Abstract
In many microorganisms helical structures are important for motility, e.g., bacterial flagella and kink propagation in Spiroplasma eriocheiris. Motile archaea also form a helical-shaped filament called the ‘archaellum’ that is functionally equivalent to the bacterial flagellum, but structurally resembles type IV pili. The archaellum motor consists of 6–8 proteins called fla accessory genes, and the filament assembly is driven by ATP hydrolysis at catalytic sites in FlaI. Remarkably, previous research using a dark-field microscopy showed that right-handed filaments propelled archaeal cells forwards or backwards by clockwise or counterclockwise rotation, respectively. However, the shape and rotational rate of the archaellum during swimming remained unclear, due to the low signal and lack of temporal resolution. Additionally, the structure and the motor properties of the archaellum and bacterial flagellum have not been precisely determined during swimming because they move freely in three-dimensional space. Recently, we developed an advanced method called “cross-kymography analysis”, which enables us to be a long-term observation and simultaneously quantify the function and morphology of helical structures using a total internal reflection fluorescence microscope. In this review, we introduce the basic idea of this analysis, and summarize the latest information in structural and functional characterization of the archaellum motor.
Keywords: archaea, archaellum, bacterial flagellum, TIRFM, cross-kymography analysis
Significance.
Motile archaea and bacteria move freely towards a better environment in three-dimensional space by rotating a helical filament called the archaellum and bacterial flagellum, respectively. Long-term observation systems of the motility have been gradually developed; however, they are specialized for quantification of functional parameters, not structures. Recently, we developed an advanced method called “cross-kymography analysis”, which enables us to simultaneously quantify the function and morphology of helical structures using a total internal reflection fluorescence microscope. We expect that this technique will be a breakthrough not only for the study of archaeal motility, but also for bacterial motility research.
The motility of organisms is driven by nano-sized molecular machines that convert chemical energy into mechanical work. Eukaryotes (whether myosin, kinesin or dynein) use the free energy of ATP hydrolysis and are responsible for intracellular work such as muscle contraction and cell division [1]. A representative example of bacterial motility is the flagellar rotation of Escherichia coli and Salmonella enterica serovar Typhimurium, which consists of about 30 different kinds of proteins and is attached to the helical filament via the hook structure [2]. The flagellar filament exits outside the bacterial cell body, allowing the cell to swim by rotating the filaments like a screw using the free energy of the ion gradient.
The ‘conventional’ motile systems of these molecular motors have been studied extensively over the last several decades using various approaches such as crystal structure, gene manipulation, and functional analysis with single-molecular techniques. Recently, forms of motility in bacteria without flagella have also been clarified, such as pili that pull [3–5], Mycoplasma legs that walk [6–8], and internal structures that contort [9,10]. In this review, we highlight the motility of the third domain, ‘Archaea’; i.e., the swimming motility driven by archaellar rotation [11]. Additionally, we describe a novel assay called ‘cross-kymography analysis’, which we use to quantify the morphology and function of helical structure in real time [11,12].
Archaea
Archaea were initially regarded as extreme-environment microorganisms. This was because halophilic archaea such as Halobacterium salinarum and Haloferax volcanii in Euryarchaeota require a few M of salt for cultivation, and the highly thermophilic archaeon Sulfolobus acidocaldarius in Crenarchaeota needs more than 60°C to grow [13]. However, archaea have recently been found in normal environments such as sea and soil [14,15]. The shape of archaea is very similar to that of bacteria, and archaea were regarded as identical to bacteria by the end of the 20th century. In 1977, Woese and Fox showed that life forms could be divided into more than prokaryotes and eukaryotes, based on the results of 16 S ribosomal RNA analysis [16]. In 1990, Woese et al. proposed that the living world consists of three domains: eukaryote, bacteria, and archaea [17].
Archaellum
The swimming archaea form a helical filament on the cell surface like swimming bacteria (Fig. 1a and b). In 1984, Alam and Oesterhelt used a dark-field microscope to demonstrate that right-handed filaments propel cells forwards or backwards by rotating filaments in clockwise and counterclockwise directions, respectively [18]. A motility machine of archaea had been thought to look similar to the bacterial flagellum, not only in function but also in structure. However, DNA sequence analyses have shown that archaea lack genes encoding the ring structures, the rod, and the hook, which are part of the bacterial flagellum. Interestingly, archaea do have operons which are a similar in sequence to archaeal and bacterial type IV pili, which serve many purposes such as twitching motility, adhesion, biofilm formation and DNA uptake [19,20]. Some of the motor components of the archaeal flagellum show homologies to components of the type IV pilus assembly apparatus [19,20]. Despite being functionally the same, the gene sequence of archaeal flagellum is completely different from that of bacterial flagellum. To prevent confusion and distinguish the bacterial and archaeal flagellum, Jarrell and Albers proposed that archaeal flagellum should be renamed “archaellum” [20]. This name was initially controversial [21,22], but has gradually been accepted; therefore, we use the archaellum in this review.
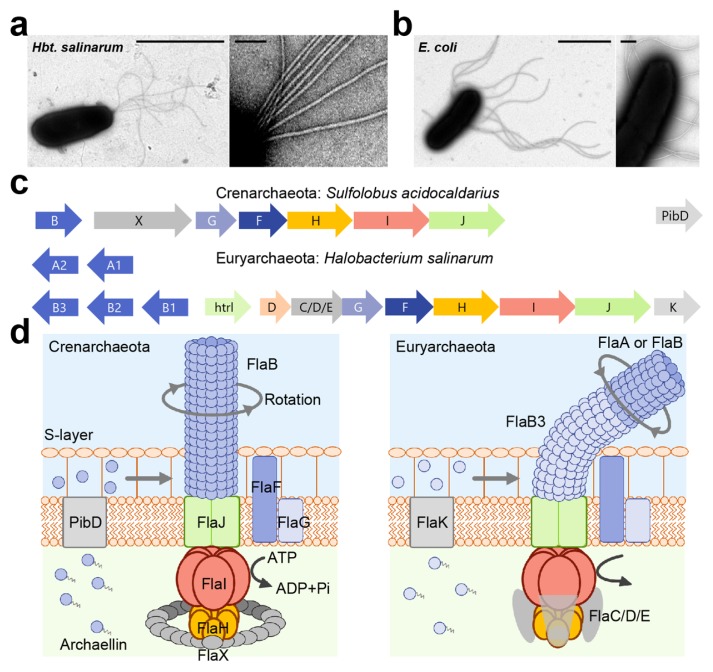
Figure 1.
Motility structure of archaea, archaellum (a) Electron micrograph of Halobacterium salinarum cell (left) and magnified image of archaella (right). Scale bars, 2 μm (left) and 0.1 μm (right). (b) Electron micrograph of Escherichia coli cell (left) and magnified image of flagella (right). Scale bars, 2 μm (left) and 0.2 μm (right). (c) The organization of archaella operon. Homologous genes are represented in the same colors. (d) The current model of archaellar motor. Pre-archaellin has a short signal sequence, and the polymerization proceed after removing this signal sequence by the signal peptidase FlaK/PibD in Euryarchaeota and PibD for Crenarchaeota. In Crenarchaeota, archaellar filament is composed of a single archaellin, FlaB, whereas Euryarchaeota possess the several archaellin genes, flaA or flaB. FlaB3 might play role of hook in Euryarchaeota. Motor is composed of a conserved the membrane protein (FlaJ), the hexametric ATPase (FlaI) and the regulator of ATPase activity (FlaH). The C-terminal domain of FlaI contains the Walker A and B motif for ATP-binding and hydrolysis, and its ATPase activity is essential for not only archaellum assembly and rotation. Nine to ten FlaH molecules assemble in a ring complex and regulates the ATPase activity of FlaI. In addition, Euryarchaeota form FlaC/D/E for motor complex, whereas Crenarchaeota possess FlaX instead. FlaC/D/E play a role for the control of rotational direction by interacting with Che proteins.
Filament assembly
The archaellum motor consists of 6 different kinds of proteins in Crenarchaeota and 8 proteins in Euryarchaeota called fla accessory genes; it is attached to the helical archaellar filament (Fig. 1b and c). Electron microscopic observation revealed that some Euryarchaeota, though not all, formed the curved hook, and that the helical filament is connected with a motor, as seen in bacterial flagella [23].
The archaellin, the filament protein, has a short signal sequence, and is assembled upon signal peptide removal by the signal peptidase FlaK/PibD in Euryarchaeota, and PibD in Crenarchaeota [19]. In Euryarchaeota the filament can consist of several archaellins as seen in many kinds of flagellated bacteria such as Caulobacter crescentus, Vibrio fischeri, and Helicobacter pylori, etc [24–27]. The archaellum of Methanococcus voltae consists of the major archaellins flaB1 and flaB2 and a minor archaellin called flaA; additionally, flaB3 plays the role of hook. The deletion mutant of flaB3 formed archaellar filaments, which did not build a hook, suggesting that the hook proteins are assembled after filament polymerization [28]. This property is one of characteristics that sets the archaellum apart from the bacterial flagellum [2]. Furthermore, a recent study showed that the swimming motility of Haloferax volcanii was improved by the deletion of the minor archaellin flgA2, suggesting that flgA2 regulates the motility and function of the major archaellin flgA1 [29]. Thus far, the reason why archaea possess several archaellins remains elusive, but it is theorized that archaea might form multiple archaellins to stabilize the archaellar structure and function in adapting to extreme environments [30].
Motor complex
All archaella operons consist of FlaF and FlaG, a transmembrane protein FlaJ, the hexametric ATPase FlaI, and the regulator of ATPase activity FlaH [19,20]. FlaF is assumed to act as stator of the archaeal motor, as it was found to interact with the S-layer proteins which are the archaeal outermost cell envelope [31]. FlaJ, a homolog of PilC in type IV pili, is an integral membrane protein and thought to form the archaeal motor platform [32]. The AAA+ ATPase FlaI possesses a Walker A, Walker B motif and a P-loop region, and has a significant homology to PilB in type IV pili and to GspE in the type II secretion system [33]. The hexametric ring structure of FlaI had a diameter of 14 nm, and the difference of the height between the ATP-bound and ADP-bound states was 1 nm. The ΔflaI mutant did not form filaments at the cell surface, indicating that FlaI is essential for the filament assembly [34,35]. Furthermore, a deletion of the first 29 amino acids of FlaI leads to a non-motile phenotype although the archaella are formed on the cell body, indicating that the N-terminal region of FlaI is responsible for archaellar motor rotation [29]. FlaH possesses conserved Walker A motifs, and interacts with FlaI. Remarkably, the RecA domain of FlaH is similar to the clock protein KaiC, suggesting that FlaH might regulate the timing of rotation and archaellar polymerization [36].
Additionally, Euryarchaeota express FlaC/D/E which possibly interact with the motor complex, whereas Crenarchaeota possesses FlaX instead [19]. FlaC/D/E plays a role in the control of rotational direction by interacting with Che proteins [37]. The purified FlaX from Sulfolobus acidocaldarius forms a 30-nm diameter ring structure, and might be essential for stabilization of FlaH, FlaI, and FlaJ [38]. Indeed, 9–10 FlaH molecules assemble inside a FlaX ring complex in vitro [36].
Swimming motility
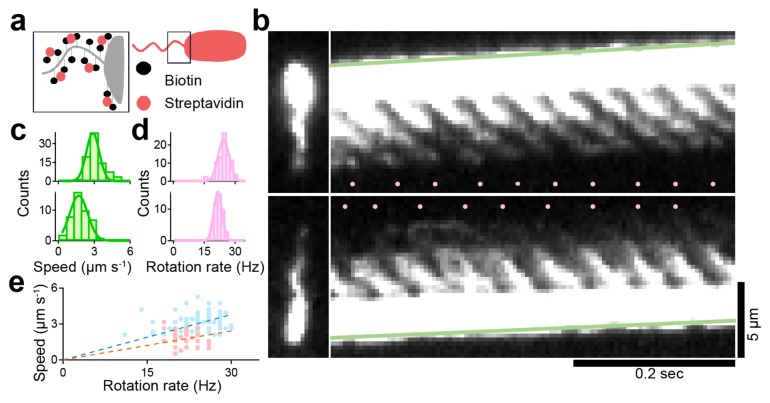
Archaellar rotation was first reported in 1984, but the shape and rotational rate of the archaellum during swimming remained unclear due to the low signal and lack of temporal resolution by a dark-field microscopy [18]. To overcome these problems, we stained the archaellar filaments with a fluorescent dye, Cy3, by biotin-avidin interaction (Fig. 2a). In this study, we used biotin with a long linker between the biotin head that recognizes avidin and the reactive group that recognizes the amino group. We succeeded in visualizing archaellar filaments with a high spatiotemporal resolution up to 2 ms. Remarkably, we also observed the two previously reported swimming modes of archaellar filament(s): pushing and pulling of the cell body [18]. Using kymograph analysis, the swimming speed of cells and rotational rate of archaellar filaments were quantified from the green slope and number of pink dots (Fig. 2b). In pushing and pulling archaella, the ‘swimming speed/rotation rate’ were 2.9±0.5 μm s−1/24.4±2.7 Hz (n=90) and 1.7±0.7 μm s−1/21.7±2.0 Hz (n=44), respectively (Fig. 2c and d). The ratios of swimming speed to archaellar rotation in pushing and pulling were thus calculated to be 0.126 and 0.081 μm (Fig. 2e), respectively, indicating that propulsion distance per rotation of archaellum was almost similar to that of bacterial flagellum [12,39].
Figure 2.
Visualization of archaellar rotation during swimming (a) Experimental-setup. Biotinylated cell is modified with streptavidin-conjugated Cy3. (b) Micrographs (left) and kymographs (right) of examples where archaella rotation pushed (upper) and pulled (lower) the cell body. The green line was drawn at the tip of the cell, so the slope of the line directly represents the swimming speed of the cell. Pink dots in each kymograph represent the moment when intensity spots appeared at the joint between the cell body and the root of the archaella, which corresponded to the rotational rate. (c) Histograms of swimming speed of pusher (upper), puller (lower). The solid lines represent the Gaussian fitting, where the peaks and SDs are 2.9±0.5 in puller and 1.7±0.7 in pusher respectively (puller, n=90; pusher n=44). (d) Histograms of rotation rate of archaellar filaments. The peaks and SDs are 24.4±2.7 in puller and 21.7±2.0 in pusher respectively. (e) Relationship between swimming speed and rotation rate. Blue and pink dots indicate the motility with archaella pushing and pulling the cell body, respectively. Dashed lines represent a linear fitting, with slopes of 0.126 and 0.081 μm per revolution. The figure was reused with permission from Kinosita et al., 2016 with modifications.
Quantification of the rotational rate and morphology of archaellum under TIRFM
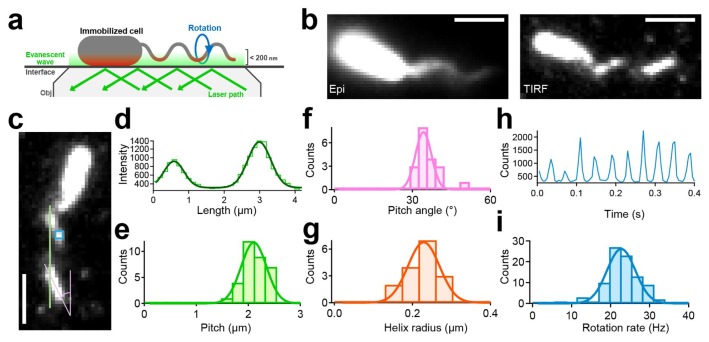
To what can the two different motility modes be attributed? There are two possibilities: (i) the rotational direction of the archaella is opposite; and (ii) the helicity of the filament is opposite. Note that the conventional observation system is not suitable for addressing these possibilities. Because swimming archaea and bacteria move freely in three-dimensional space, the fluorescent signal of filaments often moves out of focus, hindering precise determination of their helix handedness. To overcome this problem, we constructed an advanced assay using a total internal reflection fluorescence microscopy (TIRFM), which has been used for single-molecular measurements (Fig. 3a). With this technique, only the fluorescent signals at <200-nm depth from the glass surface were detected; therefore, the helicity of archaella could be determined by the orientation of the near-surface segment of a filament (Fig. 3b).
Figure 3.
Quantification of the structural and functional parameters of archaellum under TIRFM (a) Schematics of the experimental setup. (b) Fluorescent micrograph taken by epi illumination (left) and TIRF illumination (right). Scale bars, 2 μm. (c) Fluorescent micrograph to explain how to measure the pitch, the helix angle, and the rotation rate. (d) An example of intensity profile of labeled archaella along the green line in c. (e) Histograms of the pitch. Histograms of pitches in archaella structures during CW rotation (n=33) Solid line represents the Gaussian fitting. The peak and SD were 2.11±0.24 μm. (f) Histograms of pitch angle (n=20). The peak and SD were 34±5°. (g) Histograms of helix radii (n=20). The peak and SDs were 0.24±0.04 μm. (h) The typical example of the intensity changes in a fixed area positioned on archaella as shown in c. (i) Histograms of rotation rate. The peak and SD were 22.5±4.5 Hz (n=109). The figure was reused with permission from Kinosita et al., 2016 with modifications.
For the analyses, we defined the configuration as in Figure 3c, with archaella aligned parallel to the north–south orientation, and the cell body located at the north. The east–west orientation depends on the optical system. In conventional settings, a camera is set on the left-side camera port, and images are always mirror images, because the totally reflecting prism inside the inverted microscope reflects, just once, the light coming through the objective (see Fig. S4 in Kinosita et al., 2016). The portion of archaellar filaments in contact with the evanescent field was visible as short lines aligned northwest-southeast relative to the major axis of the cell body, indicating right-handed helicity. In our assay, only cells with right-handed helix were detected, without exception.
In addition to determining archaellar helicity, our assay enabled us to quantify structural parameters such as the archallar pitch, pitch angle, and helix radius from the still image. By drawing the green line along the archaellar filaments in Fig. 3c, multiple peaks could be detected corresponding to the archaellar pitch (Fig. 3d). The pitch angle was directly determined from the still image as the shallow angle of the archaella (pink line in Fig. 3c). With these values, the helix radius could be estimated with the following equation: r=1/2π×λ×tan θ, where r is the helix radius, λ is pitch, and θ is the pitch angle. From this analysis, the pitch, pitch angle, and helix radius were quantified to be 2.1±0.2 μm, 34±5°, and 0.22±0.03 μm, respectively (Fig. 3e–g). Furthermore, our assay could quantify the rotation rate precisely. By fixing on the pixels where the archaella were rotating (blue square in Fig. 3c), oscilation of the intensity could be detected due to the archaellar oscilation (Fig. 3h). Because a diameter of archaellar filaments is ~400 nm, which is a few times wider than the penetration depth of the evanescent field. Using a Fourier transform analysis, the rotational rate of archaellar filaments was estimated to be 22.5±4.5 Hz (Fig. 3i).
Cross-kymography analysis using TIRFM
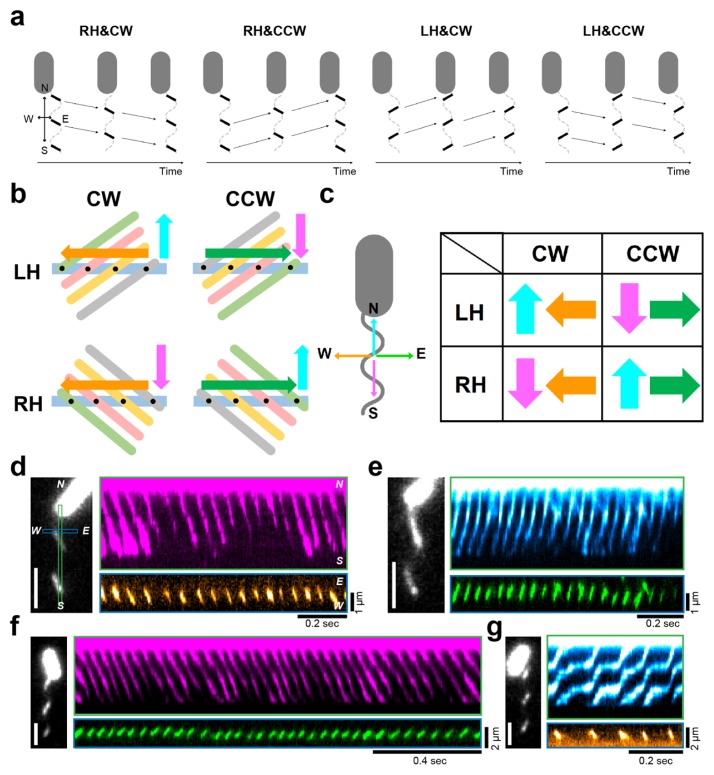
We thus developed a unique method to simultaneously quantify the function and morphology of archaellar filaments; however, rotational direction and handedness might change frequently, as in bacterial flagella. Therefore, we had to delicately check the rotational direction and helicity of the archaella from the obtained movies. To conserve time and effort, we constructed a method we call ‘cross-kymography analysis’. As shown in Figure 3c, we first defined the configuration as archaella aligned parallel to the north–south orientation with the cell body located at the north; additionally, the rotational direction was defined as the observer looking at the archaella from the protrusion outside the cell body. We then created two kinds of kymographs, north-south and east-west. In the north-south kymograph, the right-handed filamentous wave propagates away and towards the cell body, which guaranteed CW and CCW rotation, respectively (Fig. 4a). Meanwhile, the left-handed filamentous wave also propagates towards and away the cell body, which guaranteed CW and CCW rotation, respectively.
Figure 4.
Simultaneous determination of the helicity and rotational direction of archaellum by cross-kymography analysis (a) The schematics of relationships between the direction of rotation (clockwise, CW; counterclockwise, CCW) and helicity (right-handed, RH; left-handed, LH). We first defined the configuration as archaella aligned parallel to the north–south orientation and the cell body located at the north; additionally, the rotation direction is defined as the observer looking at the archaella from the protrusion direction outside the cell body. Note that a camera is set on the left-side camera port, and images are always mirror images because the totally reflecting prism inside the inverted microscope reflects, just once, the light coming through the objective. The portion of filaments that made contacts with the evanescent field was visible as short lines aligned northwest-southeast relative to the major axis of the cell body, indicating the right-handed helicity. On the contrary, the orientation of filaments is northeast-southwest, indicating the left-handed helicity. (b) The schematics to explain how to determine the helicity from the east-west kymograph. Light-blue rectangle represents the location to make the east-west kymograph. Cyan and pink arrow indicated the direction of wave propagation. Gray, orange, magenta and green ellipse represents the time course of wave propagation. Therefore, we could detect the transition of fluorescent signal at the rectangle as shown by orange or green arrows. (c) Left: definition of axes. Right: matrix of combinations to determine the directional rotation and helicity of helical structure from cross-kymography, which is the analysis of two kymographs from two different lines in the same image that perpendicularly align with each other in the region of interest. (d–g) Left: Fluorescent micrograph. Right: two kymographs taken from orthogonally oriented lines on left micrograph. In north-south kymograph, the right-handed archaellar wave propagates away (d) and towards (e) the cell body, which guaranteed the CW and CCW rotation, respectively. On the contrary, the left-handed flagellar wave propagates away (f) and towards (g) the cell body, which guaranteed the CCW and CW rotation, respectively. In the east-west kymograph, the helicity could be uniformly determine as mentioned above. Pseudo-colors of kymographs indicate the direction of spot propagation, which correspond to c. The figure was reused with permission from Kinosita et al., 2016 with modifications.
We could then uniformly determine helicity from the east-west kymograph. To explain the mechanism, we drew diagrams with four possible relationships between helicity and rotational direction; i.e., right-handed/CW, right-handed/ CCW, left-handed/CW, and left-handed/CCW (Fig. 4b). If the right-handed filament rotates in the CW direction, the wave propagates from east to west in the east-west kymograph analysis; if the wave propagates from west to east, the right-handed filament rotates in the CCW direction. Similarly, we could determine the direction of wave propagation when left-handed filaments rotated in either direction. We schematically summarized the relationship between rotational direction and helicity (Fig. 4c); consequently, the relationship can be uniformly determined from four patterns. Using this technique, we observed that the right-handed filamentous of Hbt. salinarum rotated in CW direction (Fig. 4d) and CCW direction (Fig. 4e); additionally, the left-handed filamentous of S. typhimurium also rotated in CCW direction (Fig. 4f) and in CW direction (Fig. 4g). Furthermore, we first demonstrated that the helicity of the archaellum is right-handed, even if the motor switches the direction of rotation, which was in stark contrast to the feature of bacterial flagellar filament.
Future prospects
We constructed the cross-kymography analysis under TIRFM, and expect that this technique will be a breakthrough not only for the study of archaeal motility, but also for bacterial motility research [11–12]. Flagellated bacteria show motility by swimming towards a better environment, changing rotational direction and transforming flagellar helicity [2]. Long-term observation systems have been gradually developed; however, these methods are specialized for quantification of functional parameters such as rotational rate of filaments, not flagellar structures [40–42]. Our method could reveal “change of rotational direction of flagella” and “transformation of flagellar shape by switching” simultaneously in real time and also verify the model that the flagellar filament modulates the switching frequency of the motor [43].
There are many spiral-shaped bacteria in nature; e.g., Spiroplasma eriocheiris. It shows a swimming motility in highly viscous environments by propagation of kink pairs along the cell body from front to back [44]. This propagation is considered to be driven by rotation, but there are unclear points on how the rotation is generated and transforms the helical structure of the cell body [45]. Our method will be helpful in clarifying the novel motility mechanisms of unconventional bacteria by simultaneously monitoring the function and morphology of helical structures. Finally, microscopic measurements using an optical microscope have gradually clarified the function of the archaeal motor. However, many big mysteries remain, e.g., why archaellar filaments maintain right-handedness, and how motor switch and torque is generated at a molecular level. We expect that biophysicists will join this new research field and tackle these problems with various approaches, including crystal structure, cryo-electron microscope, and the single-molecular approach.
Acknowledgements
We thank Sonja-Verena Albers for preparing the manuscript and N. Uchida and D. Nakane for helping the original experiment. This work was supported in part by the Funding Program for Next-Generation World-Leading Researchers Grant LR033 (to T. N.) from the Japan Society for the Promotion of Science, by a Grant-in-Aid for Scientific Research on Innovative Areas “Harmonized Supramolecular Motility Machinery and Its Diversity” (to T. N.) and “Fluctuation & Structure” (Grant No. 26103527 to T. N.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. Y. K was recipient of JSPS Fellowship for Japan Junior Scientists and postdoctoral fellowship for research abroad from JSPS.
Footnotes
Conflicts of interest
The authors declare no competing financial interests.
Author contribution
Y. K and T. N wrote the paper.
References
- 1.Veigel C, Schmidt CF. Moving into the cell: single-molecule studies of molecular motors in complex environments. Nat Rev Mol Cell Biol. 2011;12:163–176. doi: 10.1038/nrm3062. [DOI] [PubMed] [Google Scholar]
- 2.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 3.Maier B, Potter L, So M, Long CD, Seifert HS, Sheetz MP. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci USA. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 5.Nakane D, Nishizaka T. Asymmetric distribution of type IV pili triggered by directional light in unicellular cyanobacteria. Proc Natl Acad Sci USA. 2017;114:6593–6598. doi: 10.1073/pnas.1702395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinosita Y, Nakane D, Sugawa M, Masaike T, Mizutani K, Miyata M, et al. Unitary step of gliding machinery in Mycoplasma mobile. Proc Natl Acad Sci USA. 2014;111:8601–8606. doi: 10.1073/pnas.1310355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyata M. Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol. 2010;64:519–537. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 8.Miyata M, Hamaguchi T. Prospects for the gliding mechanism of Mycoplasma mobile. Curr Opin Microbiol. 2016;29:15–21. doi: 10.1016/j.mib.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Faure LM, Fiche JB, Espinosa L, Ducret A, Anantharaman V, Luciano J, et al. The mechanism of force transmission at bacterial focal adhesion complexes. Nature. 2016;539:530–535. doi: 10.1038/nature20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinosita Y, Uchida N, Nakane D, Nishizaka T. Direct observation of rotation and steps of the archaellum in the swimming halophilic archaeon Halobacterium salinarum. Nat Microbiol. 2016;1:16148. doi: 10.1038/nmicrobiol.2016.148. [DOI] [PubMed] [Google Scholar]
- 12.Kinosita Y, Kikuchi Y, Mikami N, Nakane D, Nishizaka T. Unforeseen swimming and gliding mode of an insect gut symbiont, Burkholderia sp. RPE64, with wrapping of the flagella around its cell body. ISME J. 2018;12:838–848. doi: 10.1038/s41396-017-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herzog B, Wirth R. Swimming behavior of selected species of Archaea. Appl Environ Microbiol. 2012;78:1670–1674. doi: 10.1128/AEM.06723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 15.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 16.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alam M, Oesterhelt D. Morphology, function and isolation of halobacterial flagella. J Mol Biol. 1984;176:459–475. doi: 10.1016/0022-2836(84)90172-4. [DOI] [PubMed] [Google Scholar]
- 19.Albers SV, Jarrell KF. The archaellum: how Archaea swim. Front Microbiol. 2015;6:23. doi: 10.3389/fmicb.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarrell KF, Albers SV. The archaellum: an old motility structure with a new name. Trends Microbiol. 2012;20:307–312. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Eichler J. Response to Jarrell and Albers: the name says it all. Trends Microbiol. 2012;20:512–513. doi: 10.1016/j.tim.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Wirth R. Response to Jarrell and Albers: seven letters less does not say more. Trends Microbiol. 2012;20:511–512. doi: 10.1016/j.tim.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Bardy SL, Mori T, Komoriya K, Aizawa S, Jarrell KF. Identification and localization of flagellins FlaA and FlaB3 within flagella of Methanococcus voltae. J Bacteriol. 2002;184:5223–5233. doi: 10.1128/JB.184.19.5223-5233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ely B, Ely TW, Crymes WB, Jr., Minnich SA. A family of six flagellin genes contributes to the Caulobacter crescentus flagellar filament. J Bacteriol. 2000;182:5001–5004. doi: 10.1128/jb.182.17.5001-5004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faulds-Pain A, Birchall C, Aldridge C, Smith WD, Grimaldi G, Nakamura S, et al. Flagellin redundancy in Caulobacter crescentus and its implications for flagellar filament assembly. J Bacteriol. 2011;193:2695–2707. doi: 10.1128/JB.01172-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostrzynska M, Betts JD, Austin JW, Trust TJ. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J Bacteriol. 1991;173:937–946. doi: 10.1128/jb.173.3.937-946.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millikan DS, Ruby EG. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol. 2004;186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaban B, Ng SY, Kanbe M, Saltzman I, Nimmo G, Aizawa S, et al. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol Microbiol. 2007;66:596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- 29.Tripepi M, Esquivel RN, Wirth R, Pohlschroder M. Haloferax volcanii cells lacking the flagellin FlgA2 are hypermotile. Microbiology. 2013;159:2249–2258. doi: 10.1099/mic.0.069617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Syutkin AS, Pyatibratov MG, Galzitskaya OV, Rodriguez-Valera F, Fedorov OV. Haloarcula marismortui archaellin genes as ecoparalogs. Extremophiles. 2014;18:341–349. doi: 10.1007/s00792-013-0619-4. [DOI] [PubMed] [Google Scholar]
- 31.Banerjee A, Tsai CL, Chaudhury P, Tripp P, Arvai AS, Ishida JP, et al. FlaF Is a beta-Sandwich Protein that Anchors the Archaellum in the Archaeal Cell Envelope by Binding the S-Layer Protein. Structure. 2015;23:863–872. doi: 10.1016/j.str.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- 33.Reindl S, Ghosh A, Williams GJ, Lassak K, Neiner T, Henche AL, et al. Insights into FlaI functions in archaeal motor assembly and motility from structures, conformations, and genetics. Mol Cell. 2013;49:1069–1082. doi: 10.1016/j.molcel.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patenge N, Berendes A, Engelhardt H, Schuster SC, Oesterhelt D. The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum. Mol Microbiol. 2001;41:653–663. doi: 10.1046/j.1365-2958.2001.02542.x. [DOI] [PubMed] [Google Scholar]
- 35.Thomas NA, Mueller S, Klein A, Jarrell KF. Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. Mol Microbiol. 2002;46:879–887. doi: 10.1046/j.1365-2958.2002.03220.x. [DOI] [PubMed] [Google Scholar]
- 36.Chaudhury P, Neiner T, D’Imprima E, Banerjee A, Reindl S, Ghosh A, et al. The nucleotide-dependent interaction of FlaH and FlaI is essential for assembly and function of the archaellum motor. Mol Microbiol. 2016;99:674–685. doi: 10.1111/mmi.13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlesner M, Miller A, Streif S, Staudinger WF, Muller J, Scheffer B, et al. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol. 2009;9:56. doi: 10.1186/1471-2180-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banerjee A, Ghosh A, Mills DJ, Kahnt J, Vonck J, Albers SV. FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI. J Biol Chem. 2012;287:43322–43330. doi: 10.1074/jbc.M112.414383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magariyama Y, Sugiyama S, Kudo S. Bacterial swimming speed and rotation rate of bundled flagella. FEMS Microbiol Lett. 2001;199:125–129. doi: 10.1111/j.1574-6968.2001.tb10662.x. [DOI] [PubMed] [Google Scholar]
- 40.Chattopadhyay S, Moldovan R, Yeung C, Wu XL. Swimming efficiency of bacterium Escherichia coli. Proc Natl Acad Sci USA. 2006;103:13712–13717. doi: 10.1073/pnas.0602043103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mears PJ, Koirala S, Rao CV, Golding I, Chemla YR. Escherichia coli swimming is robust against variations in flagellar number. Elife. 2014;3:e01916. doi: 10.7554/eLife.01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min TL, Mears PJ, Chubiz LM, Rao CV, Golding I, Chemla YR. High-resolution, long-term characterization of bacterial motility using optical tweezers. Nat Methods. 2009;6:831–835. doi: 10.1038/nmeth.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Albada SB, Tanase-Nicola S, ten Wolde PR. The switching dynamics of the bacterial flagellar motor. Mol Syst Biol. 2009;5:316. doi: 10.1038/msb.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaevitz JW, Lee JY, Fletcher DA. Spiroplasma swim by a processive change in body helicity. Cell. 2005;122:941–945. doi: 10.1016/j.cell.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Wada H, Netz RR. Model for self-propulsive helical filaments: kink-pair propagation. Phys Rev Lett. 2007;99:108102. doi: 10.1103/PhysRevLett.99.108102. [DOI] [PubMed] [Google Scholar]