Abstract
Background
Data on the expression of RCC tissues from the GEO database and patient survival data from TCGA were used to explore the prognostic significance of long noncoding RNA SNHG1. SNHG1 has been reported to participate in the development of several cancers, but, the underlying mechanism of SNHG1 in renal cell carcinoma (RCC) has not been reported. The purpose of our study was to investigate the potential function of SNHG1 in RCC.
Material/Methods
The expression of SNHG1 in 40 cases of RCC and adjacent normal tissues and 5 cell lines was detected by qRT-PCR. Cell proliferation, Transwell assay, and Western blotting assay were carried out to investigate the biological function of SNHG1. A rescue experiment was performed to verify that miR-137 can partly impede the effect of SNHG1 on renal cancer cells.
Results
SNHG1 was identified to be overexpressed in RCC tissues and RCC cell lines. High levels of SNHG1 were correlated with poor prognosis of RCC patients. Knockdown of SNHG1 suppressed the proliferation, invasion, and EMT capacity in RCC. Moreover, miR-137 abrogated the effect of SNHG1 on RCC.
Conclusions
SNHG1 is significantly upregulated in RCC and renal cancer cell lines. Overexpression of SNHG1 participates in RCC tumorigenesis by regulating miR-137.
MeSH Keywords: Carcinoma, Renal Cell; MicroRNAs; Nephrology; RNA, Long Noncoding
Background
Renal cell carcinoma (RCC) is one of the most common malignant urogenital neoplasms due to space-occupying lesions; it accounts for ~3% of all malignancies and ~90 000 patients die from RCC each year [1]. In addition, the 5-year survival rate of RCC is ≤40%. RCC is derived from renal proximal tubule cells and has clear-cell, papillary, and chromophobe subtypes [2–4]. Despite progress in therapeutic and biological approaches, current treatments are not considered to be effective [5,6]. Although surgical tumor resection is the optimal treatment strategy at present, the metastasis rate of RCC remains high (~10%) [7]. Therefore, it is necessary to find an effective molecular biomarker to predict early metastasis and develop new therapeutic approaches for RCC.
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs longer than 200 nucleotides with limited protein-coding capacity [8]. In the past, these long noncoding transcripts were regarded as transcriptional “noise” or cloning artifacts [9], but increasing evidence reveals that lncRNAs play important roles in various cell processes, including transcriptional regulation, cell proliferation, and nuclear import [10]. These findings have demonstrated the potential of lncRNAs as a promising therapeutic target. Recent studies have shown the biological functions of lncRNAs in various human cancers, including RCC [11,12].
LncRNA small nucleolar RNA host gene 1 (SNHG1) located on 11q12.3 and was verified to be upregulated in various types of cancer and was confirmed as an independent prognostic factor [13–16]. In addition, studies showed that lncRNA SNHG1 functions as an oncogene in lung cancer development, and high levels of SNHG1 are associated with tumor size, TNM stage, and poor prognosis of lung cancer patients [17]. Liu et al. reported that SNHG1 suppresses tumor progression of nasopharyngeal carcinoma and functions as a ceRNA to antagonize the effect of miR-145a-5p [18]. Wang et al. reported that SNHG1 is overexpressed in prostate cancer and negatively regulates miR-199a-3p expression [19].
The present study shows that SNHG1 is overexpressed in RCC tissues and cell lines and SNHG1 negatively influences the effect of miR-137 and promotes cell proliferation and invasion and induces EMT. The lncRNA SNHG1 might be potential therapeutic approach for the treatment of RCC by direct interaction with miR-137.
Material and Methods
Cell lines and clinical tissues
Human RCC cell lines A-498, ACHN, 786-O, and Caki-1 were purchased from the Chinese Academy of Sciences (Shanghai, China). Human Kidney cells (HK-2) were purchased from the American Type Cell Culture Collection (ATCC; Rockville, MD, USA). These cells were incubated in RPMI-1640 (Gibco, Los Angeles, CA, USA) supplemented with 10% fetal bovine serum (Gibco) and maintained at 37°C in an atmosphere containing 5% CO2.
Human renal cancer tissues and adjacent normal tissues were collected from 40 patients with histologically confirmed renal cancer who underwent radical nephrectomy at the Second Hospital of Jilin University between May 2011 and December 2016. All RCC cases were confirmed by a senior pathologist and were staged based on the 2011 Union for International Cancer Control TNM classification of malignant tumors. No patients had received any anticancer treatment. All tissues were pathologically confirmed and immediately snap-frozen in liquid nitrogen and stored at −80ºC until RNA extraction. Written informed consent for research purposes was obtained from each patient. All procedures conformed to the Declaration of Helsinki and all applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The study was approved by the Institutional Review Board of the Second Affiliated Hospital of Jilin University (Changchun, China).
Gene set enrichment analysis with SNHG1 expression
Data on 3 RCC tissues and 3 matched normal samples was downloaded from the Gene Expression Omnibus (GEO) database (GSE100666, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100666). Clinical data were obtained from the Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov). Gene Ontology (GO) term analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8. Significantly enriched gene sets were investigated, which produced a nominal P-value 0.05.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was collected from clinical tissues and cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). qRT-PCR was displayed 3 times in an ABI 7500HT fast real-time PCR System (Applied Biosystems) according to the manufacturer’s protocols. To verify the expression of SNHG1 and miR-137, endogenous mRNA was generated with a lightcycler-480 (Roche) using the SYBR Green PCR Master Mix kit (Applied Biosystems). Relative gene expression levels were calculated by comparing them to the expression level internal standard using 2−ΔΔCt method.
Plasmids, oligonucleotides, and cell transfection
SNHG1 siRNA, miR-137 mimics, miR-137 inhibitor, or negative control (NC) were obtained from Genechem (Shanghai, China). Transfection into cells was conducted using Lipofectamine 2000™ (Invitrogen) in 6-well plates. The human SNHG1 Luc-reporter was used in the ligation of the SHNG1 3′-untranslated region (UTR) PCR product. The psiCHECK2 vector (Genechem, Shanghai, China) was used to construct the SNHG1 3′UTR containing reporter.
Dual-luciferase reporter assay
O cells were co-transfected with psiCHECK2-SNHG1 wild-type (WT) or mutant-type (MUT), miR-137, and NC mimics. After 48 h, luciferase activity was detected using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instruction. Renilla luciferase activity was utilized as an internal control. Luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega Corporation).
Cell viability and colony formation assay
The number of RCC cells was evaluated using the Cell Counting Kit (CCK-8) (Dojindo, Japan) and detected at a wavelength of 490 nm, then the optical density was calculated. For colony formation assays, transfected cells were seeded onto 6-well plates (200 cells per well) and cultured for a further 14 days. Then, cells were fixed with formalin and stained with Giemsa (Sigma-Aldrich). Subsequently, the colonies (>50 cells) were counted.
Cell invasion assays
Transfected cells were seeded into the Matrigel membrane (Costa, Corning, NY, USA) in the upper chamber in serum-free medium. In the lower chamber, RPMI-1640 medium with 20% FBS was applied for 24 h, then the medium in the upper chamber was removed and the non-invading cells were wiped away. To cells in the lower chamber, we added 4% paraformaldehyde, then we stained them with 0.1% crystal violet and counted them under a microscope.
Western blot analysis
Total protein was lysed with RIPA buffer (Pierce; Thermo Fisher Scientific, Waltham, MA, USA) containing protease inhibitors cocktail (Roche, USA). Protein concentration were measured with the BCA protein assay kit (Bio-Rad Laboratories, CA, USA). We added 10% SDS-PAGE to separate equal amounts of protein (50 μg) and blotted them onto polyvinylidene fluoride membranes (PVDF, EMD Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat milk (w/v) at room temperature for 1 h and incubated with the specific rabbit anti-human antibodies E-cadherin (1: 1000 dilution, Cell Signaling Technology, USA), N-cadherin (1: 1000 dilution, Cell Signaling Technology, USA), Vimentin (1: 1000 dilution, Cell Signaling Technology, USA), and mouse anti-human GAPDH (1: 500, Santa Cruz, USA) at 4°C overnight. Immunodetection was visualized on a Gel Doc 2000 imaging system (Bio-Rad Laboratories, CA, USA).
Immunofluorescence staining
We added 4% paraformaldehyde to cells for 20 min and 0.1% Triton x-100 for 5 min. The samples were washed and blocked with 5% BSA in PBS for 1 h, then incubated with primary antibodies against Vimentin (1: 100 dilution, Cell Signaling Technology, USA) at 4°C overnight, then fixed them with fluorescence-labeled rabbit secondary antibody (1: 100, ProteinTech) for 1 h at room temperature. The nuclei were counterstained with DAPI for 10 min and assessed using fluorescence microscopy (Nikon).
Statistical analysis
All results are presented as the mean ±SD of 3 independent experiments. Statistics were analyzed using the SPSS Graduate Pack, version 19.0, statistical software (SPSS, Chicago, IL, USA). Pearson’s rank correlation analysis was used to analyze correlations between SNHG1 and miR-137. Comparisons between groups were determined using the two-tailed t test or one-way ANOVA. P<0.05 was considered to be statistically significant.
Results
SNHG1 was upregulated in RCC tissue and correlated with poor prognosis
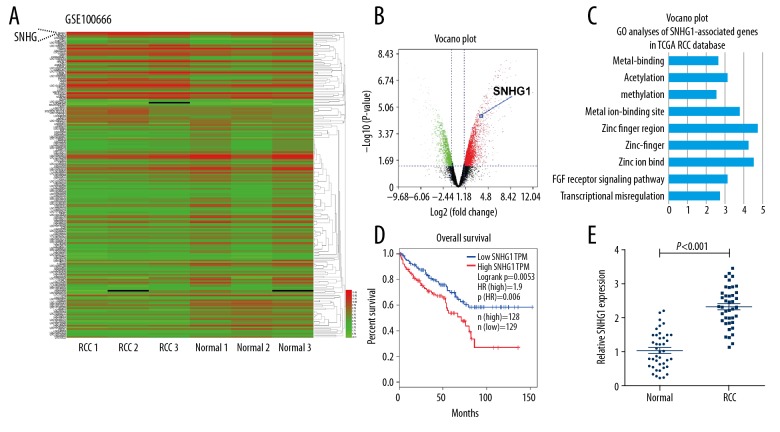
To investigate whether SNHG1 was involved in the development of RCC, we explored SNHG1 expression by using microarray data downloaded from Gene Expression Omnibus (GEO; GSE100666) and found that SNHG1 level was significantly overexpressed in RCC tissues compared with that in adjacent normal tissues (Figure 1A). The volcano plot showed that SNHG1 has over 2-fold case-control differences with a p<0.01 (Figure 1B). To further elucidate the involved function activated by SNHG1, we detected the associated genes expression profiles using data collected from the Gene Ontology (GO) database. The most prominent GO biological processes included cell migration, metal ion-binding site, and zinc ion binding (Figure 1C). Analysis of the TCGA database indicated that higher SNHG1 expression in RCC was correlated with shorter overall survival (Figure 1D; P<0.01). Then, we examined SNHG1 expression levels in 40 CRC tissues and adjacent normal tissues by employing quantitative real-time PCR (qRT-PCR) analysis. The result showed that SNGH1 expression was significantly upregulated in RCC tissues (P<0.001) (Figure 1E). These findings show that miR-137 may function as an oncogene in RCC genesis.
Figure 1.
SNHG1 is overexpressed in human RCC tissue and is correlated with poor prognosis. (A) Hierarchical clustering analysis showing lncRNAs that are differentially expressed in paired renal cell carcinoma (n=3) versus matched normal tissue (n=3) samples (fold change >2.0, P<0.05). (B) Differentially expressed lncRNAs in RCCs compared to normal tissues were determined by volcano plot. (C) Top-ranked genes in GO term analysis using DAVID, indicating that genes related to the cell invasion and migration were enriched among those affected by SNHG1 introduction. (D) Kaplan-Meier survival curve of RCC patients (TCGA) based on the levels of SNHG1 expression (log-rank test, P=0.0053). (E) Upregulated level of SNHG1 was measured in RCC tissues compared with adjacent normal tissues. (GAPDH was used as normalized control, P<0.001) with the t test. Data are presented as mean ± standard error based on at least 3 independent experiments.
Effect of miR-137 on RCC cell viability and invasion
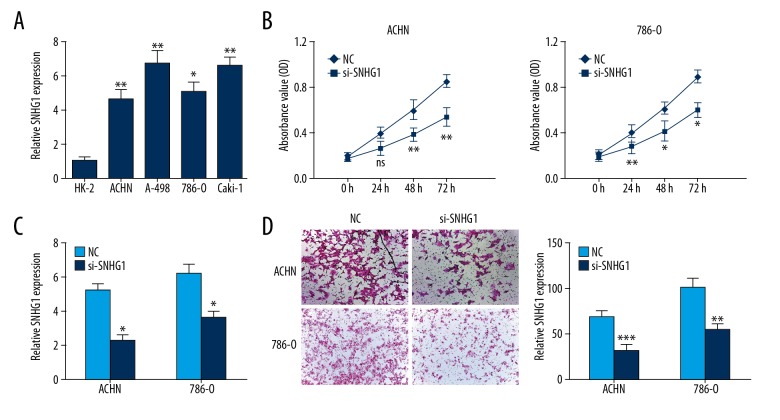
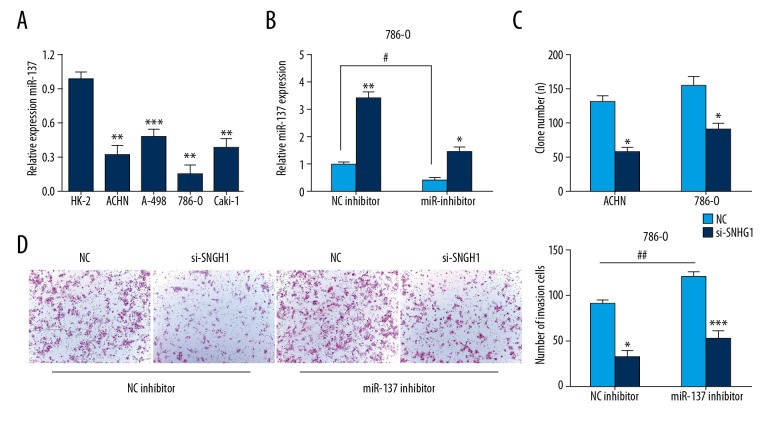
Increased SNHG1 expression was also investigated in RCC cells, including HK-2, ACHN, A-498, 786-O, and Caki-1 (P<0.05) (Figure 2A). To identify the biological function of SNHG1 in the progression of RCC, ACHN and 786-O cells were transfected with si-SNHG1 and NC, then cell growth vitality and invasion were explored. The result showed that SNHG1 silencing reduced RCC cell proliferation (Figure 2B). Colony formation assay indicated that knockdown of SNHG1 decreased the clone numbers in ACHN and 786-O cells (Figure 2C). Then, we assessed the effect of SNHG1 silencing in reducing RCC invasion capacity (Figure 2D). These results suggested the suppressive function of SNHG1 silencing on RCC cells in vitro.
Figure 2.
Knockdown of SNHG1 suppressed the proliferation and invasion of RCC cells in vitro. (A) SNHG1 expression level in RCC cell lines (ACHN, A-498, 786-0, and Caki-1) and HK-2 cells were conducted using qRT-PCR analyses. (B) RCC cell viability was analyzed using CCK-8 assay. (C) RCC cells treated with Negative Control (NC) or si-SNHG1 were cultured for 14 days, and colony formation assay was conducted. (D) RCC cell invasion ability was explored by Transwell assays. Data are presented as mean ± standard error based on at least 3 independent experiments. * P<0.05, ** P<0.01 and *** P<0.001.
Downregulation of SNHG1 regulated epithelial-mesenchymal transition (EMT) in RCC
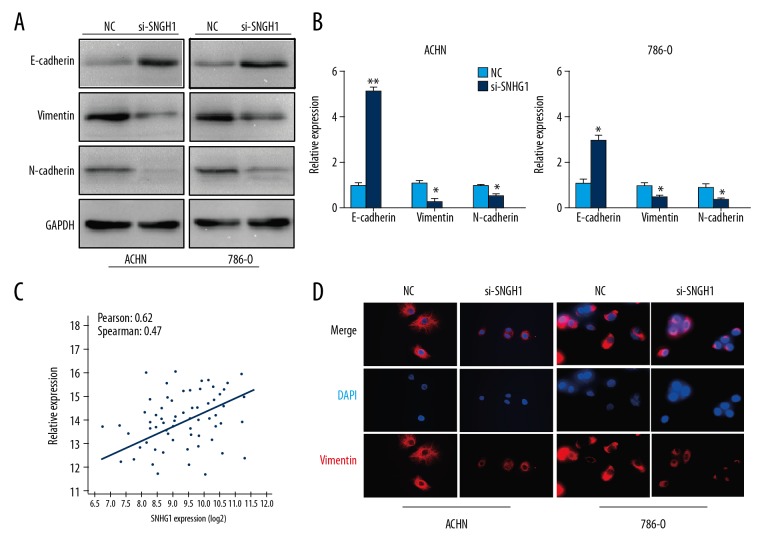
Invasion and metastasis play an important role in tumorigenesis. It is a complex process and the epithelial-mesenchymal transition (EMT) has become the classical and integral tumor invasion and metastasis component in recent years. To determine the function of SNHG1 silencing on RCC metastasis, the expression of E-cadherin, Vimentin, and N-cadherin were detected using Western blot and qRT-PCR. Data showed that SNHG1 silencing significantly increased the expression of E-cadherin, while the expressions of Vimentin and N-cadherin were downregulated (Figure 3A, 3B). The correlations of the SNHG1 expression level with Vimentin were further studied in the cohort of RCC samples from the TCGA database. The results indicated that SNHG1 expression was significantly correlated with the expression of Vimentin (R=0.47, P<0.05) (Figure 3C, 3D). Taken together, these data suggest that SNHG1 is associated with EMT and tumor metastasis in RCC.
Figure 3.
Expression of EMT markers in RCC cells. (A, B) Western blot and qRT-PCR analyses of the EMT-related proteins, E-cadherin, Vimentin, and N-cadherin were detected. (C) Correlation between SNHG1 and miR-137 expression in TCGA database (R=−0.4962, P=0.0011). (D) Immunofluorescent staining assay revealed a downregulated expression of Vimentin in si-SNHG1 cells (magnification ×400). Scale bars=100 μm. Data are presented as mean ± standard error based on at least 3 independent experiments. * P<0.05 and ** P<0.01.
SNHG1 targeted with miR-137 at 3′UTR
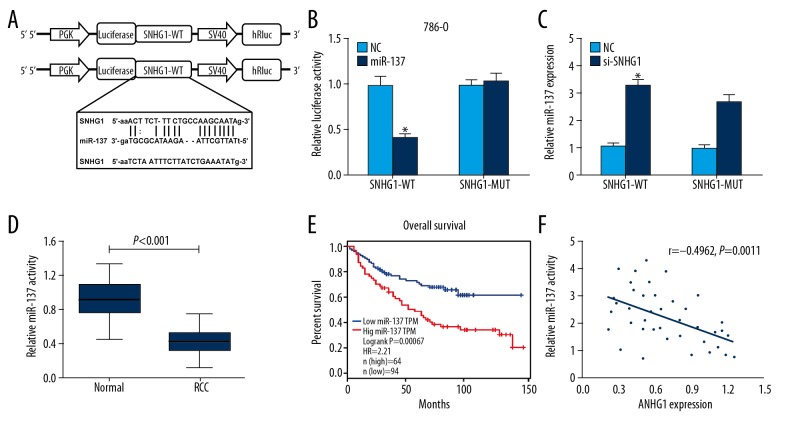
Using miRDB and Starbase database blast prediction, we found that SNHG1 has a putative miR-137 targeting site, which was validated by luciferase reporter assay (Figure 4A). We found that restoration of miR-137 expression significantly decreased the luciferase activity of WT-SNHG1-3′UTR, but not MUT-SNHG1-3′UTR in 786-O cells (Figure 4B). We then explored whether the mRNA levels of miR-137 in RCC cells are regulated by SNHG1. The results revealed that SNHG1 silencing increased the mRNA expression of miR-137 in RCC cells (Figure 4C). In addition, miR-137 expression was substantially reduced compared to the 40 RCC tissues with the pair-matched normal tissues (P<0.001) (Figure 4D). Analysis of the TCGA database indicated that low levels of miR-137 in RCC were correlated with better prognosis (Figure 4E). A significant inverse correlation between SNHG1 and miR-137 was identified at the mRNA level (R=−0.4962, P=0.0011) (Figure 4F). Taken together, these results demonstrate that miR-137 targeted SNHG1 at 3′UTR and miR-137 was negatively correlated with SNHG1.
Figure 4.
SNHG1 was targeted by miR-137 at 3′UTR. (A) The predicted sequence of miR-137 in the 3′UTR of SNHG1 is verified. (B) Relative luciferase report assay in 786-O cells co-transfected with miR-137 or NC and wild-type or mutant SNHG1 3′UTR. (C) miR-137 expression was measured by qRT-PCR assay following transfection with si-SNHG1 and NC. (D) Expression of miR-137 was identified in RCC tissues compared with adjacent normal tissues (P<0.001). (E) Patients with low level of miR-137 had better prognosis compared with those with high level in TCGA database (log-rank test, P=0.00067). (F) The correlation between SNHG1 and miR-137 levels was detected. Data are presented as mean ± standard error based on at least 3 independent experiments. * P<0.05 and ** P<0.01.
miR-137 abrogated the function of SNHG1 in 786-O cells
It had been demonstrated that miR-137 is inversely correlated with SNHG1 in RCC tissues. The expression of miR-137 was significantly decreased in RCC cells, and 786-O cells were selected for use in the rescue experiment (Figure 5A). The results indicated downregulation of miR-137 decreased the expression of miR-137 induced by SNHG1 siRNA (Figure 5B). Colony formation assay showed that miR-137 inhibitor suppressed the proliferation capacity induced by silencing the expression of SNHG1 (Figure 5C). Transwell assay showed miR-137 inhibitor suppressed the invasion ability induced by silencing the expression of SNHG1 (Figure 5D). Overall, these results reveal that miR-137 reverses the biological effect of SNHG1 on RCC cells.
Figure 5.
miR-137 reversed the effect of SNHG1 in RCC cells. (A) Expression of miR-137 in RCC cell line was displayed by qRT-PCR assay. (B) Expression of miR-137 in 786-O cells co-transfected with si-SNHG1 and miR-137 inhibitor. (C) Colony formation assay was conducted. (D) Invasion ability was determined by Transwell assays. Data are presented as mean ± standard error based on at least 3 independent experiments. * P<0.05, ** P<0.01 and *** P<0.001.
Discussion
RCC is a highly invasive cancer and despite current treatments, the 5-year survival rate is still poor. Previous studies demonstrated that aberrantly expressed molecules in tumors play important roles in tumorigenesis and predict patient survival outcomes. Cancer-specific lncRNAs have been verified to contribute in tumor progression and its underlying mechanism has attracted research attention [20]. However, the relationship between lncRNAs and RCC tumorigenesis is still unclear.
The present research is the first to investigate the expression of SNHG1 in RCC, indicating the high level of SNHG1 and the low expression of miR-137 in RCC tissue and cell lines.
The present study reveals that high levels of SNHG1 predict poor prognosis for RCC patients. We found that downregulation of SNHG1, as an oncogene in RCC, suppresses RCC cell proliferation and invasion capacity in vitro and inhibits the EMT process. Our research showed that SNHG1 exerted its effect on RCC mainly by directly targeting miR-137. To support our theory, we further investigated miR-137 tumor suppressor function, showing that transfection of miR-inhibitor could impede si-SNHG1 tumor suppressive effect on RCC growth and invasion capacity. These findings confirmed the synergistic relationship between SNHG1 and miR-137 in modulating tumor progression of RCC. This study revealed that overexpression of SNHG1 conferred a potent oncogenic signal in RCC and may be a novel therapeutic target in renal cancer.
The competing endogenous RNA (ceRNA) theory is the most important theory regarding lncRNA and lncRNA has been shown to be a sponge for regulating the expression and function of miRNA [21]. Lung adenocarcinoma transcript 1 (MALAT1) was identified to be an oncogene and may exert its effect by targeting with its P53 and PC2 [22]. The lncRNA MIAT (myocardial infarction-associated transcript) regulates cancer cell biological function by the MIAT/miR-29a-3p/HDAC4 axis, which may play a vital role in the diagnosis of gastric cancer [23]. In lung cancer, SNH1 was overexpressed in tissues and cell lines and was related to tumor size and TNM stages [24]. Jiang et al. revealed that SNHG1 contributed to the progression of osteosarcoma and miR-577 could act as a ceRNA of SNHG1. The linc-SNHG1/miR-577/WNT2B axis regulatory network might provide a novel therapeutic approach [25]. Zhang et al. confirmed that SNHG1 exacerbated hepatocellular carcinoma through suppressing miR-195 [26].
Based on this evidence, we conclude that SNHG1 function as a ceRNA of miR-137 and we have confirmed that the expression of miR-137 in RCC was significantly lower than in normal tissues and normal kidney cells. Sang et al. reported that miR-137 was downregulated in RCC and could suppress cell migration and invasion capacity [27]. Using Pearson’s correlation in the TCGA database, we found that the level of miR-137 was negatively correlated with SNHG1 mRNA expression. Then, we explored the putative binding site between SNHG1 and miR-137 using luciferase reporter assay. These results suggested that SNHG1 functions as a ceRNA by sequestering miR-137.
Conclusions
Our results demonstrate that SNHG1 functions as an oncogene by sponging miR-137 or acting as its ceRNA. It can promote proliferation, metastasis, and invasion of RCC cells, which is upregulated in RCC tissues and cells. These findings show that SNHG1 may be a potential therapeutic target in the treatment of RCC.
Footnotes
Conflicts of interest
None.
Source of support: This work was supported by a grant from the National Natural Science Foundation of China (81170669)
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics. 2012. Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 3.Meloni-Ehrig AM. Renal cancer: Cytogenetic and molecular genetic aspects. Am J Med Genet. 2002;115:164–72. doi: 10.1002/ajmg.10697. [DOI] [PubMed] [Google Scholar]
- 4.Youssef YM, White NM, Grigull J, et al. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur Urol. 2011;59:721–30. doi: 10.1016/j.eururo.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Ma W, Tao L, Wang X, et al. Sorafenib inhibits renal fibrosis induced by unilateral ureteral obstruction via inhibition of macrophage infiltration. Cell Physiol Biochem. 2016;39:1837–49. doi: 10.1159/000447883. [DOI] [PubMed] [Google Scholar]
- 6.Tian X, Dai S, Sun J, et al. Inhibition of MDM2 re-sensitizes rapamycin resistant renal cancer cells via the activation of p53. Cell Physiol Biochem. 2016;39:2088–98. doi: 10.1159/000447904. [DOI] [PubMed] [Google Scholar]
- 7.Krambeck AE, Dong H, Thompson RH, et al. Survivin and b7-h1 are collaborative predictors of survival and represent potential therapeutic targets for patients with renal cell carcinoma. Clin Cancer Res. 2007;13:1749–56. doi: 10.1158/1078-0432.CCR-06-2129. [DOI] [PubMed] [Google Scholar]
- 8.Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep. 2012;45:604–11. doi: 10.5483/BMBRep.2012.45.11.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spizzo R, Almeida MI, Colombatti A, et al. Long noncoding RNAs and cancer: A new frontier of translational research. Oncogene. 2012;31:4577–87. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Ma P, Zhao Y, et al. Biological function and mechanism of MALAT-1 in renal cell carcinoma proliferation and apoptosis: Role of the MALAT-1-livin protein interaction. J Physiol Sci. 2016;7:77–85. doi: 10.1007/s12576-016-0486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Y, Fan Q, Wang L, et al. LncRNA Snhg1, a non-degradable sponge for miR-338, promotes expression of proto-oncogene CST3 in primary esophageal cancer cells. Oncotarget. 2017;8:35750–60. doi: 10.18632/oncotarget.16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Zhang Z, Xiong L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487:146–52. doi: 10.1016/j.bbrc.2017.03.169. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Li Q, Zhou P, et al. Upregulation of the long noncoding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed Pharmacother. 2017;91:906–11. doi: 10.1016/j.biopha.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Cui Y, Zhang F, Zhu C, et al. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/b-catenin signaling pathway. Oncotarget. 2017;8:17785–94. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang HY, Yang W, Zheng FS, et al. Long noncoding RNA SNHG1 regulates zinc finger E-box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in lung squamous cell carcinoma. Biomed Pharmacother. 2017;90:650–58. doi: 10.1016/j.biopha.2017.03.104. [DOI] [PubMed] [Google Scholar]
- 18.Lan X, Liu X. LncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell Mol Med. 2018 doi: 10.1111/jcmm.13497. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Zhang Z, Xiong L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Bioph Res Co. 2017;1:146–52. doi: 10.1016/j.bbrc.2017.03.169. [DOI] [PubMed] [Google Scholar]
- 20.Gibb EA, Brown CJ, Lam WL. The functional role of long noncoding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jalali S, Bhartiya D, Lalwani MK, et al. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8:e53823. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;3:1180–89. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Wang K, Wei Y, et al. lncRNA-MIAT regulates cell biological behaviors in gastric cancer through a mechanism involving the miR-29a-3p/HDAC4 axis. Oncol Rep. 2017;6:3465–72. doi: 10.3892/or.2017.6020. [DOI] [PubMed] [Google Scholar]
- 24.Cui Y, Zhang F, Zhu C, et al. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8:17785–94. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Z, Jiang C, Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin pathway. Biochem Bioph Res Co. 2018;1:238–45. doi: 10.1016/j.bbrc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Hui Z, Dong Z, Ying M, et al. Expression of long noncoding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Med Sci Monit. 2016;22:4820–29. doi: 10.12659/MSM.898574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Sang L, Liu T, Liu H, et al. MicroRNA-137 suppresses cell migration and invasion in renal cell carcinoma by targeting PIK3R3. Int J Clin Exp Med. 2016;4:7160–67. [Google Scholar]