Haemophilus influenzae type b (Hib) conjugate vaccines have led to dramatic reductions in Hib disease among young children worldwide. Nontypeable H. influenzae (NTHi) is now the major cause of invasive H. influenzae infections.
KEYWORDS: nontypeable Haemophilus influenzae, infection, invasive disease, epidemiology, children, Japan
ABSTRACT
Haemophilus influenzae type b (Hib) conjugate vaccines have led to dramatic reductions in Hib disease among young children worldwide. Nontypeable H. influenzae (NTHi) is now the major cause of invasive H. influenzae infections. We investigated the clinical characteristics of invasive NTHi diseases among children in Japan, to clarify the pathogenicity of isolated NTHi strains. The mortality rate was 10.7%, with deaths occurring mainly among children with underlying comorbidities. Biotypes II and III were the most common, and most strains (64.3%) had multiple amino acid substitutions at the Asp-350, Ser-357, Ser-385, and/or Met-377 sites of penicillin-binding protein 3. Two strains were β-lactamase positive and ampicillin-clavulanate resistant. Biofilm indices varied widely, and IS1016 was detected in 10.7% of the strains tested. Moreover, there was wide variation in the characteristics of invasive NTHi strains. NTHi strains, showing great genetic diversity, are responsible for most invasive H. influenzae infections in children in the postvaccine era. Continuous monitoring of NTHi strains responsible for invasive diseases in children is important to detect changes in the epidemiology of invasive H. influenzae infections in the postvaccine era.
INTRODUCTION
Haemophilus influenzae is a small Gram-negative coccobacillus that commonly colonizes the human upper respiratory tract and causes both invasive and noninvasive diseases in children (1). It is classified into 7 groups, i.e., 6 chemically distinct polysaccharide capsular groups (serotypes a to f) and 1 group called nonencapsulated or nontypeable H. influenzae (NTHi). Among the encapsulated strains, H. influenzae type b (Hib) is the most virulent and causes most cases of invasive H. influenzae disease (2). In contrast, NTHi strains mainly cause noninvasive diseases (otitis media, bronchitis, and pneumonia) in healthy children, although they can also cause invasive disease, mostly in older adults and people with comorbidities (3). A substantial burden of perinatal invasive NTHi infections has been reported. Although rare, these infections can result in serious morbidity and death among pregnant women and neonates (4). Among neonates, NTHi infections are 10 times more common than Hib infections; most cases are associated with preterm birth and occur in the first week of life (5).
The introduction of Hib conjugate vaccines has dramatically decreased the incidence of invasive Hib disease globally (2, 3, 6, 7). Consequently, invasive H. influenzae infections are now mainly caused by NTHi (4, 7–9). Seven surveillance studies of H. influenzae infections in the post-Hib-vaccine era have shown clear increases in the numbers and incidence rates of invasive NTHi infections (10). Therefore, we should be aware of the virulence potentials of these strains.
In Japan, the Hib vaccine has been available on an optional basis since December 2008, and it was included in the routine immunization program for children under 5 years of age in April 2013. Since the subsidization of the Hib vaccine, the number of invasive Hib disease cases among children has decreased dramatically (11). However, invasive disease caused by NTHi, although infrequent, has been reported. The bacteriologic and epidemiologic characteristics of the NTHi strains responsible for invasive disease are poorly defined. Our study aimed to clarify the clinical impact of invasive NTHi disease in children and to determine the pathogenicity of NTHi strains isolated from patients with invasive disease.
MATERIALS AND METHODS
Bacterial isolates.
Between 2008 and 2015, a total of 28 H. influenzae strains, isolated from invasive sources such as blood and cerebrospinal fluid (CSF), were collected and sent to the National Institute of Infectious Diseases and Chiba University Hospital from hospitals in 8 distantly located prefectures in Japan. Of those strains, 21 were collected via a population-based surveillance study of invasive H. influenzae disease performed in 10 prefectures in Japan, including Chiba prefecture (12). Strains were identified as H. influenzae on the basis of colony morphology and dependence on X and V growth factors that do not lyse horse blood cells. The strains also underwent matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) at Chiba University Hospital (MALDI Biotyper Microflex LT; Bruker Daltonics, Bremen, Germany), further confirming them to be H. influenzae (13). All strains were stored at −80°C and, prior to analysis, were cultivated overnight at 37°C in 5% CO2 on BY chocolate agar (Wako Pure Chemical Industries, Osaka, Japan) supplemented with 1% IsoVitalX with 10 μg/ml hemin and 10 μg/ml β-NAD (Nippon Becton Dickinson Company, Ltd., Tokyo, Japan). DNA templates of these strains were prepared using a QIAamp DNA minikit (Qiagen, Hilden, Germany), unless otherwise noted.
Clinical data.
Patients' clinical data, collected from the treating doctors in each hospital, were deidentified at each hospital before further analysis. The investigation findings required to confirm diagnoses were as follows: (i) meningitis, isolation of H. influenzae from CSF samples; (ii) pneumonia, features of consolidation on chest radiographic images and isolation of H. influenzae from blood samples; (iii) sinusitis, opacification evident on paranasal sinus radiographic images and isolation of H. influenzae from blood samples; (iv) peritonitis, isolation of H. influenzae from both blood and ascitic fluid samples. When H. influenzae was isolated from a blood sample but the clinical symptoms, examination findings, and investigation results were unable to reveal the site of infection, the patient was categorized as having bacteremia.
Serotyping and biotyping.
Serotypes of the H. influenzae isolates were determined by slide agglutination testing using antisera (Denka Seiken Co., Ltd., Tokyo, Japan) to the bacterial polysaccharide antigens to identify capsular types a, b, c, d, e, and f. We confirmed the results using a PCR assay, according to the method reported by Falla et al. (14). For these analyses, ATCC strains such as ATCC 9327 (serotype a), ATCC 9334 (serotype b), ATCC 9007 (serotype c), ATCC 9332 (serotype d), ATCC 8142 (serotype e), and ATCC 9833 (serotype f) were used as controls for encapsulated H. influenzae. ATCC 35056 was used as a control for NTHi. Biotype analysis was performed using in-house media for urease, indole, and ornithine decarboxylase reactions, as described previously (15). These results were confirmed using a commercial kit (ID Test HN-20 Rapid kit; Nissui Pharmaceutical Company, Ltd., Tokyo, Japan).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed for all H. influenzae strains using the broth microdilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (16). β-Lactamase production was detected using a disc impregnated with nitrocefin (Cefinase paper discs; Nippon Becton Dickinson Company, Ltd., Tokyo, Japan).
The genetic profiles of penicillin-binding protein 3 (PBP-3) were investigated to identify mutations, using PCR and sequence analysis. Briefly, a single colony was used to extract DNA, and PCR was performed using the primers ftsl1F (GTTTCCCAGTCACGACGTTGTAGTTAATGCGTAACCGTGCAATTAC) and ftsl1R (TTGTGAGCGGATAACAATTTCACCACTAATGCATAACGAGGATC). The PCR products were then purified and subjected to sequence analysis using the primers SeqftsF (GTTTTCCCAGTCACGACGTTGTA) and SeqftsR (TTGTGAGCGGATAACAATTTC), using an ABI Prism 3130 genetic analyzer.
The β-lactamase gene TEM-1 was detected using a commercially available kit (Wakunaga Seiyaku Kabushikigaisha, Hiroshima, Japan). Strain Rd KW20, which is known to have no alteration in PBP-3, was used as a control. Genetically β-lactamase-negative ampicillin-resistant (BLNAR) strains were divided into subgroups, i.e., group I/II, with R517H/N526K substitutions, and group III, with M377I, S385T, and L389F substitutions in addition to N526K substitutions. Group II was further divided into subgroups a to d, based on substitutions at amino acids 502 and/or 449. Strains with M377I, S385T, and L389F substitutions and an R517H substitution rather than an N526K substitution were categorized as group III-like, based on the findings of previous studies (17, 18).
Microtiter biofilm assay.
The ability to form biofilm was assessed for all 28 strains using a quantitative biofilm assay in a microtiter polystyrene plate, as described previously (19). Briefly, the bacterial cells were grown overnight at 37°C in brain heart infusion broth with 10 μg/ml hemin and 10 μg/ml β-NAD. Overnight cultures (5 μl for each strain) were inoculated into a 96-well, flat-bottom, microtiter polystyrene plate (Becton Dickinson, Franklin Lakes, NJ) containing 200 μl of brain heart infusion broth with hemin and β-NAD, at 37°C. After 18 h of incubation, each well was washed with distilled water. Biofilm formation was visualized by staining with 0.5% crystal violet for 5 min. Two hundred microliters of 95% ethanol was added, and the biofilm was quantified in an enzyme-linked immunosorbent assay plate reader (iMark microplate reader; Bio-Rad) at 570 nm. The biofilm index was defined using the average optical density at 570 nm (OD570). All strains were tested in triplicate, and the results are reported as the averages of results from three different experiments.
Detection of adhesin genes.
Using PCR assays, NTHi strains were evaluated for possession of the following five adhesin genes: hmw1A/hmw2A, hia, hap, hifA, and p5 (20). The primer sequences used and the PCR conditions are shown in Table 1. Amplification reactions were performed using a TProfessional Basic thermocycler (Biometra GmbH, Göttingen, Germany). The products were separated on a 1.5% agarose gel, stained with ethidium bromide, and visualized with UV transillumination. Primers were designed from reference strains (as shown in Table 1) by using software (NCBI Primer-BLAST), and the primer sequences were compared with the genome sequences of approximately 10 NTHi strains registered in the NCBI database to confirm that no mutations were found at the primer sites. For the hmw, hia/hsf, hap, and p5 genes, PCR results, both positive and negative, were confirmed using another set of primers at different sites of the targeted genes (data not shown). R2866 was used as a positive control for the hia/hsf, hap, hifA, and p5 genes, and R2846 was used as a positive control for hmw. Purified water was used as a negative control.
TABLE 1.
PCR primers used for amplification of adhesin genes
| Target gene or locus | GenBank accession number(s) | Primer namea | Sequence (5′ to 3′) | Size of amplicon (bp) | Reference or source |
|---|---|---|---|---|---|
| hifA | AF020909-1 | hifA-F | ATGAAAAAAACACTWCTTGGTAGC | 624–642 | Clemans et al. (49) |
| hifA-R | TTATYCGTAAGCAATTKGGAAATC | ||||
| hmw1A/hmw2A | U08876, AY497552 | hmw-F | ATGAACAAGATATATCGTCTC | 602 | This study |
| hmw-R | CCGTGATTCACAATTTCAGC | ||||
| hia/hsf | U38617 | hia/hsf-F | TCCACCGATGCGATTAACGG | 315 | This study |
| hia/hsf-R | TGCAACGCCTGTTTTACCTTG | ||||
| Hia | NC_000907 | Hia-F | CCGAAAGCACAATGGATATGGACG | 6,235 | Satola et al. (33) |
| Hia-R | GATAAATCCTGACCTCGCTCTC | ||||
| Hap | U11024 | hap-F | GATGATGTCGGGTTTTGCCC | 532 | This study |
| hap-R | CACGCCCACATTTTGCTGTT | ||||
| Fimbrial gene | L08448 | Fimb-F | GAATACCAATGGCTAACTCGCG | 512 | This study |
| Fimb-R | ATTTCTACACGACGGTCTGGAG |
F and R correspond to the forward and reverse directions, respectively.
Multilocus sequence typing.
PCR of seven housekeeping genes (adk, atpG, frdB, fucK, mdh, pgi, and recA) was performed as described previously (21). The PCR products were then sequenced using an ABI Prism 3130 genetic analyzer, and the multilocus sequence typing (MLST) website (www.mlst.net) was used to determine the allele number and sequence type (ST) of each isolate. New allelic profiles were submitted to the database for assignment. The relatedness of the isolates was determined by constructing a gene tree with the neighbor-joining method, using MEGA7. The eBURST v3 program was used to determine the clonal complex (CC). Strains were considered to be in the same CC when at least five of the seven loci matched.
Screening for the insertion element IS1016.
PCR was performed for all 28 strains as described previously (22). Amplification reactions were performed using a MyCycler thermocycler (Bio-Rad).
Whole-genome sequencing and sequence analysis.
Fourteen draft genome sequences were generated using a MiSeq system (Illumina, San Diego, CA, USA). Genomic DNA was purified with phenol-chloroform extraction. The Nextera DNA sample preparation kit (Illumina) was used to prepare fragmented genomic DNA libraries. The quantities of the libraries were validated with an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and a Quant-iT Pico Green double-stranded DNA assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing was performed with a MiSeq system (Illumina), using MiSeq reagent kit v2 (500 cycles; Illumina), according to the manufacturer's instructions. All Illumina data sets were cleaned using Trimmomatic v0.33 (23). The draft genome sequences were assembled using Platanus v1.2.4 (24), with default parameters. Prokka v1.11 software was used for gene prediction and annotation (25). Homologous genes for IS1016, adhesin genes (hia/hsf and hifA), cap genes (capa to capf), and bexDCBA in H. influenzae were searched for using BLASTN (v2.2.28+) or BLASTP (26).
Statistical analysis.
Statistical analysis of biofilm index values was performed using the two-tailed chi-square test for independence and the Mann-Whitney U test, using SPSS 17.0 (SPSS Japan Inc.). P values of <0.05 were considered significant.
Ethical considerations.
This study was approved by the Chiba University Ethics Committee (approval no. 666). The invasive H. influenzae disease surveillance study involving the 10 prefectures was reviewed and approved by the Mie Hospital Ethics Committee and was conducted according to the principles expressed in the Declaration of Helsinki (12).
Accession number(s).
Sequence reads were assembled as contigs by de novo assembly. All draft genome sequences were deposited in the DNA Data Bank of Japan under accession numbers BGMN01000001 to BGMN01000048, BGMO01000001 to BGMO01000058, BGMP01000001 to BGMP01000045, BGMQ01000001 to BGMQ01000053, BGMR01000001 to BGMR01000043, BGMS01000001 to BGMS01000024, BGMT01000001 to BGMT01000039, BGMU01000001 to BGMU01000034, BGMV01000001 to BGMV01000048, BGMW01000001 to BGMW01000083, BGMX01000001 to BGMX01000061, BGMY01000001 to BGMY01000061, BGMZ01000001 to BGMZ01000051, and BGNA01000001 to BGNA01000050.
RESULTS
Clinical characteristics.
The patients' clinical characteristics are shown in Table 2. Four patients were neonates, and the other 24 ranged in age from 7 months to 14 years. Excluding the neonates, 33.3% of the patients (8/24 patients) had underlying diseases. Ten patients (35.7%) had pneumonia and only 1 patient, from Kagoshima prefecture, had meningitis. Three (10.7%) patients died, as follows. One patient suffered cardiac arrest on arrival at the hospital. The second patient was known to have long QT syndrome, but it was unclear whether the cause of death was arrhythmia or invasive NTHi infection. The third patient had a fever and depressed level of consciousness on admission; the patient suffered cardiac arrest soon after treatment was started. The other 25 patients, including the patient with meningitis, survived without any complications. Except for the NTHi strain isolated from the CSF, all strains were isolated from blood samples. The strains were isolated from more than one hospital in each prefecture. Some prefectures (for example, Chiba and Tokyo prefectures) are located next to each other, while others are distantly located. We could not find any specific characteristics depending on the area of isolation. Excluding 4 neonates and 4 patients with missing information, 75% of the patients (15/20 patients) were known to use childcare services or were schoolchildren.
TABLE 2.
Clinical backgrounds of children with invasive nontypeable Haemophilus influenzae diseases in Japan
| Strain no. | Yr of isolation | Patient age | Prefecture | Underlying disease(s)a | Hib vaccination (no. of doses)b | Clinical diagnosis | Disease outcome |
|---|---|---|---|---|---|---|---|
| 1 | 2008 | 1 yr | Kagoshima | None | 0 | Bacteremia | No complications |
| 2 | 2008 | 10 yr | Kochi | Reye's syndrome | 0 | Pneumonia | No complications |
| 3 | 2010 | 0 mo | Chiba | None | 0 | Pneumonia | No complications |
| 4 | 2010 | 2 yr | Fukuoka | Seckel syndrome | 0 | Bacteremia | Died |
| 5 | 2010 | 0 mo | Fukuoka | Preterm, LBW | 0 | Bacteremia | No complications |
| 6 | 2010 | 14 yr | Chiba | None | 0 | Bacteremia | No complications |
| 7 | 2010 | 1 yr | Chiba | None | 0 | CPAOA | Died |
| 8 | 2010 | 4 yr | Fukuoka | None | 0 | Bacteremia | No complications |
| 9 | 2011 | 1 mo | Chiba | None | 0 | Bacteremia | No complications |
| 10 | 2011 | 5 yr | Kagoshima | Acute lymphocytic leukemia | 0 | Bacteremia | No complications |
| 11 | 2011 | 1 yr | Chiba | None | 1 | Pneumonia | No complications |
| 12 | 2012 | 1 yr | Nagasaki | Long QT syndrome | 3 | Bacteremia | Died |
| 13 | 2012 | 7 mo | Fukuoka | Infant hepatitis, PA | 2 | Pneumonia | No complications |
| 14 | 2012 | 4 yr | Fukuoka | PA, VSD, major aortopulmonary collateral artery | 1 | Pneumonia | No complications |
| 15 | 2012 | 1 yr | Kagoshima | None | 1 | Meningitis | No complications |
| 16 | 2012 | 7 mo | Fukuoka | None | 3 | Pneumonia | No complications |
| 17 | 2012 | 2 yr | Chiba | None | 0 | Sinusitis | No complications |
| 18 | 2012 | 8 mo | Fukuoka | None | 0 | Bacteremia | No complications |
| 19 | 2012 | 10 yr | Chiba | None | 0 | Sinusitis | No complications |
| 20 | 2012 | 1 yr | Okinawa | None | 1 | Bacteremia | No complications |
| 21 | 2012 | 1 yr | Okinawa | None | 4 | Bacteremia | No complications |
| 22 | 2013 | 8 yr | Chiba | Chromosomal abnormality, hydrocephalus | 1 | Peritonitis | No complications |
| 23 | 2013 | 0 mo | Tokyo | Extremely LBW | 0 | Bacteremia | No complications |
| 24 | 2013 | 11 mo | Tokyo | None | 3 | Bacteremia | No complications |
| 25 | 2014 | 1 yr | Shizuoka | None | 4 | Pneumonia | No complications |
| 26 | 2014 | 4 yr | Chiba | None | 1 | Pneumonia | No complications |
| 27 | 2015 | 3 yr | Chiba | Preterm, LBW, jawless, tracheotomy | 4 | Pneumonia | No complications |
| 28 | 2015 | 11 mo | Chiba | None | 3 | Pneumonia | No complications |
LBW, low birth weight; PA, pulmonary atresia; VSD, ventricular septal defect; CPAOA, cardiac pulmonary arrest on arrival.
Number of doses received.
Serotyping and biotyping.
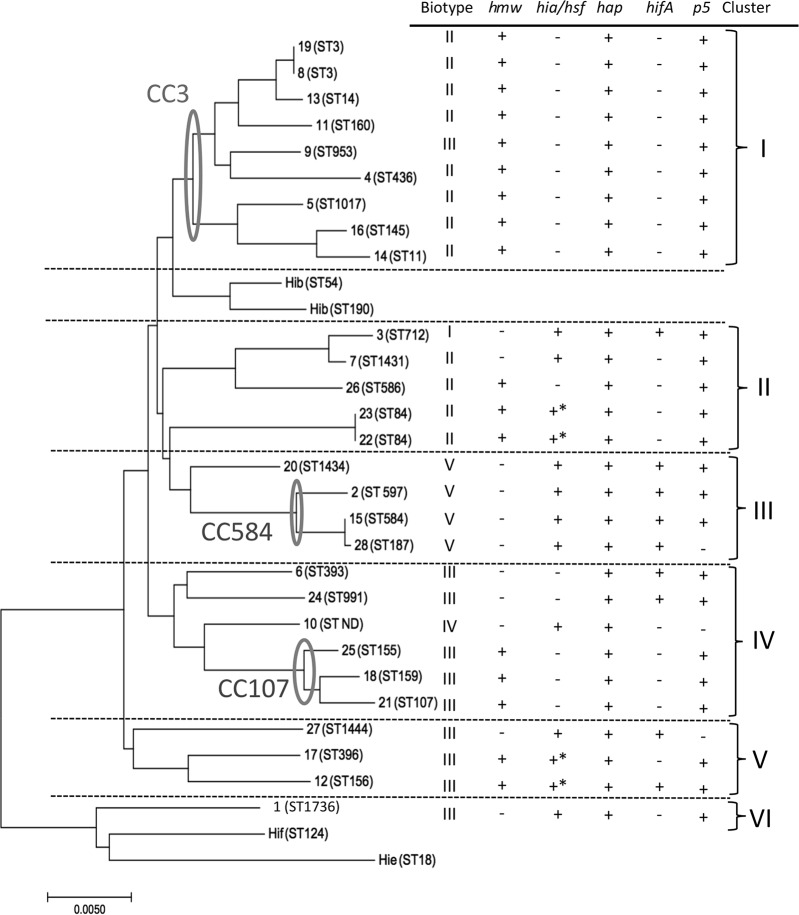
None of the 28 isolates demonstrated agglutination with capsule-typing antisera. Moreover, all isolates analyzed by PCR lacked both bexA and capsule-specific genes (defined to be NTHi in this study). Among the 4 NTHi strains isolated from neonates, 2 belonged to biotype II and the others were biotypes I and III. Of the other 24 isolates, 10 (41.7%) were biotype II, 9 (37.5%) were biotype III, 4 (16.7%) were biotype V, and 1 (4%) was biotype IV (see Fig. 2).
FIG 2.
Genetic relatedness among invasive nontypeable H. influenzae isolates. The dendrogram shows the relationships of the sequence types of the 28 invasive NTHi strains. Including the biotypes and PCR results for the adhesin genes, the strains were divided into six clusters. The PCR products of strains 12, 17, 22, and 23 were shorter than expected. BLAST results were used to confirm that those strains had only partial segments of hia/hsf.
Antimicrobial susceptibility.
The results of antimicrobial susceptibility testing are shown in Table 3. Two strains that showed high levels of aminobenzylpenicillin (ABPC) resistance (MICs of ≥16 μg/ml) were β-lactamase-producing strains. PCR analysis showed that both strains were positive for TEM-1 (data not shown). These strains also showed multiple substitutions in PBP-3 and high resistance to amoxicillin-clavulanate; they were defined as β-lactamase-positive ampicillin-clavulanate-resistant (BLPACR) strains (Table 3). Only 2 strains, strains 11 and 18, were BLNAR strains (MICs of ≥4 μg/ml). Most strains with ABPC MICs of ≥1 μg/ml had multiple amino acid substitutions in PBP-3 at Asp-350, Ser-357, Ser-385, and/or Met-377, which categorized them in group III or III-like gBLNAR strains (except for strain 17). Strain 17 also had an Ile-449-Val substitution in PBP-3, which categorized it in group IId. Group III and III-like strains had slightly higher MICs for cephalosporin antibiotics than did strains without any substitutions, but none of the strains met the criteria for resistance to cefditoren-pivoxil, cefotaxime (CTX), or ceftriaxone (CRO), as determined by the CLSI (MICs of ≥2 μg/ml). Strains with ABPC MICs of ≤0.25 μg/ml showed no PBP-3 amino acid substitutions. All strains were sensitive to meropenem and levofloxacin. Twenty-three strains (85.7%) were sensitive to clarithromycin (MICs of ≤8 μg/ml) (16).
TABLE 3.
Antimicrobial susceptibilities and β-lactamase production of invasive nontypeable Haemophilus influenzae strains isolated from children
| Strain no. | MIC (μg/ml) ofa: |
β-Lactamse productionb | Amino acid substitutions in PBP-3 |
Group | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABPC | AMC | CTX | CRO | MEM | CLR | LVX | D350 | S357 | M377 | S385 | L389 | R517 | N526 | |||
| 1 | 1 | 1/0.5 | 0.5 | 0.25 | ≤0.06 | 8 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 2 | 1 | 0.5/0.25 | 0.25 | ≤0.06 | ≤0.06 | 4 | ≤0.06 | − | N | N | T | H | III-like | |||
| 3 | 0.25 | 0.25/0.12 | ≤0.06 | ≤0.06 | ≤0.06 | 4 | ≤0.06 | − | ||||||||
| 4 | 1 | 1/0.5 | 0.5 | 0.25 | 0.12 | 1 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 5 | 0.25 | 0.25/0.12 | ≤0.06 | ≤0.06 | ≤0.06 | 8 | ≤0.06 | − | ||||||||
| 6 | ≤0.12 | 0.25/0.12 | ≤0.06 | ≤0.06 | ≤0.06 | 4 | ≤0.06 | − | ||||||||
| 7 | 0.25 | 0.5/0.25 | ≤0.06 | ≤0.06 | ≤0.06 | 8 | ≤0.06 | − | ||||||||
| 8 | ≤0.12 | 0.25/0.12 | ≤0.06 | ≤0.06 | ≤0.06 | 8 | ≤0.06 | − | ||||||||
| 9 | ≥16 | 8/4 | 0.5 | 0.25 | 0.25 | 16 | ≤0.06 | + | N | N | I | T | F | K | III | |
| 10 | 2 | 2/1 | 0.5 | 0.25 | 0.25 | 8 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 11 | 4 | 8/4 | 1 | 0.25 | 0.25 | 8 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 12 | 2 | 2/1 | 1 | 0.25 | ≤0.06 | 16 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 13 | 1 | 1/0.5 | 0.25 | 0.12 | 0.12 | 8 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 14 | 1 | 1/0.5 | 0.5 | 0.12 | ≤0.06 | 8 | ≤0.06 | − | N | N | I | T | F | H | III-like | |
| 15 | 0.25 | 0.5/0.25 | ≤0.06 | ≤0.06 | ≤0.06 | 4 | ≤0.06 | − | ||||||||
| 16 | 0.25 | 0.5/0.25 | ≤0.06 | ≤0.06 | ≤0.06 | 8 | ≤0.06 | − | ||||||||
| 17 | 1 | 1/0.5 | ≤0.06 | ≤0.06 | 0.12 | 16 | ≤0.06 | − | K | II d | ||||||
| 18 | 4 | 4/2 | 1 | 0.25 | 0.5 | 32 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 19 | 0.25 | 0.25 | ≤0.06 | ≤0.06 | ≤0.06 | 8 | ≤0.06 | − | ||||||||
| 20 | 2 | 2/1 | 0.5 | 0.25 | 0.5 | 8 | 0.12 | − | N | N | I | T | F | K | III | |
| 21 | 2 | 2/1 | 0.5 | 0.12 | ≤0.06 | 8 | ≤0.06 | − | N | N | I | T | F | H | III-like | |
| 22 | 1 | 1/0.5 | 0.12 | ≤0.06 | 0.25 | 4 | ≤0.06 | − | N | N | I | T | H | III-like | ||
| 23 | 1 | 1/0.5 | 0.12 | ≤0.06 | 0.25 | 8 | ≤0.06 | − | N | N | I | T | H | III-like | ||
| 24 | 1 | 1/0.5 | 0.25 | ≤0.06 | ≤0.06 | 4 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 25 | 0.25 | 0.25/0.12 | ≤0.06 | ≤0.06 | ≤0.06 | 16 | ≤0.06 | − | ||||||||
| 26 | 1 | 0.5/0.25 | 0.5 | 0.25 | ≤0.06 | 4 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 27 | 2 | 2/1 | 0.5 | 0.25 | 0.25 | 8 | ≤0.06 | − | N | N | I | T | F | K | III | |
| 28 | 16 | 16/8 | 0.5 | 0.25 | 0.12 | 8 | ≤0.06 | + | N | N | I | T | F | K | III | |
ABPC, aminobenzylpenicillin; AMC, amoxicillin-clavulanic acid; CTX, cefotaxime; CRO, ceftriaxone; MEM, meropenem; CLR, clarithromycin; LVX, levofloxacin.
−, negative; +, positive.
Except for the patient with strain 7, who experienced cardiac arrest before arrival at the hospital for initiation of treatment, all of the patients were treated with antibiotics. The majority of the patients were treated with ABPC, ABPC-sulbactam (SBT), CRO, or CTX, which are antibiotics recommended in the Japanese guidelines for bacteremia (50). The patients with BLPACR strains (strains 9 and 28) were both successfully treated with CTX.
Biofilm formation.
The biofilm index values of the NTHi strains isolated from patients with invasive diseases are shown in Fig. 1. Heterogeneity was observed, and the biofilm indices varied widely, from 0.006 to 1.592. The average biofilm formation of invasive strains was greater than that of strains isolated from sputum and nasal cavities of patients with noninvasive, community-acquired pneumonia; however, this difference was not statistically significant (Fig. 1).
FIG 1.
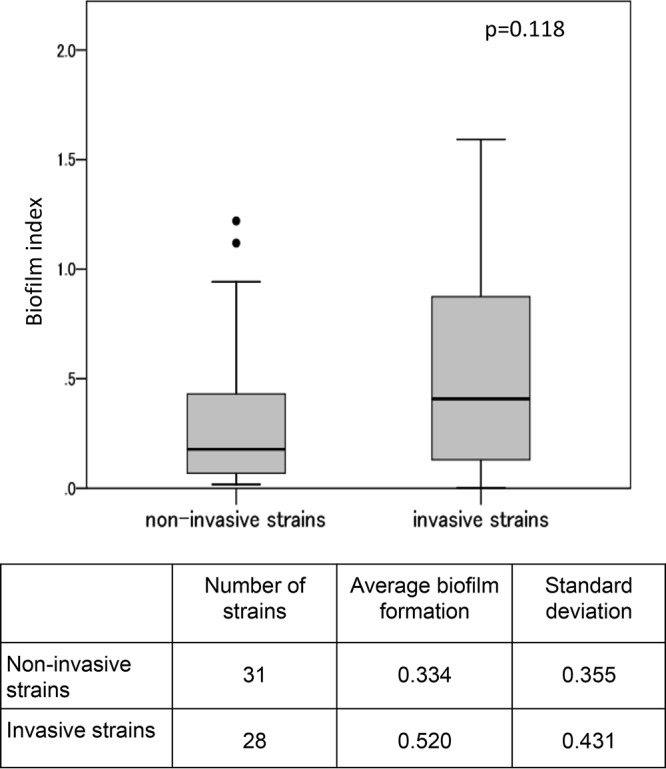
Comparison of the biofilm indices of invasive and noninvasive nontypeable H. influenzae strains. Thirty-one noninvasive NTHi strains were collected from nasal cavities or sputum of children with respiratory infections (these strains were used only for biofilm assays). The boxplot shows the medians and 10th, 25th, 75th, and 90th percentiles; outliers are plotted as individual points. The biofilm index was defined using the average OD570 value.
Prevalence of adhesin genes.
The PCR results showed 17 strains as being positive for hmw (60.7%) and 13 strains as being positive for hia/hsf (46.4%). The PCR products of strains 12, 17, 22, and 23 were shorter than expected. BLAST results were used to confirm that those strains had only partial segments of hia/hsf. In terms of the fimbrial gene, most strains (89.3%) possessed the p5 gene, while only 9 (32.1%) harbored hifA (Fig. 2).
MLST.
Molecular typing using MLST showed wide variety among the 28 isolates, which had 26 different STs (Table 4). Strains 1, 3, 7, 20, and 27 had new STs, which we submitted as ST-1736, ST-712, ST-1431, ST-1434, and ST-1444, respectively. Strain 10 lacked fuculokinase (fucK) and so could not be genotyped. eBURST analysis distributed the 28 isolates into 3 clonal groups, with 11 singletons. The largest clonal group, CC-3, included 9 isolates. CC-3 is also the largest CC of NTHi isolates in the database. Within CC-3, there were 8 STs, including 2 isolates of ST-3 and 1 isolate each of ST-11, ST-14, ST-145, ST-160, ST-436, ST-953, and ST-1017. The other 2 groups included 3 strains each, i.e., CC-107 (ST-107, ST-155, and ST-159) and CC-584 (ST-187, ST-584, and ST-597). Among the 11 singletons, the most frequent ST was ST-84, with 2 isolates. A phylogenic tree based on MLST relatedness is shown in Fig. 2.
TABLE 4.
Sequence types and allelic profiles of the invasive nontypeable Haemophilus influenzae isolates
| Strain no. | Sequence typea | Allele |
||||||
|---|---|---|---|---|---|---|---|---|
| adk | atpG | frdB | fucK | mdh | pgi | recA | ||
| 1 | 1736 | 68 | 43 | 38 | 7 | 95 | 165 | 18 |
| 2 | 597 | 11 | 33 | 7 | 1 | 7 | 41 | 29 |
| 3 | 712 | 63 | 54 | 102 | 1 | 17 | 31 | 10 |
| 4 | 436 | 40 | 1 | 1 | 10 | 15 | 1 | 5 |
| 5 | 1017 | 1 | 1 | 1 | 14 | 9 | 7 | 13 |
| 6 | 393 | 42 | 2 | 38 | 41 | 104 | 55 | 10 |
| 7 | 1431 | 63 | 54 | 165 | 1 | 17 | 31 | 10 |
| 8 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 5 |
| 9 | 953 | 153 | 11 | 18 | 18 | 62 | 1 | 5 |
| 10 | ND | 139 | 2 | 16 | ND | 62 | 40 | 4 |
| 11 | 160 | 40 | 1 | 1 | 14 | 1 | 59 | 3 |
| 12 | 156 | 26 | 2 | 15 | 7 | 22 | 56 | 3 |
| 13 | 14 | 5 | 1 | 1 | 1 | 1 | 2 | 5 |
| 14 | 11 | 1 | 8 | 1 | 14 | 9 | 14 | 13 |
| 15 | 584 | 5 | 33 | 7 | 1 | 26 | 41 | 29 |
| 16 | 145 | 1 | 8 | 1 | 14 | 22 | 14 | 13 |
| 17 | 396 | 10 | 2 | 15 | 8 | 26 | 61 | 3 |
| 18 | 159 | 33 | 8 | 16 | 16 | 17 | 2 | 29 |
| 19 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 5 |
| 20 | 1434 | 35 | 1 | 22 | 36 | 7 | 14 | 29 |
| 21 | 107 | 33 | 8 | 16 | 16 | 49 | 2 | 3 |
| 22 | 84 | 29 | 7 | 13 | 1 | 45 | 13 | 1 |
| 23 | 84 | 29 | 7 | 13 | 1 | 45 | 13 | 1 |
| 24 | 991 | 63 | 50 | 22 | 15 | 10 | 40 | 3 |
| 25 | 155 | 16 | 8 | 16 | 16 | 30 | 1 | 3 |
| 26 | 586 | 116 | 1 | 1 | 13 | 17 | 25 | 16 |
| 27 | 1444 | 26 | 11 | 104 | 7 | 26 | 5 | 42 |
| 28 | 187 | 5 | 33 | 7 | 32 | 26 | 41 | 29 |
ND, not determined.
Insertion element IS1016.
PCR was performed for all 28 strains; only 3 strains had the insertion element IS1016. In addition, whole-genome sequencing, which was performed for 14 isolates, was used to analyze the existence of IS1016. Of the 14 strains, 2 were positive for IS1016 and were also PCR positive. All 14 strains were negative for the cap and bex genes.
DISCUSSION
NTHi display wide genetic diversity (27–29). To date, attempts to identify a potential virulence factor that is present in NTHi strains isolated from patients with invasive infections but not from those with noninvasive NTHi infections have been unsuccessful. It is important to understand the clinical characteristics of patients with such infections, to determine the virulence potential of NTHi strains isolated from invasive diseases. Cases of NTHi meningitis are rare in children, but bacteremia and bacteremic pneumonia are more common (29). Our study confirmed that most invasive NTHi infections were bacteremia without a focus of infection, followed by bacteremic pneumonia.
Three children (10.7%) in our study died, probably from NTHi infections; however, most children without underlying disease(s), including the patient with meningitis, recovered from invasive NTHi infections without complications. Although neonates with perinatal NTHi infections often have serious comorbidities, those children also recovered well. This could be because of easy access to hospitals in Japan and the early initiation of appropriate antibiotic treatment. NTHi strains are known to be a cause of sudden unexpected infant death (30). Strain 7 was cultured from postmortem samples. Because the NTHi isolate was the only bacterium isolated from that patient, NTHi infection was thought to be the cause of death in that case.
Biotyping is a way to categorize the phenotypes of H. influenzae strains by their enzyme activity. Biotypes II and III, the two major NTHi biotypes found in a healthy respiratory tract, accounted for 79.2% of the NTHi isolates in our study. These results were similar to those of several studies from Japan that showed that 77 to 95% of the H. influenzae strains isolated mainly from nonsterile sites of respiratory tract infections were either biotype II or biotype III (31, 32). All of the IS1016-positive strains were biotype III, a unique finding, compared with those that have been reported (33). Furthermore, although NTHi biotype IV is most commonly isolated from neonates (34), other biotypes were detected in our study.
There are two known mechanisms by which H. influenzae acquires resistance to aminopenicillins, namely, production of β-lactamase and a change in PBP-3. Among ABPC-resistant strains, β-lactamase production (TEM-1 or ROB-1) is common in other countries (35–37); however, Japan is known to have a high percentage of gBLNAR strains with PBP-3 substitutions that are encoded by the ftsI gene (38, 39). The ftsI gene encodes an essential transpeptidase that catalyzes cross-linking of the peptidoglycan septal cell wall during cell division (40). Only 2 strains (6.7%) were BLNAR, which was relatively low, compared to other studies from Japan that showed that 30 to 40% of H. influenzae strains isolated mainly from nonsterile sites were BLNAR (38, 41). In our study, any mutation in the ftsI gene yielded 4- to 8-fold resistance to ABPC. PBP-3 substitutions are categorized into several groups. Strains with low-level resistance possess the R517H (group I) or N526K (group II) substitution. Group III is defined as having M377I, S385T, and L389F substitutions, in addition to N526K; this is clinically important, as this group shows higher levels of resistance to extended-spectrum cephalosporins (42). In our study, strains with PBP-3 substitutions were mainly categorized as group III (Table 3), which is a distinctive feature of gBLNAR strains in Japan (38, 39). The higher levels of resistance to cephalosporins among these strains should be taken into account when choosing antibiotics to treat patients with invasive H. influenzae infections in Japan. Another study showed that resistance to cephalosporins is dependent on the PBP-3 substitutions R517H, N526K, S385T, and L389F (43). In our study, strains with S385T, L389F, and R517H or N526K substitutions showed more than 2- to 4-fold resistance to cephalosporins. Strains 14 and 21, both with L389F, showed higher levels of cephalosporin resistance than did strains 22 and 23, which did not have this mutation. None of the patients, including those who died (patients 4 and 7), were considered to have been treated with ineffective antibiotics, to which the isolated NTHi strains were resistant.
MLST of the isolates from our study showed wide diversity, except for ST-3 and ST-84, which had 2 strains each. We found no relationship between ST and the geographic region or clinical presentation of the patients. However, using the phylogenic tree and including biotypes and PCR results for the adhesin genes, these strains were divided into six clusters. Cluster I strains were CC-3 and mainly biotype II, with positive hmw and negative hia/hsf results for all strains. The hmw gene is known to be highly expressed in NTHi strains but is generally absent in encapsulated strains, while hia is found in many non-Hib encapsulated strains and NTHi strains that lack hmw (20). In contrast, isolates in cluster III were all positive for hia/hsf and negative for hmw, also with biotype V, an uncommon biotype for NTHi strains, which may be one of the characteristics of invasive NTHi strains in children (33). None of the isolates belonged to CC-6, which is known to contain Hib strains and is detected worldwide (http://haemophilus.mlst.net). Several studies have indicated that ST-14 has a PBP-3 type A pattern (D350N, M377I, A502V, N526K, V547I, and N569S) (18). In our study, however, our ST-14 strain (strain 13) had S385T. Moreover, our study showed a high frequency of group III gBLNAR strains, despite their STs. Interestingly, strain 10 lacked fucK. Fuculokinase is one of several enzymes involved in the fucose pathway; fucose is used as an energy and carbon source in bacteria. H. influenzae isolates that lack fucK have been reported (44). In the report, all isolates were NTHi; the prevalence of fucK-negative strains was 2%. The strains were obtained from sterile sites (blood and CSF) and belonged to various biotypes. For encapsulated H. influenzae strains, the capsule and the genes surrounding the cap locus are thought to be the major determinants of virulence. In division I Hib strains, the cap locus is flanked by direct repeats of an insertion element known as IS1016 (45). Whole-genome analysis revealed that 2 strains encoded IS1016. Including the other 14 strains that underwent PCR analysis to investigate the existence of IS1016, 3 strains (10.7%) were positive for IS1016. Although capa to capf and bexA to bexD were completely absent in these strains, the existence of IS1016 suggests that these strains are genetically more closely related to encapsulated strains.
Many microorganisms form biofilms as protection against bactericidal agents, bacteriophages, and host clearance mechanisms (46). We used a microtiter biofilm assay as an indicator of biofilm formation. We did not visually observe biofilm formation, but the data suggest the potential of each strain for biofilm formation (47). A previous report showed increased biofilm formation among the NTHi strains collected from patients with invasive disease or otitis media, compared with NTHi isolates from patients with community-acquired pneumonia or chronic obstructive airway disease or from healthy colonized subjects (48). We also found higher biofilm index values for isolates from patients with invasive diseases than for isolates from patients with respiratory tract infections, but the difference was not statistically significant.
We collected strains from 8 (of 47) prefectures in Japan and included data from the nationwide population-based surveillance study on invasive H. influenzae disease in Japan, which includes 10 prefectures (Hokkaido, Fukushima, Niigata, Chiba, Mie, Okayama, Kouchi, Fukuoka, Kagoshima, and Okinawa) (12); only 6 prefectures had cases of invasive NTHi infections during the study period. The populations of the 10 prefectures account for approximately 20% of the Japanese population. This is the only study to have evaluated the trends in invasive H. influenzae infections in Japanese children after introduction of the Hib vaccine.
In conclusion, we revealed the bacteriologic characteristics of NTHi strains responsible for invasive H. influenzae diseases among children in Japan, and we described the clinical characteristics of children with such infections. Although most patients recovered without complications, the mortality rate of invasive NTHi disease was 10.7%. Invasive NTHi strains in Japan have peculiar PBP-3 substitutions that result in low penicillin and cephalosporin sensitivity, suggesting the need for a new vaccine to prevent invasive NTHi diseases or suggesting that other antibiotic classes that are effective against ABPC-resistant or BLNAR strains be used to treat such infections in Japan. Moreover, continuous monitoring of the NTHi strains responsible for invasive diseases in children is important to detect changes in the epidemiology of invasive H. influenzae infections in the post-Hib vaccine era.
ACKNOWLEDGMENTS
We gratefully acknowledge the dedicated efforts of the staff of the bacteriological examination room for isolating and storing the bacterial isolates. We truly appreciate Arnold L. Smith providing us with the NTHi strains R2866 and R2846. We also express our appreciation to the pediatricians in Japan who sent the bacterial isolates for analysis and to Hideki Akeda, Kenji Okada, Yuho Horikoshi, Ichiro Ohgawara, Tetsuya Sato, and Hiroyuki Moriuchi for their contributions to this study.
This study was financially supported by a grant from the Japanese Ministry of Health, Labor, and Welfare under the category of research on emerging and re-emerging infectious diseases (H-Shinko-Ippan-00). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We report no potential conflicts of interest.
REFERENCES
- 1.Agrawal A, Murphy TF. 2011. Haemophilus influenzae infections in the H. influenzae type b conjugate vaccine era. J Clin Microbiol 49:3728–3732. doi: 10.1128/JCM.05476-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peltola H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin Microbiol Rev 13:302–317. doi: 10.1128/CMR.13.2.302-317.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ladhani S, Slack MP, Heath PT, von Gottberg A, Chandra M, Ramsay ME. 2010. Invasive Haemophilus influenzae disease, Europe, 1996–2006. Emerg Infect Dis 16:455–463. doi: 10.3201/eid1603.090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins S, Litt DJ, Flynn S, Ramsay ME, Slack MP, Ladhani SN. 2015. Neonatal invasive Haemophilus influenzae disease in England and Wales: epidemiology, clinical characteristics, and outcome. Clin Infect Dis 60:1786–1792. doi: 10.1093/cid/civ194. [DOI] [PubMed] [Google Scholar]
- 5.Falla TJ, Dobson SR, Crook DW, Kraak WA, Nichols WW, Anderson EC, Jordens JZ, Slack MP, Mayon-White D, Moxon ER. 1993. Population-based study of non-typable Haemophilus influenzae invasive disease in children and neonates. Lancet 341:851–854. doi: 10.1016/0140-6736(93)93059-A. [DOI] [PubMed] [Google Scholar]
- 6.Adams WG, Deaver KA, Cochi SL, Plikaytis BD, Zell ER, Broome CV, Wenger JD. 1993. Decline of childhood Haemophilus influenzae type b (Hib) disease in the Hib vaccine era. JAMA 269:221–226. doi: 10.1001/jama.1993.03500020055031. [DOI] [PubMed] [Google Scholar]
- 7.Slack MP, Azzopardi HJ, Hargreaves RM, Ramsay ME. 1998. Enhanced surveillance of invasive Haemophilus influenzae disease in England, 1990 to 1996: impact of conjugate vaccines. Pediatr Infect Dis J 17(Suppl):S204–S207. [DOI] [PubMed] [Google Scholar]
- 8.Campos J, Hernando M, Román F, Pérez-Vázquez M, Aracil B, Oteo J, Lázaro E, de Abajo F. 2004. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J Clin Microbiol 42:524–529. doi: 10.1128/JCM.42.2.524-529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berndsen MR, Erlendsdóttir H, Gottfredsson M. 2012. Evolving epidemiology of invasive Haemophilus infections in the post-vaccination era: results from a long-term population-based study. Clin Microbiol Infect 18:918–923. doi: 10.1111/j.1469-0691.2011.03700.x. [DOI] [PubMed] [Google Scholar]
- 10.Langereis JD, de Jonge MI. 2015. Invasive disease caused by nontypeable Haemophilus influenzae. Emerg Infect Dis 21:1711–1718. doi: 10.3201/eid2110.150004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishiwada N, Hishiki H, Nagasawa K, Naito S, Sato Y, Chang B, Sasaki Y, Kimura K, Ohnishi M, Shibayama K. 2014. The incidence of pediatric invasive Haemophilus influenzae and pneumococcal disease in Chiba prefecture, Japan, before and after the introduction of conjugate vaccines. Vaccine 32:5425–5431. doi: 10.1016/j.vaccine.2014.07.100. [DOI] [PubMed] [Google Scholar]
- 12.Suga S, Chang B, Asada K, Akeda H, Nishi J, Okada K, Wakiguchi H, Maeda A, Oda M, Ishiwada N, Saitoh A, Oishi T, Hosoya M, Togashi T, Oishi K, Ihara T. 2015. Nationwide population-based surveillance of invasive pneumococcal disease in Japanese children: effects of the seven-valent pneumococcal conjugate vaccine. Vaccine 33:6054–6060. doi: 10.1016/j.vaccine.2015.07.069. [DOI] [PubMed] [Google Scholar]
- 13.Zhu B, Xiao D, Zhang H, Zhang Y, Gao Y, Xu L, Lv J, Wang Y, Zhang J, Shao Z. 2013. MALDI-TOF MS distinctly differentiates nontypable Haemophilus influenzae from Haemophilus haemolyticus. PLoS One 8:e56139. doi: 10.1371/journal.pone.0056139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. 1994. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol 32:2382–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Killian M. 2005. Genus III: Haemophilus, p 883–904. In Garrity GM, Brenner DJ, Kreig NR, Staley JT (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2, part B Springer, New York, NY. [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing; 26th informational supplement. CLSI M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.Hotomi M, Fujihara K, Billal DS, Suzuki K, Nishimura T, Baba S, Yamanaka N. 2007. Genetic characteristics and clonal dissemination of β-lactamase-negative ampicillin-resistant Haemophilus influenzae strains isolated from the upper respiratory tract of patients in Japan. Antimicrob Agents Chemother 51:3969–3976. doi: 10.1128/AAC.00422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skaare D, Anthonisen IL, Caugant DA, Jenkins A, Steinbakk M, Strand L, Sundsfjord A, Tveten Y, Kristiansen BE. 2014. Multilocus sequence typing and ftsI sequencing: a powerful tool for surveillance of penicillin-binding protein 3-mediated beta-lactam resistance in nontypeable Haemophilus influenzae. BMC Microbiol 14:131. doi: 10.1186/1471-2180-14-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy TF, Kirkham C. 2002. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol 2:7. doi: 10.1186/1471-2180-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.St Geme JM., III 2002. Molecular and cellular determinants of non-typeable Haemophilus influenzae adherence and invasion. Cell Microbiol 4:191–200. doi: 10.1046/j.1462-5822.2002.00180.x. [DOI] [PubMed] [Google Scholar]
- 21.Meats E, Feil EJ, Stinger S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG. 2003. Characterization of encapsulated and nonencapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohkusu K, Nash KA, Inderlied CB. 2005. Molecular characterisation of Haemophilus influenzae type a and untypeable strains isolated simultaneously from cerebrospinal fluid and blood: novel use of quantitative real-time PCR based on the cap copy number to determine virulence. Clin Microbiol Infect 11:637–643. doi: 10.1111/j.1469-0691.2005.01203.x. [DOI] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, Kohara Y, Fujiyama A, Hayashi T, Itoh T. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerquetti M, Ciofi degli Atti ML, Renna G, Tozzi AE, Garlaschi ML, Mastrantonio P. 2000. Characterization of non-type B Haemophilus influenzae strains isolated from patients with invasive disease. J Clin Microbiol 38:4649–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erwin AL, Nelson KL, Mhlanga-Mutangadura T, Bonthuis PJ, Geelhood JL, Morlin G, Unrath WC, Campos J, Crook DW, Farley MM, Henderson FW, Jacobs RF, Mühlemann K, Satola SW, van Alphen L, Golomb M, Smith AL. 2005. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect Immun 73:5853–5863. doi: 10.1128/IAI.73.9.5853-5863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardines R, Giufrè M, Mastrantonio P, Ciofi degli Atti ML, Cerquetti M. 2007. Nontypeable Haemophilus influenzae meningitis in children: phenotypic and genotypic characterization of isolates. Pediatr Infect Dis J 26:577–582. doi: 10.1097/INF.0b013e3180616715. [DOI] [PubMed] [Google Scholar]
- 30.Prtak L, Al-Adnani M, Fenton P, Kudesia G, Cohen MC. 2010. Contribution of bacteriology and virology in sudden unexpected death in infancy. Arch Dis Child 95:371–376. doi: 10.1136/adc.2009.162792. [DOI] [PubMed] [Google Scholar]
- 31.Kaneko M, Bando Y, Fujita T, Hirose Y, Suganuma E, Ishii M, Takahashi T. 2017. Encapsulated pleural effusion due to Haemophilus influenzae biotype II in a child with trisomy 21: a case report and literature review. IDCases 10:93–96. doi: 10.1016/j.idcr.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada F, Ando Y, Tanoue S, Ishii R, Matsushita S, Ono A. 2012. Radiological findings in acute Haemophilus influenzae pulmonary infection. Br J Radiol 85:121–126. doi: 10.1259/bjr/48077494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satola SW, Napier B, Farley MM. 2008. Association of IS1016 with the hia adhesin gene and biotypes V and I in invasive nontypeable Haemophilus influenzae. Infect Immun 76:5221–5227. doi: 10.1128/IAI.00672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quentin R, Musser JM, Mellouet M, Sizaret PY, Selander RK, Goudeau A. 1989. Typing of urogenital, maternal, and neonatal isolates of Haemophilus influenzae and Haemophilus parainfluenzae in correlation with clinical source of isolation and evidence for a genital specificity of H. influenzae biotype IV. J Clin Microbiol 27:2286–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doern GV, Brueggemann AB, Pierce G, Holley HP Jr, Rauch A. 1997. Antibiotic resistance among clinical isolates of Haemophilus influenzae in the United States in 1994 and 1995 and detection of β-lactamase-positive strains resistant to amoxicillin-clavulanate: results of a national multicenter surveillance study. Antimicrob Agents Chemother 41:292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lâm TT, Claus H, Elias J, Frosch M, Vogel U. 2015. Ampicillin resistance of invasive Haemophilus influenzae isolates in Germany 2009–2012. Int J Med Microbiol 305:748–755. doi: 10.1016/j.ijmm.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 37.Jansen WT, Verel A, Beitsma M, Verhoef J, Milatovic D. 2008. Surveillance study of the susceptibility of Haemophilus influenzae to various antibacterial agents in Europe and Canada. Curr Med Res Opin 24:2853–2861. doi: 10.1185/03007990802381505. [DOI] [PubMed] [Google Scholar]
- 38.Hoshino T, Sato Y, Toyonaga Y, Hanaki H, Sunakawa K. 2013. Nationwide survey of the development of drug resistance in the pediatric field in 2007 and 2010: drug sensitivity of Haemophilus influenzae in Japan (second report). J Infect Chemother 19:495–503. doi: 10.1007/s10156-013-0591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ubukata K, Shibasaki Y, Yamamoto K, Chiba N, Hasegawa K, Takeuchi Y, Sunakawa K, Inoue M, Konno M. 2001. Association of amino acid substitutions in penicillin-binding protein 3 with β-lactam resistance in β-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob Agents Chemother 45:1693–1699. doi: 10.1128/AAC.45.6.1693-1699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahata S, Ida T, Senju N, Sanbongi Y, Miyata A, Maebashi K, Hoshiko S. 2007. Horizontal gene transfer of ftsI, encoding penicillin-binding protein 3, in Haemophilus influenzae. Antimicrob Agents Chemother 51:1589–1595. doi: 10.1128/AAC.01545-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morozumi M, Chiba N, Okada T, Sakata H, Matsubara K, Iwata S, Ubukata K. 2013. Antibiotic susceptibility in relation to genotype of Streptococcus pneumoniae, Haemophilus influenzae, and Mycoplasma pneumoniae responsible for community-acquired pneumonia in children. J Infect Chemother 19:432–440. doi: 10.1007/s10156-012-0500-x. [DOI] [PubMed] [Google Scholar]
- 42.Dabernat H, Delmas C, Seguy M, Pelissier R, Faucon G, Bennamani S, Pasquier C. 2002. Diversity of β-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae. Antimicrob Agents Chemother 46:2208–2218. doi: 10.1128/AAC.46.7.2208-2218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shuel ML, Karlowsky KE, Law DK, Tsang RS. 2011. Nonencapsulated or nontypeable Haemophilus influenzae are more likely than their encapsulated or serotypeable counterparts to have mutations in their fucose operon. Can J Microbiol 57:982–986. doi: 10.1139/w11-017. [DOI] [PubMed] [Google Scholar]
- 44.Ridderberg W, Fenger MG, Norskov-Lauritsen N. 2010. Haemophilus influenzae may be untypable by the multilocus sequence typing scheme due to a complete deletion of the fucose operon. J Med Microbiol 59:740–742. doi: 10.1099/jmm.0.018424-0. [DOI] [PubMed] [Google Scholar]
- 45.Satola SW, Collins JT, Napier R, Farley MM. 2007. Capsule gene analysis of invasive Haemophilus influenzae: accuracy of serotyping and prevalence of IS1016 among nontypeable isolates. J Clin Microbiol 45:3230–3238. doi: 10.1128/JCM.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin L, Kida Y, Ishiwada N, Ohkusu K, Kaji C, Sakai Y, Watanabe K, Furumoto A, Ichinose A, Watanabe H. 2014. The relationship between biofilm formations and capsule in Haemophilus influenzae. J Infect Chemother 20:151–156. doi: 10.1016/j.jiac.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Puig C, Domenech A, Garmendia J, Langereis JD, Mayer P, Calatayud L, Liñares J, Ardanuy C, Marti S. 2014. Increased biofilm formation by nontypeable Haemophilus influenzae isolates from patients with invasive disease or otitis media versus strains recovered from cases of respiratory infections. Appl Environ Microbiol 80:7088–7095. doi: 10.1128/AEM.02544-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clemans DL, Marrs CF, Patel M, Duncan M, Gilsdorf JR. 1998. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect Immun 66:656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arakawa S, Kasai M, Kawai S, Sakata H, Mayumi T. 2018. The JAID/JSC guide to clinical management of infectious diseases 2017: sepsis and catheter related blood stream infection. Jpn J Chemother 66:82–117. [Google Scholar]