ABSTRACT
Conjugative transfer of plasmids in enterococci is promoted by intercellular communication using peptide pheromones. The regulatory mechanisms that control transfer have been extensively studied in vitro. However, the complicated systems that regulate the spread of these plasmids did not evolve in the laboratory test tube, and remarkably little is known about this form of signaling in the intestinal tract, the primary niche of these organisms. Because the evolution of Enterococcus faecalis strains and their coresident pheromone-inducible plasmids, such as pCF10, have occurred in the gastrointestinal (GI) tract, it is important to consider the functions controlled by pheromones in light of this ecology. This review summarizes our current understanding of the pCF10-encoded pheromone response. We consider how selective pressures in the natural environment may have selected for the complex and very tightly regulated systems controlling conjugation, and we pay special attention to the ecology of enterococci and the pCF10 plasmid as a gut commensal. We summarize the results of recent studies of the pheromone response at the single-cell level, as well as those of the first experiments demonstrating a role for pheromone signaling in plasmid transfer and in GI tract competitive fitness. These results will serve as a foundation for further in vivo studies that could lead to novel interventions to reduce opportunistic infections and the spread of antibiotic resistance.
KEYWORDS: antibiotic resistance, nosocomial infections, intestinal microbiota, horizontal gene transfer
INTRODUCTION
The tetracycline resistance plasmid pCF10 is a member of a family of pheromone-inducible conjugative plasmids of Enterococcus faecalis; these elements frequently carry antibiotic resistance or virulence genes, which can readily be transferred horizontally by the induction of conjugation machinery. Cells containing pCF10 are potential plasmid donors, and these can be signaled by recipients secreting the chromosomally encoded peptide pheromone cCF10 to induce conjugative transfer of pCF10 (Fig. 1). Over a dozen different but related enterococcal plasmids, each responding to a distinct chromosomally encoded peptide pheromone, have been identified, as reviewed by Clewell and Dunny (1).
FIG 1.
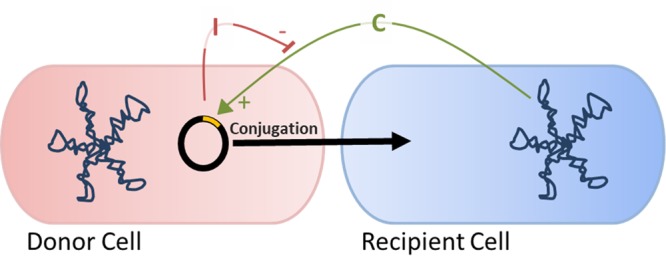
Plasmids can be transferred from donor to recipient cells in a cell-cell contact-mediated process called conjugation. For pCF10, conjugation is induced by the cCF10 (C) peptide pheromone produced from the chromosome of potential recipient cells. A pCF10-encoded counteracting peptide, iCF10 (I), inhibits this response; I functions to block spontaneous self-induction in donors and is critical for shutdown of the donor response to C following an induction cycle.
Pheromone-inducible conjugation was first discovered during experiments examining the kinetics of plasmid transfer and how the alteration of growth conditions impacted transfer frequency (2). Important observations that eventually led to the conclusion that intercellular signaling regulated enterococcal conjugation included the following: (i) initial rates of plasmid transfer were very low upon mixing recipients and donors, but after about 60 to 90 min of coculture, transfer rates showed a sharp increase significantly higher than the growth rate, (ii) in mixed cultures of donors and recipients, clumps, or large aggregates, were visible to the naked eye when plasmid transfer frequencies were high, and (iii) preincubation of donor cells with cell-free recipient culture supernatants dramatically increased transfer frequencies upon the addition of recipient cells to the donor cells, and the conditioned medium from recipients also induced clumping of donor cultures whether or not recipient cells were present. Subsequent follow-up studies demonstrated that cell aggregation and efficient plasmid transfer were induced by the same clumping-inducing agent (CIA) peptide. The primary determinant of clumping is a large cell wall-anchored surface protein generically termed “aggregation substance” and is highly conserved among several different pheromone-responsive plasmids, as exemplified by the PrgB protein of pCF10 and the Asa1 protein of pAD1.
We now know that for pCF10 and several other plasmids, the genes encoding aggregation substances are a part of a long inducible operon composed of over 25 genes. These gene products collectively mediate donor/recipient attachment, formation of a mating channel between the attached cells, and processing of the plasmid DNA for transfer to the recipient via the channel (3).
REGULATION OF CONJUGATION
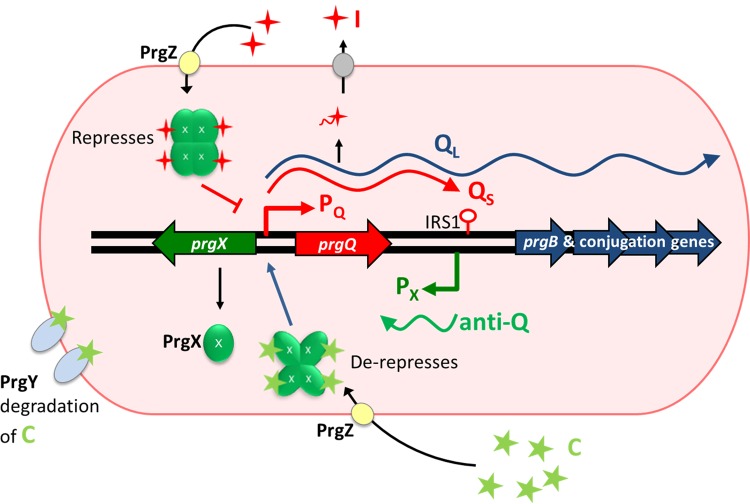
The intricate system controlling pCF10 conjugation involves secretion and signaling by two short peptides, cCF10 (LVTLVFV) and iCF10 (AITLIFI) (abbreviated C and I, respectively); each of these binds the cytoplasmic pheromone receptor PrgX and modulates the transcription of prgQ and the downstream genes (Fig. 2). Potential pCF10 recipients signal to pCF10-containing donor cells by releasing cCF10. In the absence of recipients, donor self-induction is prevented by pCF10-encoded iCF10 and reduction of donor-produced cCF10 by plasmid-encoded PrgY (3). The import of cCF10 is enhanced by PrgZ, a member of the OppA family (4, 5). It is interesting that PrgX, PrgY, and PrgZ, which all specifically bind cCF10, have independently evolved their cCF10 binding functions from distinct ancestor proteins (5).
FIG 2.
The pCF10 regulatory region. Induction and expression of the conjugation machinery from pCF10 are responsive to signaling by the peptide pheromone cCF10 (C; sequence, LVTLVFV) in association with PrgX tetramers which decrease repression the prgQ promoter. PrgX, in complex with iCF10 (I; sequence, AITLIFI) enhances PrgX repression of prgQ and reduces downstream conjugation machinery transcription. Antisense interactions between small RNAs (sRNAs; QS and anti-Q) from the 5′ ends of the prgQ and prgX operons mediate reciprocal negative-regulatory functions, amplifying the effects of peptides and PrgX in controlling prgQ transcription extension through the entire operon. See the text for more details.
As indicated in Fig. 2, PrgX is the master transcriptional regulator that interacts with the cCF10 and iCF10 peptide pheromones and that controls the expression of pCF10 conjugation genes by regulating the initiation of transcription from PQ, the promoter for the prgQ conjugation operon. It is well known that PrgX is a negative regulator of PQ and that cCF10 reduces PrgX repression, while iCF10 enhances repression. The current model for peptide-mediated modulation of PrgX activity posits that PrgX-peptide complexes, with either cCF10 or iCF10, exist as tetramers that bind the PQ region via two operator sites. Subtle structural differences between the peptide-PrgX-DNA complexes account for the increased ability of PrgX-I complexes to block RNA polymerase from binding to PQ, thereby reducing prgQ expression. Changes in the induction state of a donor cell result from replacement of one type of PrgX DNA-bound complex with the opposing type, which in turn is controlled by the relative concentrations of the two peptides in each donor cell. A detailed description of this model, along with recent experimental evidence supporting its refinement from earlier versions, was recently published by Chen et al. (6).
While the PrgX-peptide-related genetic switch comprises the core circuit controlling conjugation, there are multiple downstream posttranscriptional mechanisms that also operate to amplify the effects of pheromone induction and completely shut down the expression of most conjugation genes in uninduced cells. The most important of these mechanisms involves the control of transcription termination at IRS1, a stem-loop structure about 385 nucleotides (nt) downstream from the transcription start site of the prgQ operon (Fig. 2). Repression of the PQ promoter is never complete, and exponentially growing donor cells constitutively express a 385-nt “short Q” (QS) transcript that terminates at IRS1. Induced cells make increased levels of QS, as well as extended “long Q” (QL) transcripts that can reach the 3′ end of the operon. The decision to either terminate at IRS1 or extend transcription is modulated by antisense interactions between nascent prgQ transcripts and “anti-Q,” an opposing antisense RNA that corresponds to the first 102 nucleotides of prgX mRNA (Fig. 2) (2). Binding of anti-Q to nascent Q mRNA shifts the secondary structure of QS to a “terminator” conformation, whereas unbound QS assumes an “antiterminator” fold, reading through IRS1 and downstream genes. In uninduced cells, the levels of anti-Q are sufficient to bind all the nascent prgQ transcripts, such that cells exclusively produce QS. However, pheromone induction greatly increases QS production, which titrates the anti-Q pool and allows for increased levels of QL. We also note that prgQ, the first functional open reading frame (ORF) at the 5′ end of the operon fully contained within QS, actually encodes the iCF10 peptide. This provides a built-in mechanism for a timed shutdown of the induction response to exogenous cCF10, providing a limited time window in which the conjugation apparatus can be generated.
Basal QS expression in uninduced donor cultures leads to a cell density-dependent increase in iCF10 levels in the culture medium during growth. Since iCF10 inhibits the response to cCF10, donor cells at high density are poorly induced for conjugation even in the presence of large recipient populations. The iCF10 peptide thus functions as a self-sensing quorum signal to limit induction at high donor population densities and to mediate shutdown after induction. In contrast, cCF10 functions as a mate-sensing signal (7).
Taken together, these mechanisms function as a dose-dependent extremely tightly regulated expression system with potential applications for basic genetic studies and biotechnology. This is illustrated by a recent study in which toxin genes were successfully cloned under the control of the PQ pheromone-responsive promoter from pCF10 (8). In the absence of cCF10, the clones grew normally, but in response to exogenously added cCF10, cells exhibited dose-dependent growth inhibition. In previous work, these toxin genes could only be cloned in cells expressing their cognate antitoxin genes.
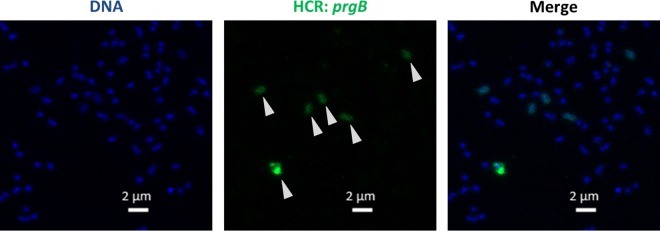
Initially, mathematical modeling suggested that the pCF10 regulatory system would behave as a bistable switch (9), but follow-up using a stochastic model predicted that there could be more variability in the induced response within individual cells ranging from uninduced, to slightly induced, to very induced. In order to resolve these discrepancies, it was necessary to develop experimental approaches to quantify the pheromone response at the single-cell level (10). The importance of single-cell analysis to fully understand the biology of the pheromone response is also suggested by previous work demonstrating that biofilm growth can impact plasmid copy number, plasmid-mediated antibiotic resistance, and the dynamics of the pheromone response (7, 11, 12). Our recent studies used a combination of protein reporters and fluorescence in situ hybridization chain reaction (HCR)-mediated transcript labeling to assess the response of individual donor cells to cCF10; we found that the response to induction was heterogeneous and most likely stochastically driven (10). The HCR technique (demonstrated in Fig. 3) is highly sensitive and may enable future interrogation of the pheromone response in complex environments, like the gastrointestinal tract.
FIG 3.
Induction of pCF10-carrying E. faecalis cells is heterogeneous, as observed using fluorescence in situ HCR. Expression of prgB from pCF10 can be observed in a subset of E. faecalis cells containing pCF10 exposed to 2.5 ng · ml−1 cCF10. Green, Alexa Fluor 488 HCR-labeled prgB; blue, Hoechst 33342 labeling cell DNA. Arrowheads indicate individual cells expressing different levels of prgB mRNA.
BENEFITS AND RISKS OF PHEROMONE INDUCTION
Beyond plasmid transfer (and spread of potentially beneficial genes carried by the plasmid, such as antibiotic resistance), there may be other fitness benefits to pheromone induction. Because pCF10 carries nearly 60 protein-coding genes, including uvr orthologs and genes for determinants for numerous secreted proteins, predicted enzymes, and transcription factors (13), it would not be surprising if carriage of the plasmid impacted the fitness of the host bacterium in the GI tract. For example, there is substantial evidence for a functional role of aggregation substance in the interaction of E. faecalis with its mammalian host. Surface expression of aggregation substance increases adherence to host tissues and also modulates the innate immune response to enterococcal infection, thus increasing virulence in multiple models of opportunistic infection (14–19). The formation of biofilms in vitro is also greatly enhanced by aggregation substance (20), and this adhesin could also promote intestinal colonization, as discussed below.
On the other hand, enhancement of plasmid transfer and host colonization by pheromone-induced expression of aggregation substance is not without serious costs to the donor cell. In liquid cultures, heavily aggregated cells show reduced growth, probably as a result of impaired diffusion of nutrients to cells enmeshed within large aggregates. Recent evidence has also suggested that in highly induced cultures of pCF10-containing donor cells, expression and secretion of the PrgB protein (and possibly other surface proteins) are associated with lysis and release of extracellular DNA (eDNA) by a subpopulation of the induced cells (20). This eDNA probably enhances aggregate formation, conjugative plasmid transfer, and biofilm development, providing a fitness benefit to the unlysed plasmid-carrying cells and enabling the spread of the plasmid to new hosts. Another pheromone-induced gene product, PrgU, a small predicted RNA binding protein, plays a critical role in modulating the pheromone response and limiting the toxic effects of prgB overexpression (21). Thus, the carriage of a conjugative pheromone-inducible plasmid by E. faecalis essentially forces the host cell to balance the potential fitness benefits of plasmid carriage and inducible surface adhesin expression, with the potential lethal effects of an overly vigorous response to pheromone. This situation likely contributed to the evolution of the complex, and extremely tightly controlled, regulatory system associated with pCF10 and other pheromone plasmids. To fully investigate the genetic determinants of pCF10 affecting fitness, an unbiased comprehensive genetic screen is a major goal of our future research.
PHYSIOLOGY AND ECOLOGY OF ENTEROCOCCI IN THE NATIVE NICHE: PATHOGEN VERSUS COMMENSAL
E. faecalis is a leading cause of nosocomial infections and endocarditis, and enterococcal endocarditis correlates with mortality rates of 11 to 22% (22, 23). Along with high levels of inherent antibiotic resistance of enterococci growing in biofilms, hospital-acquired infections by strains of vancomycin- and multidrug-resistant Enterococcus spp. are becoming especially problematic (24, 25). Antibiotic resistance spread in the opportunistic pathogen E. faecalis is particularly troubling due to its ability to rapidly transfer plasmids containing genes for antibiotic resistance to other E. faecalis cells via conjugative transfer of plasmids, like pCF10.
There are some “epidemic” strains of enterococci responsible for opportunistic nosocomial infections, for example, MMH594 (26), that may be better competitors in the GI tract against commensal strains. Epidemic strains could also thrive in the ecological environment of a gut that has been disrupted by antibiotic or cancer treatment. However, most enterococci are commensal and cause few issues for healthy individuals. Even in the case of epidemic patient-to-patient spread, opportunistic infections, such as bloodstream or wound infections, constitute an “evolutionary dead-end” for the causative strain. This is because patient-to-patient spread occurs via the fecal-oral route, and as a result, the primary determinant of fitness is the ability to colonize the GI tract of the hospital patient. Therefore, it is likely that most enterococci have evolved to have a commensal lifestyle, and competition for colonization and persistence in the gut is a primary driver of evolution. It is therefore of interest to determine whether pheromone signaling impacts both plasmid transfer and competitive fitness in the GI tract.
STUDY OF PHEROMONE SIGNALING IN THE NATIVE NICHE
Although in vitro study of pCF10 has revealed extensive details about the regulation and kinetics of plasmid transfer, detailed in vivo characterization of the plasmid's transfer dynamics and ecology in natural environments is more challenging. Most relevant animal studies have relied on antibiotic suppression of the resident microbiota to establish enterococcal strains of interest in sufficient numbers for analysis. Although these protocols might be considered an extreme disruption of the niche, they could resemble common clinical situations where overgrowth of enterococci in the intestinal tract is triggered by preceding antibiotic treatment (27). Plasmid transfer was reported at high frequency in animals in which it was detected (28, 29), indicating that transfer, possibly facilitated by pheromone signaling between donors and recipients, occurs in vivo. However, the nature of the animal models and the markers carried on the plasmids chosen in these studies did not allow investigators to distinguish between the role of the pheromone signaling in vivo and the potential selective advantages of plasmid genes unrelated to the pheromone response.
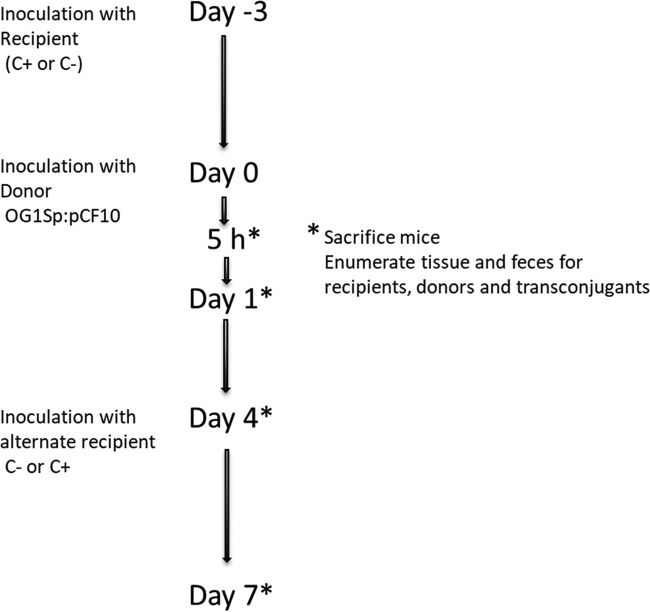
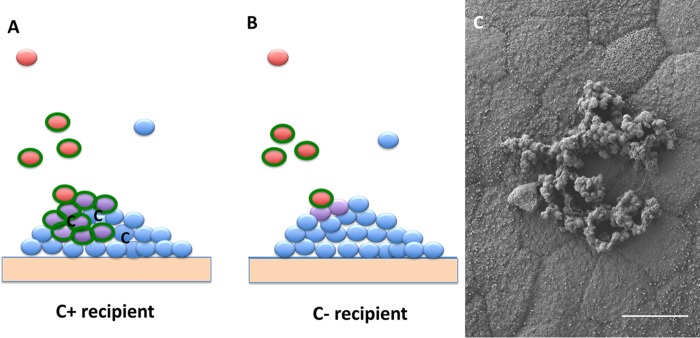
We recently used a germfree mouse model to begin elucidation of the role of cell-cell communication between donors and recipients in vivo (Hirt et al. [30]). The potential recipient strain was first established in the mice, followed by introduction of the donor strain (Fig. 4). To demonstrate the effect of the peptides in vivo, we compared the recipient ability of a plasmid-free wild-type strain (C+) to that of an isogenic C− recipient strain engineered by a single TA→CG base pair change in the chromosomal ccfA gene to produce a peptide (LATLVFV) closely related to cCF10 but lacking inducing activity for pCF10 (31). While plasmid transfer to this C− recipient did occur, frequencies were 100-fold lower than those observed with C+ recipients, suggesting a requirement of cCF10 for high-efficiency plasmid transfer. In spite of the reduced frequencies observed with the C− recipient, plasmid transfer occurred at modest levels within 24 h of donor inoculation. Previous studies (31, 32) demonstrated induction by one or more host factors (probably albumin-lipid complexes) in serum. These host factors appear to act by sequestering I, which leads to self-induction of donors by endogenously produced C. We hypothesize that the transfer into the C− recipient in the GI tract results from low-level induction of donors by a similar mechanism, which then allows donor and recipient interaction on recipient colonies on the intestinal wall (Fig. 5A and B). Microcolonies formed by C+ recipients contain C at high levels, and the plasmid can penetrate the colony. In a C− colony, plasmid transfer would be most likely restricted to the surface of the colony or very shallow penetration into the colony due to spatial restrictions and the lack of C production within the colony. This model is based in part on recent observations that plasmid-free E. faecalis can form abundant microcolonies on the mucosal surface throughout the GI tract when introduced into germfree mice (33) (also shown in Fig. 5C). To test the model, we are engineering a C− donor strain and will examine pCF10 transfer in vivo to a C− recipient; if the model is correct, we expect to see an extremely low level of transfer.
FIG 4.
Timeline for conjugation experiments in germfree mice colonized with E. faecalis donors and recipients. Mice were gavaged with 2.9 × 107 to 4.3 × 107 recipients and, after 3 to 4 days, with 2 × 108 to 4.4 × 108 donors. In experiments with C+ as the primary recipient, the C− recipient was introduced on day 4; alternatively, mice initially colonized with C− recipients were later inoculated with C+ recipients on day 4. Mice were sacrificed at various time points ranging from 5 h to 7 days after donor inoculation, as depicted. For experiments with a competing microbiota, recipients were inoculated at day −4 before donor inoculation, followed by the competing microbiota on day −3 (30).
FIG 5.
Model for in vivo plasmid transfer of E. faecalis plasmid pCF10. (A) The wild-type recipient (blue) is established as a microcolony on the intestinal wall. C is produced in the colony but not in sufficient quantity to induce donors in the lumen. Incoming donor cells (red) can be induced at low levels by a host factor to express AS (green) allowing cell-cell contact with the microcolony. Plasmid transfer occurs, establishing transconjugants (purple) that can spread the plasmid through the colony due to the presence of high concentrations of C. (B) C− recipients (blue) establish microcolonies on the intestinal wall, similar to what is shown in panel A, and donors are induced by a host factor and bind to the colony. Limited plasmid transfer occurs but cannot proceed into the core of the colony due to the lack of C production by the recipients in the microcolony. (C) Low-voltage field-emission scanning electron microscopy (FE-SEM) micrograph illustrating E. faecalis strain OG1RF colonization and microcolony formation in the proximal colon of a germfree Swiss-Webster mouse (scale bar = 5 μm). SEM provided by Aaron Barnes; see the work of Barnes et al. (33) for further description of microcolony formation in the intestinal tract.
Another significant finding relevant to the model shown in Fig. 5 was that the presence of a coresident C+ strain in the GI tract did not significantly increase pCF10 transfer into a coresident C− recipient in vivo, even though extracellular rescue of the recipient defect of C− strains by a coresident C+ recipient is readily observed in vitro. The failure of C+ recipients to restore high levels of transfer to coresident C− recipients by extracellular complementation suggests that, even in mice colonized by C+ recipients, recipient-produced pheromone is not readily available in the lumen of the intestine at sufficient concentrations to impact transfer frequencies.
We detected plasmid transfer 5 h after donor inoculation in the intestinal tract, before either donor or transconjugants could be isolated from feces (30). This demonstrates that the intestinal tract is the actual location for plasmid transfer, ruling out mechanisms involving transfer in expelled feces followed by coprophagic reingestion.
We increased the complexity of the system by introducing a competing defined community of five common intestinal microbes. Plasmid transfer was modestly reduced in the lower intestinal tract where the members of the introduced community reach the highest density. However, transfer in other regions of the GI tract was not significantly reduced. In summary, the early results of the germfree mouse model established the importance of cCF10 for high-efficiency transfer and pointed to the necessity for close contact between donor and recipient for pheromone induction of plasmid transfer.
Our initial studies of pCF10 transfer in vivo also revealed a contribution of the plasmid to competitive fitness in the GI tract that was not related to antibiotic selection or plasmid-encoded bacteriocin production. This effect was not detected in previous suppressed microbiota models (28, 29). A strong candidate for the factor increasing fitness is aggregation substance (AS), which could offer donors protection from immune system responses (18), provide the ability to attach to host structures (34), or allow the colonization of already-established recipient microcolonies, as described above. AS may be the major factor on the plasmid for increased fitness, but other factors cannot be excluded. The impact of AS on plasmid transfer in the germfree mouse model will be assessed in future experiments.
Ultimately, direct visualization of pheromone induction and plasmid transfer in vivo will be required to validate the model discussed above (Fig. 5). For this purpose, we have developed a variety of fluorescently labeled strains that allow us to distinguish between recipients, donors, and transconjugants. Although plasmid transfer is highly efficient, detection of the event in vivo might be challenging, with only 1 in 1,000 recipients receiving the plasmid. On the other hand, if our model is correct, transconjugants should be concentrated on the outer edges of adherent microcolonies, together with donors. Thus, high-resolution imaging may also allow identification of colonies that were initiated by recipients that could possibly benefit from harboring pCF10 and therefore expand their ecological niche. Alternatively, labeling of transcripts in vivo with fluorescence in situ HCR will allow expansion of the investigation of pCF10 gene expression under in vivo conditions (10) and allow more quantitative analysis of gene expression at the single-cell level (Fig. 3).
CONCLUSION
The in vivo biology and ecology of the microbial inhabitants of the GI tract are complex, and very little is known about the maintenance of commensal microbes that can also become opportunistic pathogens. Enterococcus faecalis is one such inhabitant of this niche, and exploration of the biology of pCF10 maintenance and spread in this environment has just begun. A better understanding of the fitness benefits conferred by pheromone plasmids in vivo may facilitate the development of novel approaches to control antibiotic-resistant hospital infections.
ACKNOWLEDGMENTS
We acknowledge Aaron Barnes for provision of the SEM micrograph in Fig. 5C.
This work, was supported by HHS National Institutes of Health (NIH) grants AI122742 and GM118079 to G.M.D. R.J.B. was supported in part by a predoctoral traineeship under T32-GM008347.
REFERENCES
- 1.Clewell DB, Dunny GM. 2002. Conjugation and genetic exchange in enterococci, p 265–300. In Gilmore M, Clewell D, Courvalin P, Dunny G, Murray B, Rice L (ed), The enterococci. ASM Press, Washington, DC. [Google Scholar]
- 2.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A 75:3479–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunny GM. 2013. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47:457–482. doi: 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- 4.Leonard BA, Podbielski A, Hedberg PJ, Dunny GM. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci U S A 93:260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunny GM, Berntsson RPA. 2016. Enterococcal sex pheromones: evolutionary pathways to complex, two-signal systems. J Bacteriol 198:1556–1562. doi: 10.1128/JB.00128-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Bandyopadhyay A, Kozlowicz BK, Haemig HAH, Tai A, Hu W-S, Dunny GM. 2017. Mechanisms of peptide sex pheromone regulation of conjugation in Enterococcus faecalis. Microbiologyopen 6:e492. doi: 10.1002/mbo3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee A, Cook LCC, Shu C-C, Chen Y, Manias DA, Ramkrishna D, Dunny GM, Hu W-S. 2013. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc Natl Acad Sci U S A 110:7086–7090. doi: 10.1073/pnas.1212256110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver KE, Chen Y, Miiller EM, Johnson JN, Dangler AA, Manias DA, Clem AM, Schjodt DJ, Dunny GM. 2017. Examination of Enterococcus faecalis toxin-antitoxin system toxin Fst function utilizing a pheromone-inducible expression vector with tight repression and broad dynamic range. J Bacteriol 199:e00065-17. doi: 10.1128/JB.00065-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee A, Johnson CM, Shu C-C, Kaznessis YN, Ramkrishna D, Dunny GM, Hu W-S. 2011. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc Natl Acad Sci U S A 108:9721–9726. doi: 10.1073/pnas.1101569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breuer RJ, Bandyopadhyay A, O'Brien SA, Barnes AMT, Hunter RC, Hu W-S, Dunny GM. 2017. Stochasticity in the enterococcal sex pheromone response revealed by quantitative analysis of transcription in single cells. PLoS Genet 13:e1006878. doi: 10.1371/journal.pgen.1006878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook LC, Dunny GM. 2013. Effects of biofilm growth on plasmid copy number and expression of antibiotic resistance genes in Enterococcus faecalis. Antimicrob Agents Chemother 57:1850–1856. doi: 10.1128/AAC.02010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook L, Chatterjee A, Barnes A, Yarwood J, Hu W-S, Dunny G. 2011. Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol Microbiol 81:1499–1510. doi: 10.1111/j.1365-2958.2011.07786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirt H, Manias DA, Bryan EM, Marklund JK, Staddon JH, Paustian ML, Kapur V, Dunny GM. 2005. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol 187:1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, Clewell DB, Zervos MJ. 1993. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother 37:2474–2477. doi: 10.1128/AAC.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlievert PM, Gahr PJ, Assimacopoulos AP, Dinges MM, Stoehr JA, Harmala JW, Hirt H, Dunny GM. 1998. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect Immun 66:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlievert PM, Chuang-Smith ON, Peterson ML, Cook LCC, Dunny GM. 2010. Enterococcus faecalis endocarditis severity in rabbits is reduced by IfG Fabs interfering with aggregation substance. PLoS One 5:e13194. doi: 10.1371/journal.pone.0013194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCormick JK, Hirt H, Dunny GM, Schlievert PM. 2000. Pathogenic mechanisms of enterococcal endocarditis. Curr Infect Dis Rep 2:315–321. doi: 10.1007/s11908-000-0009-9. [DOI] [PubMed] [Google Scholar]
- 18.Rakita RM, Vanek NN, Jacques-Palaz K, Mee M, Mariscalco MM, Dunny GM, Snuggs M, Van Winkle WB, Simon SI. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun 67:6067–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang ON, Schlievert PM, Wells CL, Manias DA, Tripp TJ, Dunny GM. 2009. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect Immun 77:539–548. doi: 10.1128/IAI.01034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatty M, Cruz MR, Frank KL, Laverde Gomez JA, Andrade F, Garsin DA, Dunny GM, Kaplan HB, Christie PJ. 2015. Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance), and PrgC contribute to plasmid transfer, biofilm formation, and virulence. Mol Microbiol 95:660–677. doi: 10.1111/mmi.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatty M, Camacho MI, Gonzalez-Rivera C, Frank KL, Dale JL, Manias DA, Dunny GM, Christie PJ. 2016. PrgU: a suppressor of sex pheromone toxicity in Enterococcus faecalis. Mol Microbiol 103:398–412. doi: 10.1111/mmi.13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards MJ, Edwards JR, Culver DH, Gaynes RP. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect Control Hosp Epidemiol 21:510–515. doi: 10.1086/501795. [DOI] [PubMed] [Google Scholar]
- 23.McDonald JR, Olaison L, Anderson DJ, Hoen B, Miro JM, Eykyn S, Abrutyn E, Fowler VG, Habib G, Selton-Suty C, Pappas PA, Cabell CH, Corey GR, Marco F, Sexton DJ. 2005. Enterococcal endocarditis: 107 cases from the international collaboration on endocarditis merged database. Am J Med 118:759–766. doi: 10.1016/j.amjmed.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 24.Miller W, Munita JM, Arias CA. 2014. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 12:1221–1236. doi: 10.1586/14787210.2014.956092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibrahim GM, Sundaramoorthy S, Madhavan R. 2015. Enterococcus: an emerging global superbug. Int J Sci Res 4:1539–1543. [Google Scholar]
- 26.Huycke MM, Spiegel CA, Gilmore MS. 1991. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 35:1626–1634. doi: 10.1128/AAC.35.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakticová V, Hutton-Thomas R, Meyer M, Gurkan E, Rice LB. 2006. Antibiotic-induced enterococcal expansion in the mouse intestine occurs throughout the small bowel and correlates poorly with suppression of competing flora. Antimicrob Agents Chemother 50:3117–3123. doi: 10.1128/AAC.00125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licht TR, Laugesen D, Jensen LB, Jacobsen BL. 2002. Transfer of the pheromone-inducible plasmid pCF10 among Enterococcus faecalis microorganisms colonizing the intestine of mini-pigs. Appl Environ Microbiol 68:187–193. doi: 10.1128/AEM.68.1.187-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huycke MM, Gilmore MS, Jett BD, Booth JL. 1992. Transfer of pheromone-inducible plasmids between Enterococcus faecalis in the Syrian hamster gastrointestinal tract. J Infect Dis 166:1188–1191. doi: 10.1093/infdis/166.5.1188. [DOI] [PubMed] [Google Scholar]
- 30.Hirt H, Greenwood Quaintance KE, Karau MJ, Till LM, Kashyap PC, Patel R, Dunny GM. Enterococcus faecalis sex pheromone cCF10 enhances conjugative plasmid transfer in vivo. mBio 9:e00037-18. doi: 10.1128/mBio.00037-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandler JR, Hirt H, Dunny GM. 2005. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci U S A 102:15617–15622. doi: 10.1073/pnas.0505545102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirt H, Schlievert PM, Dunny GM. 2002. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect Immun 70:716–723. doi: 10.1128/IAI.70.2.716-723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnes AMT, Dale JL, Chen Y, Manias DA, Greenwood Quaintance KE, Karau MK, Kashyap PC, Patel R, Wells CL, Dunny GM. 2017. Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model. Virulence 8:282–296. doi: 10.1080/21505594.2016.1208890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chuang-Smith ON, Wells CL, Henry-Stanley MJ, Dunny GM. 2010. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLoS One 5:e15798. doi: 10.1371/journal.pone.0015798. [DOI] [PMC free article] [PubMed] [Google Scholar]