ABSTRACT
Streptococcus gallolyticus subsp. gallolyticus, a member of the group D streptococci, is normally found in the bovine rumen and human gut. It is an opportunistic pathogen that was recently determined to be a bacterial driver of colorectal cancer, in addition to causing other diseases, such as infective endocarditis, bacteremia, neonatal meningitis, and septicemia. As an emerging pathogen, not much is known about this bacterium, its virulence mechanisms, or its virulence regulatory pathways. Previous studies suggest that S. gallolyticus subsp. gallolyticus uses a ComRS pathway, one of many Streptococcus quorum-sensing circuitries, for competence. However, thus far, the ubiquitous ComABCDE pathway has not been studied, nor has its regulatory role in S. gallolyticus subsp. gallolyticus. We therefore sought to study the S. gallolyticus subsp. gallolyticus ComABCDE quorum-sensing pathway and have identified its peptide pheromone, which is termed the competence-stimulating peptide (CSP). We further determined that this peptide regulates the production of bacteriocin-like inhibitory substances (BLISs), a phenotype that has been linked with the ComABCDE pathway in both Streptococcus pneumoniae and Streptococcus mutans. Our data show that S. gallolyticus subsp. gallolyticus TX20005 produces a 21-mer CSP signal, which differs from CSP signals of other Streptococcus species in that its active form begins three residues after the double-glycine leader signal of the ComC precursor peptide. Additionally, our data suggest that this peptide might not be related to competence induction, as opposed to CSP signaling peptides in other Streptococcus species. This study provides the first evidence that S. gallolyticus subsp. gallolyticus utilizes quorum sensing to eliminate competitors, presenting a potential pathway to target this emerging human pathogen.
IMPORTANCE Streptococcus gallolyticus subsp. gallolyticus is an emerging human pathogen known as a causative agent of infective endocarditis, and recently, of colorectal cancer. In this work, we revealed a functional quorum-sensing circuitry in S. gallolyticus subsp. gallolyticus, including the identification of the central signaling peptide pheromone, competence-stimulating peptide (CSP), and the regulatory role of this circuitry in the production of bacteriocin-like inhibitory substances (BLISs). This work uncovered a mechanism by which this bacterium outcompetes other bacterial species and thus provides a potential tool to study this opportunistic pathogen.
KEYWORDS: S. gallolyticus, competence-stimulating peptide, quorum sensing
INTRODUCTION
Streptococcus gallolyticus subsp. gallolyticus, a member of the group D streptococci belonging to the bovis group, was originally classified as Streptococcus bovis biotype I, until it was later changed to Streptococcus gallolyticus, which consists of three subspecies, S. gallolyticus subsp. gallolyticus (biotype I), S. gallolyticus subsp. pasteurianus (biotype II.2), and S. gallolyticus subsp. macedonicus (1–4). Since the late 1970s, a strong association has been established between patients suffering from bacteremia or endocarditis caused by S. gallolyticus subsp. gallolyticus and colorectal cancer (CRC) (5–11). Recently, S. gallolyticus subsp. gallolyticus has been shown to promote colon cancer cell proliferation and tumor growth, establishing that S. gallolyticus subsp. gallolyticus is a bacterial driver of CRC; however, the exact mechanism remains unknown (12, 13).
The competence quorum-sensing (QS) regulon was first discovered in Streptococcus pneumoniae, where it was shown that a signaling peptide, termed competence-stimulating peptide (CSP), induces S. pneumoniae to become competent (14–16). This QS circuitry consists of five components, collectively termed ComABCDE, and is centered on the CSP signaling peptide. The CSP signaling peptide is derived from its precursor peptide, ComC, and contains an N-terminal double glycine leader signal sequence that gets cleaved by the ABC transporter, ComAB, prior to being excreted into the environment (17). Upon reaching a threshold concentration, the CSP activates a two-component signal-transduction system (TCSTS) involving a histidine kinase (ComD) and a DNA-binding transcriptional response regulator (ComE). ComD activation leads to phosphorylation of ComE, resulting in the expression of QS genes (comABCDE) and the alternative sigma factor, SigX/ComX, a master regulator of competence genes (Fig. 1) (17, 18). This competence circuitry (ComABCDE) has been shown to be conserved in 12 Streptococcus species that are naturally transformable, with variations in the CSP sequence between species and, in a few cases, within different strains of the same species (pherotypes) (16, 19–21). The competence regulon in S. pneumoniae and Streptococcus mutans was also found to be linked to bacteriocin production (17). A different QS circuit, ComRS, was discovered in two species of the salivarius group, Streptococcus thermophilus and Streptococcus salivarius (22). In these species, a propeptide pheromone, ComS, gets secreted into the environment and cleaved either during or after its secretion to produce a 7-amino-acid signaling peptide, termed sigma inducing peptide (XIP) (23). XIP is imported into the intracellular environment, where it binds to a transcriptional regulator, ComR. The binding of XIP to ComR then upregulates the expression of ComX, leading to induction of competence. This pathway was found to exist in some species of the mutans, pyogenic, and bovis groups, suggesting that this alternative QS circuitry is also conserved among streptococci (24).
FIG 1.
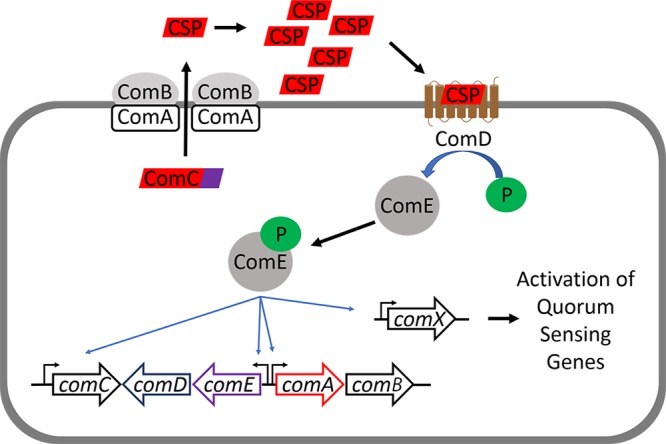
Streptococcus ComABCDE QS pathway. ComC is processed intracellularly and secreted by ComAB as the mature CSP. Upon reaching a threshold concentration, CSP binds and activates its cognate receptor, ComD. Activation of ComD leads to ComE phosphorylation, which results in the autoinduction of the QS circuitry (ComABCDE) and the expression of comX, the master regulator of QS-dependent phenotypes.
The genomes of many bovis group species, including those of a few strains of S. gallolyticus subsp. gallolyticus, are publicly available, and analysis of the S. gallolyticus subsp. gallolyticus genome suggests that it contains both comABCDE and comRS operons. Sequencing of the S. gallolyticus subsp. gallolyticus genome showed that it contains a comABCDE operon; however, the comC gene was previously claimed to be an inactive allele, with no further justification provided on why the authors concluded that this gene is inactive (25). Nonetheless, no further studies were conducted to confirm the presence of these operons or identify their functions (26–28). It was suggested through genomic analysis that the bovis group uses the ComRS circuitry for competence. This hypothesis was later confirmed experimentally in two species, Streptococcus infantarius and Streptococcus macedonicus (25). Additionally, it was shown that the recombinant Strep II-tagged ComR of S. gallolyticus subsp. gallolyticus strain UNC34 can bind DNA probes containing the promoter regions of both comS and comX in the presence of its cognate XIP, suggesting that S. gallolyticus subsp. gallolyticus probably uses the ComRS pathway for competence (24). However, no studies to date were able to demonstrate competence in S. gallolyticus subsp. gallolyticus. The purpose of this work was to study the ComABCDE circuitry in S. gallolyticus subsp. gallolyticus by predicting the CSP sequence, isolating the CSP, and determining its regulatory role. Our results show that S. gallolyticus subsp. gallolyticus uses a 21-mer CSP that is involved in QS-dependent production of bacteriocin-like inhibitory substances (BLISs).
RESULTS AND DISCUSSION
Multiple sequence alignments of ComC sequences.
A prediction of the S. gallolyticus subsp. gallolyticus CSP was made by performing a multiple-sequence alignment (MSA) of ComC sequences of several S. gallolyticus subsp. gallolyticus strains, using the ComC protein sequences of S. pneumoniae (strains D39 and TIGR4) and Streptococcus intermedius strain NCDO 2227 as reference sequences. The MSA predicted S. gallolyticus subsp. gallolyticus to have a 24-mer CSP containing a positively charged, N-terminal amino acid that differs from the negatively charged, N-terminal amino acids of the reference sequences (see Fig. S1 in the supplemental material). After removing the double glycine leader signal from the original MSA, we performed another alignment to look for any similarities between the predicted S. gallolyticus subsp. gallolyticus CSP and the reference CSP sequences. This analysis indicated that the fourth residue of the predicted S. gallolyticus subsp. gallolyticus CSP is likely the first residue of the mature CSP, as it follows the trend in the reference CSPs of having a negatively charged, N-terminal amino acid (Fig. 2). We therefore hypothesized that S. gallolyticus subsp. gallolyticus produces and utilizes a 21-mer CSP. Lastly, we noticed that the S. gallolyticus subsp. gallolyticus strain DSM 16831 sequence differs from those of the other strains in eight positions and is one amino acid longer (22 mer), suggesting the presence of pherotypes in this species, similarly to those of S. pneumoniae (16, 29). To support our hypothesis, we designed primers to amplify the intergenic region in front of the comC gene and the 3′ coding strand region of the comD gene from S. gallolyticus subsp. gallolyticus TX20005. We PCR amplified and sequenced the comC gene, which matched with the results of the uploaded genome of S. gallolyticus subsp. gallolyticus TX20005 (GL397173.1). Furthermore, the translated sequence matched the ComC sequences of S. gallolyticus subsp. gallolyticus TX20005 (EFM28801.1) and other S. gallolyticus subsp. gallolyticus strains (Fig. 2).
FIG 2.
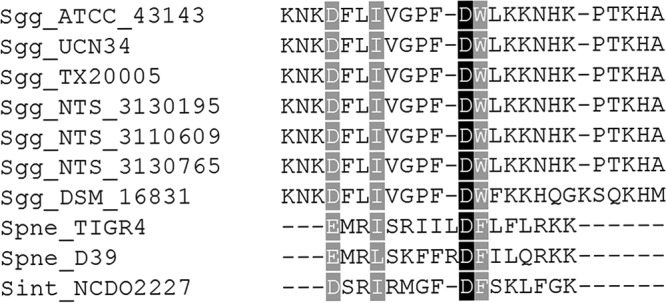
Multiple-sequence alignment of ComC. The ComC sequences of several S. gallolyticus subsp. gallolyticus strains, with double-glycine leader signal removed, were aligned. The ComC sequences of S. pneumoniae D39, S. pneumoniae TIGR4, and S. intermedius NCDO2227 were used as reference sequences.
Isolation of predicted CSP peptide from spent, cell-free supernatants.
Our next step in identifying the S. gallolyticus subsp. gallolyticus CSP was to isolate it from bacterial supernatants. Following precipitation of the excreted crude peptide mixture from cell-free supernatants of S. gallolyticus subsp. gallolyticus TX20005, we purified various peptides/proteins using reversed-phase high-performance liquid chromatography (RP-HPLC). We observed an intense peak eluting between 29.5 to 30.5 min that contained a mass of 2,492.8 Da, as determined by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Fig. 3). This mass corresponds to the predicted 21-mer CSP sequence (mass without the first three N-terminal residues, 2,491.9 Da). This analysis provided the first support for our hypothesis that the mature CSP in S. gallolyticus subsp. gallolyticus is a 21-mer peptide bearing a negatively charged residue at the N terminus. Overall, we were able to recover approximately 2.5 mg of this peptide from 50 ml of overnight-grown S. gallolyticus subsp. gallolyticus spent supernatants, corresponding to a 20 μM concentration.
FIG 3.
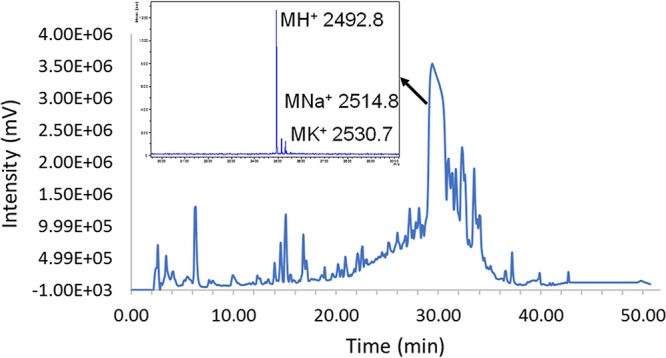
Isolation and identification of S. gallolyticus subsp. gallolyticus CSP. RP-HPLC chromatogram trace of crude protein extract from S. gallolyticus subsp. gallolyticus cell-free supernatants and MALDI-TOF MS of a fraction collected between 29.5 to 30.5 min (major peak).
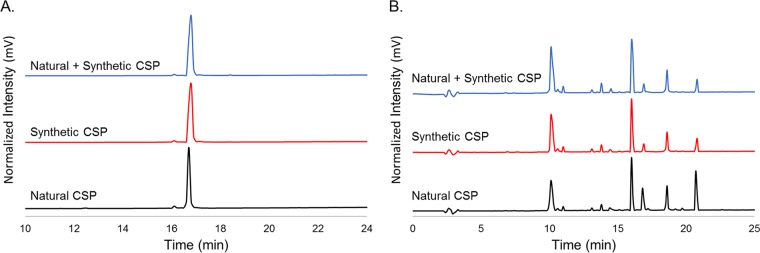
Chemical synthesis of predicted CSP and comparison to the naturally isolated CSP.
We decided to synthesize the predicted CSP sequence using microwave-assisted 9-fluorenyl methoxycarbonyl (Fmoc)-based solid-phase peptide synthesis and obtained 2 mg of purified peptide (3.8% yield of purified peptide, >95% purity). We compared the synthetic peptide to the peptide isolated from cell-free supernatants to confirm the identity of the S. gallolyticus subsp. gallolyticus CSP. HPLC analysis of both peptides revealed a single peak with the same retention time (Fig. 4A; see also Fig. S2 and S3 in the supplemental material). Furthermore, a combined fraction of both the synthetic and isolated peptides resulted in a single peak in the HPLC with the same retention time (Fig. 4A; see also Fig. S4 in the supplemental material). In addition, positive electrospray ionization (ESI+) high-resolution mass spectrometry (HRMS) of both peptides exhibited similar ionization trends (+2, +3, and +4 charged states; all exact masses [EM] within 5 ppm error) (see Fig. S8 and S9 in the supplemental material). We then submitted both the naturally isolated and synthetic peptides to the Nevada Proteomics Center for tandem MS analysis. This analysis confirmed that the amino acid sequence is DFLIVGPFDWLKKNHKPTKHA; however, uncertainty remained in regard to the order of the first two amino acids (see Fig. S10 to S13 in the supplemental material). Thus, we treated both samples with chymotrypsin, a protease that selectively cleaves the amide bonds following an aromatic residue (phenylalanine, tyrosine, or tryptophan), with the aim of confirming the order of the first two residues by obtaining either an LIVGPF fragment (DF) or a DLIVGPF fragment (FD). After treating both naturally isolated and synthetic S. gallolyticus subsp. gallolyticus CSPs with chymotrypsin, we observed similar chromatograms by analytical RP-HPLC analysis (Fig. 4B; see also Fig. S14 and S15 in the supplemental material), and we observed a mass of 645.39 Da using ESI+ HRMS for both peptides, corresponding to an amino acid sequence of LIVGPF. Together, these results confirmed that the CSP sequence of S. gallolyticus subsp. gallolyticus is DFLIVGPFDWLKKNHKPTKHA. Lastly, we performed structural analysis of both peptides using circular dichroism and found that S. gallolyticus subsp. gallolyticus CSP has a weak α-helical propensity in membrane-mimicking conditions (approximately 10% helicity) (see Fig. S16 and S17 in the supplemental material) (30).
FIG 4.
Comparison of synthetic and isolated CSPs. (A) Overlay with offsets of analytical RP-HPLC chromatograms of purified natural, synthetic, and natural and synthetic CSP. (B) Overlay with offset of analytical RP-HPLC chromatograms of chymotrypsin digestion of natural, synthetic, and natural and synthetic CSP.
Phenotypic assays.
After confirming the CSP sequence in S. gallolyticus subsp. gallolyticus, we set out to determine its regulatory role. Since these peptides are termed competence-stimulating peptides and have been shown to induce competence in several Streptococcus species (14, 21), we first attempted to induce competence in S. gallolyticus subsp. gallolyticus using the synthetic CSP and pALH122, a plasmid that contains an erythromycin-resistance gene. All our attempts to induce competence were unsuccessful. We tried plating the mixtures grown in Todd-Hewitt agar supplemented with 0.5% yeast extract (THY) containing CSP and plasmid, using various concentrations of synthetic CSP (100 nM, 500 nM, and 10 μM) at different time points (30 min, 1 h, 2 h, 3 h, 4 h, and 16 h) with no transformants. We decided to try the transformation of S. gallolyticus subsp. gallolyticus using a chemically defined medium to help induce competence with the synthetic CSP, as was shown for S. mutans (31–33). We repeated the experiment, growing S. gallolyticus subsp. gallolyticus in a chemically defined medium, M9 supplemented with 1% glucose and 1% biotin, prior to plating, and again we observed no transformants.
After our attempts to induce competence failed, we turned to assessing biofilm formation, as this phenotype was also correlated to the competence regulon in S. pneumoniae, S. intermedius, S. gordonii, and S. mutans (34–38). To this end, we performed a crystal violet biofilm quantification assay. Our results show no significant difference in biofilm formation with various concentrations of CSP (10 μM, 1 μM, and 0.1 μM) compared to our negative control (dimethyl sulfoxide [DMSO]) (data not shown). These results do not necessarily rule out any role of this circuitry in regulating these phenotypes; rather, they indicate that for the conditions we tested, this QS circuitry failed to play a significant role.
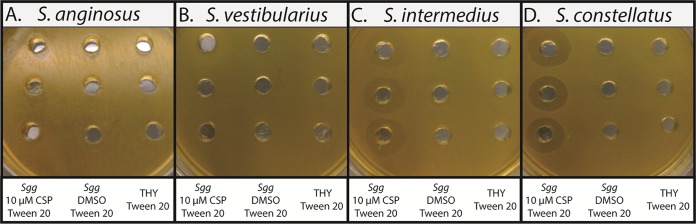
We next tested the role of the competence regulon in bacteriocin production. To this end, we performed an interspecies inhibition assay, where we tested the ability of S. gallolyticus subsp. gallolyticus to inhibit the growth of several Streptococcus species, namely, Streptococcus intermedius F0413, Streptococcus vestibularius F0396, Streptococcus mutans ATCC 25175, Streptococcus anginosus ATCC 33397, Streptococcus constellatus ATCC 27823 and Streptococcus agalactiae MNZ938, following treatment of S. gallolyticus subsp. gallolyticus with either CSP (10 μM or 100 nM) or DMSO (negative control). Our results revealed similar CSP-mediated inhibition for both concentrations of CSP against S. intermedius F0413, S. anginosus ATCC 33397, S. constellatus ATCC 27823, and S. vestibularius F0396, whereas neither self-inhibition nor inhibition against S. agalactiae MNZ938 or S. mutans ATCC 25175 was observed (see Fig. S18 in the supplemental material). Moreover, treatment with only S. gallolyticus subsp. gallolyticus CSP (no bacteria) did not induce any growth inhibition against any of the species, indicating that S. gallolyticus subsp. gallolyticus CSP is not toxic to the bacteria tested and that the CSP-mediated inhibition was due to a regulatory role it possesses. To verify the removal of all S. gallolyticus subsp. gallolyticus cells, we attempted the experiment again, using filtered supernatants, and observed no inhibition. We performed another experiment, where we grew S. gallolyticus subsp. gallolyticus without CSP and only added it to the centrifuged supernatants prior to placing the supernatants in the agar wells; in this case, we observed inhibition zones. Lastly, we tested the centrifuged supernatants for the presence of S. gallolyticus subsp. gallolyticus cells and found that the supernatants contained a few bacterial cells. Together, these experiments confirm that the observed inhibition is CSP-dependent. Moreover, the results indicate that either the inhibitory agent (presumably bacteriocin) is cell associated or that the observed inhibition is mediated by a different mechanism requiring direct cell contact. In a previous study, Mantovani et al. showed that a Streptococcus bovis bacteriocin is cell associated and requires detergents to be released from the cell surface (39). We therefore repeated the interspecies inhibition assay against the four species that were found to be inhibited by S. gallolyticus subsp. gallolyticus (S. intermedius F0413, S. anginosus ATCC 33397, S. constellatus ATCC 27823, and S. vestibularius F0396), using only one CSP concentration (10 μM), but this time, following centrifugation, we washed the overnight cells with phosphate-buffered saline (PBS) containing Tween 20 to remove any cell-associated bacteriocin-like inhibitory substances (BLISs) (40), prior to treatment with either CSP or DMSO. Following the 4 h of incubation, we added Tween 20 prior to sterile-filtration of the supernatants and addition to the agar plates containing the Streptococcus species. Interestingly, CSP-dependent inhibition was observed against only two species (S. intermedius F0413 and S. constellatus ATCC 27823), while the growth of the other two species (S. anginosus ATCC 33397 and S. vestibularius F0396) was not affected (Fig. 5). We concluded that S. gallolyticus subsp. gallolyticus utilizes two distinct mechanisms to outcompete other Streptococcus species and that both mechanisms are CSP-dependent; S. intermedius F0413 and S. constellatus ATCC 27823 are inhibited through a secreted BLIS, while S. anginosus ATCC 33397 and S. vestibularius F0396 are inhibited through an unknown mechanism that involves direct cell contact. Finally, to further characterize the BLIS, we washed CSP-treated cells with PBS, added 0.1% Tween 20, filter-sterilized the solution, and treated with ammonium sulfate to precipitate peptide and protein contents. Then, following resuspension of the precipitate, we observed interspecies inhibition, suggesting that the BLIS is peptide/protein based. We isolated the BLIS using RP-HPLC (see Fig. S19 in the supplemental material) and found that only one fraction exhibited interspecies inhibition against S. intermedius F0413 and S. constellatus ATCC 27823, suggesting that one BLIS is responsible for the observed inhibition.
FIG 5.
Images comparing results from S. gallolyticus subsp. gallolyticus interspecies inhibition assay, where S. gallolyticus subsp. gallolyticus is incubated with CSP (10 μM) or DMSO, followed by addition of Tween 20, sterile filtration, and testing of the supernatants against S. anginosus ATCC 33397 (A), S. vestibularius F0396 (B), S. intermedius F0413 (C), and S. constellatus ATCC 27823 (D). Inhibition was observed in panels C and D but not in panels A and B.
There is a continuous debate regarding the role of the ComABCDE circuitry in both competence and bacteriocin production. Our results provide supporting evidence that the ComABCDE pathway is likely related to the production of a BLIS in S. gallolyticus subsp. gallolyticus. Our data suggest that this BLIS is a bacteriocin; however, additional studies are needed to determine the identity of the secreted agent/s that are responsible for the CSP-mediated growth inhibition. These studies are ongoing in our lab and will be reported in due course.
In summary, we demonstrated that S. gallolyticus subsp. gallolyticus has a functional comC gene that produces a 21-mer CSP. Furthermore, our results indicate that the CSP precursor peptide is unique compared to other known Streptococcus species CSP precursor peptides, in that its active sequence begins three residues after the double glycine leader signal, a phenomenon that has never been observed in other Streptococcus species (16). These findings suggest that either the ComAB differs in S. gallolyticus subsp. gallolyticus compared to those of other streptococci or that S. gallolyticus subsp. gallolyticus possesses an extracellular protease, such as SepM, which was found to process the C termini of CSPs in other streptococci prior to binding to the ComD receptor (41). Additionally, our bioinformatics analysis suggests that S. gallolyticus subsp. gallolyticus strains might be divided into two pherotypes with distinct CSP sequences. Finally, we found that S. gallolyticus subsp. gallolyticus is capable of inhibiting the growth of other Streptococcus species through two distinct CSP-dependent mechanisms: (i) the production of a BLIS and (ii) direct cell contact. This observation could explain the ability of S. gallolyticus subsp. gallolyticus to outcompete other bacteria and occupy particular niches related to infective endocarditis and/or CRC.
MATERIALS AND METHODS
All chemical reagents and solvents were purchased from Sigma-Aldrich or Chem-Impex and used without further purification. Double-distilled water (ddH2O) was purified using a Millipore Analyzer feed system. Solid-phase 2-(4-(chloromethyl)phenoxy)acetamide (Cl-MPA) ProTide resin was purchased from CEM Corporation. 9-Fluorenyl methoxycarbonyl (Fmoc)-protected l-α-amino acids were purchased from Advanced ChemTech.
Reversed-phase high-performance liquid chromatography (RP-HPLC) was performed using a Shimadzu ultra fast liquid chromatography (UFLC) system equipped with a CBM-20A communications bus module, two LC-20AT pumps, an SIL-20A autosampler, an SPD-20A UV-visible (UV-Vis) detector, a CTO-20A column oven, and an FRC-10A fraction collector. All RP-HPLC solvents (ddH2O and HPLC-grade acetonitrile [ACN]) contained 0.1% trifluoroacetic acid (TFA). Preparative RP-HPLC was performed using a Phenomenex Kinetex 5-μm, 100-Å C18 column (250 × 10 mm), whereas analytical RP-HPLC was performed using a Phenomenx Kinetex 5-μm, 100-Å C18 column (250 by 4.6 mm). Fmoc solid-phase peptide synthesis was performed on a Discover Microwave and a Liberty1 automated peptide synthesizer (CEM Corporation). PCR amplification was performed in an Eppendorf Mastercycler gradient 5331 PCR machine (Eppendorf). Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) data were obtained by mixing 0.75 μl of sample with 0.75 μl of matrix solution (α-cyano-4-hydroxycinnamic acid dissolved in ddH2O-ACN [1:1] with 0.1% TFA) on an MSP 96 polished steel target plate (Bruker Daltonics) and allowing it to air dry. Data were obtained using a Bruker Microflex spectrometer equipped with a 60-Hz (337-nm wavelength) nitrogen laser and a reflectron. MALDI-TOF MS data were obtained using the reflectron's positive ion mode with the following settings: ion source 1, 19 kV; ion source 2, 15.9 kV; lens, 8.75 kV; reflector, 20 kV; up to 300-Da matrix suppression; 200 laser shots per sample; and detector gain, 1,594 V. Exact mass (EM) data were obtained on an Agilent Technologies 6230 time of flight mass spectrometer (an HRMS system) with the following settings for positive electrospray ionization (ESI+) mode: capillary voltage, 3,500 V; fragmentor voltage, 175 V; skimmer voltage, 65 V; Oct 1 RF Vpp, 750 V; gas temperature, 325°C; drying gas flow rate = 3 liters/min; and nebulizer, 25 lb/in2.
Bacterial growth conditions.
S. gallolyticus subsp. gallolyticus TX20005, S. intermedius F0413, S. vestibularius F0396, and S. agalactiae MNZ938 freezer stock cultures were obtained from the Biodefense and Emerging Infections Research Resources Repository (BEI Resources), while S. mutans ATCC 25175, S. anginosus ATCC 33397, and S. constellatus ATCC 27823 freezer stock cultures were obtained from the Streptococcus Lab at the Centers for Disease Control and Prevention. The following procedure was followed for each obtained isolate: a freezer stock was streaked onto a plate of Todd-Hewitt agar supplemented with 0.5% yeast extract (THY). The plate was incubated for 12 to 24 h in a CO2 incubator (37°C with 5% CO2). Fresh colonies were picked and Gram-stained to ensure purity. After purity was confirmed, colonies (2 to 5) were picked and inoculated into a sterilized culture tube containing 3 ml of sterile THY broth and incubated statically in a CO2 incubator for 8 to 24 h.
PCR amplification of RNA polymerase B and the comC gene.
PCR amplification was conducted using established protocols (42, 43); see the supplemental material for details.
Sequencing and verification of PCR-amplified rpoB and comC genes.
Sequencing and verification of PCR products were conducted at the Nevada Genomics Center, using established protocols (44, 45). The sequenced partial rpoB gene sequences were used to confirm the identity of our isolates to the species level; see the supplemental material for details.
Predicting the S. gallolyticus subsp. gallolyticus competence-stimulating peptide sequence.
The following ComC protein sequences were downloaded from the NCBI database for various S. gallolyticus subsp. gallolyticus strains and for S. pneumoniae and S. intermedius for multiple-sequence alignment: WP_009854999.1, AQP43123.1, CBI14504.1, EFM28801.1, OCW49321.1, KJE98669.1, OAV84346.1, WP_000799689.1, AAK76284.1, and CAA04354.1. The sequences were aligned in Molecular Evolutionary Genetics Analysis (MEGA) 7.0 software, using ClustalW with the default settings. The aligned sequences were then trimmed from the N terminus to the first residue after the double glycine leader signal, and another alignment was performed under the same conditions. This alignment was used to predict the possible CSP sequence. Multiple sequence alignments were then exported and edited using Boxshade 3.21 (https://www.ch.embnet.org/software/BOX_form.html).
Isolation of crude peptides from bacterial supernatants.
Overnight S. gallolyticus subsp. gallolyticus culture (25 ml) was centrifuged at 4,600 × g for 10 min. The supernatants were then filtered through a sterile 0.22-μm polyethersulfone (PES) filter into a sterile, empty 50-ml centrifuge tube. Ammonium sulfate was added to the filtered supernatants to give a 20% (wt/vol) concentration and mixed by inversion until all of the ammonium sulfate went into solution. The solution was stored at 4°C for 45 min, followed by centrifugation at 4,600 × g for 15 min. The supernatants were discarded and the remaining pellet was dissolved in 10 ml of ddH2O-ACN (1:1) and freeze-dried. The lyophilized material was purified by RP-HPLC.
Bacteriocin-like inhibitory substance extraction and isolation.
Overnight S. gallolyticus subsp. gallolyticus culture (500 ml) was centrifuged at 4,600 × g for 10 min. The supernatants were discarded, whereas the cell pellet was resuspended in 25 ml of sterile PBS. The resuspended cells were centrifuged at 4,600 × g for 10 min, and the supernatants were discarded. This process was repeated 3 times. The washed cells were resuspended in 10 ml of sterile PBS containing 10 μl Tween 20 and shaken vigorously for 30 min. The mixture was centrifuged at 4,600 × g for 10 min, and the supernatants were filtered through a 0.22-μm PES filter. Ammonium sulfate was added (40% [wt/vol]) and the mixture was stored at 4°C for 1 h and centrifuged at 4,600 × g for 30 min. The supernatants were discarded, and the remaining pellet was dissolved in 5 ml of H2O and freeze-dried. RP-HPLC purification was performed, and the various fractions were freeze-dried prior to being resuspended in 250 μl of sterile water and tested in the interspecies inhibition assay against S. intermedius F0413 and S. constellatus ATCC 27823.
Peptide purification.
Peptide purification was conducted using RP-HPLC and established procedures (46); see the supplemental material for details.
Solid-phase peptide synthesis.
Peptide synthesis was conducted using established protocols (47–49); see the supplemental material for details.
Peptide mapping.
Samples of synthetic S. gallolyticus subsp. gallolyticus CSP and naturally isolated S. gallolyticus subsp. gallolyticus CSP were prepared in a 20 μM concentration in ddH2O and provided to the Nevada Proteomics Center for peptide mapping by ESI-tandem MS. The amino acid sequences of multiple charge states were processed using Scaffold 4 software.
Chymotrypsin digestion.
Chymotrypsin (Worthington Biochemical Corporation) was dissolved in 1 mM HCl solution to a final concentration of 1 μg/ml. Both naturally isolated and synthetic S. gallolyticus subsp. gallolyticus CSPs were dissolved to a final concentration of 1 mg/ml in PBS (pH 7.6) containing 1 mM CaCl2. A mixture containing 1:20 chymotrypsin-S. gallolyticus subsp. gallolyticus CSP was prepared and incubated at 37°C for 12 to 16 h. Samples were analyzed by analytical HPLC to confirm cleavage of the peptide. The chymotrypsin-S. gallolyticus subsp. gallolyticus CSP mixtures were purified using RP-HPLC to remove buffer salts and freeze-dried prior to being resuspended in ddH2O for fragment analysis by ESI+ HRMS to determine the correct sequence of the S. gallolyticus subsp. gallolyticus CSP.
Circular dichroism spectroscopy.
Circular dichroism spectra were acquired as previously described (46); see the supplemental material for details.
Crystal violet biofilm assay.
Biofilm quantification using crystal violet was performed as previously described (50); see the supplemental material for details.
Plasmid extraction.
Plasmid extraction and quantification were conducted using established protocols (51); see the supplemental material for details.
Transformation assay.
A 1:10 dilution of overnight culture was made in sterilized THY and incubated statically in a CO2 incubator (37°C with 5% CO2) for 4 to 6 h (optical density at 600 nm [OD600], ∼0.25). Six sterile microcentrifuge tubes for each experimental time point (30 min, 1 h, 2 h, 3 h, 4 h, and 16 h) received 154 μl of sterile medium (THY or M9 with 1% glucose and 1% biotin) and 40 μl of the fresh culture (for a total of 36 tubes). The tubes were incubated at 37°C with shaking at 50 rpm for 30 min prior to addition of CSP, plasmid, or DMSO. For each time point, 2 tubes (one THY and one M9) received 2 μl of 1 mM CSP with 4 μl of sterile water, 2 other tubes (one THY and one M9) were treated with 4 μl of pALH122 (40 ng/μl) with 2 μl of DMSO, and the final 2 tubes (one THY and one M9) were treated with 2 μl of 1 mM CSP and 4 μl of pALH122. The microcentrifuge tubes were incubated at 37°C with shaking at 50 rpm. Six tubes (three THY and three M9), consisting of CSP only, plasmid only, and CSP plus plasmid conditions were removed at the appropriate time point, and the content of each tube was spread plated onto two agar plates (100 μl for each plate), one plate of THY agar and one plate of THY agar with erythromycin (10 ng/μl final concentration). The plates were incubated for 24 to 96 h in a CO2 incubator (37°C with 5% CO2) prior to being inspected for colonies. The procedure was repeated using working stocks of 50 μM and 10 μM CSP to give final concentrations of 500 nM and 100 nM CSP, respectively.
Interspecies inhibition assay.
Three sterile microcentrifuge tubes received 445 μl of sterile THY and 50 μl of S. gallolyticus subsp. gallolyticus overnight culture. A fourth sterile microcentrifuge tube received 495 μl of sterile THY and 5 μl of CSP (10 μM final concentration). The three S. gallolyticus subsp. gallolyticus-containing microcentrifuge tubes were incubated aerobically for 30 min at 37°C with shaking at 50 rpm. Each tube then received either 5 μl of CSP (10 μM or 100 nM final concentration) or DMSO (negative control), followed by 4 h of incubation in a CO2 incubator (37°C with 5% CO2). The tubes were then centrifuged at 14,000 × g for 5 min to pellet as many cells as possible. The supernatants were then transferred to a sterile microcentrifuge tube and centrifuged again at 14,000 × g for 5 min. The process was repeated one more time, and the supernatants were used for the interspecies inhibition assay.
For each isolate tested against S. gallolyticus subsp. gallolyticus, 250 μl of overnight culture was spread plated onto a THY agar plate, and the plate was allowed to dry for 10 min. After the plate was dry, wells were made by using the larger diameter of a sterile 200 μl pipette tip, and the agar plugs were picked out using the smaller-diameter tip. Wells were made in triplicates for each of the following conditions: S. gallolyticus subsp. gallolyticus supernatants and DMSO, THY and 10 μM CSP, S. gallolyticus subsp. gallolyticus supernatants and 10 μM CSP, and S. gallolyticus subsp. gallolyticus supernatants and 100 nM CSP. A volume of 85 μl was transferred to three wells for each condition and incubated overnight in a CO2 incubator (37°C with 5% CO2). After overnight incubation, the plates were inspected for zones of inhibition around the wells. The experiment was repeated for 3 consecutive days. The same process was repeated, with the exception that the supernatants were filtered with a 0.45-μm polytetrafluoroethylene (PTFE) filter for one trial and a 0.22-μm PES filter for another trial, prior to being placed in the wells. These experiments were also repeated for 3 consecutive days. In a different trial, S. gallolyticus subsp. gallolyticus cells were grown under the same conditions, but with no added CSP during the incubation; only after 3 consecutive high-speed centrifugations or filtration using a 0.22-μm PES filter, were 5 μl of CSP added to reach the final concentration of 10 μM. A volume of 85 μl of these two supernatants was transferred to three wells and incubated overnight in a CO2 incubator (37°C with 5% CO2), prior to inspection for zones of inhibition. These experiments were also repeated for 3 consecutive days. Finally, the same procedure was repeated, using either 10 μM final concentration CSP or DMSO, with the exception that at the end of the 4 h of incubation, 0.5 μl of Tween 20 was added to the supernatants and shaken vigorously for 10 min, followed by centrifugation at 14,000 × g. The supernatants were filtered through a sterile 0.22-μm PES filter and used for the interspecies inhibition assay. These experiments were also repeated for 3 consecutive days.
Accession number(s).
The comC nucleotide and protein sequences of S. gallolyticus subsp. gallolyticus TX20005 were deposited in GenBank under the assigned accession numbers MF964227 and AWK22889.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Nevada INBRE through a grant from the National Institute of General Medical Sciences (grant GM103440).
The S. agalactiae strain MNZ938 (NR-43897) was obtained through BEI Resources, NIAID, and NIH, while the following strains were obtained as part of the Human Microbiome Project: S. vestibularis strain F0396 (HM-561), S. intermedius F0413 (HM-368), and S. gallolyticus subsp. gallolyticus strain TX20005 (HM-272). We also thank Lesley McGee from the CDC Streptococcus Lab for providing S. mutans ATCC 25175, S. anginosus ATCC 33397, and S. constellatus ATCC 27823. Additionally, we thank Dennis G. Cvitkovitch from the University of Toronto for providing plasmid pALH122 and Matthew J. Tucker from the University of Nevada, Reno, for the use of the CD spectrometer.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00709-17.
REFERENCES
- 1.Beck M, Frodl R, Funke G. 2008. Comprehensive study of strains previously designated Streptococcus bovis consecutively isolated from human blood cultures and emended description of Streptococcus gallolyticus and Streptococcus infantarius subsp. coli. J Clin Microbiol 46:2966–2972. doi: 10.1128/JCM.00078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlegel L, Grimont F, Ageron E, Grimont PA, Bouvet A. 2003. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int J Syst Evol Microbiol 53:631–645. [DOI] [PubMed] [Google Scholar]
- 3.Lapage SP, Sneath PHA, Lessel EF, Skerman VBD, Seeliger HPR, Clark WA (ed). 1992. International code of nomenclature of bacteria. American Society for Microbiology, Washington, DC. [PubMed] [Google Scholar]
- 4.Dekker JP, Lau AF. 2016. An update on the Streptococcus bovis group: classification, identification, and disease associations. J Clin Microbiol 54:1694–1699. doi: 10.1128/JCM.02977-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein RS, Recco RA, Catalano MT, Edberg SC, Casey JI, Steigbigel NH. 1977. Association of Streptococcus bovis with carcinoma of the colon. N Engl J Med 297:800–802. doi: 10.1056/NEJM197710132971503. [DOI] [PubMed] [Google Scholar]
- 6.Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. 2011. Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis 53:870–878. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- 7.Boleij A, Muytjens CM, Bukhari SI, Cayet N, Glaser P, Hermans PW, Swinkels DW, Bolhuis A, Tjalsma H. 2011. Novel clues on the specific association of Streptococcus gallolyticus subsp gallolyticus with colorectal cancer. J Infect Dis 203:1101–1109. doi: 10.1093/infdis/jiq169. [DOI] [PubMed] [Google Scholar]
- 8.Boleij A, Tjalsma H. 2012. Gut bacteria in health and disease: a survey on the interface between intestinal microbiology and colorectal cancer. Biol Rev 87:701–730. doi: 10.1111/j.1469-185X.2012.00218.x. [DOI] [PubMed] [Google Scholar]
- 9.Corredoira J, Grau I, Garcia-Rodriguez JF, Alonso-Garcia P, Garcia-Pais M, Rabuñal R, Garcia-Garrote F, Ardanuy C, Coira A, Lopez-Alvarez M. 2015. The clinical epidemiology and malignancies associated with Streptococcus bovis biotypes in 506 cases of bloodstream infections. J Infect 71:317–325. doi: 10.1016/j.jinf.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Gupta A, Madani R, Mukhtar H. 2010. Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis 12:164–171. doi: 10.1111/j.1463-1318.2009.01814.x. [DOI] [PubMed] [Google Scholar]
- 11.Waisberg J, Matheus CdO, Pimenta J. 2002. Infectious endocarditis from Streptococcus bovis associated with colonic carcinoma: case report and literature review. Arq Gastroenterol 39:177–180. doi: 10.1590/S0004-28032002000300008. [DOI] [PubMed] [Google Scholar]
- 12.Kumar R, Herold JL, Schady D, Davis J, Kopetz S, Martinez-Moczygemba M, Murray BE, Han F, Li Y, Callaway E. 2017. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog 13:e1006440. doi: 10.1371/journal.ppat.1006440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng J, Song Q, Tang X, Liang X, Fan H, Peng H, Guo Q, Zhang Z. 2014. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog 6:26. doi: 10.1186/1757-4749-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Håvarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomasz A. 1966. Model for the mechanism controlling the expression of competent state in pneumococcus cultures. J Bacteriol 91:1050–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Håvarstein LS, Hakenbeck R, Gaustad P. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J Bacteriol 179:6589–6594. doi: 10.1128/jb.179.21.6589-6594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanker E, Federle MJ. 2017. Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes 8:E15. doi: 10.3390/genes8010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo P, Li H, Morrison DA. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol Microbiol 50:623–633. doi: 10.1046/j.1365-2958.2003.03714.x. [DOI] [PubMed] [Google Scholar]
- 19.Berg KH, Biørnstad TJ, Johnsborg O, Håvarstein LS. 2012. Properties and biological role of streptococcal fratricins. Appl Environ Microbiol 78:3515–3522. doi: 10.1128/AEM.00098-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y-H, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salvadori G, Junges R, Khan R, Åmdal HA, Morrison DA, Petersen FC. 2017. Natural transformation of oral streptococci by use of synthetic pheromones. Methods Mol Biol 1537:219–232. doi: 10.1007/978-1-4939-6685-1_13. [DOI] [PubMed] [Google Scholar]
- 22.Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol 192:1444–1454. doi: 10.1128/JB.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haustenne L, Bastin G, Hols P, Fontaine L. 2015. Modeling of the ComRS signaling pathway reveals the limiting factors controlling competence in Streptococcus thermophilus. Front Microbiol 6:1413. doi: 10.3389/fmicb.2015.01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N, Chambellon E, Gardan R, Hols P. 2013. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol Microbiol 87:1113–1132. doi: 10.1111/mmi.12157. [DOI] [PubMed] [Google Scholar]
- 25.Morrison DA, Guédon E, Renault P. 2013. Competence for natural genetic transformation in the Streptococcus bovis group streptococci S. infantarius and S. macedonicus. J Bacteriol 195:2612–2620. doi: 10.1128/JB.00230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinse D, Vollmer T, Rückert C, Blom J, Kalinowski J, Knabbe C, Dreier J. 2011. Complete genome and comparative analysis of Streptococcus gallolyticus subsp. gallolyticus, an emerging pathogen of infective endocarditis. BMC Genomics 12:400. doi: 10.1186/1471-2164-12-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin I-H, Liu T-T, Teng Y-T, Wu H-L, Liu Y-M, Wu K-M, Chang C-H, Hsu M-T. 2011. Sequencing and comparative genome analysis of two pathogenic Streptococcus gallolyticus subspecies: genome plasticity, adaptation and virulence. PLoS One 6:e20519. doi: 10.1371/journal.pone.0020519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rusniok C, Couvé E, Da Cunha V, El Gana R, Zidane N, Bouchier C, Poyart C, Leclercq R, Trieu-Cuot P, Glaser P. 2010. Genome sequence of Streptococcus gallolyticus: insights into its adaptation to the bovine rumen and its ability to cause endocarditis. J Bacteriol 192:2266–2276. doi: 10.1128/JB.01659-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iannelli F, Oggioni MR, Pozzi G. 2005. Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptoccoccus pneumoniae. FEMS Microbiol Lett 252:321–326. doi: 10.1016/j.femsle.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Luo P, Baldwin RL. 1997. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry 36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 31.Shah GR, Caufield P. 1993. Enhanced transformation of Streptococcus mutans by modifications in culture conditions. Anal Biochem 214:343–346. doi: 10.1006/abio.1993.1503. [DOI] [PubMed] [Google Scholar]
- 32.Lemme A, Gröbe L, Reck M, Tomasch J, Wagner-Döbler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol 193:1863–1877. doi: 10.1128/JB.01363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. 2012. Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J Bacteriol 194:3774–3780. doi: 10.1128/JB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loo C, Corliss D, Ganeshkumar N. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol 182:1374–1382. doi: 10.1128/JB.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y-H, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen FC, Pecharki D, Scheie AA. 2004. Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J Bacteriol 186:6327–6331. doi: 10.1128/JB.186.18.6327-6331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romao S, Memmi G, Oggioni MR, Trombe M-C. 2006. LuxS impacts on LytA-dependent autolysis and on competence in Streptococcus pneumoniae. Microbiology 152:333–341. doi: 10.1099/mic.0.28406-0. [DOI] [PubMed] [Google Scholar]
- 38.Vidal JE, Ludewick HP, Kunkel RM, Zähner D, Klugman KP. 2011. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun 79:4050–4060. doi: 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantovani HC, Hu H, Worobo RW, Russell JB. 2002. Bovicin HC5, a bacteriocin from Streptococcus bovis HC5. Microbiology 148:3347–3352. doi: 10.1099/00221287-148-11-3347. [DOI] [PubMed] [Google Scholar]
- 40.Riley MA, Chavan MA. 2007. Bacteriocins: ecology and evolution. Springer-Verlag, Berlin, Germany [Google Scholar]
- 41.Biswas S, Cao L, Kim A, Biswas I. 2016. SepM, a streptococcal protease involved in quorum sensing, displays strict substrate specificity. J Bacteriol 198:436–447. doi: 10.1128/JB.00708-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kellogg D, Rybalkin I, Chen S, Mukhamedova N, Vlasik T, Siebert P, Chenchik A. 1994. TaqStart antibody: “hot start” PCR facilitated by a neutralizing monoclonal antibody directed against Taq DNA polymerase. Biotechniques 16:1134–1137. [PubMed] [Google Scholar]
- 43.Saris PEJ, Paulin LG, Uhlén M. 1990. Direct amplication of DNA from colonies of Bacillus subtilis and Escherichia coli by the polymerase chain reaction. J Microbiol Methods 11:121–126. doi: 10.1016/0167-7012(90)90012-U. [DOI] [Google Scholar]
- 44.Kheterpal I, Mathies RA. 1999. Capillary array electrophoresis DNA sequencing. Anal Chem 71:31A–37A. doi: 10.1021/ac990099w. [DOI] [PubMed] [Google Scholar]
- 45.Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res 8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, Koirala B, Sanchez LA, Phillips NR, Hamry SR, Tal-Gan Y. 2017. Structure–activity relationships of the competence stimulating peptides (CSPs) in Streptococcus pneumoniae reveal motifs critical for intra-group and cross-group ComD receptor activation. ACS Chem Biol 12:1141–1151. doi: 10.1021/acschembio.7b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collins JM, Porter KA, Singh SK, Vanier GS. 2014. High-efficiency solid phase peptide synthesis (HE-SPPS). Org Lett 16:940–943. doi: 10.1021/ol4036825. [DOI] [PubMed] [Google Scholar]
- 48.Kates SA, Solé NA, Beyermann M, Barany G, Albericio F. 1996. Optimized preparation of deca(l-alanyl)-l-valinamide by 9-fluorenylmethyloxycarbonyl (Fmoc) solid-phase synthesis on polyethylene glycol-polystyrene (PEG-PS) graft supports, with 1,8-diazobicyclo [5.4. 0]-undec-7-ene (DBU) deprotection. Pept Res 9:106–113. [PubMed] [Google Scholar]
- 49.Sandhya K, Ravindranath B. 2008. A protocol for racemization-free loading of Fmoc-amino acids to Wang resin. Tetrahedron Lett 49:2435–2437. doi: 10.1016/j.tetlet.2008.02.055. [DOI] [Google Scholar]
- 50.Kratochvil MJ, Tal-Gan Y, Yang T, Blackwell HE, Lynn DM. 2015. Nanoporous superhydrophobic coatings that promote the extended release of water-labile quorum sensing inhibitors and enable long-term modulation of quorum sensing in Staphylococcus aureus. ACS Biomater Sci Eng 1:1039–1049. doi: 10.1021/acsbiomaterials.5b00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Branovic K, Forcic D, Ivancic J, Strancar A, Barut M, Gulija TK, Zgorelec R, Mazuran R. 2004. Application of short monolithic columns for fast purification of plasmid DNA. J Chromatogr B Analyt Technol Biomed Life Sci 801:331–337. doi: 10.1016/j.jchromb.2003.11.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.