and azole cross-resistance rates were notably high in C. tropicalis and C. glabrata, and their fluconazole resistance rates increased significantly over the 5 years. Less common Candida species also exhibited high fluconazole resistance rates, and molecular or mass spectrum methods were essential for the identification of uncommon species. Antifungal resistance has become a threat, and continued surveillance is still warranted.
ACKNOWLEDGMENTS
The co-principal investigators in the 65 hospitals participating in the China Hospital Invasive Fungal Surveillance Net (CHIF-NET) Study Group (CHIF-NET 2010 to 2014) are as follows: (1) Ying-Chun Xu and He Wang, Peking Union Medical College Hospital, Beijing; (2) Mei Kang and Yu-Ling Xiao, West China Hospital of Sichuan University, Chengdu, Sichuan Province; (3) Zi-Yong Sun and Zhong-Ju Chen, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei Province; (4) Kang Liao and Peng-Hao Guo, The First Affiliated Hospital of Zhongshan University, Guangzhou, Guangdong Province; (5) Yan-Ping Luo and Li-Yan Ye, The General Hospital of People's Liberation Army, Beijing; (6) Zhi-Dong Hu and Na Yue, General Hospital of Tianjin Medical University, Tianjin; (7) Ya-Ning Mei and Gen-Yan Liu, Jiangsu Province Hospital, Nanjing, Jiangsu Province; (8) Da-Wen Guo and Shu-Lan Chen, The First Clinic College of Harbin Medical University, Harbin, Heilongjiang Province; (9) Ruo-Yu Li and Zhe Wan, Peking University First Hospital, Beijing; (10) Yu-Hong Pan and Lan-Mei Gao, Fujian Medical University Union Hospital, Fuzhou, Fujian Province; (11) Yun-Zhuo Chu and Fu-Shun Li, First Hospital of China Medical University, Shenyang, Liaoning Province; (12) Yun-Song Yu and Jie Lin, Sir Run Run Shaw Hospital, Hangzhou, Zhejiang Province; (13) Xian-Ju Feng and Hui Xu, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan Province; (14) Qing Yang, The First Affiliated Hospital of the Zhejiang University School of Medicine, Hangzhou, Zhejiang Province; (15) Hai-Feng Shao, General Hospital of Nanjing Military Area Command, Nanjing, Jiangsu Province; (16) Wen-En Liu and Hong-Ling Li, Xiangya Hospital, Central South University, Changsha, Hunan Province; (17) Huo-Xiang Lv and Qu-Hao Wei, Zhejiang Province People's Hospital, Hangzhou, Zhejiang Province; (18) Yong Wang and Yan Jin, Shandong Provincial Hospital, Qingdao, Shandong Province; (19) Li-Wen Liu, The People's Hospital of Liaoning Province, Shenyang, Liaoning Province; (20) Dan-Hong Su, The First Affiliated Hospital of Guangzhou Medical Collage, Guangzhou, Guangdong Province; (21) Yu-Xing Ni, Shanghai Ruijin Hospital, Shanghai; (22) Gui-Ling Zou and Xue-Fei Du, The Fouth Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province; (23) Xin-Lan Hu and Ning Li, Fujian Provincial Hospital, Fuzhou, Fujian Province; (24) Ling Ma and Shuai-Xian Du, Union Hospital, Tongji Medical College of Huazhong University of Science and Technology, Wuhan, Hubei Province; (25) Xiu-Lan Song, The First Hospital of Jiaxing, Jiaxing, Zhejiang Province; (26) Hua Yu and Xiang-Ning Huang, Sichuan Academy of Medical Sciences and Sichuan Provincial People's Hospital, Chengdu, Sichuan Province; (27) Tie-Li Zhou and Qing Wu, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang Province; (28) Wei-Jia and Gang Li, The General Hospital Affiliated to Ningxia Medical University, Yinchuan, Ningxia Province; (29) Qiang-Qiang Zhang, Huashan Hospital, Fudan University, Shanghai; (30) Zhi-Jie Zhang, The Second Hospital Affiliated to China Medical University, Shenyang, Liaoning Province; (31) Zhi-Yong Zhang, Southwest Hospital Affiliated to the Third Military Medical University, Chongqing; (32) Rong Zhang and Hong-Wei Zhou, The Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, Zhejiang Province; (33) Xiu-Li Xu and Xiao Chen, Xijing Hospital, Fourth Military Medical University, Xi'an, Shaanxi Province; (34) Li-Ping Zhang and Li Yan, The First Affiliated Hospital of Chongqing Medical Hospital, Chongqing; (35) Xue-Song Xu and Wei Li, China-Japan Union Hospital of Jilin University, Changchun, Jilin Province; (36) Tie-Ying Hou and Li-Yan Zhang, Guangdong Provincial People's Hospital, Guangzhou, Guangdong Province; (37) Lin-Qiang Deng and Hui Chen, Jiangxi Province People's Hospital, Nanchang, Jiangxi Province; (38) Ke-Cheng Li and Fei Xia, Third Affiliated Hospital of Wenzhou Medical College, Wenzhou, Zhejiang Province; (39) Wei Song and Yong-Xin Shi, The Affiliated Qingdao Municipal Hospital of Qingdao University Medical College, Qingdao, Shandong Province; (40) Yuan-Hong Xu and Ji-Lu Shen, The First Affiliated Hospital of AnHui University of Science and Technology, Hefei, Anhui Province; (41) Xiao-Min Xu, Ningbo Second Hospital, Ningbo, Zhejiang Province; (42) Guo-Xiong Li and Hui Ding, Central Hospital of Lishui City, Lishui, Zhejiang Province; (43) Rong Tang and Xing Ding, Shanghai First People's Hospital, Shanghai; (44) Jian-Hong Zhao and Dong-Yan Shi, The Second Hospital of Hebei Medical University, Shijiazhuang, Hebei Province; (45) Jing Wang and Xiao-Guang Xiao, First Affiliated Hospital of Dalian Medical University, Dalian, Liaoning Province; (46) Ling Meng, Second Affiliated Hospital of Lanzhou University, Lanzhou, Gansu Province; (47) Xiao-Ming Wang and Xu-Feng Ji, The First Hospital of Jilin University, Changchun, Jilin Province; (48) Su-Fei Yu and Chun-Yan Xu, Zhejiang Taizhou Hospital, Taizhou, Zhejiang Province; (49) Qiong Zhang and Ping Ji, The First Hospital of Xinjiang Medical University, Urumchi, Xinjiang Province; (50) Long-Hua Hu and Bai-Ling Zhang, The Second Affiliated Hospital of Nanchang University, Nanchang, Jiangxi Province; (51) Bin Yang and Yu-Lan Lin, First Affiliated Hospital of Fujian Medical University, Fuzhou, Fujian Province; (52) Jin-E Lei, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, Shaanxi Province; (53) Hai-Bin Wang and Jing Zhu, First Affiliated Hospital of PLA General Hospital, Beijing; (54) Hong-Jie Liang, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Province; (55) Xiao-Ling Ma and Huai-Wei Lu, Anhui Provincial Hospital, Hefei, Anhui Province; (56) Wen-Cheng Xue, General Hospital of Shenyang Military Area, Shenyang, Liaoning Province; (57) Bin Shan and Yan Du, The First Affiliated Hospital, Kunming Medical University, Kunming, Yunnan Province; (58) Xiang-Yang Chen, People's Hospital of Zhengzhou, Zhengzhou, Henan Province; (59) Run-Mei Zhang and Jian-Bang Kang, The Second Hospital of Shanxi Medical University, Taiyuan, Shanxi Province; (60) Jian-Lei Zhang, Tianjin First Centre Hospital, Tianjin; (61) Wei Cao, The Second Xiangya Hospital of Central South University, Changsha, Hunan Province; (62) Jun-Lin Zhang and Quang Fu, The Affiliated Hospital of Inner Mongolia Medical University, Hohhot, Inner Mongolia Province; (63) Yan-Ping Fan, Dalian Municipal Central Hospital, Dalian, Liaoning Province; (64) Lian-Hua Wei and Feng-Mei Zou, Gansu Province People's Hospital, Lanzhou, Gansu Province; and (65) Yan-Yan Guo, Tangshan Gongren Hospital, Tangshan, Hebei Province.
This work was supported by the Peking Union Medical College Hospital Out-standing Young Talents Program (JQ201703), the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2016-I2M-1-014), and the Beijing Innovation Base Cultivation and Development Special Fund (Z171100002217068).
We declare no conflict of interests.
Contributor Information
on behalf of the China Hospital Invasive Fungal Surveillance Net (CHIF-NET) Study Group:
Ying-Chun Xu, He Wang, Mei Kang, Yu-Ling Xiao, Zi-Yong Sun, Zhong-Ju Chen, Kang Liao, Peng-Hao Guo, Yan-Ping Luo, Li-Yan Ye, Zhi-Dong Hu, Na Yue, Ya-Ning Mei, Gen-Yan Liu, Da-Wen Guo, Shu-Lan Chen, Ruo-Yu Li, Zhe Wan, Yu-Hong Pan, Lan-Mei Gao, Yun-Zhuo Chu, Fu-Shun Li, Yun-Song Yu, Jie Lin, Xian-Ju Feng, Hui Xu, Qing Yang, Hai-Feng Shao, Wen-En Liu, Hong-Ling Li, Lv Huo-Xiang, Qu-Hao Wei, Yong Wang, Yan Jin, Li-Wen Liu, Dan-Hong Su, Yu-Xing Ni, Gui-Ling Zou, Xue-Fei Du, Xin-Lan Hu, Ning Li, Ling Ma, Shuai-Xian Du, Xiu-Lan Song, Hua Yu, Xiang-Ning Huang, Tie-Li Zhou, Qing Wu, Wei-Jia, Gang Li, Qiang-Qiang Zhang, Zhi-Jie Zhang, Zhi-Yong Zhang, Rong Zhang, Hong-Wei Zhou, Xiu-Li Xu, Xiao Chen, Li-Ping Zhang, Li Yan, Xue-Song Xu, Wei Li, Tie-Ying Hou, Li-Yan Zhang, Lin-Qiang Deng, Hui Chen, Ke-Cheng Li, Fei Xia, Wei Song, Yong-Xin Shi, Yuan-Hong Xu, Ji-Lu Shen, Xiao-Min Xu, Guo-Xiong Li, Hui Ding, Rong Tang, Xing Ding, Jian-Hong Zhao, Dong-Yan Shi, Jing Wang, Xiao-Guang Xiao, Ling Meng, Xiao-Ming Wang, Xu-Feng Ji, Su-Fei Yu, Chun-Yan Xu, Qiong Zhang, Ping Ji, Long-Hua Hu, Bai-Ling Zhang, Bin Yang, Yu-Lan Lin, Jin-E Lei, Hai-Bin Wang, Jing Zhu, Hong-Jie Liang, Xiao-Ling Ma, Huai-Wei Lu, Wen-Cheng Xue, Bin Shan, Yan Du, Xiang-Yang Chen, Run-Mei Zhang, Jian-Bang Kang, Jian-Lei Zhang, Wei Cao, Jun-Lin Zhang, Quang Fu, Yan-Ping Fan, Lian-Hua Wei, Feng-Mei Zou, and Yan-Yan Guo
REFERENCES
- 1.Kullberg BJ, Arendrup MC. 2015. Invasive candidiasis. N Engl J Med 373:1445–1456. doi: 10.1056/NEJMra1315399. [DOI] [PubMed] [Google Scholar]
- 2.Arendrup MC, Patterson TF. 2017. Multidrug-resistant Candida: epidemiology, molecular mechanisms, and treatment. J Infect Dis 216:S445–S451. doi: 10.1093/infdis/jix131. [DOI] [PubMed] [Google Scholar]
- 3.Castanheira M, Messer SA, Rhomberg PR, Pfaller MA. 2016. Antifungal susceptibility patterns of a global collection of fungal isolates: results of the SENTRY antifungal surveillance program (2013). Diagn Microbiol Infect Dis 85:200–204. doi: 10.1016/j.diagmicrobio.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. 2017. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis 17:e383–e392. doi: 10.1016/S1473-3099(17)30316-X. [DOI] [PubMed] [Google Scholar]
- 5.Grim SA, Berger K, Teng C, Gupta S, Layden JE, Janda WM, Clark NM. 2012. Timing of susceptibility-based antifungal drug administration in patients with Candida bloodstream infection: correlation with outcomes. J Antimicrob Chemother 67:707–714. doi: 10.1093/jac/dkr511. [DOI] [PubMed] [Google Scholar]
- 6.Kollef M, Micek S, Hampton N, Doherty JA, Kumar A. 2012. Septic shock attributed to Candida infection: importance of empiric therapy and source control. Clin Infect Dis 54:1739–1746. doi: 10.1093/cid/cis305. [DOI] [PubMed] [Google Scholar]
- 7.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. doi: 10.1093/cid/civ1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Xiao M, Chen SC, Kong F, Sun ZY, Liao K, Lu J, Shao HF, Yan Y, Fan H, Hu ZD, Chu YZ, Hu TS, Ni YX, Zou GL, Xu YC. 2012. In vitro susceptibilities of yeast species to fluconazole and voriconazole as determined by the 2010 National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study. J Clin Microbiol 50:3952–3959. doi: 10.1128/JCM.01130-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao M, Fan X, Chen SC, Wang H, Sun ZY, Liao K, Chen SL, Yan Y, Kang M, Hu ZD, Chu YZ, Hu TS, Ni YX, Zou GL, Kong F, Xu YC. 2015. Antifungal susceptibilities of Candida glabrata species complex, Candida krusei, Candida parapsilosis species complex and Candida tropicalis causing invasive candidiasis in China: 3 year national surveillance. J Antimicrob Chemother 70:802–810. doi: 10.1093/jac/dku460. [DOI] [PubMed] [Google Scholar]
- 10.Cheng JW, Yu SY, Xiao M, Wang H, Kudinha T, Kong F, Xu YC. 2016. Identification and antifungal susceptibility profile of Candida guilliermondii and Candida fermentati from a multicenter study in China. J Clin Microbiol 54:2187–2189. doi: 10.1128/JCM.00938-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou X, Xiao M, Chen SC, Wang H, Cheng JW, Chen XX, Xu ZP, Fan X, Kong F, Xu YC. 2016. Identification and antifungal susceptibility profiles of Candida haemulonii species complex clinical isolates from a multicenter study in China. J Clin Microbiol 54:2676–2680. doi: 10.1128/JCM.01492-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Zhang L, Kudinha T, Kong F, Ma XJ, Chu YZ, Kang M, Sun ZY, Li RY, Liao K, Lu J, Zou GL, Xiao M, Fan X, Xu YC. 2016. Investigation of an unrecognized large-scale outbreak of Candida parapsilosis sensu stricto fungaemia in a tertiary-care hospital in China. Sci Rep 6:27099. doi: 10.1038/srep27099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan X, Xiao M, Liao K, Kudinha T, Wang H, Zhang L, Hou X, Kong F, Xu YC. 2017. Notable increasing trend in azole non-susceptible Candida tropicalis causing invasive candidiasis in China (August 2009 to July 2014): molecular epidemiology and clinical azole consumption. Front Microbiol 8:464. doi: 10.3389/fmicb.2017.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou X, Xiao M, Chen SC, Kong F, Wang H, Chu YZ, Kang M, Sun ZY, Hu ZD, Li RY, Lu J, Liao K, Hu TS, Ni YX, Zou GL, Zhang G, Fan X, Zhao YP, Xu YC. 2017. Molecular epidemiology and antifungal susceptibility of Candida glabrata in China (August 2009 to July 2014): a multi-center study. Front Microbiol 8:880. doi: 10.3389/fmicb.2017.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Xiao M, Wang H, Gao R, Fan X, Brown M, Gray TJ, Kong F, Xu YC. 2014. Yeast identification algorithm based on use of the Vitek MS system selectively supplemented with ribosomal DNA sequencing: proposal of a reference assay for invasive fungal surveillance programs in China. J Clin Microbiol 52:572–577. doi: 10.1128/JCM.02543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. 2009. M44-A2. Method for antifungal disk diffusion susceptibility testing of yeasts, 2nd ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2018. M60. Performance standards for antifungal susceptibility testing of yeasts, 1st ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2011. M44-S3. Zone diameter interpretive standards, corresponding minimal inhibitory concentration (MIC) interpretive breakpoints, and quality control limits for antifungal disk diffusion susceptibility testing of yeasts; 3rd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Pfaller MA, Castanheira M, Messer SA, Moet GJ, Jones RN. 2011. Echinocandin and triazole antifungal susceptibility profiles for Candida spp., Cryptococcus neoformans, and Aspergillus fumigatus: application of new CLSI clinical breakpoints and epidemiologic cutoff values to characterize resistance in the SENTRY antimicrobial surveillance program (2009). Diagn Microbiol Infect Dis 69:45–50. doi: 10.1016/j.diagmicrobio.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Pfaller MA, Messer SA, Woosley LN, Jones RN, Castanheira M. 2013. Echinocandin and triazole antifungal susceptibility profiles for clinical opportunistic yeast and mold isolates collected from 2010 to 2011: application of new CLSI clinical breakpoints and epidemiological cutoff values for characterization of geographic and temporal trends of antifungal resistance. J Clin Microbiol 51:2571–2581. doi: 10.1128/JCM.00308-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaller MA, Rhomberg PR, Messer SA, Jones RN, Castanheira M. 2015. Isavuconazole, micafungin, and 8 comparator antifungal agents' susceptibility profiles for common and uncommon opportunistic fungi collected in 2013: temporal analysis of antifungal drug resistance using CLSI species-specific clinical breakpoints and proposed epidemiological cutoff values. Diagn Microbiol Infect Dis 82:303–313. doi: 10.1016/j.diagmicrobio.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Pfaller MA, Messer SA, Jones RN, Castanheira M. 2015. Antifungal susceptibilities of Candida, Cryptococcus neoformans and Aspergillus fumigatus from the Asia and Western Pacific region: data from the SENTRY antifungal surveillance program (2010-2012). J Antibiot (Tokyo) 68:556–561. doi: 10.1038/ja.2015.29. [DOI] [PubMed] [Google Scholar]
- 23.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, Rodloff A, Fu W, Ling TA, Global Antifungal Surveillance Group. 2010. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol 48:1366–1377. doi: 10.1128/JCM.02117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. 2012. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev 36:288–305. doi: 10.1111/j.1574-6976.2011.00278.x. [DOI] [PubMed] [Google Scholar]
- 25.Lockhart SR, Messer SA, Pfaller MA, Diekema DJ. 2008. Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis. J Clin Microbiol 46:2659–2664. doi: 10.1128/JCM.00803-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YC, Lin YH, Chen KW, Lii J, Teng HJ, Li SY. 2010. Molecular epidemiology and antifungal susceptibility of Candida parapsilosis sensu stricto, Candida orthopsilosis, and Candida metapsilosis in Taiwan. Diagn Microbiol Infect Dis 68:284–292. doi: 10.1016/j.diagmicrobio.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Lockhart SR, Iqbal N, Cleveland AA, Farley MM, Harrison LH, Bolden CB, Baughman W, Stein B, Hollick R, Park BJ, Chiller T. 2012. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J Clin Microbiol 50:3435–3442. doi: 10.1128/JCM.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan TY, Hsu LY, Alejandria MM, Chaiwarith R, Chinniah T, Chayakulkeeree M, Choudhury S, Chen YH, Shin JH, Kiratisin P, Mendoza M, Prabhu K, Supparatpinyo K, Tan AL, Phan XT, Tran TT, Nguyen GB, Doan MP, Huynh VA, Nguyen SM, Tran TB, Van Pham H. 2016. Antifungal susceptibility of invasive Candida bloodstream isolates from the Asia-Pacific region. Med Mycol 54:471–477. doi: 10.1093/mmy/myv114. [DOI] [PubMed] [Google Scholar]
- 29.Berkow EL, Lockhart SR. 2017. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist 10:237–245. doi: 10.2147/IDR.S118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andes DR, Safdar N, Baddley JW, Playford G, Reboli AC, Rex JH, Sobel JD, Pappas PG, Kullberg BJ, Mycoses Study Group. 2012. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin Infect Dis 54:1110–1122. doi: 10.1093/cid/cis021. [DOI] [PubMed] [Google Scholar]
- 31.Xiao M, Wang H, Lu J, Chen SC, Kong F, Ma XJ, Xu YC. 2014. Three clustered cases of candidemia caused by Candida quercitrusa and mycological characteristics of this novel species. J Clin Microbiol 52:3044–3048. doi: 10.1128/JCM.00246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cassagne C, Normand AC, L'Ollivier C, Ranque S, Piarroux R. 2016. Performance of MALDI-TOF MS platforms for fungal identification. Mycoses 59:678–690. doi: 10.1111/myc.12506. [DOI] [PubMed] [Google Scholar]
- 33.Posteraro B, De Carolis E, Vella A, Sanguinetti M. 2013. MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Expert Rev Proteomics 10:151–164. doi: 10.1586/epr.13.8. [DOI] [PubMed] [Google Scholar]
- 34.Xiao M, Fan X, Chen XX, Wang H, Zhang L, Xu ZP, Kudinha T, Kong F, Xu YC. 2016. Misidentification of a rare species, Cryptococcus laurentii, by commonly used commercial biochemical methods and matrix-assisted laser desorption ionization–time of flight mass spectrometry systems: challenges for clinical mycology laboratories. J Clin Microbiol 54:226–229. doi: 10.1128/JCM.02830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo LN, Xiao M, Cao B, Qu F, Zhan YL, Hu YJ, Wang XR, Liang GW, Gu HT, Qi J, Yuan H, Min R, Wang FY, Liu LJ, Wang HB, Jiang W, Duan XG, Xu WJ, Yu YH, Su JR, Zhang JZ, Nong JQ, Liu SM, Li J, Liu JT, Yue ZG, Yang D, Guo J, Zhao R, Zhang YN, Yang XM, Liu XQ, Hsueh PR, Xu YC. 2017. Epidemiology and antifungal susceptibilities of yeast isolates causing invasive infections across urban Beijing, China. Future Microbiol 12:1075–1086. doi: 10.2217/fmb-2017-0036. [DOI] [PubMed] [Google Scholar]