Abstract
Echinoderms exhibit several fascinating evolutionary innovations that are rarely seen in the animal kingdom, but how these animals attained such features is not well understood. Here we report the sequencing and analysis of the genome and extensive transcriptomes of the sea cucumber Apostichopus japonicus, a species from a special echinoderm group with extraordinary potential for saponin synthesis, aestivation and organ regeneration. The sea cucumber does not possess a reorganized Hox cluster as previously assumed for all echinoderms, and the spatial expression of Hox7 and Hox11/13b potentially guides the embryo-to-larva axial transformation. Contrary to the typical production of lanosterol in animal cholesterol synthesis, the oxidosqualene cyclase of sea cucumber produces parkeol for saponin synthesis and has “plant-like” motifs suggestive of convergent evolution. The transcriptional factors Klf2 and Egr1 are identified as key regulators of aestivation, probably exerting their effects through a clock gene-controlled process. Intestinal hypometabolism during aestivation is driven by the DNA hypermethylation of various metabolic gene pathways, whereas the transcriptional network of intestine regeneration involves diverse signaling pathways, including Wnt, Hippo and FGF. Decoding the sea cucumber genome provides a new avenue for an in-depth understanding of the extraordinary features of sea cucumbers and other echinoderms.
Introduction
Echinoderms, which first appeared in the early Cambrian period1, represent the second largest group of deuterostomes, and together with their sister phylum Hemichordata, they occupy a critical phylogenetic position for understanding the evolutionary origin of chordates2. The radiation of echinoderms was believed to be responsible for the Mesozoic Marine Revolution3, and they can adapt to various oceanic environments, even to the biotic desert of the deep sea. Their superb adaptation attributes include several fascinating evolutionary innovations that are rarely seen in the animal kingdom such as a pentaradial body plan with rigid calcitic skeletons4, biosynthesis of saponins5, and extraordinary potential for regeneration6. However, our understanding of their biology and evolution is hindered by poor sampling of their genomes7–10, and their relatively large, highly heterozygous genomes generally challenge our ability to obtain high-quality assemblies8,11,12.
Sea cucumbers (Supplementary Figure S1), praised as the “jewel of the seabed”13, are a special echinoderm group that possesses the well-known striking features of echinoderms and are more distinctive than other echinoderm groups in aspects of saponin biosynthesis, aestivation and visceral regeneration. Sea cucumbers can produce saponins (also known as holothurins), which act as chemical defense agents against predators and parasites14,15 and are also highly valued bioactive compounds with many appealing biomedical properties, such as anti-cancer, anti-inflammatory, anti-bacterial and immunomodulatory effects (sea cucumbers are sometimes referred to as “Marine Ginseng” in Asian countries)16–18. Saponins are widespread in plants but are rarely found in the animal kingdom19,20, and how sea cucumbers gained the ability to synthesize saponins remains enigmatic. In response to high temperatures, sea cucumbers can protect themselves by entering a physiological state called aestivation, which is a process that can last for up to 4 months21 and is characterized by inactivity, feeding cessation, intestine degeneration and metabolic rate depression22,23. Sea cucumbers also possess another defensive mechanism called evisceration, with which they can expel internal organs (e.g., intestine and respiratory tree) out of their body when they get stressed, and the missing organs can be regenerated concurrently within 7 days24,25. Although the aestivation and regeneration properties of sea cucumbers have received increasing research attention23, the molecular regulatory mechanisms underlying these interesting phenomena remain obscure.
The sea cucumber Apostichopus japonicus (Selenka 1867), represents one of the most dominant and economically important species in the Western Pacific Ocean along the coasts of China, Japan, Korea and Russia, with considerable edible and medicinal value26. It is also among the best molecularly characterized sea cucumber species27–30, making it a good candidate for whole-genome sequencing. Recent sequencing efforts have been devoted to understanding its morphological evolution and visceral regeneration10,12, but their saponin synthesis and aestivation abilities remain poorly explored at whole genome levels. Here, we report sequencing of the genome and extensive transcriptomes of A. japonicus. Our analysis revealed novel genomic features and molecular changes that may contribute to the evolutionary innovations of sea cucumber or echinoderm-characteristic adaptive traits, providing insights into saponin synthesis and aestivation regulation.
Results
Genome sequencing, assembly and characterization
Like other marine invertebrates31–34, echinoderm genomes are generally difficult to sequence and assemble due to their relatively large genomes and/or remarkably high genome heterozygosity8. By taking advantage of both Illumina short-read and PacBio long-read sequencing technologies, we performed whole-genome shotgun sequencing of a wild individual of A. japonicus, which produced 352 Gb of clean data, corresponding to an average genome coverage of 370× (Supplementary Table S1). The final genome assembly was 952 Mb with a contig N50 of 45 kb and a scaffold N50 of 196 kb, and over 90% of the assembly was covered by the longest 4,784 scaffolds (>63 kb) (Supplementary Tables S2, S3). The total assembly length was close to the estimates from k-mer analysis (~1.0 Gb; Supplementary Figure S2) and flow cytometry analysis35 (~0.9 Gb; Supplementary Figure S3). The integrity of the assembly was demonstrated by mapping 97.4–99.3% of the transcriptome data sets (Supplementary Table S4). The assembly was further anchored to chromosomes based on a high-density genetic linkage map28 by assigning 1,949 scaffolds to 22 linkage groups (Fig. 1; Supplementary Table S5).
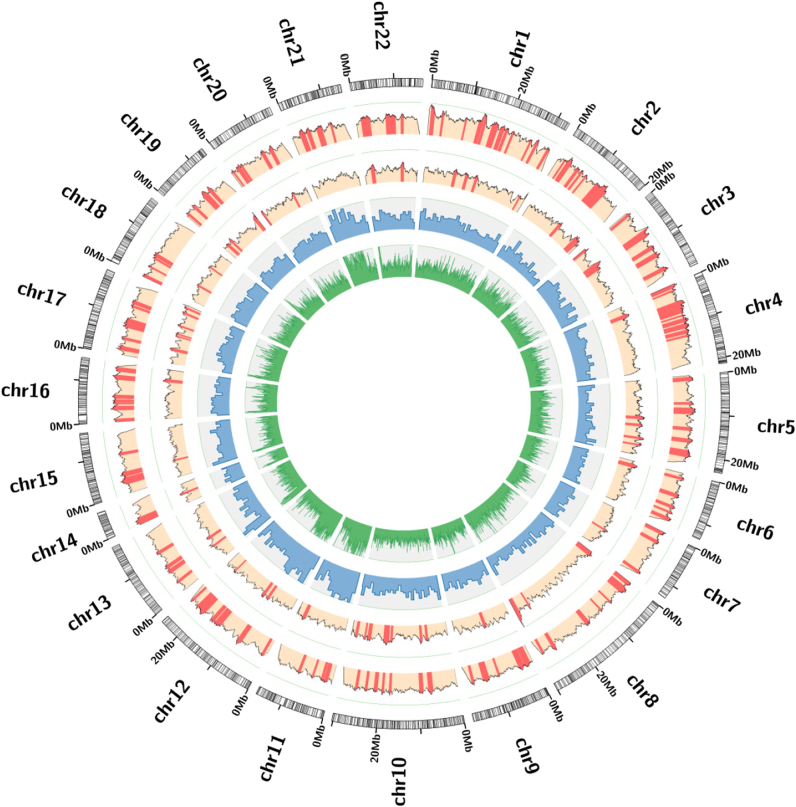
Fig. 1. Genome landscape and polymorphism analysis of the sea cucumber Apostichopus japonicus.
From outer to inner circles: I, marker distribution on 22 chromosomes at the Mb scale; II and III, single nucleotide polymorphism (SNP) density across genome (II) or CDS (III) drawn in 1-Mb sliding windows with a 50-kb step (yellow columns), and polymorphism hotspot regions (p < 1e–4) are colored red; IV and V, gene density and repeat density across the genome, respectively, drawn in 1-Mb non-overlapping windows
The sea cucumber genome contains 29,451 protein-coding genes supported by known protein sequences and/or transcriptomic data (Supplementary Table S6). Functional analysis via comparison with various public protein databases annotated all of the predicted genes (Supplementary Figure S4; Supplementary Tables S7, S8). The sea cucumber genome contains 254 Mb of repetitive sequences accounting for 26% of the genome. This percentage is lower than those of other echinoderm genomes (26-41%; Supplementary Table S9). DNA transposons represent the most abundant repeat type (3.5%), followed by long interspersed elements (2.1%) and tandem repeats (2.0%) (Supplementary Table S9; Supplementary Figure S5).
We conducted a comprehensive comparison of our assembly with the two A. japonicus assemblies generated in previous studies10,12. In terms of assembly quality, ours and Zhang’s assembly is much better than Jo’s version (Supplementary Table S10). Although Zhang’s assembly has higher contiguity than ours, gene completeness is comparable between the two assemblies (Supplementary Table S10), suggesting that the quality of both assemblies is sufficiently high for many downstream analyses. The majority of genes and pathways mentioned by Zhang et al.10 relating to morphological evolution and visceral regeneration could be found in our assembly with largely consistent biological and/or evolutionary characteristics (Supplementary Tables S11–S14).
Polymorphism analysis identified 2.43 million single-nucleotide polymorphisms (SNPs) in the assembled individual. A genome-wide scan of polymorphism based on the assembled individual and 8 additional resequenced individuals (Supplementary Table S15) identified 110 highly polymorphic genomic regions (≥500 kb) in the genome (Fig. 1). SNP density in coding sequences (CDSs) varies dramatically among genes, ranging from 0 to 222 SNPs per kb (Fig. 1). We identified 3,241 highly polymorphic genes (HPGs) that are statistically significant relative to the chromosomal background (Supplementary Table S16). Functional enrichment analysis of these HPGs reveals that they are involved in diverse cellular functions or biological processes such as catabolic processes, reproductive processes, regulation of homeostatic processes, and regulation of signaling (Supplementary Table S17). These HPGs may contribute to the sea cucumber’s superb adaptation by enhancing the plasticity of its coding repertoire.
Hox/ParaHox clusters and body plan evolution
Echinoderms, albeit evolving from the same bilaterian ancestor that gave rise to chordates, display a variety of highly derived, Cambrian-derived body architectures36 for which the molecular driving force(s) remain poorly understood37. Understanding the organization of echinoderm Hox clusters is of particular interest, as Hox genes play a key role in animal body plan determination38. Much of our previous knowledge came from the sea urchin Strongylocentrotus purpuratus, leading to a prevailing hypothesis that the echinoderm pentameral body plan was a consequence of a reorganized Hox cluster39,40. This hypothesis was, however, recently challenged by the finding of an intact Hox cluster in the sea star Acanthaster planci37,41. Coinciding with the new report by Zhang et al.10, we identified a Hox cluster and a ParaHox cluster in the sea cucumber genome, and the gene composition and orientation of both clusters was highly consistent with those of the sea star Hox and ParaHox clusters (Fig. 2a; Supplementary Figure S6). The finding of typical Hox clusters in both sea cucumber and sea star suggests that it was unlikely that the ancestor of echinoderms possessed a reorganized Hox cluster as found in sea urchin, thus invalidating the previous hypothesis correlating the derived pentameral body plan with the reorganized Hox cluster40. Despite having a typical Hox cluster, the sea cucumber lacks Hox4 and Hox6, and the latter is also absent in Echinoidea, Asteroidea and Ophiuroidea, implicating that the loss of Hox6 might have already occurred before the split of Echinozoa and Asterozoa.
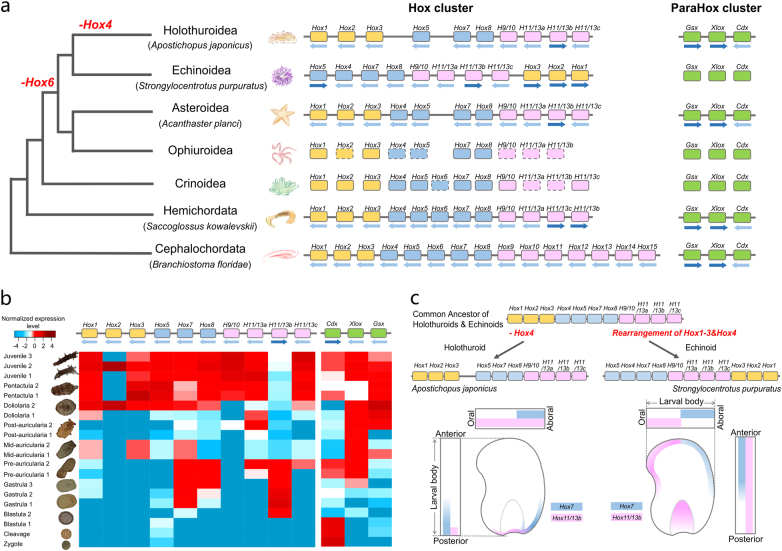
Fig. 2. The cluster organization and tempo-spatial expression of Hox genes during the development of A. japonicus.
a Cluster organization of the Hox and ParaHox genes of the sea cucumber A. japonicus and other echinoderms37,41,112. The sea cucumber has a typical Hox cluster similar to that of the sea star A. planci, invalidating the previous hypothesis that Hox clusters of all echinoderms are reorganized39,40. The genes whose identity existed ambiguously are shown with rectangles in dashed lines. b Temporal expression of the sea cucumber Hox and ParaHox cluster genes. Contrary to their ParaHox counterpart, expression of the sea cucumber Hox cluster during development does not exhibit temporal colinearity as typically found in chordate Hox clusters. Compared with other Hox genes, Hox7 and Hox11/13b show prominent expression during gastrulation, likely participating in determination of the larval body plan. c Inferred Hox cluster evolution and spatial expression of Hox7 and Hox11/13b in sea cucumber and sea urchins. Presumably the common ancestor of holothuroids and echinoids contained a typical Hox cluster without Hox6, which is largely preserved in the sea cucumber lineage (except the loss of Hox4) but had undergone a few rearrangements in the sea urchin lineage. The spatial expression of Hox7 and Hox11/13b shows colinearity during gastrulationalong the anterior/posterior (A/P) axis for the sea cucumber A. japonicus45 and along the oral/aboral (O/A) axis for sea urchins46–48. This finding probably suggests the important roles of the two Hox genes in guiding axial transformation from the embryonic to larval stage in echinozoans
During development, most echinoderms rearrange their body plans through drastic axial transformation, e.g., from embryonic anterior/posterior (A/P) axis to larval/adult oral/aboral (O/A) axis, with changes from 0° in holothurians, 90° in echinoids to 180° in crinoids42. To gain insights into this poorly understood process, we investigated the expression dynamics of all Hox genes during sea cucumber embryonic and larval stages. We found that contrary to their ParaHox counterpart, the expression of the sea cucumber Hox cluster during development did not exhibit temporal colinearity as typically found in chordate Hox clusters43 (Fig. 2b), which is possibly related to the derived body plan. Compared with other Hox genes that are mostly expressed after metamorphosis, sea cucumber Hox7 and Hox11/13b show prominent expression during gastrulation (Fig. 2b), and they are also the only two genes significantly expressed in sea urchin embryos44. Further investigation of the spatial expression of Hox7 and Hox11/13b in sea cucumber45 and sea urchins46–48 revealed spatial colinearity during gastrulation but in different axial directions, i.e., A/P axis for sea cucumber and O/A axis for sea urchin (Fig. 2c), corresponding to their larval/adult axial directions (i.e., no axial change in sea cucumber, whereas there was a 90° change in sea urchin). This interesting finding suggests that Hox7 and Hox11/13b might play an important role in guiding axial transformation from the embryonic to larval stage. Considering that axial transformation before larval metamorphosis only occurs in the sea urchin lineage, it is likely that such a unique transformation in sea urchin could be related to the reorganization of its Hox cluster (i.e., the translocation of Hox1-Hox3; Fig. 2c).
Saponin biosynthesis and convergent evolution
Saponins are secondary metabolites that are commonly present in the plant kingdom but are only found in a few animal lineages, including sea cucumber19. Sea cucumber saponins (also known as holothurins) belong to the triterpene glycoside family of natural products16. Current knowledge about saponin biosynthesis has come mostly from plant studies19,20, and how sea cucumbers gained the ability to synthesize saponins is a source of intrigue. As both sterols and triterpenes are synthesized via the mevalonate (MVA) pathway49, we first investigated the integrity of the MVA pathway in sea cucumber for the route of cholesterol synthesis in animals, and we found that the two genes Cyp51 and Dhcr7 were absent in the sea cucumber genome (Fig. 3a; Supplementary Table S18). This finding, in contrast to the observation of full gene sets in sea urchin S. purpuratus and starfish A. planci (Fig. 3a; Supplementary Table S18), suggests that the sea cucumber A. japonicus might have lost the ability to synthesize cholesterol de novo, which is consistent with previous observations of extremely low cholesterol levels in sea cucumber50. In addition, the absence of Cyp51 (i.e., C-14 sterol demethylase) in the sea cucumber genome also supports the previous speculation that the blockage of C-14 demethylation leads to the accumulation of 14α-methylated Δ9(11)-sterols in cell membranes of sea cucumber, contributing to resistance to their own toxins15.
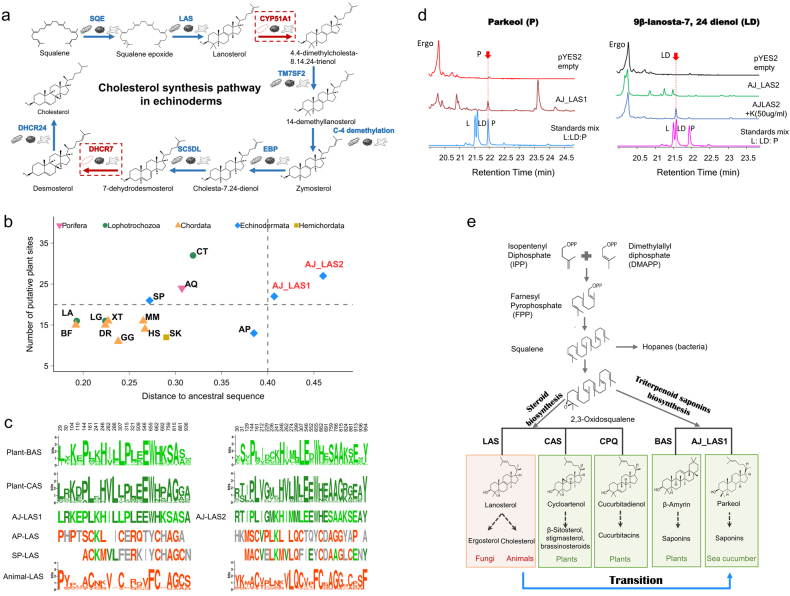
Fig. 3. Saponin biosynthesis and convergent evolution of LAS genes in A. japonicus.
a Gene representation of the canonical animal cholesterol synthesis pathway in the sea cucumber genome. The sea cucumber lacks two genes, Cyp51 and Dhcr7, suggesting that it might have lost its de novo cholesterol synthesis ability, consistent with the previous observation of extremely low cholesterol content in sea cucumber50. b Evolutionary analysis of the lanosterol synthase (LAS) genes in sea cucumber and other animals. Compared to the inferred ancestral bilaterian LAS sequence (see Methods for details), LAS1 and LAS2 in sea cucumber show the highest sequence divergence and possess more putative plant sites than most other animals. c Overview of plant-like motifs in the sea cucumber LAS sequences and comparison with those of the animal consensus LAS sequence and plant consensus BAS and CAS sequences. The plant-like motifs in the sea cucumber LAS sequences are not present in sea urchin and starfish, suggesting the de novo acquirement of these motifs in the sea cucumber lineage. d Product determination by yeast expression of sea cucumber LAS1 and LAS2. Contrary to the general expectation that animal LAS produces lanosterol, neither sea cucumber LAS1 nor LAS2 produces lanosterol. Sea cucumber LAS1 produces parkeol (previously identified as the triterpene precursor of sea cucumber saponins;51), whereas LAS2 produces 9β-lanosta-7, 24 dienol. e Summary of the pathways leading to saponin biosynthesis and steroid biosynthesis. In contrast to the plant kingdom, saponin biosynthesis is rarely found in the animal kingdom19,20. The extraordinary ability of saponin synthesis in sea cucumber is enabled by its modification of lanosterol synthase, which possibly occurred through convergent evolution
Sterols and triterpenes share the common biosynthetic precursor 2,3-oxidosqualene, which can be cyclized by different oxidosqualene cyclases (OSCs) to produce sterols [lanosterol synthase (LAS) in fungi and animals and cycloartenol synthase (CAS)/cucurbitadienol synthase (CPQ) in plants] or triterpenes [e.g., β-amyrin synthase (BAS) in plants]20 (Fig. 3e). We found two predicted OSC genes (named in this study as LAS1 and LAS2) in the sea cucumber genome (Supplementary Figures S7, S8). Evolutionary analysis suggests that the sea cucumber LAS genes show high evolutionary rates compared with diverse animal groups, and contain many plant-like motifs that are not present in sea urchin and starfish (Fig. 3b, c; Supplementary Figure S9). Contrary to the general expectation that cyclization of 2,3-oxidosqualene by animal LAS produces lanosterol, functional analysis of yeast expressing A. japanicus LAS1 and LAS2 revealed that neither produces lanosterol. Instead the main products were identified as parkeol (LAS1) and 9β-lanosta-7, 24-dienol (LAS2) (Fig. 3d; Supplementary Figures S10-S12). Parkeol has previously been suggested to be the triterpene precursor of saponins in sea cucumbers51, although whether 9β-lanosta-7, 24 dienol is also a saponin precursor remains to be determined. Previous studies suggested that the LASs of different eukaryotic lineages emerged from an ancestral plant-like CAS that is capable of producing both parkeol and cycloartenol52. A switch of LAS product specificity from lanosterol to parkeol may reflect the convergent evolution of sea cucumber LAS (prone to plant-like CAS; Fig. 3e), enabling the evolutionary appearance of effective chemical tools of defense in sea cucumbers.
Key regulators and transcriptional network of aestivation
Studies on aestivation to date have focused primarily on terrestrial animals such as amphibians and land snails53, with relatively little attention paid to understanding animal aestivation in marine environments23,54. To understand the regulatory mechanism of sea cucumber aestivation (Supplementary Figure S13), we conducted large-scale transcriptome sequencing by generating 39 data sets (Supplementary Table S20) from multiple organs (body wall, muscle, respiratory tree and intestine) across different aestivation states. Body wall showed the most differential expressed genes (DEGs) and differentially expressed transcriptional factors (DE-TFs), followed by muscle, respiratory tree and intestine (Fig. 4a; Supplementary Figures S14, S15; Supplementary Tables S21–S25), suggesting that body wall may be more sensitive in response to thermal stress than other organs. Nine TFs showed differential expression during aestivation in all four organs, of which Klf2 and Egr1 were the most significant TFs especially in body wall (Fig. 4b). Egr1 and members of the Klf family are known to participate in biorhythm regulation55–58, e. g., Egr1 can regulate the clock gene Cry155, the key repressor gene that can propel the animal into sleep phase (through the inhibition of key activators Clock and Bmal159). We therefore investigated the expression patterns of clock-related genes during the aestivation of sea cucumber. To ensure a reliable evaluation and inference, two analytical approaches (RNAseq and quantitative PCR) were adopted, both of which produced largely consistent results (Fig. 4c; Supplementary Figure S16). Expression patterns of Cry1 in body wall coincide with the aestivation states of sea cucumber, with increased expression during early and deep aestivation and decreased expression after aestivation (Fig. 4c; Supplementary Figure S16). The expression of key activators Clock and Bmal1 is suppressed during deep aestivation (corresponding to the enhanced role of the repressor Cry1; Fig. 4c; Supplementary Figure S16), and is re-activated with emerging arousal from aestivation (corresponding to the decreasing effect of Cry1; Fig. 4c; Supplementary Figure S16). These observations suggest that sea cucumber aestivation, such as other types of dormancy60,61, might be a clock gene-controlled process with an extended sleep phase possibly triggered by Egr1 and/or Klf2 (Fig. 4c).
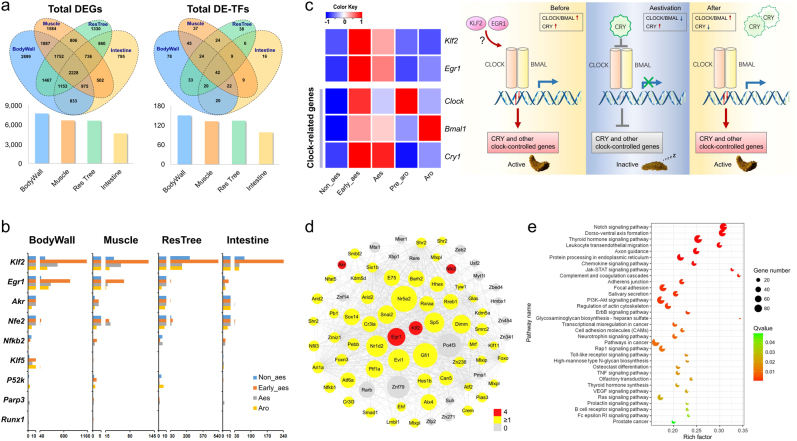
Fig. 4. Key regulators and transcriptional network of aestivation in A. japonicus.
a Identification of differentially expressed genes (DEGs) and differentially expressed transcriptional factors (DE-TFs) in four organs (body wall, muscle, respiratory tree and intestine) during different states of aestivation. Venn diagrams and histograms show the shared gene numbers between organs and the absolute gene numbers in each organ. Among the four organs, body wall shows the most DEGs and DE-TFs, representing the most responsive organ during sea cucumber aestivation. b Expression profiles of nine TFs showing differential expression during aestivation in all four organs. Compared with other TFs, Klf2 and Egr1 are the most significant TFs, especially in body wall, likely playing important roles in the regulation of aestivation. Aestivation states: non-aestivation (Non_aes); early aestivation (Early_aes); deep aestivation (aes); and arousal from aestivation (Aro). c Expression heatmap of Klf2, Egr1 and clock-related genes during different aestivation states (according to quantitative PCR results), and the inferred clock gene-controlled regulation model. Klf2 and Egr1 may trigger the upregulation of Cry1 (either directly or indirectly through Clock and Bmal1) during sea cucumber aestivation, which propels the animal into an extended sleep phase, and decreased Cry1 expression makes the animal awaken from aestivation. Aestivation states are the same as depicted in (b) except Pre_aro representing initial arousal from aestivation. d Co-expression TF network of the aestivation-responsive model AM7. Klf2 and Egr1 are recognized as hub transcription factors in the network. The TFs showing differential expression in all organs are labeled in red, whereas for the remaining DE-TFs showing differential expression in at least one organ are labeled in yellow. e KEGG enrichment analysis of the AM7 module. The AM7 module governs diverse gene pathways, including those participating in cell proliferation and differentiation, seasonal rhythmicity and immune responses, suggesting the complex mechanism of molecular regulation during sea cucumber aestivation. The circle size and filled portion represent the gene numbers (from the AM7 module) and percentage of differentially expressed genes (DGEs) in a given pathway, respectively. The statistical significance is colored according to Q values
To understand the gene regulatory network of aestivation, we constructed a gene co-expression network using the 39 transcriptome data sets, and identified 6 aestivation-related modules, with AM7 as the most significant module across three organs (body wall, muscle and respiratory tree; Supplementary Figure S17; Supplementary Table S27). In particular, the transcription factors Klf2 and Egr1 were recognized as hub transcription factors in the AM7 network (Fig. 4d; Supplementary Table S28), further supporting their role as key regulators of aestivation. Pathway enrichment analysis revealed that the AM7 module governs complex molecular regulation involving with diverse gene pathways (Fig. 4e; Supplementary Table S29), including Notch, Jak-STAT and PI3K-Akt signaling pathways for cell proliferation, differentiation and apoptosis62, the thyroid hormone signaling pathway for seasonal rhythmicity63, the neurotrophin signaling pathway for neuronal survival, growth and differentiation64, and chemokine, complement and Toll-like receptor signaling pathways for immune responses65. Taken together, our study presents the first step to identify the key regulators and gene network for sea cucumber’s aestivation, and our novel findings provide insights into how sea cucumber optimizes its physiological states for long-term viability during aestivation.
Regulation of intestine hypometabolism and regeneration
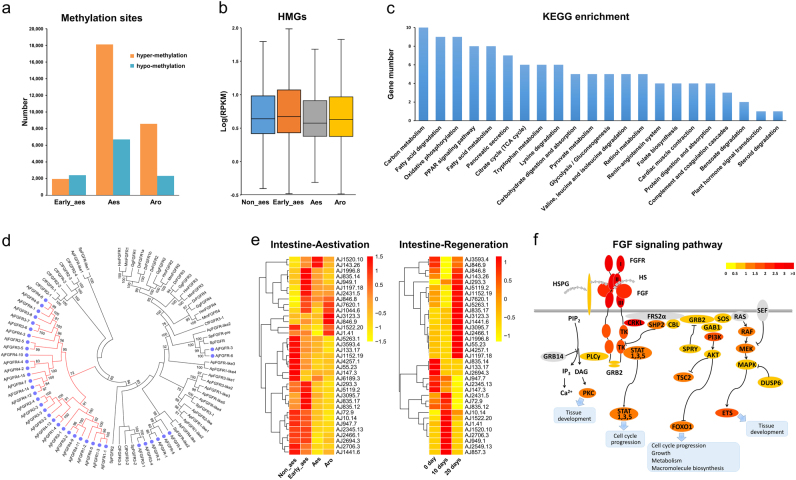
The intestine of sea cucumber has long attracted great research interests due to its extraordinary potential for degeneration and regeneration23. During aestivation, sea cucumber experiences intestine atrophy and depresses its global metabolic rate, and such intestine degeneration can fully recover after arousal from aestivation. Previous preliminary studies suggested that DNA methylation may play an important role during sea cucumber aestivation66,67, but to date there is still lack of direct evidence linking DNA methylation with hypometabolism and potential targeted genes remain to be uncovered. We conducted DNA methylome profiling of intestine samples from normal and different aestivation states using the MethylRAD technique68 and found that a remarkable increase in DNA methylation level during aestivation (Fig. 5a). We identified 411 hypermethylated genes (HMGs) during aestivation (Supplementary Table S30), 64% of which showed transcriptional suppression during aestivation (Fig. 5b). Functional enrichment analysis of the identified HMGs revealed that they are involved in numerous metabolic pathways such as carbon metabolism, fatty acid metabolism, pyruvate metabolism and retinol metabolism (Fig. 5c). This finding suggests that the phenomenon of intestine hypometabolism in sea cucumber results from transcriptional suppression of various metabolic pathways mediated through DNA hypermethylation.
Fig. 5. Epigenetic regulation of intestine hypometabolism and participation of the expanded Fgfr family in intestine regeneration of A. japonicus.
a Identification of differentially methylated sites during different aestivation states. The intestine of sea cucumber shows prominent hypermethylation during aestivation. b Expression profiles of significantly hypermethylated genes (HMGs), showing the overall transcriptional suppression of these HMGs. c KEGG enrichment analysis of HMGs. HMGs are involved in numerous metabolic pathways, suggesting that intestine hypometabolism is caused by transcriptional suppression of metabolic pathways mediated through DNA hypermethylation. d The phylogeny of the Fgfr gene family in sea cucumber and other animals. The Fgfr gene family shows significant expansion in the sea cucumber genome (38 in contrast to 4–13 in other echinoderms or chordates). The expanded gene members in sea cucumber mostly form a separate clade (indicated by the red cluster). Numbers above the branches are support percentages for 1000 bootstrap replicates. The accession numbers or IDs of corresponding genes displayed in the tree are provided in Supplementary Table S37. e Expression heat maps of the Fgfr genes in sea cucumber intestine during different stages of aestivation and regeneration. Expression of the Fgfr genes is mostly suppressed during aestivation (corresponding to intestine atrophy), whereas it is activated during the regeneration process. f Expression of the FGF signaling pathway72 during intestine regeneration in sea cucumber. The potential roles of the FGF signaling pathway in intestine regeneration are supported by observation of the activation of various downstream cascades during the intestine regeneration process in sea cucumber. Fold-change (regeneration stages vs. the control stage) is color-coded
To understand the transcriptional regulation of intestine regeneration, we conducted transcriptome profiling of normal and regenerated intestines at 10 and 20 dpe (days post-evisceration) (Supplementary Figure S18; Supplementary Table S31). We identified 6,511 DGEs (Supplementary Tables S32, S33; Supplementary Figure S19), and co-expression network analysis identified two regeneration-related modules that involved diverse signaling pathways, including Wnt and Hippo, which are well-known for participating in intestine regeneration in sea cucumber and other animals69,70 (Supplementary Figures S20-S24; Supplementary Tables S34-S36). Among the identified DGEs, the significant expansion of the fibroblast growth factor receptor (Fgfr) gene family in the sea cucumber genome (38 in contrast to 4–13 in other echinoderms or chordates; Fig. 5d) is of particular interest, as this family is known to be crucial for animal organogenesis and regeneration by mediating the FGF signaling pathway71,72. Expression analysis of Fgfr genes suggests that their transcription is mostly suppressed during aestivation (corresponding to intestine atrophy), whereas it is activated during the regeneration process (Fig. 5e). Their potential roles in mediating the FGF signaling pathway for regeneration are further supported by observation of the activation of various downstream cascades of this pathway during the intestine regeneration process (Fig. 5f).
Discussion
Echinoderms exhibit several fascinating evolutionary innovations that are rarely seen in the animal kingdom, and how they attained such extraordinary biological features through evolution has been a long-standing unsolved mystery. Here we present a high quality of the sea cucumber A. japonicus genome assembly and extensive transcriptomes, with the assembly quality comparable or superior to many previously published echinoderm genomes8,11,12. Our analysis of the sea cucumber genome revealed novel genomic features and molecular changes that may contribute to the evolutionary appearance of striking biological features. The sea cucumber has a typical Hox cluster similar to that of sea star A. planci, invalidating the previous hypothesis that Hox clusters of all echinoderms are reorganized39,40. We also revealed that the spatial expression patterns of Hox7 and Hox11/13b were potentially responsible for embryo-to-larva axial transformation in echinoderms. The extraordinary ability of sea cucumbers to make saponins is enabled by its modification of lanosterol synthase, which possibly occurred through convergent evolution. This implies that even for very complex metabolic pathways, modifying just one key gene could lead to the generation of a new adaptive trait in an organism. Klf2 and Egr1 were identified as putative key regulators during sea cucumber aestivation, probably exerting their effects through a clock gene-controlled process. While circadian regulation has been extensively studied in many organisms, the exact ways of how these key regulators exert their effects in sea cucumber’s aestivation and whether in the same way as in circadian regulation remain to be thoroughly explored. We found that intestine hypometabolism during aestivation is driven by the DNA hypermethylation-mediated expression suppression of various metabolic gene pathways, providing direct evidence linking DNA methylation with hypometabolism. The intestine regeneration involves diverse signaling pathways including Wnt, Hippo and FGF. The outstanding expansion of the Fgfr gene family may promote regeneration potential and contribute to the extraordinary regeneration capacity of sea cucumber. The sea cucumber genome, together with extensive transcriptomes, represents an invaluable resource and provides a new avenue for understanding the evolutionary appearance and molecular regulation of these extraordinary biological features in sea cucumbers and other echinoderms.
Materials and methods
Genome sequencing and genome size estimation
Genomic DNA was extracted from the gonad tissue of a wild female sea cucumber individual using the conventional phenol/chloroform extraction method73. To address the challenge of high genome heterozygosity in echinoderms8, we used a combinational sequencing strategy both with traditional paired-end/mate pair sequencing and long read sequencing. Consequently, we constructed short-insert paired-end libraries (180 bp, 350 bp, 450 bp, 500 bp and 550 bp) using the Illumina standard protocol (San Diego, USA) and long-insert mate-pair libraries (5 Kb, 10 Kb, 15 Kb and 20 Kb) following the Cre-lox recombination-based protocol74. Then these libraries were sequenced using an Illumina HiSeq 2000 platform, which produced 349.34 Gb of raw data. Long read sequencing was done by a Pacific Biosciences (PacBio) system, which generated 23.22 Gb of raw data with an average read length of ~9 Kb. After quality filtering, 352.38 Gb of high-quality data were used for genome assembly.
The genome size of A. japonicus was estimated using flow cytometry75 and k-mer analysis76. Flow cytometry analysis of A. japonicus was conducted as previously described75 using the scallop Chlamys farreri as an internal reference standard. The distribution of 19-mers was calculated based on paired-end reads derived from the DNA libraries with insert sizes of 180 bp, 350 bp and 500 bp. The genome size was estimated using the following formula:77 genome size = (total number of 19-mers) / (position of peak length).
Genome assembly and genome-quality assessment
To conquer high genome heterozygosity, we adopted a hybrid assembly strategy to generate long genome contigs. The genome assembly backbone was first constructed using Illumina short-insert paired-end reads by MaSuRCA with its default parameters78, and the long PacBio sequencing reads were recruited to link and extend the backbones by SSpace-long79 with parameters –k 1 –l 1 and PBJelly280. A homemade Perl script was used to recover part of abandoned sspace-LongRead links due to strict internal settings, which led to an improved assembly. Due to the unsatisfactory scaffolding efficiency of PacBio long reads for the sea cucumber genome8, we also adopted a classic strategy using traditional mate pair sequencing in the scaffolding step. In total, 140.85 Gb of Illumina mate pair sequencing reads with large insert lengths ranging from 5 kb to 20 kb were used to connect and integrate the assembled contigs into scaffolds through SSpace-standard with parameters −k 2 −a 0.7 −x 0 −m 5079. A total of 1,238,063 assembled mRNAs contigs were also used to evaluate the gene coverage of the genome assembly.
Linkage map-based chromosome anchoring
A total of 3,063 2b-RAD81 marker sequences were obtained from a high-density linkage map of A. japonicus28 and aligned back to the assembly using SOAP82 with the parameter settings of -M 4 –r 0 –v 2. Only markers with a unique location were used for anchoring and orienting scaffolds to different linkage groups. Scaffolds in conflict with the genetic map (such as markers from a different linkage on the same scaffold) were checked manually with 10 kb mate-paired reads.
Genome annotation
Repeat sequences were predicted via two approaches (homology-based method and de novo prediction). RepeatMasker was used for homology-based transposable element (TE) prediction: we ran searches against RepBase sequences with default parameters83. De novo TEs in the genome were detected by RepeatMasker based on a de novo repeat library, constructed by RepeatModeller (http://www.repeatmasker.org/RepeatModeler.html). These unclassified elements were further classified using the following strategy: (a) Unknown repeat sequences were aligned to Repbase using blastn; (b) Remaining unknown repeats were aligned to RepBase using tblastx; (c) Remaining unknown repeats were aligned to Uniprot database; and (d) Remaining repeats were classified by TE-class using multiple classification methods. The results from the above strategies were combined to generate final repeat database.
To predict the genes in the A. japonicus genome, three approaches (de novo, transcriptome-based and homolog-based predictions) were employed. For de novo predictions, we used three ab initio gene prediction tools—FGENESH using settings of printmRNA = yes, printExons = yes, Organism = Sea Urchin / S. purpuratus84, GENEMARK with --ES parameter85 and Augustus86, to predict coding genes. A primary model predicted by CEGMA was used as an Augustus training set. To improve the prediction accuracy, 7,739 A. japonicus expressed sequence tags (ESTs) with a total length of 1.171 Mb were obtained from NCBI. These ESTs were integrated with unigenes assembled by Trinity87 and then used to train a prediction model in PASA88 with parameters of -C -R --ALIGNERS blat. Protein sequences from twelve sequenced genomes (Branchiostoma floridae, Caenorhabditis elegans, Crassostrea gigas, Ciona intestinalis, Drosophila melanogaster, Danio rerio, H. robusta, Helobdella sapiens, Mus musculus, Nematostella vectensis, Patinopecten yessoensis, and S. purpuratus) were downloaded from Ensembl (Release 85) or JGI or EchinoBase. The protein sequences of the selected species were aligned to the repeat-masked sea cucumber genome using the Exonerate89 tool with parameters of --model protein2genome --refineboundary 1000 --showvulgar no --showalignment no --showquerygff no --showtargetgff yes.
The non-redundant consensus set of gene structures was integrated in EVidenceModeler (EVM)90 using all the gene evidence predicted above with parameters of --segmentSize 100000 --overlapSize 10000. The locations of the untranslated regions (UTRs) were added using PASA88 based on transcripts alignment. Next, we removed the gene models that were not supported by protein or transcript alignment, or were not supported by more than 2 de novo prediction methods. In total, 29,451 protein-coding genes were retained, constituting the final gene set of A. japonicus.
Functional annotation of protein-coding genes was first performed by means of BLASTP (E value threshold: 1e–05) against the protein databases SwissProt and Nr. Annotation information from the best BLASTP hits was retained for the sea cucumber gene set. Protein domains were annotated by searching the InterPro (v29.0) database. Gene Ontology (GO) terms for each gene were retrieved from the corresponding InterPro entry. The sea cucumber gene set was also mapped to the KEGG pathway database (release 53) to identify the best match for each gene.
Transcriptome analysis of developmental stages
Embryos (zygote, blastulae and gastrulae), larvae (auricularia, doliolaria, and pentactula) and juveniles of A. japonicus were collected based on artificial fertilization of sex-matured adults and larval cultivation according to Zhang et al.91. Total mRNA was extracted from each sample ( > 1000 embryos/larvae per developmental stage) by following the protocol described by Du et al.27. All RNA-seq libraries were constructed using the NEB Next mRNA Library Prep Kit by following the manufacturer’s instructions and then were subjected to paired-end 100-bp (PE100) sequencing on the Illumina HiSeq 2000 platform. Sequencing reads were aligned to the A. japonicus genome using STAR aligner92 with its default parameters. Gene expression levels in terms of RPKM were estimated by HTseq93 and custom Perl scripts.
Polymorphism analysis
To characterize the nucleotide polymorphism in the A. japonicus genome, reads from eight resequenced individuals were aligned to the assembled genome using BWA software94 with the settings of -n 15 -o 1 -e 10. Afterwards, SAMtools95 was used to sort alignments and filter PCR duplicates. SNPs were called using SAMtools mpileup and bcftools with a minimal mapping quality of 50, and sites with a read depth lower than 4 or higher than 4 times the average sequencing depth were filtered out.
SNP density was defined as the number of SNP sites per unit region among all 6 sequenced individuals. SNP density across the genome was calculated in 1-Mb sliding windows with a step size of 50 kb, and SNP density in the CDS region of each gene was also estimated. Genomic regions or CDSs with high SNP density were subjected to one-sided Fisher’s exact test compared to the corresponding chromosomal background, and the cutoff p-value was set to 1e–4. The distribution of SNP density in chromosomes or genes was visualized using Circos software96.
Homeobox gene analysis
To identify the Hox and ParaHox genes, homeodomains were searched in the A. japonicus genome using BLAST with an E value threshold of 1e−5 against all homeodomain sequences retrieved from the HomeoDB database (http://homeodb.zoo.ox.ac.uk/)97 and were further confirmed by comparing the results to the Conserved Domains Database (http://www.ncbi.nlm.nih.gov/cdd). Homeobox genes were classified based on BLAST results, molecular phylogeny and manual inspection of conserved residues. The homeodomain regions of Hox genes were used to construct the phylogenetic tree using the Neighbor-Joining method98 in MEGA799. Evolutionary distances were computed using the p-distance method100. All positions containing gaps and missing data were eliminated and the robustness of the resulting phylogenies was tested by a reanalysis of 1,000 bootstrap replicates101. Heat maps of Hox and ParaHox gene expression were drawn using custom R scripts that call the heatmap.2 function of gplots.
Evolutionary analysis of MVA pathway genes in echinoderms
Known genes participating in the human MVA pathway were downloaded from NCBI protein database and aligned to the full gene sets of three echinoderms (A. japonicus, S. purpuratus and A. planci) by BLASTP with 1e-5. BLAST hits in every echinoderm species were further checked by domain searching to ensure the presence of expected protein domains.
OSC (BAS, CAS, LAS) protein sequences in animals and plants were first aligned using MEGA 7.099, and positions with more than two-thirds gaps were removed. The amino acid percentage in every remaining position was calculated for plant BAS genes, plant CAS genes and animal LAS genes. The amino acids found in an animal lineage with a percentage smaller than 20% in animal LAS genes and larger than 55% in plant BAS or CAS genes, were defined as plant-like sites. The ancestral animal sequence of the LAS gene was deduced based on the following criterion: the amino acid was the predominant type in at least two of three animal groups (non-bilaterian, protostomia and deuterostomia). The distance between the LAS gene in every animal species and the ancestral LAS sequence was calculated using MEGA 7.099 with the parameters of bootstrap method (1000 replications), Poisson model and pairwise deletion.
Functional analysis of AJ-LAS genes using yeast expression system
Gene synthesis
For the two OSC sequences (AJ-LAS1 and AJ-LAS2) identified in the A. japonicus genome, we opted to synthesize them as gblocks from IDT (Integrated DNA Technologies) since their sequences were of good quality and appropriate length. Each of the OSCs was divided into two ~1.2 Kb fragments with an overlapping region of 50 bp in the middle (Supplementary Table S19). We employed this strategy to allow homologous recombination of these two fragments along with an empty vector resulting in the construction of an in-frame expression vector in the yeast cell.
Yeast cloning
All cloning and expression analysis was carried out in the yeast strain GIL77 (gal2 hem3-6 erg7 ura3-167)102. Expression vectors were constructed using in vivo homologous recombination in yeast. The open reading frames (ORFs) of OSCs were amplified from gblocks using the oligonucleotides listed in Supplementary Table S19. Each primer contained a region that overlapped with the pYES2 vector sequences (the 5′ end of the forward primer overlapped with the GAL1 promoter sequence, and the 5′ of the reverse primer overlapped with the CYC1 terminator sequence). The 3′ ends of the primers matched the beginning and end of the respective OSCs gblock fragments. The gblocks of each of the OSCs were amplified using the primers listed in Supplementary Table S19. The obtained PCR fragments were co-transformed into the GIL77 strain along with XbaI/HindIII-linearized pYES2 vector. Yeast transformation was performed using standard protocols (Yeastmaker™ Yeast transformation system 2, Clontech Laboratories). This resulted in vivo recombination between the pYES2 vector and the gblock OSC fragments. Plasmids were recovered from yeast transformants, transformed back into E. coli and sequence verified by sequencing the whole gene using three different primers spanning the entire length of the gene. The sequencing primers are listed in the Supplementary Table S19.
Yeast expression
For expression analysis, yeast strains were grown at 28 °C in 5 ml cultures in selective medium (SD-URA + 2% glucose + supplements) until saturation (~2 days). The supplements included the following: ergosterol (Fluka), 20 µg/ml; hemin (Sigma-Aldrich), 13 µg/ml; and Tween- 80 (Sigma-Aldrich), 5 mg/ml. Cells were then pelleted, washed in ddH2O, transferred to induction medium (SD-URA + 2% Galactose) and incubated for an additional 2 days for the accumulation of triterpenes. They were then pelleted and washed once with ddH2O before triterpene extraction.
Triterpene extraction and GC-MS analysis
The yeast pellets were mixed with 0.5 ml saponification reagent (20% KOH in 50% ethanol) and incubated at 65 °C for 2 h before extraction with an equal volume of hexane. The extraction step was repeated two additional times to maximize triterpene recovery. The extract was then dried down and the residue was dissolved in 500 µl of hexane. For rapid qualitative analysis, extracts were run on TLC plates (Silica gel on Al foil, 10 cm × 5 cm, FLUKA, Cat #70644) using a hexane: ethyl acetate (6:1) solvent system. Compounds were visualized by spraying the plates with acetic acid: H2SO4: p-anisaldehyde (48:1:1 v/v) and heating to 120 °C for 5 min on a TLC plate heater. For GC-MS analysis, 100 µl aliquots of hexane extract were transferred to 150 µl inserts and subjected to GC-MS analysis. Samples were run on a HP-5MS column (30 m × 0.25 mm i.d., 0.25 µm film) (Agilent). The injector port, source and transfer line temperatures were set at 250 °C and an oven temperature program from 80 °C (2 min) to 290 °C (30 min) at 20 °C/min was used. The carrier gas was helium and the flow rate was 1.2 ml/min. Samples were injected in splitless mode with 3 µl sample volume. The output was used to search the NISTv8 library to assign identity to peaks in the GC-MS traces. Product abundance was calculated as the percentage of total cyclic products using integrated peak areas. All experiments were repeated to confirm the reproducibility of triterpene profiles.
Inhibition of endogenous lanosterol-14α demethylase activity
Initial LAS1 or LAS2 expression experiments indicated a high degree of modification of the LAS1 products by endogenous sterol pathway enzymes. Here we hypothesized that the step immediately downstream of lanosterol synthase in yeast involves lanosterol-14α demethylase (CYP51, erg11), which might be a limiting step in further downstream modifications. Inhibition of erg11 might lead to an accumulation of LAS1 products without further modifications. Initial optimization experiments revealed that 50 µg/ml ketoconazole was sufficient for the complete inhibition of endogenous erg11 activity. Ketoconazole was dissolved in DMSO and applied to yeast cultures during the induction phase of the culturing process.
Triterpene standards
Lanosterol (Cat# L5768) was purchased from Sigma-Aldrich, whereas parkeol and lanostadienol were purified and characterized according to published literatures103,104. The standards were dissolved and diluted to 0.5 mg/ml in hexane and used in GC-MS. Ketoconazole (Catlog # K1003) was purchased from Sigma dissolved in DMSO and used in yeast media.
Transcriptome analysis of sea cucumber aestivation
Adult sea cucumbers (80–120 g body weight) were collected from the coast of Liaoning, China (121°33′47″E, 38°51′55″N). The animals were acclimated in seawater aquaria (~500 l) at 15 °C for one week before use and were fed mixed feed once a day during this period. The feed ingredients included fresh sea mud (40%), Sargassum thunbergii (30%), and sea cucumber compound feed (30%, An-yuan company, China). Several individuals were maintained as the non-aestivation control group, and the other were slowly induced into aestivation by increasing the water temperature from 15 °C to 25 °C at a rate of 0.5 °C per day. Animals in this state were labeled as the early aestivation group. The rest were maintained at 25 °C for 15 days, after which some of the animals were sampled as the deep-aestivation group. The remaining animals were subjected to a decrease in water temperature back to 18 °C (at a rate of 0.5 °C per day), held at 18 °C for 2 days and then sampled as the arousal-from-aestivation group105. For each state, transcriptome sequencing was independently conducted for two to three individuals (i.e., biological replicates) to ensure reliable quantification of gene expression. Total mRNA was extracted from four organs (body wall, muscle, respiratory tree and intestine) following the protocol described by Du et al27. All RNA-seq libraries were constructed using the NEB Next mRNA Library Prep Kit following the manufacturer’s instructions and then were subjected to paired-end 100-bp (PE100) sequencing on the Illumina HiSeq 2000 platform. Sequencing reads were aligned to the A. japonicus genome using STAR aligner92 with its default parameters. Gene expression levels in terms of RPKM were estimated by HTseq93 and custom Perl scripts.
The transcription factor (TF) sequences of 8 selected animals (H. sapiens, M. musculus, Gallus gallus, Xenopus tropicalis, D. rerio, D. melanogaster, C. elegans and C. intestinalis) were downloaded from AnimalTFDB 2.0 (http://bioinfo.life.hust.edu.cn/AnimalTFDB/) and Ensembl (http://asia.ensembl.org/index.html), and the homologous proteins in sea cucumber were identified by comparing these known TF sequences against the A. japonicus genome using the BLAST algorithm with an E value threshold of 1e-10. The obtained candidate genes were further verified based on their annotations. Differentially expressed genes (DEGs) and differentially expressed TFs (DE-TFs) were detected using the edgeR package (p < 0.05)106.
The co-expression gene network was constructed by WGCNA software107 using 39 gene expression data sets from four organs (body wall, muscle, respiratory tree and intestine) across four different states (non-aestivation; early aestivation; deep aestivation; and arousal from aestivation), with the parameters of minimum module size = 300, cutting height = 0.997, and deepSplit = F. Cytoscape108 was employed for visualization of the co-expression network. To identify the aestivation-related module, over-representation analysis of the aestivation DEGs was performed for each module using a hypergeometric test with p values adjusted by the Benjamini–Hochberg method for multiple-test correction. The hubness of a gene in the aestivation-related module (AM7) was measured by its connection strength with other genes in the module and was determined by intra-modular connectivity (Kwithin)109. KEGG enrichment analysis of AM7 was performed using the EnrichPipeline110. Directed acycline graphs (DAG) of GO terms corresponding to biological process were generated using OmicShare tools (www.omicshare.com/tools).
Quantitative PCR validation
To further verify the expression patterns of aestivation-related genes (Klf2, Egr1, Clock, Bmal1 and Cry1), quantitative PCR (qPCR) analyses were conducted. As we ran out of original samples for RNAseq analyses, another set of aestivation-related samples was adopted for qPCR validation. The adult sea cucumbers were acclimated in seawater aquaria (~500 l) at 15 °C for 1 week before use and were fed mixed feed once a day during this period. Several individuals were maintained at 15 °C as the control non-aestivation group, and the others were slowly induced into aestivation by increasing the water temperature from 15 °C to 22 °C at a rate of 1 °C per day and held at 22 °C for 1 week. According to the degree of intestine degeneration, animals in this state were labeled as the early aestivation group. The rest were maintained in water with the temperature increased to 28 °C at a rate of 1 °C per day and held at 28 °C for 1 week, and some of the animals in this state with complete degeneration of intestines were sampled as the deep-aestivation group. The remaining animals were subjected to a decrease in water temperature back to 22 °C (at a rate of 1 °C per day) and held at 22 °C for 1 week, and animals in this state were sampled as the pre-arousal group according to the degree of intestine restoration. The water temperature was continuously decreased to 15 °C at a rate of 1 °C per day and held at 15 °C for 1 week, and then, sea cucumbers with complete restoration of intestines were sampled as arousal from aestivation animals. For each state, total mRNA was extracted from body wall using an RNeasy Lipid Tissue Mini Kit (QIAGEN). Real-time PCR was conducted using the SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) on an ABI7500 real-time PCR System. The running program was as follows: 30 s at 95 °C, followed by 40 cycles of 15 s at 95 °C, 35 s at 55 °C and 25 s at 72 °C. Dpolm and Grb2 were used as an endogenous control for the normalization of gene expression111. Gene-specific primers were designed using Primer Premier 5.0, and the primer sequences are listed in Supplementary Table S26. For each gene of interest, 12 samples per stage were assayed. All PCR reactions were conducted in triplicate. Melt curve analysis was performed at the end of each PCR to confirm PCR specificity. The mRNA expression of each gene was quantified relative to that of reference genes Dpolm and Grb2 using the ΔΔCt method. Grubbs test was used for the detection of outliers. Statistical analyses of the data were performed with the SPSS (version 16.0) statistical software package using independent t-tests. Differences were considered significant if p < 0.05.
DNA methylation analysis of intestine during aestivation
Genomic DNA was extracted from intestine (three samples at each aestivation state) using the standard phenol/chloroform extraction method. MethylRAD libraries were constructed by following the protocol described by Wang et al.68 and were subjected to single end sequencing (1 × 50 bp) on an Illumina HiSeq2500 sequencer. Raw reads were first preprocessed to remove any sequences with ambiguous basecalls (N), long homopolymer regions (>10 bp) and excessive low-quality positions (>20% positions with quality score < 10). Then, high-quality reads were mapped to the target MspJI sites extracted from the A. japonicus genome using the SOAP program (parameters: -r 0 –v 2 –M 4). The sites detected in at least one group of three samples with coverage > 2 were used for differential DNA methylation analysis. The sum of all methylation sites in one gene was used to define its methylation level. Differential DNA methylation analysis between groups was conducted based on the method described by Wang et al68. Hyper-methylation genes with an average RPKM > 2 were used for KEGG enrichment analysis by EnrichPipeline110.
Transcriptome analysis of intestine regeneration
Adult sea cucumbers (70–100 g) were collected from the coast of Liaoning, China (121°33′47″E, 38°51′55″N) and acclimated in seawater aquaria (~500 l) at 15 °C for 2 weeks prior to treatment, while being fed mixed feed once a day. After acclimation, the regeneration group (30 sea cucumbers) was treated to induce evisceration by intra-coelomic injection of 0.35 M KCl. The animals were left undisturbed in aquaria to allow regeneration for 10 days, which corresponded to cell dedifferentiation stages for intestine regeneration. Intestine tissues of two or three individuals from the non-injured group, the 10-day regeneration group and the 20-day regeneration group were collected for transcriptome sequencing. Total mRNA was extracted following the protocol described by Du et al27. All RNA-seq libraries were constructed using the NEB Next mRNA Library Prep Kit following the manufacturer’s instructions and then were subjected to paired-end 100-bp (PE100) sequencing on the Illumina HiSeq 2000 platform. Sequencing reads were aligned to the A. japonicus genome using STAR aligner92 with its default parameters. Gene expression levels in terms of RPKM were estimated by HTseq93 and custom Perl scripts.
Differentially expressed genes (DEGs) in intestine regeneration were detected according to the procedure described in the edgeR package (p < 0.05)106. GO and KEGG enrichment analysis of DEGs was performed using the EnrichPipeline110. The genes in all stages of samples with sum (RPKM) > 2 were used for network construction by WGCNA107 with the parameters of minimum module size = 300, cutting height = 0.99, and deepSplit = F. To identify the regeneration -related module, over-representation analysis of the regeneration DEGs was performed for each module using a hypergeometric test. Cytoscape108 was employed for visualization of the co-expression networks. KEGG enrichment analysis was performed using the EnrichPipeline110.
Phylogenetic analysis of Fgfr genes
To understand the phylogeny of Fgfr genes of A. japonicus, the homologs from several selected animals including sea urchin (S. purpuratus), scallop (C. farreri), human (H. sapiens), mouse (M. musculus), chicken (G. gallus) and zebrafish (D. rerio), were retrieved from NCBI (http://www.ncbi.nlm.nih.gov) and Ensembl (http://useast.ensembl.org). All the retrieved FGFR amino acid sequences were used for phylogenetic analysis with the A. japonicus FGFRs. Phylogenetic trees were constructed using MEGA7 with the Neighbor-Joining method98,99. Bootstrapping with 1000 replications was conducted to evaluate the robustness of the phylogenetic tree.
Data availability
This genome project has been registered in NCBI under the BioProject accession no. PRJNA413998. The sequencing data of the genome, transcriptomes and methylomes have been deposited in NCBI Sequence Read Archive under the accession numbers. SRX3311475, SRX3311477, SRX3311479-SRX3311482, SRX3311484, SRX3311485, SRX3311487, SRX3311488 (genome); SRX3302890-SRX3302909, SRX3299141-SRX3299148 (transcriptomes) and SRX3311539 (methylomes).
Electronic supplementary material
Acknowledgements
We acknowledge the grant support from the National High Technology Research and Development Program of China (863 program; 2012AA10A412), the National Natural Science Foundation of China (31672688, 31772849 and 31502165), the Fundamental Research Funds for the Central Universities (201841001), the Taishan Scholar Project Fund of Shandong Province of China, Chang Jiang Scholars Program of Ministry of Education, the AoShan Talents Program of Qingdao National Laboratory for Marine Science and Technology (2015ASTP-ES02 and 2016ASKJ14), the National Infrastructure of Fishery Germplasm Resources (2017DKA30470), and a grant for Chinese Outstanding Talents in Agricultural Scientific Research (to Y.C.). A.O. and R.T. would like to acknowledge the funding support from European Union grant KBBE-2013-7 (TriForC), the Biotechnological and Biological Sciences Research Council Institute Strategic Programme Grant ‘Molecules from Nature’ (BB/P012523/1) and the John Innes Foundation.
Author Contributions
Z.B., S.W., Y.C. and Zu.Z. conceived the study and designed major scientific objectives. Z.B., S.W., Y.C., Zu.Z., R.W., Yul.L. and J.D. coordinated the whole project. J.D., J.J., and Ze.Z. participated in sea cucumber cultivation and sample preparation. C.M. conducted DNA extraction and library preparation for genome sequencing. Yuq.L. conducted library preparation for MethylRAD sequencing. Lih.Z., H.L. and Jia.W. conducted mRNA extraction and library preparation for transcriptome sequencing. R.W., T.L., H.S. and X.M. participated in initial genome analysis. R.W., L.B. and T.L. participated in genome sequencing, assembly and annotation. Yul.L., Jin.L and L.Y. participated in genome polymorphism analysis. S.W., X.X., and L.B. participated in Hox gene analysis. S.W., Yul.L., and R.W. participated in saponin synthesis pathway and evolutionary analysis. R.T. and A.O. conducted the yeast expression of sea cucumber LAS genes. S.W., Yul.L., Jin.W., Jia.L and X.X. participated in aestivation analysis. Yul.L., Jin.W., S.W. and Y.X. participated in regeneration analysis. Jin.W. and Lin.Z. participated in gene network analysis. Xl.H., W.J., S.L., Y.Z., Xt.H., Q.X. and W.L. participated in discussions and provided suggestions for manuscript improvement. Y.W. provided computational services and technical support. S.W., Yul.L., R.W. and Z.B. did most of the writing with input from other authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Yuli Li, Ruijia Wang, Xiaogang Xun, Jing Wang, Lisui Bao, Ramesha Thimmappa, Jun Ding, Jingwei Jiang.
Electronic supplementary material
Supplementary Information accompanies the paper at (10.1038/s41421-018-0030-5).
Contributor Information
Zunchun Zhou, Phone: +86 411 84678047, Email: zunchunz@hotmail.com.
Yaqing Chang, Phone: +86 411 84762652, Email: yqchang@dlou.edu.cn.
Zhenmin Bao, Phone: +86 532 82031960, Email: zmbao@ouc.edu.cn.
Shi Wang, Phone: +86 532 82031969, Email: swang@ouc.edu.cn.
References
- 1.Bottjer DJ, Davidson EH, Peterson KJ, Cameron RA. Paleogenomics of echinoderms. Science. 2006;314:956–960. doi: 10.1126/science.1132310. [DOI] [PubMed] [Google Scholar]
- 2.Lowe CJ, Clarke DN, Medeiros DM, Rokhsar DS, Gerhart J. The deuterostome context of chordate origins. Nature. 2015;520:456–465. doi: 10.1038/nature14434. [DOI] [PubMed] [Google Scholar]
- 3.Signor PW, Brett CE. The mid-paleozoic precursor to the mesozoic marine revolution. Paleobiology. 1984;10:229–245. doi: 10.1017/S0094837300008174. [DOI] [Google Scholar]
- 4.Lowe CJ, Wray GA. Radical alterations in the roles of homeobox genes during echinoderm evolution. Nature. 1997;389:718–721. doi: 10.1038/39580. [DOI] [PubMed] [Google Scholar]
- 5.Kelly MS. Echinoderms: Their culture and bioactive compounds. Prog. Mol. SubC. Biol. 2005;39:139–165. doi: 10.1007/3-540-27683-1_7. [DOI] [PubMed] [Google Scholar]
- 6.Carnevali, M. D. C. & Burighel, P. Regeneration in echinoderms and ascidians. eLS10.1002/9780470015902.a0022102 (2010).
- 7.Consortium SUGS, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2009;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron RA, Kudtarkar P, Gordon SM, Worley KC, Gibbs RA. Do echinoderm genomes measure up? Mar. Genom. 2015;22:1–9. doi: 10.1016/j.margen.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall MR, et al. The crown-of-thorns starfish genome as a guide for biocontrol of this coral reef pest. Nature. 2017;544:231–234. doi: 10.1038/nature22033. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, et al. The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLoS Biol. 2017;15:e2003790. doi: 10.1371/journal.pbio.2003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long KA, Nossa CW, Sewell MA, Putnam NH, Ryan JF. Low coverage sequencing of three echinoderm genomes: The brittle star Ophionereis fasciata, the sea star Patiriella regularis, and the sea cucumber Australostichopus mollis. Gigascience. 2016;5:20. doi: 10.1186/s13742-016-0125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo J, et al. Draft genome of the sea cucumber Apostichopus japonicus and genetic polymorphism among color variants. Gigascience. 2017;6:1–6. doi: 10.1093/gigascience/giw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiew PL, Don MM. Jewel of the seabed: Sea cucumbers as nutritional and drug candidates. Int. J. Food Sci. Nutr. 2012;63:616–636. doi: 10.3109/09637486.2011.641944. [DOI] [PubMed] [Google Scholar]
- 14.Bakus GJ. Defensive mechanisms and ecology of some tropical holothurians. Mar. Biol. 1968;2:23–32. doi: 10.1007/BF00351634. [DOI] [Google Scholar]
- 15.Stonik VA, Kalinin VI, Avilov SA. Toxins from sea cucumbers (holothuroids): chemical structures, properties, taxonomic distribution, biosynthesis and evolution. J. Nat. Toxins. 1999;8:235–248. [PubMed] [Google Scholar]
- 16.Kim SK, Himaya SW. Triterpene glycosides from sea cucumbers and their biological activities. Adv. Food Nutr. Res. 2012;65:297–319. doi: 10.1016/B978-0-12-416003-3.00020-2. [DOI] [PubMed] [Google Scholar]
- 17.Kwak JY. Relationships between chemical structures and functions of triterpene glycosides isolated from sea cucumbers. Front. Chem. 2014;2:77. doi: 10.3389/fchem.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahrami Y, Franco CM. Acetylated triterpene glycosides and their biological activity from holothuroidea reported in the past six decades. Mar. Drugs. 2016;14:E147. doi: 10.3390/md14080147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osbourn A, Goss RJM, Field RA. The saponins - polar isoprenoids with important and diverse biological activities. Nat. Prod. Rep. 2011;28:1261–1268. doi: 10.1039/c1np00015b. [DOI] [PubMed] [Google Scholar]
- 20.Thimmappa R, Geisler K, Louveau T, O’Maille P, Osbourn A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 2014;65:225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- 21.Li F, et al. Study on aestivating habit of sea cucumber Apostichopus japonicus selenka: the factors relating to aestivating. J. Fish. China. 1996;3:49–57. [Google Scholar]
- 22.Yang, H., Hamel, J. F. & Mercier, A. (eds) The Sea CucumberApostichopus japonicus: History, Biology And Aquaculture. (Academic Press, United States, 2015).
- 23.Wang, T., Sun, L. & Chen, M. in: The Sea Cucumber Apostichopus japonicus: History, Biology And Aquaculture (eds Yang, H., Hamel, J. F. & Mercier, A.) Ch. 11 (Academic Press, United States, 2015).
- 24.Bai MM. Regeneration in the holothurian, holothuria scabra jager. Indian J. Exp. Biol. 1971;9:467–471. [PubMed] [Google Scholar]
- 25.Shukalyuk AI, Dolmatov IY. Regeneration of the digestive tube in the holothurian Apostichopus japonicusafter evisceration. Russ. J. Mar. Biol. 2001;27:168–173. doi: 10.1023/A:1016717502616. [DOI] [Google Scholar]
- 26.Toralgranda V, Lovatelli A, Vasconcellos M. Sea cucumbers. a global review of fisheries and trade. Fao Fish. Aquacult. Tech. Pap. 2008;516:1–317. [Google Scholar]
- 27.Du H, et al. Transcriptome sequencing and characterization for the sea cucumber Apostichopus japonicus (selenka, 1867) PLoS ONE. 2012;7:e33311. doi: 10.1371/journal.pone.0033311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian M, et al. Construction of a high-density genetic map and quantitative trait locus mapping in the sea cucumber Apostichopus japonicus. Sci. Rep. 2015;5:14852. doi: 10.1038/srep14852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen M, et al. Understanding mechanism of sea cucumber Apostichopus japonicus aestivation: Insights from tmt-based proteomic study. Comp. Biochem. Phys. D. 2016;19:78–89. doi: 10.1016/j.cbd.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Sun L, et al. Metabolic responses to intestine regeneration in sea cucumbers Apostichopus japonicus. Comp. Biochem. Phys. D. 2017;22:32–38. doi: 10.1016/j.cbd.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Small KS, Brudno M, Hill MM, Sidow A. Extreme genomic variation in a natural population. Proc. Natl Acad. Sci. USA. 2007;104:5698–5703. doi: 10.1073/pnas.0700890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 2017;1:120. doi: 10.1038/s41559-017-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, et al. Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat. Commun. 2017;8:1721. doi: 10.1038/s41467-017-01927-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grohme MA, et al. The genome of schmidtea mediterranea and the evolution of core cellular mechanisms. Nature. 2018;554:56–61. doi: 10.1038/nature25473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, et al. Genome size determination of sea cucumber (Apostichopus japonicus) J. Fish. China. 2012;36:8. [Google Scholar]
- 36.Smith AB, Zamora S, Alvaro JJ. The oldest echinoderm faunas from gondwana show that echinoderm body plan diversification was rapid. Nat. Commun. 2013;4:1385. doi: 10.1038/ncomms2391. [DOI] [PubMed] [Google Scholar]
- 37.Byrne M, Martinez P, Morris V. Evolution of a pentameral body plan was not linked to translocation of anterior Hox genes: The echinoderm Hox cluster revisited. Evol. Dev. 2016;18:137–143. doi: 10.1111/ede.12172. [DOI] [PubMed] [Google Scholar]
- 38.Pearson JC, Lemons D, Mcginnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 39.Mooi R, David B. Radial symmetry, the anterior/posterior axis, and echinoderm Hox genes. Annu. Rev. Ecol. Evol. Syst. 2008;39:43–62. doi: 10.1146/annurev.ecolsys.39.110707.173521. [DOI] [Google Scholar]
- 40.David B, Mooi R. How Hox genes can shed light on the place of echinoderms among the deuterostomes. EvoDevo. 2014;5:1–19. doi: 10.1186/2041-9139-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baughman KW, et al. Genomic organization of Hox and ParaHox clusters in the echinoderm Acanthaster planci. Genesis. 2014;52:952–958. doi: 10.1002/dvg.22840. [DOI] [PubMed] [Google Scholar]
- 42.Peterson KJ, Arenas-Mena C, Davidson EH. The a/p axis in echinoderm ontogeny and evolution: evidence from fossils and molecules. Evol. Dev. 2000;2:93–101. doi: 10.1046/j.1525-142x.2000.00042.x. [DOI] [PubMed] [Google Scholar]
- 43.Tschopp P, Duboule D. A regulatory ‘landscape effect’ over the Hoxd cluster. Dev. Biol. 2011;351:288–295. doi: 10.1016/j.ydbio.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 44.Arenas-Mena C, Martinez P, Cameron RA, Davidson EH. Expression of the Hox gene complex in the indirect development of a sea urchin. Proc. Natl Acad. Sci. USA. 1998;95:13062–13067. doi: 10.1073/pnas.95.22.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikuchi M, Omori A, Kurokawa D, Akasaka K. Patterning of anteroposterior body axis displayed in the expression of Hox genes in sea cucumber Apostichopus japonicus. Dev. Genes Evol. 2015;225:275–286. doi: 10.1007/s00427-015-0510-7. [DOI] [PubMed] [Google Scholar]
- 46.Angerer LM, et al. Progressively restricted expression of a homeo box gene within the aboral ectoderm of developing sea urchin embryos. Genes Dev. 1989;3:370–383. doi: 10.1101/gad.3.3.370. [DOI] [PubMed] [Google Scholar]
- 47.Dobias SL, Zhao AZ, Tan H, Bell JR. & Maxson R. Sphbox7, a new abd-b class homeobox gene from the sea urchin Strongylocentrotus purpuratus: Insights into the evolution of Hox gene expression and function. Dev. Dynam. 1996;207:450–460. doi: 10.1002/(SICI)1097-0177(199612)207:4<450::AID-AJA9>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 48.Ishii M, et al. Hbox1 and hbox7 are involved in pattern formation in sea urchin embryos. Dev. Growth Differ. 1999;41:241–252. doi: 10.1046/j.1440-169X.1999.413426.x. [DOI] [PubMed] [Google Scholar]
- 49.Chappell J. The genetics and molecular genetics of terpene and sterol origami. Curr. Opin. Plant Biol. 2002;5:151–157. doi: 10.1016/S1369-5266(02)00241-8. [DOI] [PubMed] [Google Scholar]
- 50.Svetashev VI, Levin VS, Lam CN, Nga DT. Lipid and fatty acid composition of holothurians from tropical and temperate waters. Comp. Biochem. Phys. B. 1991;98:489–494. doi: 10.1016/0305-0491(91)90242-6. [DOI] [Google Scholar]
- 51.Kerr RG, Chen Z. In vivo and in vitro biosynthesis of saponins in sea cucumbers. J. Nat. Prod. 1995;58:172–176. doi: 10.1021/np50116a002. [DOI] [PubMed] [Google Scholar]
- 52.Sawai S, et al. Plant lanosterol synthase: Divergence of the sterol and triterpene biosynthetic pathways in eukaryotes. Plant Cell Physiol. 2006;47:673–677. doi: 10.1093/pcp/pcj032. [DOI] [PubMed] [Google Scholar]
- 53.Storey KB, Storey JM. Aestivation: Signaling and hypometabolism. J. Exp. Biol. 2012;215:1425–1433. doi: 10.1242/jeb.054403. [DOI] [PubMed] [Google Scholar]
- 54.Loomis SH. Diapause and estivation in sponges. Prog. Mol. Subcell. Biol. 2010;49:231–243. doi: 10.1007/978-3-642-02421-4_11. [DOI] [PubMed] [Google Scholar]
- 55.Fustin J, et al. Egr1 involvement in evening gene regulation by melatonin. FASEB J. 2009;23:764–773. doi: 10.1096/fj.08-121467. [DOI] [PubMed] [Google Scholar]
- 56.Guillaumond F, et al. Krüppel-like factor klf10 is a link between the circadian clock and metabolism in liver. Mol. Cell. Biol. 2010;30:3059–3070. doi: 10.1128/MCB.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spörl F, et al. Krüppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc. Natl Acad. Sci. USA. 2012;109:10903–10908. doi: 10.1073/pnas.1118641109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao W, et al. Egr1 regulates hepatic clock gene amplitude by activatingper1 transcription. Sci. Rep. 2015;5:15212. doi: 10.1038/srep15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kume K, et al. Mcry1 and mcry2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 60.Meuti ME, Denlinger DL. Evolutionary links between circadian clocks and photoperiodic diapause in insects. Integr. Comp. Biol. 2013;53:131–143. doi: 10.1093/icb/ict023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo K. Signaling cross talk between tgf-β/smad and other signaling pathways. Cold Spring Harb. Perspect. Biol. 2017;9:a022137. doi: 10.1101/cshperspect.a022137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dardente H, Hazlerigg DG, Ebling FJP. Thyroid hormone and seasonal rhythmicity. Front. Endocrinol. 2014;5:19. doi: 10.3389/fendo.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitre M, Mariga A, Chao MV. Neurotrophin signalling: Novel insights into mechanisms and pathophysiology. Clin. Sci. 2017;131:13–23. doi: 10.1042/CS20160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hibino T, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 66.Zhao Y, Chen M, Storey KB, Sun L, Yang H. DNA methylation levels analysis in four tissues of sea cucumber Apostichopus japonicus based on fluorescence-labeled methylation-sensitive amplified polymorphism (f-msap) during aestivation. Comp. Biochem. Phys. B. 2015;181:26–32. doi: 10.1016/j.cbpb.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 67.Wang T, Yang H, Zhao H, Chen M, Wang B. Transcriptional changes in epigenetic modifiers associated with gene silencing in the intestine of the sea cucumber, Apostichopus japonicus (selenka), during aestivation. Chinese J. Oceanol. Limnol. 2011;29:1267–1274. doi: 10.1007/s00343-011-0143-2. [DOI] [Google Scholar]
- 68.Wang S, et al. MethylRAD: A simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes. Open Biol. 2015;5:150130. doi: 10.1098/rsob.150130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mashanov VS, García-Arrarás JE. Gut regeneration in holothurians: A snapshot of recent developments. Biol. Bull. 2011;221:93–109. doi: 10.1086/BBLv221n1p93. [DOI] [PubMed] [Google Scholar]
- 70.Zhao B, Tumaneng K, Guan KL. The hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell. Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sánchez AA, Tsonis PA. Bridging the regeneration gap: Genetic insights from diverse animal models. Nat. Rev. Genet. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- 72.Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wires Dev. Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sambrook, J., Fritsch, E. F. & Maniatis, T. E. (eds) Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor Lab. Press, United States, 1989).
- 74.Van NF, et al. Illumina mate-paired DNA sequencing-library preparation using cre-lox recombination. Nucleic Acids Res. 2012;40:e24. doi: 10.1093/nar/gkr1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang L, et al. Fosmid library construction and initial analysis of end sequences in Zhikong scallop (Chlamys farreri) Mar. Biotechnol. 2007;9:606–612. doi: 10.1007/s10126-007-9014-4. [DOI] [PubMed] [Google Scholar]
- 76.Meng Q, et al. Growth and reproductive performance of triploid yesso scallops (Patinopecten yessoensis) induced by hypotonic shock. J. Shellfish Res. 2016;31:1113–1122. doi: 10.2983/035.031.0422. [DOI] [Google Scholar]
- 77.Dolezel J, Bartos J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytom. Part. A. 2003;51:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- 78.Zimin AV, et al. The masurca genome assembler. Bioinformatics. 2013;29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boetzer M, Pirovano W. Sspace-longread: Scaffolding bacterial draft genomes using long read sequence information. BMC Bioinforma. 2014;15:211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.English AC, et al. Mind the gap: Upgrading genomes with pacific biosciences rs long-read sequencing technology. PLoS ONE. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang S, Meyer E, Mckay JK, Matz MV. 2b-RAD: A simple and flexible method for genome-wide genotyping. Nat. Methods. 2012;9:808–810. doi: 10.1038/nmeth.2023. [DOI] [PubMed] [Google Scholar]
- 82.Huang J, et al. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 2016;12:e1006433. doi: 10.1371/journal.pgen.1006433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jurka J. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/S0168-9525(00)02093-X. [DOI] [PubMed] [Google Scholar]
- 84.Solovyev V, Kosarev P, Seledsov I, Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006;7:S10.1–12. doi: 10.1186/gb-2006-7-s1-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lukashin AV, Borodovsky M. Genemark.Hmm: New solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stanke M, Steinkamp R, Waack S, Morgenstern B. Augustus: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grabherr MG, et al. Full-length transcriptome assembly from rna-seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haas BJ, et al. Improving the arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinforma. 2005;6:1–11. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haas BJ, et al. Automated eukaryotic gene structure annotation using evidencemodeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang, Q. & Liu, Y. (eds) Cultivation Techniques on Sea Cucumber and Sea Urchin. (Ocean Univ. China Press, China, 1998).
- 92.Dobin A, et al. Bioinformatics. 2013. Star: Ultrafast universal rna-seq aligner; pp. 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anders S, Pyl PT, Huber W. Htseq--a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li H, Durbin R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li H, et al. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhong YF, Butts T, Holland PW. Homeodb: A database of homeobox gene diversity. Evol. Dev. 2008;10:516–518. doi: 10.1111/j.1525-142X.2008.00266.x. [DOI] [PubMed] [Google Scholar]
- 98.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 99.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nei M, Kumar S. Molecular evolution and phylogenetics. Heredity. 2001;86:385–385. [Google Scholar]
- 101.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 102.Kushiro T, Shibuya M, Ebizuka Y. Beta-amyrin synthase--cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur. J. Biochem. 1998;256:238–244. doi: 10.1046/j.1432-1327.1998.2560238.x. [DOI] [PubMed] [Google Scholar]
- 103.Wu TK, Wang TT, Chang CH, Liu YT, Shie WS. Importance of Saccharomyces cerevisiae oxidosqualene-lanosterol cyclase tyrosine 707 residue for chair-boat bicyclic ring formation and deprotonation reactions. Org. Lett. 2008;10:4959–4962. doi: 10.1021/ol802036c. [DOI] [PubMed] [Google Scholar]
- 104.Liu YT, Hu TC, Chang CH, Shie WS, Wu TK. Protein engineering of Saccharomyces cerevisiae oxidosqualene-lanosterol cyclase into parkeol synthase. Org. Lett. 2012;14:5222–5225. doi: 10.1021/ol302341h. [DOI] [PubMed] [Google Scholar]
- 105.Zhao Y, Yang H, Storey KB, Chen M. Differential gene expression in the respiratory tree of the sea cucumber Apostichopus japonicus during aestivation. Mar. Genom. 2014;18(Pt B):173–183. doi: 10.1016/j.margen.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 106.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. 2005;4:1–45. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 108.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559–559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen S, et al. De novo analysis of transcriptome dynamics in the migratory locust during the development of phase traits. PLoS ONE. 2010;5:e15633. doi: 10.1371/journal.pone.0015633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang L, Wang S, Chang Y, Bao Z, Ding J. Selection of reference genes for quantitative real-time PCR of Apostichopus japonicus intestine during different periods of aestivation. Period. Ocean Univ. China. 2016;46:35–43. [Google Scholar]
- 112.Mito T, Endo K. PCR survey of Hox genes in the crinoid and ophiuroid: Evidence for anterior conservation and posterior expansion in the echinoderm Hox gene cluster. Mol. Phylogenet. Evol. 2000;14:375–388. doi: 10.1006/mpev.1999.0707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This genome project has been registered in NCBI under the BioProject accession no. PRJNA413998. The sequencing data of the genome, transcriptomes and methylomes have been deposited in NCBI Sequence Read Archive under the accession numbers. SRX3311475, SRX3311477, SRX3311479-SRX3311482, SRX3311484, SRX3311485, SRX3311487, SRX3311488 (genome); SRX3302890-SRX3302909, SRX3299141-SRX3299148 (transcriptomes) and SRX3311539 (methylomes).