Abstract
The protein kinase Aurora A (AurA) is essential for the formation of bipolar mitotic spindles in all eukaryotic organisms. During spindle assembly, AurA is activated through two different pathways operating at centrosomes and on spindle microtubules. Recent studies have revealed that these pathways operate quite differently at the molecular level, activating AurA through multifaceted changes to the structure and dynamics of the kinase domain. These advances provide an intimate atomic-level view of the finely tuned regulatory control operating in protein kinases, revealing mechanisms of allosteric cooperativity that provide graded levels of regulatory control, and a previously unanticipated mechanism for kinase activation by phosphorylation on the activation loop. Here, I review these advances in our understanding of AurA function, and discuss their implications for the use of allosteric small molecule inhibitors to address recently discovered roles of AurA in neuroblastoma, prostate cancer and melanoma.
Keywords: allosteric regulation, aurora kinases, protein dynamics
Introduction
The Aurora kinases are essential components of the mitotic machinery in all eukaryotic organisms. Members of this protein family were first discovered in Drosophila, where mutations in the aur-a gene cause severe mitotic defects including failure of centrosome separation and formation of monopolar spindles [1]. A closely related kinase was identified independently in budding yeast in a screen for mutants defective in chromosome segregation (Ipl1, increase-in-ploidy 1) [2]. Further studies in Drosophila, Caenorhabditis elegans and Xenopus clarified that these metazoan organisms possess two different Aurora kinases with distinct functions in mitotic cells [3,4]. The mammalian orthologues of these Aurora kinases, now called Aurora A (AurA) and Aurora B, were subsequently found to be amplified in several cancers [5,6], spurring rapid progress in dissecting the biological functions of this kinase family. AurA is localized to the poles of mitotic cells and plays a central role in the assembly of bipolar spindles. In contrast, Aurora B is found at the spindle midzone and regulates the spindle attachment of the chromosomes, as well as cytokinesis. Mammals also possess a third Aurora kinase, Aurora C; expressed predominantly in testes, its function is relatively poorly understood [7,8].
AurA first associates with centrosomes in the G2 stage of the cell cycle, where it helps drive the process of centrosome maturation, in which the pericentriolar material, such as the γ-tubulin ring complex, is recruited to the centrosomes to promote the nucleation of microtubules [9]. Centrosomal AurA also plays an important role in the G2/M transition through a pathway involving direct phosphorylation and activation of the polo-like kinase PLK1 [10,11]. Later in mitosis, in metaphase, a separate pool of AurA associates with the centrosome-proximal microtubules of the spindle, where it plays important roles in the assembly and function of the bipolar spindle [12].
The discovery that Aurora kinases are widely overexpressed in a variety of solid tumors led to considerable interest in targeting the Aurora kinases, and several inhibitors entered clinical trials starting in the late 2000s [13,14]. Although preclinical studies of many AurA inhibitors showed efficacy in various cancer models, by and large clinical response rates in patients with solid tumors have been disappointing [15]. As observed with inhibitors targeting other kinase families [16,17], poor responses in trials were probably exacerbated by a lack of an established causal link between AurA overexpression and tumorigenesis, poor understanding of the differential roles of AurA in different cancers and patient populations, and the inability to appropriately stratify patients accordingly. Recently, however, specific biochemical roles have been identified for AurA in the pathogenesis of melanoma, where a hyperactivated form of the kinase causes chromosome instability and DNA damage, and in neuroblastoma and prostate cancer, where AurA blocks degradation of the oncogenic transcription factor N-Myc [18–20]. In these cases, AurA forms multiprotein complexes with biochemical, structural and dynamic properties that are distinct from those of AurA in normal cells. These advances in understanding specific mechanistic roles of AurA in disease are catalyzing renewed clinical interest in AurA inhibitors for these patient populations. It is hoped that greater understanding of the distinctive features of disease-associated AurA, particularly its unique dynamics, may aid the design of novel therapeutics that selectively target these forms of the kinase.
The unique allosteric properties of AurA stem from the loss of the regulatory hydrophobic motif found in the closely related AGC kinases
The mammalian Aurora kinases consist of a highly conserved C-terminal kinase domain and a much less conserved N-terminal domain of varying length (Figure 1A). The function of the N-terminal domain of AurA is still poorly understood, but it includes a role in targeting the protein for ubiquitination by the anaphase promoting complex, which mediates its degradation late in mitosis [21,22]. The N-terminal domain has also been reported to play some role in restraining the catalytic activity of the kinase domain [23], although the inhibitory effects appear to be modest compared with those mediated directly through the kinase domain, and most studies have focused on the latter regulatory mechanisms. These include autophosphorylation of a conserved threonine on the activation loop of the kinase domain (T288 in human AurA, shown in blue in Figure 1A), as well as protein–protein interactions with the spindle assembly factor Tpx2 (targeting protein for Xklp2, shown in magenta in Figure 1A).
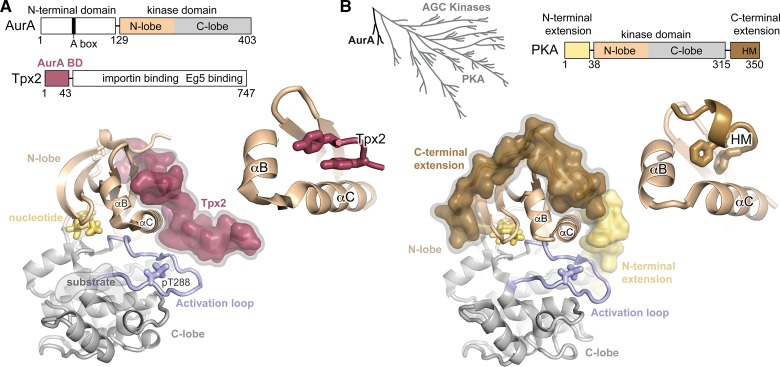
Figure 1. AurA and the AGC-family kinases share a common regulatory architecture.
(A) Top: The domain architectures of AurA and Tpx2 are shown, highlighting the N-terminal domain and kinase domain of AurA, and the AurA-binding domain (AurA BD) of Tpx2. The kinase domain of AurA is divided into the N-lobe and C-lobe. The location of the A-box in the N-terminal domain of AurA, which targets the kinase for degradation, is also shown. Bottom: X-ray structure of the AurA kinase domain bound to residues 1–43 of human Tpx2 (PDB ID: 1OL5). The N-lobe and C-lobe are shown in beige and gray, respectively, the activation loop in blue and Tpx2 in magenta. The inset shows an expanded view of the docking site of Tpx2 on the N-lobe. (B) Top: A phylogenetic tree illustrating the relationship of the Aurora kinases to the AGC kinase family is shown on the left (adapted from ref. [24]), and the domain architecture of the AGC kinase PKA is shown on the right. Bottom: Structure of PKA (PDB ID: 1L3R) with the C-terminal tail containing the HM colored brown, the N-terminal extension colored yellow, and the remainder of the kinase domain colored as in (A). The inset shows the docking of the HM to the PIF pocket of the kinase.
Although the Aurora kinases have been grouped phylogenetically into their own family outside the seven main clades of eukaryotic protein kinases (ePKs), their kinase domains are quite closely related to those of the AGC-family kinases and the polo-like kinases [24]. Indeed, the kinase domain catalytic core of AurA shares close to 40% sequence identity with several AGC kinases and with the polo-like kinase PLK4, which is similar to the degree of sequence conservation found within these two other families [24]. Many of the functional and regulatory properties of AurA can be viewed as variations on the general themes found in the AGC kinases.
All ePKs have a similar three-dimensional structure consisting of an N-terminal lobe (N-lobe) that is predominantly β-sheet, and a C-terminal lobe (C-lobe) that is mostly α-helical, with the active site cleft formed between the lobes (Figure 1A). In most kinases, the series of β-strands that comprise the N-lobe is interrupted by a single α-helix (called helix αC), which forms one wall of the active site and plays a central role in regulating kinase activity [25]. A second key regulatory structure, the activation loop, resides in the C-lobe, contains the conserved catalytic Asp-Phe-Gly (DFG) motif in the active site, and forms the binding site for substrate peptides at the base of the active site cleft (Figure 1A). The activation loop is a site of regulatory phosphorylation in most protein kinases [26].
The Aurora kinases, PLKs and AGC kinases all share an additional α-helical segment in their N-lobes, missing in most other kinase families, that immediately precedes the αC-helix, and is termed αB (Figure 1A). The rigid αB helix acts as a spacer to separate the αC-helix from the core β-sheet of the N-lobe, resulting in a shallow pocket on the surface of the N-lobe above the αC-helix. This pocket, termed the PIF pocket (PIF stands for ‘PDK1-interacting fragment'), is used as a surface for regulatory interactions. In the AGC kinases, a polypeptide motif present in the C-terminal tail, termed the hydrophobic motif (HM), binds in cis to the PIF pocket [27], stimulating kinase activity (Figure 1B). A prominent difference between the AGC kinases and the Aurora kinases is that the C-terminal HM is missing in the latter. Instead, the PIF pocket of AurA has been repurposed for protein–protein interactions in trans that serve both a localization and an allosteric activation role. The spindle assembly factor Tpx2 recruits AurA to the mitotic spindle through an interaction with its PIF pocket (Figure 1A) that closely resembles that of the AGC kinases with their HMs [28]. It appears that many of the unusual allosteric and dynamic properties of AurA may stem from the loss of the HM and the repurposing of the PIF pocket, and are best understood in the light of AGC kinase regulation.
The regulation of the AGC-family kinases has been extensively studied [29]. The canonical activation pathway for AGC kinases is the co-ordinated phosphorylation of both the HM and the activation loop [30]. Phosphorylation of a residue near the HM by an upstream kinase, usually one of the mTOR complexes [31,32], primes the HM to interact with the PIF pocket of PDK1 [33,34] which allows PDK1 to phosphorylate the activation loop of the other kinase and activate it. Docking of the phosphorylated HM back to its own kinase domain further increases kinase activity [35]. Thus, in most AGC kinases, PIF pocket engagement and activation loop phosphorylation are highly coupled events that activate the kinase in a concerted fashion. A key difference in the allosteric wiring of AurA compared with this canonical AGC model is that engagement of the PIF pocket by Tpx2, and phosphorylation of the activation loop on T288, have been partially uncoupled from one another, and that the enzyme is correspondingly more responsive to activation by either of these two regulatory inputs [36]. This seems to have allowed cells to utilize these inputs for two independent activation pathways of the kinase operating in different cellular contexts in mitosis.
Distinct activation mechanisms for two mitotic pools of AurA that co-operate in the assembly of bipolar spindles
The assembly of bipolar spindles is a critical function of mitotic cells essential for faithful segregation of the duplicated chromosomes into the two daughter cells [37]. AurA plays key roles in two distinct but complementary pathways of microtubule assembly required for the formation of bipolar spindles. These pathways were uncovered by elegant studies in Xenopus egg extracts, which showed that spindle microtubule assembly can be initiated independently by either the addition of chromatin to egg extract, or by the addition of sperm nuclei containing centrosomes, which act as microtubule organizing centers [38–40]. Although there is partial redundancy between the two pathways, in most cells both the chromatin-stimulated and centrosome-directed pathways are thought to be important for the faithful assembly of bipolar spindles.
Chromatin-stimulated microtubule assembly occurs through a RanGTP/Tpx2-dependent pathway in which release of the spindle assembly factor Tpx2 from importins around condensed chromatin by the small GTPase Ran results in the recruitment of AurA [12,41,42], which phosphorylates the γ-tubulin ring complex adaptor NEDD1 [43], and the microtubule binding protein HURP[44], promoting microtubule nucleation. Once the bipolar spindle forms, this Tpx2-associated pool of AurA redistributes along the spindle microtubules [45]. In contrast, the role of AurA in centrosome maturation and centrosome-driven spindle assembly is independent of Tpx2 [12], and instead involves recruitment of AurA to the centrosome by the centrosomal scaffolding protein Cep192 [46], as well as interactions with other proteins such as Ajuba [47]. Subsequent phosphorylation of the microtubule stabilizing protein TACC3 by activated centrosomal AurA is a key step in promoting efficient microtubule nucleation at centrosomes [48,49].
Considerable effort has been invested in dissecting the mechanisms governing activation of the two mitotic pools of AurA localized to the spindle and to the centrosome. The emerging consensus is that the centrosomal pool of AurA is activated by autophosphorylation on residue T288 in the activation loop, whereas the spindle microtubule-associated pool is instead allosterically activated by the interaction with Tpx2. Specifically, the binding of AurA to Cep192 drives AurA oligomerization, autophosphorylation and activation at centrosomes [46,50], although the molecular mechanism of autophosphorylation has been controversial [51,52]. The Cep192/AurA interaction is required for centrosomes to function as microtubule organizing centers, and Cep192 itself is essential for both centrosome maturation and spindle assembly in C. elegans and human cells [53,54]. After autophosphorylation of AurA, the interaction with Cep192 appears to become dispensable, as phosphorylated AurA can promote microtubule assembly at the centrosome even if displaced from Cep192 [46]. In contrast, Tpx2-associated AurA appears to function largely independently of phosphorylation on T288. Although Tpx2 can stimulate AurA autophosphorylation in vitro [55], RanGTP-stimulated microtubule assembly does not seem to drive efficient AurA autophosphorylation under physiological conditions [46], and a T288A mutant of AurA does not impair this specific pathway, but does block the centrosomal functions of AurA [56]. In support of this, numerous studies using phosphospecific antibodies against T288 have observed that while AurA protein is localized both to the centrosomes and to the microtubules of bipolar spindles, T288-phosphorylated AurA is only observed at the centrosomes [18,46,56,57]. The phosphatase PP6 has been shown to be important for maintaining Tpx2-associated AurA in the unphosphorylated state [58].
Collectively, these studies point to the existence of two largely independent activation pathways for AurA in mitotic cells. This view is consistent with recent work showing that AurA can be activated independently by Tpx2 or by phosphorylation on T288 by up to ∼100-fold in vitro [36,51,52,59]. Together, they support an emerging picture that the tight coupling of PIF pocket engagement and activation loop phosphorylation that operates in the AGC-family kinases has been largely lost in AurA, where these two regulatory inputs operate more independently. Surprisingly, this decoupling can be traced to a unique feature of the active site of AurA.
A water-mediated allosteric network in the active site allows AurA to be activated independently by either Tpx2 binding or phosphorylation on T288
A central feature of activated protein kinases is the assembly of an interlocked network of hydrophobic amino acids, termed the regulatory spine, that stabilizes the active conformation of the kinase domain [60,61]. The regulatory spine consists of one residue from the N-terminal β-sheet, a residue on the αC-helix, the phenylalanine residue of the catalytic Asp-Phe-Gly (DFG) motif, and an additional residue in the C-lobe (Figure 2A). The assembly of the spine locks the N- and C-lobes of the kinase together, trapping the DFG motif between them in a catalytically competent conformation in which the DFG Asp residue can co-ordinate Mg–ATP (Figure 2A, right panel). In the autoinhibited states of protein kinases, this structure is usually disassembled to block kinase activity, either by rearrangements of the DFG motif, or by movements of the αC-helix, or by both [25].
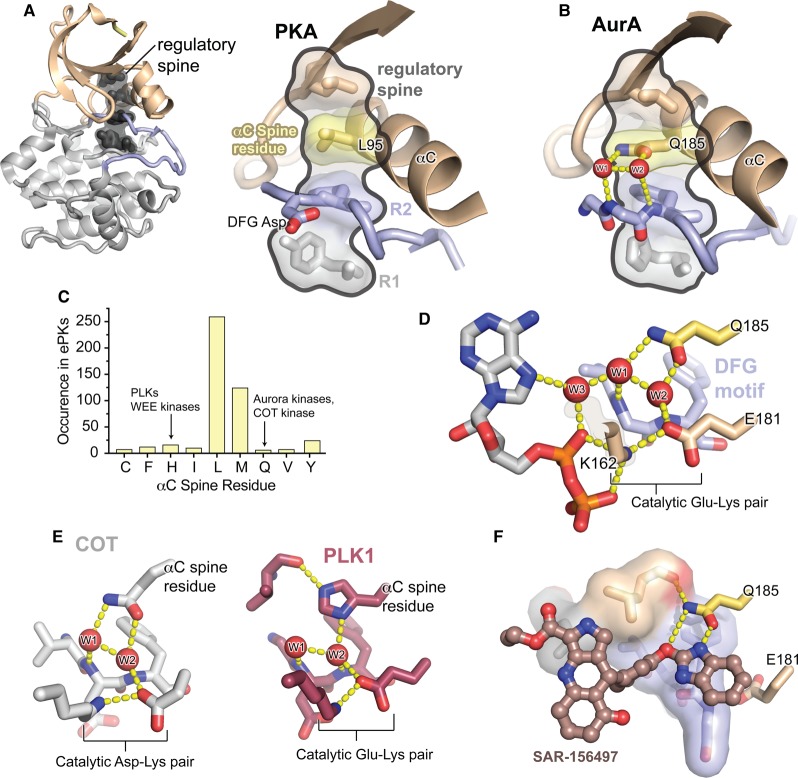
Figure 2. The unusual regulatory spine of AurA contains a functionally important water-mediated allosteric network.
(A) Left: Structure of the kinase domain core of PKA, colored as in Figure 1, with the four residues of the regulatory spine highlighted in dark gray. Right: Enlarged view of the regulatory spine of PKA, with the αC spine residue highlighted in yellow. (B) Enlarged view of the regulatory spine of AurA, colored as in (A), showing the interactions of the unusual Q185 αC spine residue with structured water molecules (red) co-ordinated to the DFG motif in the active site. (C) Histogram showing the frequency of different amino acid residues at the αC spine position across all ePKs. Values are derived from the original multiple sequence alignments published by Manning et al. [24]. (D) View of the water network in the AurA active site (PDB ID: 5G1X), showing the Q185 residue, three water molecules, the Glu-Lys salt bridge, and the bound ADP molecule. (E) Left: View of the active site of COT kinase showing the similar interactions of the glutamine αC spine residue with the active site water molecules (PDB ID: 4Y85). Right: View of the active site of PLK1 showing the water network mediated by the histidine αC spine residue (PDB ID: 2OWB). (F) Structure of SAR-156497 bound to AurA showing the interactions of the small molecule with the Q185 residue (PDB ID: 4UZD).
A salient feature of the regulatory spine is its high degree of hydrophobicity. However, in the Aurora kinases, the regulatory spine residue emanating from the αC-helix, referred to here as the αC spine residue, is instead a polar glutamine residue (Figure 2B), in contrast with the leucine or methionine residues found at this position in >75% of all ePKs (Figure 2C). This highly unusual glutamine is conserved in the Aurora kinases of all eukaryotes, including yeast Ipl1, suggesting that it plays an important functional role unique to this subfamily.
It was recently shown that the αC spine glutamine residue, Q185 in human AurA, mediates an extensive network of hydrogen-bonding interactions with ordered water molecules in the active site (Figure 2D), interconnecting the αC-helix, the DFG motif and the catalytically important Glu-Lys salt bridge [36]. Molecular dynamics simulations showed that this water network is surprisingly stable, with the same water molecules remaining bound in a similar configuration for up to 100 ns in simulations, in contrast with the picosecond correlation times typical of interfacial water molecules. Interestingly, the inclusion of a Tpx2 fragment bound to AurA in the simulations was found to stabilize both the water network and the αC-helix itself, and this effect required the Q185 sidechain. This suggested that the water network constitutes an allosteric pathway for communicating the binding of Tpx2, through the αC-helix and Q185 residue, to the active site. Indeed, in subsequent experiments, substitution of Q185 with hydrophobic residues, including replacement with the leucine and methionine residues found in most other kinases (see Figure 2C), was found to severely impair the activation of AurA by Tpx2 binding or phosphorylation. Interestingly, the activity of the mutant enzymes could be rescued by activating them with both Tpx2 and phosphorylation simultaneously. Thus, the replacement of Q185 with a canonical hydrophobic αC spine residue appears to revert AurA to a more AGC kinase-like state in which both regulatory inputs (αC-helix docking and activation loop phosphorylation) are simultaneously required for activation. This suggests that the enhanced coupling of the αC-helix to the active site mediated by Q185 may have emerged in an early ancestral Aurora kinase as a way to uncouple αC-docking and activation loop phosphorylation from one another and repurpose them for independent activation pathways.
Although polar αC spine residues are very rare in ePKs, they are not unique to Aurora kinases. At least three other ePKs have a glutamine residue at this position, and an X-ray structure of one of them, the Cancer Osaka Thyroid (COT) kinase, shows that the glutamine residue nucleates a similar water-mediated hydrogen bond network to that seen in AurA [62] (Figure 2E). In addition, the polo-like kinases, and several other ePKs, possess a histidine residue at this position, and crystal structures show that the histidine interacts with equivalent ordered water molecules in a similar geometry (Figure 2E) [63]. The polo-like kinases possess a PIF pocket-like crevice on their N-lobes, but they are not known to use this surface for protein–protein interactions. However, their activity is strictly dependent on activation loop phosphorylation — indeed, phosphorylation of the PLK1 activation loop is a key function of AurA at the centrosome [10,11]. Although it is not known whether the histidine αC spine residue is functionally important in the PLKs, one might speculate, by analogy to AurA, that the histidine residue arose in the PLKs to stabilize the αC-helix and allow the enzymes to be activated without engagement of the PIF pocket.
The presence of a polar residue in the usually hydrophobic back pocket of some kinase active sites has important implications for inhibitor specificity. Most Aurora kinase inhibitors do not extend sufficiently far into the active site to interact with Q185, but a recently described inhibitor developed by Sanofi, SAR156497, does directly recognize this residue (Figure 2F) [64]. Remarkably, this compound was reported to be an exceptionally selective inhibitor of the Aurora kinases, with IC50 values below 10 nM for AurA, B and C, but >10 µM for all other kinases in a 110-kinase panel. Such a high degree of selectivity is exceedingly rare for ATP-competitive kinase inhibitors and demonstrates that recognition of unusual polar αC spine residues may be an important and relatively unharnessed source of selectivity for future drug development. Presumably the glutamine-containing COT, and the histidine-containing polo-like kinases, which are both important therapeutic targets [65,66], could also be selectively targeted by novel inhibitors designed to form hydrogen bonds to these residues.
Phosphorylation and Tpx2 binding enhance AurA activity by triggering distinct but complementary structural rearrangements
Most protein kinases are regulated by large-scale conformational changes that serve to reorient key elements of the catalytic machinery [25]. In particular, the catalytic DFG motif and αC-helix undergo major structural movements away from their active conformations into comparatively stable autoinhibited conformations. A common rearrangement of the DFG motif is the adoption of a so-called DFG-Out state, in which a crankshaft-like motion of the backbone of the Asp and Phe residues of the DFG motif reorients the Asp sidechain to point away from the active site, preventing the coordination of Mg–ATP. This structural change, referred to as the DFG flip, also disassembles the regulatory spine by moving the Phe residue of the DFG motif out of alignment with the other spine residues. Because the activation loop immediately follows the DFG motif in the primary sequence, the DFG flip causes the activation loop to move across the active site cleft, blocking the binding site for substrate peptides (Figure 1A).
The DFG-Out state is known to be the primary autoinhibition mechanism for only a small subset of protein kinases, notably several families of tyrosine kinases [67–69], but it has been observed in X-ray structures of a much wider range of kinase families, where it is thought to be trapped by inhibitors. Indeed, because the DFG flip fundamentally alters the chemical makeup of the kinase active site, much attention has been given to how this structural change affects interactions with inhibitors, including controlling binding and release kinetics [70,71] and determining inhibitor selectivity patterns [72,73].
Whether the DFG-Out state plays a functional role in AurA was unclear until very recently. Crystal structures of AurA bound to various inhibitors have shown that this kinase can adopt a variety of different DFG-Out states (Figure 3A) [74–77]. Structures also show that AurA adopts the active DFG-In state when bound to the Tpx2 activator [28]. However, the DFG-In state is also observed in numerous structures of the kinase determined in the absence of either Tpx2 or phosphorylation on the T288 residue [78,79]. The diversity of conformations observed in crystal structures led to confusion as to the role of these conformational changes in regulating AurA, including whether the kinase is naturally regulated by a DFG flip, and which of the various DFG-Out states captured in crystal structures might represent a genuine autoinhibited state that restrains AurA activity in vivo.
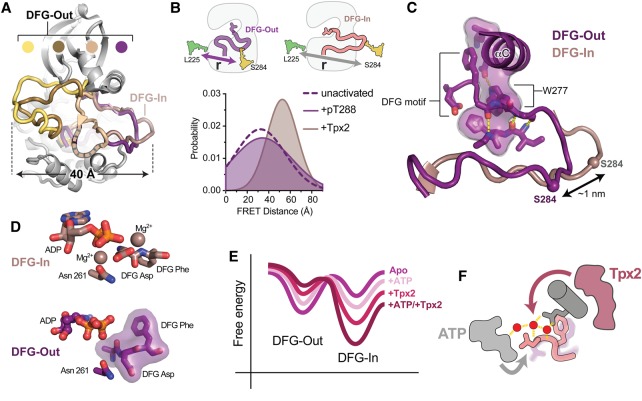
Figure 3. Tpx2 binding to AurA triggers a conformational transition from a DFG-Out state to the active DFG-In state.
(A) Selected crystal structures of AurA in which the kinase adopts DFG-Out states with widely differing positioning of the activation loop [PDB IDs: 3UOK (yellow), 2WTV (brown), 5EW9 (beige) and 5L8K (dark purple)]. The active DFG-In state is highlighted for comparison (pink, PDB ID: 1OL5). Structures were aligned on the entire kinase domain. (B) Schematics of the dye-labeling scheme used to track the position of the activation loop by FRET (top), and (bottom) the distribution of inter-dye distances determined by time-resolved FRET for inactive AurA (dashed line), AurA activated by Tpx2 (pink), and AurA activated by phosphorylation on T288 (dark purple-shaded area). The data are taken from Ruff et al. [88]. (C) Structure of the probable DFG-Out state adopted by AurA in solution (purple). The flipped DFG motif is indicated, and the partially intact regulatory spine in which the W277 residue has taken the place of the DFG phenylalanine residue, is shown as a transparent surface. The conformation of the activation loop in the active DFG-In state is shown for reference (pink). The Cα atom of the residue labeled with a fluorescent dye is shown as a sphere in both structures. (D) Top: View of the active site in the active DFG-In state showing the bound nucleotide and magnesium ions, the DFG motif and the Asn 261 residue, which, together with the DFG Asp, co-ordinates one of the magnesium ions (PDB ID: 1OL5). Bottom: View of the active site in the crystal structure of the native DFG-Out state showing bound nucleotide, the flipped DFG motif and the Asn261 residue (PDB ID 5L8K). Magnesium coordination is lost in this state. (E) Illustrative free energy landscape for the DFG-In/DFG-Out transition, showing how nucleotide and Tpx2 binding lead to graded changes in the populations of the DFG-In and DFG-Out states. (F) Schematic of the AurA active site showing the DFG motif (light pink), αC-helix and Q185 residue (dark gray), water network (red spheres and yellow hydrogen bonds) along with bound Tpx2 (magenta) and ATP (light gray). The colored arrows represent the allosteric coupling of Tpx2 to the DFG motif through the water network (magenta), and the coupling of ATP to the DFG motif through the water network and magnesium ions (gray). The faded DFG-Out state (purple) represents the promotion of the DFG-In state by nucleotide and Tpx2.
Activation of AurA by Tpx2 involves a DFG flip
These questions were recently answered in a study that used spectroscopy to track the structural movements of the kinase occurring in response to the binding of Tpx2 [36]. Using samples of AurA labeled on the activation loop and a distal surface of the kinase domain with fluorescent donor and acceptor dyes, Förster resonance energy transfer (FRET) experiments were performed that revealed a 1–2 nm movement of the activation loop upon addition of a peptide derived from Tpx2, consistent with a DFG flip coupled to a substantial structural rearrangement of the loop (Figure 3B). With the active conformation well established, the measured scale and direction of this structural change helped pinpoint the specific DFG-Out state adopted by AurA prior to activation by Tpx2. Most of the DFG-Out states captured in crystal structures of AurA bound to inhibitors have the activation loop displaced 3–4 nm across the active site cleft from its position in the active state (Figure 3A). The exception is a DFG-Out state in which a more modest movement of the DFG motif leads to a comparatively small rearrangement of the activation loop, with the labeling site predicted to move ∼1 nm (Figure 3C). In addition to several inhibitor complexes [77,80–82], this DFG-Out state is observed in a structure of AurA bound to ADP [83], suggesting compatibility with the binding of physiological substrates and products (Figure 3D). The tyrosine kinase Abl has been shown to adopt a very similar DFG-Out state bound to an ATP-peptide conjugate inhibitor [84]. An interesting feature of this DFG-Out state is that the N-terminal segment of the activation loop is anchored onto the kinase domain by a series of β-sheet hydrogen bonds (Figure 3C), preventing the larger rearrangements of the activation loop seen in other DFG-Out states of AurA (Figure 3A). The hydrogen-bonding pattern is similar to that seen in the DFG-In state, but with a register shift of one amino acid in the activation loop β-strand. The register shift results in the sidechain of W277 swapping positions with the phenylalanine of the DFG motif (F275), such that the packing of the regulatory spine is partially restored (Figure 3C), perhaps contributing to the relative stability of this DFG-Out state.
A major advantage of the spectroscopic methods employed in these studies is the ability to probe the populations of the DFG-In and DFG-Out states in solution and how they are altered by ligand binding, information that cannot be gained from crystal structures. Indeed, the data provide a rare glimpse of the nuanced way in which allostery functions in AurA, with ligands imparting relatively small population shifts between conformational states, as opposed to the wholesale switches suggested by X-ray structures (Figure 3E). For instance, nucleotide binding causes a partial population shift toward the DFG-In state, such that the populations of the DFG-In and DFG-Out states change from favoring the DFG-Out state to becoming approximately equal. This coupling is presumably mediated through coordination of Mg/ATP by the DFG Asp residue (Figure 3D), and the participation of the nucleotide in the water-mediated hydrogen bond network discussed above (Figure 2D), but the strength of the coupling is surprisingly small. This may reflect a requirement for maintaining the accessibility of the DFG-Out state amidst a high concentration of ATP in the cell.
The binding of Tpx2 to AurA causes a more pronounced shift to the DFG-In state than does nucleotide binding, with the DFG-Out state becoming only minimally populated (Figure 3E). The discovery that Tpx2 and nucleotide both promote the DFG-In state explains why these ligands bind cooperatively to AurA, with the binding of one increasing the affinity of the other by ∼5-fold [36,59]. The switch from the DFG-Out to the DFG-In state triggered by Tpx2 helps to switch the enzyme on by increasing the concentration of the catalytically competent state. However, since the kinase samples the DFG-In state appreciably when bound only to nucleotide, the further population shift induced by Tpx2 can explain at most a 2–3-fold increase in activity. The remainder of the ∼50-fold activation of AurA must be attributed to the allosteric effects of Tpx2 on the αC-helix and the hydrogen bond network mediated by Q185, which can be thought of as enhancing the intrinsic catalytic activity of the DFG-In state (Figure 3F). Thus, the DFG flip is only one part of the activation mechanism, with tuning of the DFG-In state equally if not more critical. This view is reinforced by subsequent studies of the mechanism of activation of AurA by phosphorylation, discussed below, and makes sense in the light of the close relationship of AurA to the AGC kinases, which are thought to be primarily regulated through dynamical changes to the DFG-In state [85]. Perhaps the loss of the HM in the Aurora kinases, by substantially increasing the inherent dynamics of the enzyme, made the DFG-Out state accessible for an additional level of regulatory control.
Activation of AurA by phosphorylation occurs through remodeling of the DFG-In state
Phosphorylation on T288 activates AurA in vitro to a similar degree as Tpx2 binding, giving a ∼100-fold increase in kinase activity [52,59]. Many protein kinases are activated by phosphorylation on an equivalent site in the activation loop, and it has long been thought that this post-translational modification serves to clamp the activation loop down on the C-lobe of the kinase, trapping the kinase in the active DFG-In state [86]. Surprisingly, two recent independent studies, using different spectroscopic methods to track the activation loop, revealed that this is not the case for AurA.
One study exploited the self-quenching of tetramethylrhodamine dyes to monitor transitions between active and inactive states by single-molecule fluorescence [87]. This approach revealed that T288-phosphorylated AurA dynamically transitions between active (fluorescent) and inactive (quenched) states, much like the unphosphorylated enzyme. In contrast, Tpx2 binding was observed to cause a pronounced switch to the active state. A second study used time-resolved FRET to map the distribution of conformations sampled by the activation loop with and without phosphorylation on T288 [88]. The results showed that the activation loop of both unphosphorylated and phosphorylated AurA samples a range of conformations in solution that span the DFG-Out and DFG-In states (Figure 3B, compare dashed and solid purple lines in the FRET distance distribution). In contrast, upon binding to Tpx2, the activation loop of both unphosphorylated and phosphorylated AurA adopts a more constrained conformation consistent with adoption of the DFG-In state (Figure 3B, solid pink line in the FRET distance distribution). These results were corroborated using an independent experimental method, double electron–electron resonance (DEER) spectroscopy, in which the fluorescent dyes are replaced by paramagnetic spin probes. The results of the DEER experiments were fully consistent with the FRET experiments [88], confirming that the activation loop of phosphorylated AurA adopts a range of different conformations, as observed in the unphosphorylated kinase.
These surprising results raised the question of how phosphorylation on its own activates AurA so strongly, without causing a population shift to the DFG-In state. Reasoning that the large DFG-Out subpopulation of phosphorylated AurA might be obscuring an activating structural change in the DFG-In subpopulation in their ensemble experiments, Ruff et al. [88] used a DFG-In-selective inhibitor, SNS-314, to trap AurA in the DFG-In state. DEER experiments on these samples revealed that phosphorylation on T288 indeed triggers a structural change within the DFG-In state, confirming that phosphorylation operates by altering the structure and dynamics of the DFG-In subpopulation (Figure 4A). Molecular dynamics simulations suggested a model for the structural transition triggered by phosphorylation (Figure 4B). Simulations performed on the unphosphorylated kinase lacking Tpx2, but initiated from the active DFG-In conformation, showed a strong tendency of the activation loop to refold into an alternative structure in which a short helical turn formed spanning residues P282 to R286. This effect was not observed in simulations of the kinase with phosphorylation on T288, which instead stably sample the active conformation. Because the T288 phosphorylation site lies at the C-terminus of the short helix observed in the simulations (Figure 4B, bottom panel), phosphorylation would destabilize this autoinhibited DFG-In conformation due to unfavorable interactions with the helix dipole. This structural model is consistent with the scale of the phosphorylation-driven structural change observed in the DEER experiments (Figure 4A). It would appear that AurA, in addition to the DFG-Out state, employs an autoinhibited DFG-in substate for additional regulatory control. Although this was not anticipated, many other families of protein kinases, including the AGC kinase MSK1 [89], as well as the more distantly related cyclin-dependent kinases [90] and Src-family kinases [91], are also regulated by autoinhibited DFG-In states.
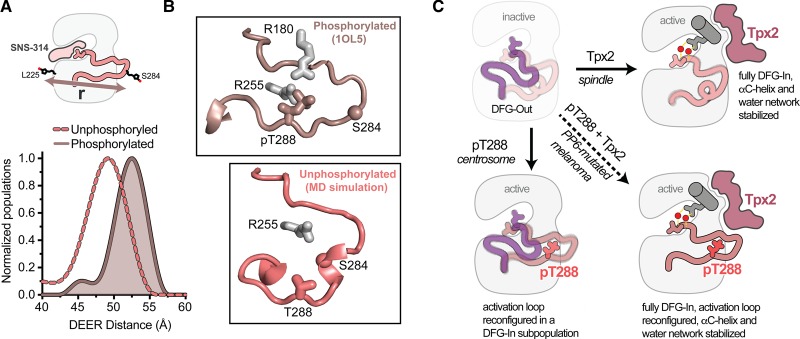
Figure 4. Phosphorylation of T288 triggers a rearrangement of the activation loop within the DFG-In state.
(A) Schematic of the spin-probe labeling scheme (top) and measured spin-spin distances (bottom) from double electron–electron resonance experiments on phosphorylated AurA (darker solid line and shading) and unphosphorylated AurA (lighter dashed line). Data are taken from Ruff et al. [88]. The inhibitor SNS-314 was used to isolate the DFG-In state. (B) Conformation of the activation loop in the active DFG-In substate promoted by phosphorylation on T288 (top, PDB ID: 1OL5), and putative structure of the autoinhibited DFG-In substate adopted in the absence of phosphorylation, identified in molecular dynamics simulations (bottom). The Cα atom of the residue on the activation loop labeled with the spin probe is shown as a sphere in both panels. (C) Schematics of the four main activation states of AurA highlighting the complementary structural and dynamic changes triggered by Tpx2 binding and phosphorylation (pT288), and the dynamically quenched state that results when these factors act together, e.g. in PP6-mutated melanoma cells.
Together these studies showed that Tpx2 and phosphorylation have quite different effects on the structure and dynamics of AurA, with Tpx2 triggering a population shift from the DFG-Out to the DFG-In state and stabilizing the αC-helix and associated water network, and phosphorylation instead altering the conformation of the activation loop in the DFG-In subpopulation. A schematic representation of these differing activation mechanisms is shown in Figure 4C. The two activation pathways are highly complementary, and when AurA is activated by both Tpx2 and phosphorylation together, the dynamics of the enzyme are quenched: the kinase homogeneously adopts the DFG-In state, the activation loop is locked in the active configuration, and the αC-helix and coupled water network are maximally stabilized. This ‘doubly-activated’ form of AurA is found on the spindles of melanoma cells bearing mutations in the AurA-directed phosphatase PP6, and contributes to their disease pathology [18].
Remarkably, phosphorylation of AurA on T288 by itself does not trigger a substantial population shift to the DFG-In state. While this runs counter to the canonical view that phosphorylation clamps the activation loop in the active state [26], a recent study of the interaction between AurA and its substrate TACC3 suggests a possible explanation. TACC3 is an important regulator of microtubule dynamics at the centrosome, and its centrosomal localization, and ability to form stabilizing cross-links between microtubules, is controlled by AurA through phosphorylation on serine 558 [48,49]. Recently, AurA was shown to use a secondary docking site on its N-lobe to efficiently recruit TACC3 and phosphorylate S558 [92]. Binding of TACC3 to this docking site also enhances the intrinsic catalytic activity of AurA, and it was postulated that this occurs through an allosteric mechanism in which TACC3 docking promotes the DFG flip. The dynamic equilibrium between DFG-Out and DFG-In states of phosphorylated AurA may be required to permit additional levels of regulatory control by substrates, potentially tuning the substrate specificity of this pool of the kinase.
The different conformational dynamics of AurA activated by either Tpx2 or phosphorylation have potential ramifications for selectively targeting the corresponding cellular pools of AurA with inhibitors. As Tpx2 enforces the DFG-In state, it might be predicted that the spindle-associated pool of AurA would be less susceptible to DFG-Out inhibitors, and more susceptible to DFG-In inhibitors. Conversely, since the phosphorylated enzyme found at the centrosome appears to still substantially sample the DFG-Out state, it is possible that this pool of AurA can be selectively targeted with DFG-Out inhibitors. Whether such selective targeting is possible with existing inhibitors remains to be seen. Further studies to dissect the extent to which particular inhibitors selectively recognize one conformational state or the other, and the way in which these preferences intersect with the differential conformational dynamics of different forms of AurA, will help address the feasibility of this goal.
A defective anchoring mechanism in the AurA activation loop may contribute to its heightened dynamics
The high degree of conformational dynamics displayed by the phosphorylated activation loop of AurA suggests that the active conformation is relatively unstable in this kinase. A comparison of the activation loops of AurA and the AGC kinases suggests that AurA is indeed missing a key structural feature that may be necessary to stabilize the active conformation.
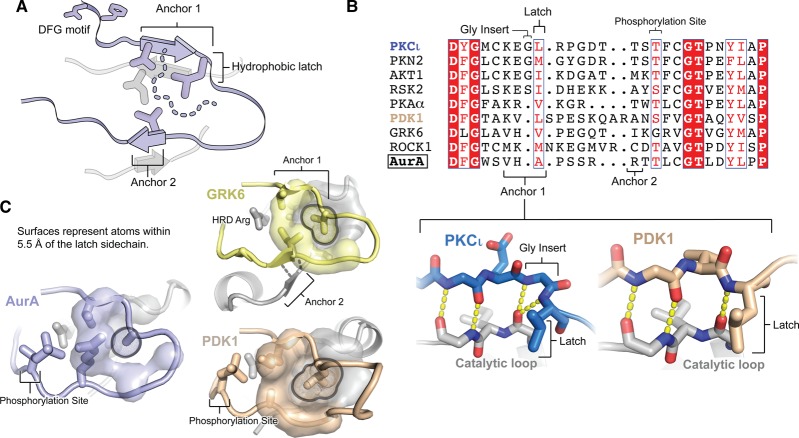
In kinases adopting the active conformation, the activation loop is anchored onto the C-lobe of the kinase domain by two short stretches of β-sheet (Figure 5A); an N-terminal anchor (anchor point 1) involves hydrogen bonds to the catalytic loop beneath it [26], and a second anchor (anchor point 2) links the C-terminal segment of the activation loop to the loop connecting the αEF and αF helices, through two or three hydrogen bonds [93]. An important feature of the anchoring mechanism, first described in the tyrosine kinases [93], is that the final residue of anchor point 1 projects a hydrophobic sidechain into a pocket formed by the C-lobe (Figure 5A). We refer to this structure as the hydrophobic latch.
Figure 5. Defective anchoring of the AurA activation loop may underlie its heightened dynamics.
(A) Schematic representation of the activation loop of an active protein kinase, showing the DFG motif at the N-terminus, the two anchor points that stabilize the N- and C-terminal halves of the loop, and the hydrophobic latch residue at the end of Anchor point 1. The sidechains of several residues located on the anchor points and the underlying catalytic loop are shown, which form part of the interaction surface (dashed line) for the hydrophobic latch residue. (B) Sequence alignment of the activation loops of AurA and members of eight AGC kinase subfamilies. The alignment was manually curated based on X-ray structures of each kinase [PDB IDs: 4DC2 (PKCι), 4CRS (PKN2), 4EKK (AKT1), 4NW6 (RSK2), 1L3R (PKA), 4RQK (PDK1), 3NYN (GRK6), 4W7P (ROCK1), 1OL5 (AurA)]. In the lower inset, a comparison is shown of the structures of Anchor point 1 in PKCι (a kinase with a glycine insert before the hydrophobic latch residue) and PDK1 (a kinase lacking the insert), highlighting the similar positioning of the leucine hydrophobic latch residue in the two cases. (C) Crystal structures are shown highlighting the anchoring of the activation loop in AurA (blue) and the AGC kinases GRK6 (yellow) and PDK1 (beige). Atoms within 5.5 Å of the sidechain of the hydrophobic latch residue are shown in a surface representation (for clarity surfaces were excluded for the residues preceding and following the latch residue).
Analysis of AGC kinase sequence alignments, combined with X-ray structures of members of eight different AGC kinase subfamilies, demonstrates that the hydrophobic latch is conserved in the AGC kinases as well, with isoleucine, leucine and valine being the most common latch residues (Figure 5B). Interestingly, there are two different classes of latch in the AGC kinases; about half the members of this family have the latch residue as the third and final residue of anchor point 1, but the remainder have a single-amino acid insertion immediately before the latch residue, usually glycine, extending the anchor point to four residues. In X-ray structures of this latter group, which includes Akt and the PKCs, the inserted residue adopts a left-handed α-helical conformation, resulting in an abrupt reversal of the direction of the polypeptide chain that positions the hydrophobic latch residue similarly to its location in the kinases lacking the insertion (Figure 5B, compare lower panels). Left-handed α-helices are energetically disfavored for residues other than glycine [94], and the conservation of glycine at this position suggests that evolutionary selective pressure has operated to maintain the positioning of the hydrophobic latch residue in this kinase family.
Unlike the AGC kinases, AurA has an alanine residue instead of a large hydrophobic residue at the latch position (Figure 5B), and the hydrophobic packing around the alanine sidechain is less extensive and less complementary than in the AGC kinases (Figure 5C). Thus, upon adoption of the active state, a considerably larger hydrophobic surface area is buried by the valine, leucine and isoleucine latches of the AGC kinases, than by the alanine latch of AurA. Considering common estimates for solvation free energies of these hydrophobic amino acids [95,96], these differences could alter the relative stabilities of the active and inactive states by as much as 1–2 kcal/mol. It is intriguing that in the other established case of a protein kinase with an alanine latch residue, the tyrosine kinase Csk, the impaired anchoring of the activation loop is responsible for the inability of the kinase to phosphorylate generic peptide substrates and the resulting specialization toward its cognate protein substrates, the Src-family kinases [93]. It is likely that the suboptimal latch residue of AurA is one of the primary reasons for the relative instability of the active conformation of the activation loop even when phosphorylated on T288, and the resulting heightened dynamics of this kinase.
Targeting disease-associated cellular pools of AurA with allosteric small molecules as novel therapeutic strategies in melanoma, neuroblastoma and prostate cancer
The studies reviewed above have highlighted the crucial importance of allostery and long-range conformational changes for the physiological regulation of AurA, begging the question of whether synthetic small molecules could be developed to exploit these allosteric properties. Indeed, the heightened dynamics of the phosphorylation-activated enzyme might render it particularly susceptible to conformational modulation with small molecules. Several recent studies have reported the identification of small molecule ligands that bind to allosteric sites on AurA and modulate kinase activity. Further development of molecules of this class could lead to novel therapies to treat several specific cancers in which AurA has recently been shown to play an important role.
Disruption of the AurA–Tpx2 interaction with small molecules
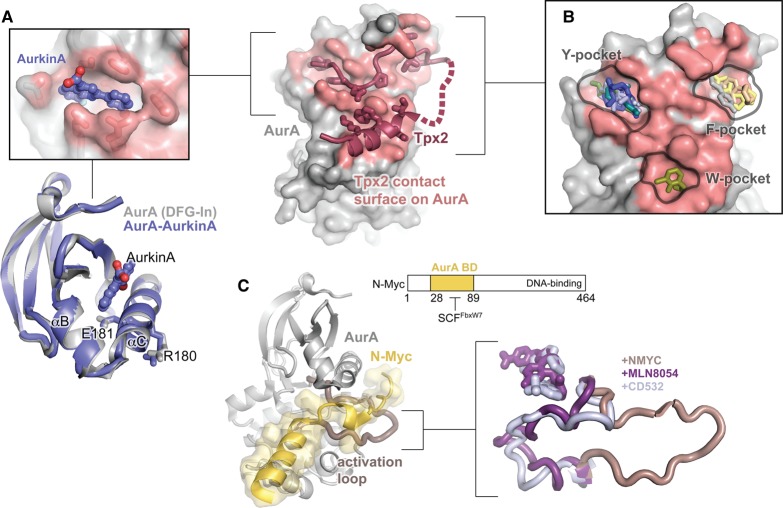
Using a high-throughput screen for molecules that displace Tpx2 from AurA, Janaček et al. identified a drug-like molecule they named AurkinA that binds to part of the Tpx2-binding surface on the N-lobe of AurA [97] (Figure 6A). AurkinA inhibits kinase activity in a non-ATP-competitive manner and mislocalizes AurA away from the mitotic spindle in cells. Interestingly, the molecule also inhibits activation of AurA by phosphorylation in vitro. At first glance, this is somewhat surprising, because the crystal structure of AurA bound to AurkinA shows only small perturbations to the structure of the N-lobe that are mostly restricted to local rearrangements of sidechains necessary to accommodate the small molecule (Figure 6A, bottom panel). In light of the recent discovery that phosphorylated AurA is unusually dynamic [87,88], a plausible alternative mechanism is that AurkinA inhibits kinase activity by restricting catalytically important motions of the enzyme.
Figure 6. Targeting AurA with allosteric small molecules.
(A) The structure of AurA bound to Tpx2 is shown on the right with the kinase domain in gray, Tpx2 shown in magenta, and the Tpx2-contact surface on AurA shown in pink. The top left panel shows an enlarged view of the structure of AurA bound to the allosteric small molecule AurkinA, showing AurkinA (blue) residing in the PIF pocket of the kinase. The same structure is shown underneath, aligned to the structure of free AurA in the DFG-In state (gray, PDB ID: 1OL7). Only the N-lobe is shown. (B) The locations of a representative subset of the small molecules found to bind to the Tpx2 binding surface by McIntyre et al. [98]. The molecules localize to three discrete pockets identified as the Y-, F-, and W-pockets based on the identity of the hydrophobic Tpx2 residues that occupy them in the AurA–Tpx2 complex. (C) Left: Structure of AurA bound to an N-terminal segment of N-Myc (residues 28–89). AurA is shown in gray with the activation loop highlighted in pink, and N-Myc is shown in yellow (PDB ID: 5G1X). The domain architecture of N-Myc is shown on the top right. The AurA-binding domain (AurA BD) is thought to overlap with the site of recognition by the ubiquitin ligase SCFFbxW7. Right: Comparison of the conformation of the AurA activation loop in structures of the kinase bound to N-Myc (pink) and the DFG-Out kinase inhibitors MLN-8054 (purple, PDB ID: 2WTV) and CD532 (blue, PDB ID: 4J8M). Structures were aligned on the entire kinase domain.
Another recent study involved the use of high-throughput crystallography to screen a fragment library for fragments that bind to AurA [98]. This unbiased approach led to the identification of >50 compounds that bind to allosteric pockets on AurA, and showed a striking preference for binding to the precise surfaces on the N-lobe that form the Tpx2-binding site (Figure 6B). The molecules cluster into three ‘hotspots’ on the surface that correspond to pockets occupied by large hydrophobic residues of Tpx2 when the latter is bound to AurA. Binding and inhibition studies showed that many of the compounds interfered with Tpx2 binding and activation of AurA, confirming an allosteric mode of action and validating the general approach of targeting the Tpx2-binding surface. Several of the compounds also inhibited the activity of phosphorylated AurA in the absence of Tpx2, as was observed with AurkinA, further confirming that ligands binding to this site, in addition to displacing Tpx2, may also allosterically modulate kinase activity. Interestingly, some of the compounds were found to increase rates of AurA autophosphorylation, highlighting that this allosteric coupling may be either positive or negative depending on the nature of the ligand.
Together these studies suggest that the AurA–Tpx2 interaction is surprisingly druggable, and that allosteric modulation of this interaction at the mitotic spindle is a potentially feasible goal. It will be interesting to see if compounds of this class have activity in tumor cells, particularly in cases where the spindle-associated pool of AurA is perturbed. A notable example is the ∼10% of melanomas that carry mutations in the gene encoding the catalytic subunit of the PP6 phosphatase [99]. Inactivation of PP6 in melanoma cells leads to hyperactivation of the spindle pool of AurA, which drives chromosome instability and DNA damage, and sensitizes cells to AurA inhibitors [18,58,100]. Allosteric inhibitors that disrupt the AurA–Tpx2 interaction represent a novel approach to targeting these melanoma tumors that may be superior to the use of conventional AurA inhibitors, as they could, in principle, spare the important functions of AurA at the centrosome. However, as discussed above, the allosteric AurA ligands discovered to date show that such an allosteric binding mode does not guarantee that the molecule will not also block activation of AurA by phosphorylation. Further progress in understanding how small molecules that target the Tpx2 binding site affect the structure and dynamics of the kinase domain may be essential to maximize the efficacy of this potential therapeutic approach.
Disruption of the AurA/N-Myc interaction in neuroblastoma and prostate cancer
In addition to its catalytic roles, AurA has recently been shown to play an essential non-catalytic scaffolding function in several cancers by stabilizing the oncogenic transcription factor N-Myc, a close homolog of the c-Myc proto-oncogene. It has long been known that the MYCN gene, which encodes N-Myc, is heavily amplified in high-risk cases of neuroblastoma [101], and it was recently reported that similar amplification of MYCN occurs in neuroendocrine prostate cancer (NEPC), a highly aggressive treatment-resistant form of prostate cancer [20]. In cells from both of these tumor types, the binding of AurA to N-Myc protein blocks degradation of the transcription factor by the ubiquitination/proteasome machinery [19,20]. Disruption of this complex by AurA knockdown or with small molecules can promote N-Myc degradation and cell death in cancer cell lines [19,102,103].
The structural basis for the interaction between AurA and N-Myc was recently determined [104], and revealed that a segment near the N-terminus of N-Myc binds across the activation loop of AurA, trapping the loop in the active conformation (Figure 6C, left). Indeed, the conformation of AurA bound to N-Myc is essentially identical with that of AurA complexed with Tpx2, and this remains the only example of a crystal structure in which AurA adopts the active conformation in the absence of Tpx2. This remarkable result explained the empirical observation that certain DFG-Out AurA inhibitors — but not DFG-In inhibitors — can promote dissociation of N-Myc from AurA [102,103], since they induce a conformational change of the activation loop that is incompatible with the binding mode of N-Myc (Figure 6C, right panel).
The discovery that DFG-Out AurA inhibitors can cause N-Myc dissociation and degradation raises the exciting possibility that such inhibitors could be used as novel treatments for patients with MYCN-amplified neuroblastoma and NEPC, patient populations that currently have limited treatment options. Unfortunately, most existing DFG-Out AurA inhibitors have limited capacity to displace N-Myc from AurA in vivo, with studies in cells demonstrating varying degrees of efficacy ranging from partial dissociation to no effect [102,103]. Although clinical trials in neuroblastoma and NEPC with some of these compounds are ongoing, results from early phases suggest that responses are limited, and in some cases actually lower for NMYC-amplified patients [105], suggesting that any efficacy is due to conventional enzymatic inhibition rather than enhanced degradation of N-Myc. Presumably most available DFG-Out AurA inhibitors, such as MLN-8054 and alisertib, have a limited energetic preference for the DFG-Out state, and so do not efficiently transduce their nanomolar binding affinity into the large conformational changes needed to block the association of N-Myc. In the case of MLN-8054, this hypothesis is supported by X-ray structures showing that the drug can bind to both the DFG-In and DFG-Out states of AurA [75], and a recent single-molecule fluorescence study, which showed that it triggers an incomplete population shift to the inactive state in solution [87]. A greater understanding of the chemical features of an inhibitor that confer strong selectivity for the DFG-Out state could be essential to pave the way for new treatments in this area. The various emerging spectroscopic approaches for studying allosteric transitions in AurA in solution are well suited to address this question by simultaneously tracking inhibitor binding and kinase conformation. Future studies in this vein are likely to shed considerable light on the interplay between small molecule binding and kinase conformational dynamics, and may accelerate the discovery of inhibitors that selectively target individual cellular pools of AurA in disease.
Outlook
AurA possesses a composite allosteric regulatory system that reflects the enzyme's multifaceted functions in mitosis. It is informative to consider AurA as a modified AGC-family kinase, in which stabilization of the αC-helix by HM docking, and stabilization of the activation loop by phosphorylation, have been partially decoupled into separate activation pathways. It appears that this decoupling was brought about in an ancestral Aurora kinase by supplementing the regulatory spine with an additional network of polar interactions mediated by an unusual polar αC spine residue, which further link the αC-helix to the DFG motif. Studies of the two activation pathways of AurA consequently provide valuable insight into the individual effects of αC-docking and phosphorylation on kinase structure and function.
Crystal structures of the active and inactive states of protein kinases have provided much insight into their regulation, but these static snapshots do not capture important dynamic effects that mediate allostery. Much of the recent progress in understanding the regulation of AurA has been driven by new spectroscopic methods that probe kinase dynamics in solution, providing information about both the structures of individual conformational states and the populations of those states. A central conclusion is that Tpx2 binding and phosphorylation activate the kinase by different, albeit complementary, mechanisms: Tpx2 promotes a population shift from the inactive DFG-Out to the active DFG-In state, while phosphorylation instead causes a change in the structure of the DFG-In subpopulation. The complementary nature of these effects, in which the dynamics of the enzyme are suppressed when activated by both phosphorylation and Tpx2, probably reflects the fact that equivalent activation pathways function concomitantly in the AGC kinases.
Protein kinases varying widely in their degree of dependence on activation loop phosphorylation. An important insight gained from the recent studies of AurA is that phosphorylation of the kinase activation loop may in some cases only marginally stabilize the active conformation. The degree of dynamics in the kinase activation loop both before and after phosphorylation may depend heavily on the intrinsic stability of the active conformation, which can be influenced by factors such as the effectiveness of the anchor points and the identity of the hydrophobic latch residue. Differences in these latter properties among kinases are likely a driving force for determining their responsiveness to activation loop phosphorylation.
The discovery that forms of AurA with unique conformational and dynamic properties are associated with specific disease states provides an excellent opportunity for applying insights into allostery and dynamics to the development of therapeutics targeting these pathological forms of the kinase. Further study of the dynamics of disease-associated forms of AurA, particularly in complex with N-Myc, is needed to bring this goal to fruition. With the advent of effective methods for studying AurA dynamics, we would appear to be at a tipping point where advances in understanding the basic biochemical mechanisms underpinning allosteric coupling in this kinase are poised to help bring about new therapies that could substantially impact cancer patients. As these methodologies for dissecting AurA dynamics are readily applicable to other systems, the resulting insights are likely to catalyze advances in our understanding of allosteric regulation in other kinase families.
Abbreviations
- AurA
Aurora A
- C-lobe
C-terminal lobe
- COT
Cancer Osaka Thyroid
- DEER
double electron–electron resonance
- ePKs
eukaryotic protein kinases
- FRET
Förster resonance energy transfer
- HM
hydrophobic motif
- Ipl1
increase-in-ploidy 1
- NEPC
neuroendocrine prostate cancer
- N-lobe
N-terminal lobe
- PIF
PDK1-interacting fragment
Competing Interests
The Author declares that there are no competing interests associated with this manuscript.
References
- 1.Glover D.M., Leibowitz M.H., McLean D.A. and Parry H. (1995) Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81, 95–105 10.1016/0092-8674(95)90374-7 [DOI] [PubMed] [Google Scholar]
- 2.Chan C.S. and Botstein D. (1993) Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135, 677–691 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roghi C., Giet R., Uzbekov R., Morin N., Chartrain I., Le Guellec R. et al. (1998) The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J. Cell Sci. 111(Pt 5), 557–572 PMID: [DOI] [PubMed] [Google Scholar]
- 4.Schumacher J.M., Golden A. and Donovan P.J. (1998) AIR-2: an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143, 1635–1646 10.1083/jcb.143.6.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff J.R., Anderson L., Zhu Y., Mossie K., Ng L., Souza B. et al. (1998) A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17, 3052–3065 10.1093/emboj/17.11.3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H., Kuang J., Zhong L., Kuo W.-l., Gray J., Sahin A. et al. (1998) Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 20, 189–193 10.1038/2496 [DOI] [PubMed] [Google Scholar]
- 7.Kimura M., Matsuda Y., Yoshioka T. and Okano Y. (1999) Cell cycle-dependent expression and centrosome localization of a third human aurora/Ipl1-related protein kinase, AIK3. J. Biol. Chem. 274, 7334–7340 10.1074/jbc.274.11.7334 [DOI] [PubMed] [Google Scholar]
- 8.Carmena M. and Earnshaw W.C. (2003) The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4, 842–854 10.1038/nrm1245 [DOI] [PubMed] [Google Scholar]
- 9.Hannak E., Kirkham M., Hyman A.A. and Oegema K. (2001) Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 155, 1109–1116 10.1083/jcb.200108051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seki A., Coppinger J.A., Jang C.-Y., Yates J.R. and Fang G. (2008) Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 320, 1655–1658 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macůrek L., Lindqvist A., Lim D., Lampson M.A., Klompmaker R., Freire R. et al. (2008) Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- 12.Kufer T.A., Silljé H.H.W., Körner R., Gruss O.J., Meraldi P. and Nigg E.A. (2002) Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158, 617–623 10.1083/jcb.200204155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung C.H.A., Coumar M.S., Hsieh H.-P. and Chang J.-Y. (2009) Aurora kinase inhibitors in preclinical and clinical testing. Expert. Opin. Investig. Drugs 18, 379–398 10.1517/13543780902806392 [DOI] [PubMed] [Google Scholar]
- 14.Boss D.S., Beijnen J.H. and Schellens J.H.M. (2009) Clinical experience with aurora kinase inhibitors: a review. Oncologist 14, 780–793 10.1634/theoncologist.2009-0019 [DOI] [PubMed] [Google Scholar]
- 15.Bavetsias V. and Linardopoulos S. (2015) Aurora kinase inhibitors: current status and outlook. Front. Oncol. 5, 278 10.3389/fonc.2015.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S. et al. (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 17.Asghar U., Witkiewicz A.K., Turner N.C. and Knudsen E.S. (2015) The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 14, 130–146 10.1038/nrd4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond D., Zeng K., Espert A., Bastos R.N., Baron R.D., Gruneberg U. et al. (2013) Melanoma-associated mutations in protein phosphatase 6 cause chromosome instability and DNA damage owing to dysregulated Aurora-A. J. Cell Sci. 126, 3429–3440 10.1242/jcs.128397 [DOI] [PubMed] [Google Scholar]
- 19.Otto T., Horn S., Brockmann M., Eilers U., Schüttrumpf L., Popov N. et al. (2009) Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell 15, 67–78 10.1016/j.ccr.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 20.Lee J.K., Phillips J.W., Smith B.A., Park J.W., Stoyanova T., McCaffrey E.F. et al. (2016) N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell 29, 536–547 10.1016/j.ccell.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda K., Mihara H., Kato Y., Yamaguchi A., Tanaka H., Yasuda H. et al. (2000) Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene 19, 2812–2819 10.1038/sj.onc.1203609 [DOI] [PubMed] [Google Scholar]
- 22.Littlepage L.E. and Ruderman J.V. (2002) Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16, 2274–2285 10.1101/gad.1007302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Ni J., Huang Q., Ren W., Yu L. and Zhao S. (2007) Identification of the auto-inhibitory domains of Aurora-A kinase. Biochem. Biophys. Res. Commun. 357, 347–352 10.1016/j.bbrc.2007.03.129 [DOI] [PubMed] [Google Scholar]
- 24.Manning G., Whyte D.B., Martinez R., Hunter T. and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 25.Huse M. and Kuriyan J. (2002) The conformational plasticity of protein kinases. Cell 109, 275–282 10.1016/S0092-8674(02)00741-9 [DOI] [PubMed] [Google Scholar]
- 26.Nolen B., Taylor S. and Ghosh G. (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 10.1016/j.molcel.2004.08.024 [DOI] [PubMed] [Google Scholar]
- 27.Knighton D.R., Zheng J.H., Ten Eyck L.F., Ashford V.A., Xuong N.H., Taylor S.S. et al. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 10.1126/science.1862342 [DOI] [PubMed] [Google Scholar]
- 28.Bayliss R., Sardon T., Vernos I. and Conti E. (2003) Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 12, 851–862 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- 29.Pearce L.R., Komander D. and Alessi D.R. (2010) The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11, 9–22 10.1038/nrm2822 [DOI] [PubMed] [Google Scholar]
- 30.Alessi D.R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P. et al. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551 PMID: [PMC free article] [PubMed] [Google Scholar]
- 31.Sarbassov D.D., Guertin D.A., Ali S.M. and Sabatini D.M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 10.1126/science.1106148 [DOI] [PubMed] [Google Scholar]
- 32.García-Martínez J.M. and Alessi D.R. (2008) mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 416, 375–385 10.1042/BJ20081668 [DOI] [PubMed] [Google Scholar]
- 33.Biondi R.M., Kieloch A., Currie R.A., Deak M. and Alessi D.R. (2001) The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 20, 4380–4390 10.1093/emboj/20.16.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins B.J., Deak M., Arthur J.S., Armit L.J. and Alessi D.R. (2003) In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J. 22, 4202–4211 10.1093/emboj/cdg407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frödin M., Antal T.L., Dummler B.A., Jensen C.J., Deak M., Gammeltoft S. et al. (2002) A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J. 21, 5396–5407 10.1093/emboj/cdf551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cyphers S., Ruff E.F., Behr J.M., Chodera J.D. and Levinson N.M. (2017) A water-mediated allosteric network governs activation of Aurora kinase A. Nat. Chem. Biol. 13, 402–408 10.1038/nchembio.2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gadde S. and Heald R. (2004) Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14, R797–R805 10.1016/j.cub.2004.09.021 [DOI] [PubMed] [Google Scholar]
- 38.Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A. et al. (1996) Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382, 420–425 10.1038/382420a0 [DOI] [PubMed] [Google Scholar]
- 39.Heald R., Tournebize R., Habermann A., Karsenti E. and Hyman A. (1997) Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138, 615–628 10.1083/jcb.138.3.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawin K.E. and Mitchison T.J. (1991) Mitotic spindle assembly by two different pathways in vitro. J. Cell Biol. 112, 925–940 10.1083/jcb.112.5.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruss O.J., Carazo-Salas R.E., Schatz C.A., Guarguaglini G., Kast J., Wilm M. et al. (2001) Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell 104, 83–93 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- 42.Tsai M.-Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J. et al. (2003) A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5, 242–248 10.1038/ncb936 [DOI] [PubMed] [Google Scholar]
- 43.Pinyol R., Scrofani J. and Vernos I. (2013) The role of NEDD1 phosphorylation by Aurora A in chromosomal microtubule nucleation and spindle function. Curr. Biol. 23, 143–149 10.1016/j.cub.2012.11.046 [DOI] [PubMed] [Google Scholar]
- 44.Wong J., Lerrigo R., Jang C.-Y. and Fang G. (2008) Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol. Biol. Cell 19, 2083–2091 10.1091/mbc.e07-10-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma N., Tulu U.S., Ferenz N.P., Fagerstrom C., Wilde A. and Wadsworth P. (2010) Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell 21, 979–988 10.1091/mbc.e09-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joukov V., De Nicolo A., Rodriguez A., Walter J.C. and Livingston D.M. (2010) Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc. Natl Acad. Sci. U.S.A. 107, 21022–21027 10.1073/pnas.1014664107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M. et al. (2003) Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114, 585–598 10.1016/S0092-8674(03)00642-1 [DOI] [PubMed] [Google Scholar]
- 48.Kinoshita K., Noetzel T.L., Pelletier L., Mechtler K., Drechsel D.N., Schwager A. et al. (2005) Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol. 170, 1047–1055 10.1083/jcb.200503023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peset I., Seiler J., Sardon T., Bejarano L.A., Rybina S. and Vernos I. (2005) Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J. Cell Biol. 170, 1057–1066 10.1083/jcb.200504037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joukov V., Walter J.C. and De Nicolo A. (2014) The Cep192-organized aurora A-Plk1 cascade is essential for centrosome cycle and bipolar spindle assembly. Mol. Cell 55, 578–591 10.1016/j.molcel.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dodson C.A., Yeoh S., Haq T. and Bayliss R. (2013) A kinetic test characterizes kinase intramolecular and intermolecular autophosphorylation mechanisms. Sci. Signal. 6, ra54 10.1126/scisignal.2003910 [DOI] [PubMed] [Google Scholar]
- 52.Zorba A., Buosi V., Kutter S., Kern N., Pontiggia F., Cho Y.-J. et al. (2014) Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. eLife 3, e02667 10.7554/eLife.02667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomez-Ferreria M.A., Rath U., Buster D.W., Chanda S.K., Caldwell J.S., Rines D.R. et al. (2007) Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17, 1960–1966 10.1016/j.cub.2007.10.019 [DOI] [PubMed] [Google Scholar]
- 54.Kemp C.A., Kopish K.R., Zipperlen P., Ahringer J. and O'Connell K.F. (2004) Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511–523 10.1016/S1534-5807(04)00066-8 [DOI] [PubMed] [Google Scholar]
- 55.Eyers P.A., Erikson E., Chen L.G. and Maller J.L. (2003) A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol 13, 691–697 10.1016/S0960-9822(03)00166-0 [DOI] [PubMed] [Google Scholar]
- 56.Toya M., Terasawa M., Nagata K., Iida Y. and Sugimoto A. (2011) A kinase-independent role for Aurora A in the assembly of mitotic spindle microtubules in Caenorhabditis elegans embryos. Nat. Cell Biol. 13, 708–714 10.1038/ncb2242 [DOI] [PubMed] [Google Scholar]
- 57.Dutertre S., Cazales M., Quaranta M., Froment C., Trabut V., Dozier C. et al. (2004) Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J. Cell Sci. 117, 2523–2531. 10.1242/jcs.01108 [DOI] [PubMed] [Google Scholar]
- 58.Zeng K., Bastos R.N., Barr F.A. and Gruneberg U. (2010) Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J. Cell Biol. 191, 1315–1332 10.1083/jcb.201008106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dodson C.A. and Bayliss R. (2012) Activation of Aurora-A kinase by protein partner binding and phosphorylation are independent and synergistic. J. Biol. Chem. 287, 1150–1157 10.1074/jbc.M111.312090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kornev A.P., Haste N.M., Taylor S.S. and Eyck L.F. (2006) Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl Acad. Sci. U.S.A. 103, 17783–17788 10.1073/pnas.0607656103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kornev A.P., Taylor S.S. and Ten Eyck L.F. (2008) A helix scaffold for the assembly of active protein kinases. Proc. Natl Acad. Sci. U.S.A. 105, 14377–14382 10.1073/pnas.0807988105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gutmann S., Hinniger A., Fendrich G., Drückes P., Antz S., Mattes H. et al. (2015) The crystal structure of cancer Osaka thyroid kinase reveals an unexpected kinase domain fold. J. Biol. Chem. 290, 15210–15218 10.1074/jbc.M115.648097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kothe M., Kohls D., Low S., Coli R., Cheng A.C., Jacques S.L. et al. (2007) Structure of the catalytic domain of human polo-like kinase 1. Biochemistry 46, 5960–5971 10.1021/bi602474j [DOI] [PubMed] [Google Scholar]
- 64.Carry J.-C., Clerc F., Minoux H., Schio L., Mauger J., Nair A. et al. (2015) SAR156497, an exquisitely selective inhibitor of aurora kinases. J. Med. Chem. 58, 362–375 10.1021/jm501326k [DOI] [PubMed] [Google Scholar]
- 65.Johannessen C.M., Boehm J.S., Kim S.Y., Thomas S.R., Wardwell L., Johnson L.A. et al. (2010) COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972 10.1038/nature09627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matheson C.J., Backos D.S. and Reigan P. (2016) Targeting WEE1 kinase in cancer. Trends Pharmacol. Sci. 37, 872–881 10.1016/j.tips.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 67.Hubbard S.R., Wei L., Ellis L. and Hendrickson W.A. (1994) Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 372, 746–754 10.1038/372746a0 [DOI] [PubMed] [Google Scholar]
- 68.Nagar B., Hantschel O., Young M.A., Scheffzek K., Veach D., Bornmann W. et al. (2003) Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell 112, 859–871 10.1016/S0092-8674(03)00194-6 [DOI] [PubMed] [Google Scholar]
- 69.Mol C.D., Dougan D.R., Schneider T.R., Skene R.J., Kraus M.L., Scheibe D.N. et al. (2004) Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J. Biol. Chem. 279, 31655–31663 10.1074/jbc.M403319200 [DOI] [PubMed] [Google Scholar]
- 70.Shan Y., Seeliger M.A., Eastwood M.P., Frank F., Xu H., Jensen M.O. et al. (2009) A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc. Natl Acad. Sci. U.S.A. 106, 139–144 10.1073/pnas.0811223106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agafonov R.V., Wilson C., Otten R., Buosi V. and Kern D. (2014) Energetic dissection of Gleevec's selectivity toward human tyrosine kinases. Nat. Struct. Mol. Biol. 21, 848–853 10.1038/nsmb.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis M.I., Hunt J.P., Herrgard S., Ciceri P., Wodicka L.M., Pallares G. et al. (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 10.1038/nbt.1990 [DOI] [PubMed] [Google Scholar]
- 73.Zhao Z., Wu H., Wang L., Liu Y., Knapp S., Liu Q. et al. (2014) Exploration of type II binding mode: A privileged approach for kinase inhibitor focused drug discovery? ACS Chem. Biol. 9, 1230–1241. 10.1021/cb500129t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin M.P., Zhu J.-Y., Lawrence H.R., Pireddu R., Luo Y., Alam R. et al. (2012) A novel mechanism by which small molecule inhibitors induce the DFG flip in Aurora A. ACS Chem. Biol. 7, 698–706 10.1021/cb200508b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dodson C.A., Kosmopoulou M., Richards M.W., Atrash B., Bavetsias V., Blagg J. et al. (2010) Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: insights into selectivity and drug design. Biochem. J. 427, 19–28 10.1042/BJ20091530 [DOI] [PubMed] [Google Scholar]
- 76.de Groot C.O., Hsia J.E., Anzola J.V., Motamedi A., Yoon M., Wong Y.L. et al. (2015) A cell biologist's field guide to aurora kinase inhibitors. Front. Oncol. 5, 285 10.3389/fonc.2015.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fancelli D., Moll J., Varasi M., Bravo R., Artico R., Berta D. et al. (2006) 1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: identification of a potent Aurora kinase inhibitor with a favorable antitumor kinase inhibition profile. J. Med. Chem. 49, 7247–7251 10.1021/jm060897w [DOI] [PubMed] [Google Scholar]
- 78.Nowakowski J., Cronin C.N., McRee D.E., Knuth M.W., Nelson C.G., Pavletich N.P. et al. (2002) Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography. Structure 10, 1659–1667 10.1016/S0969-2126(02)00907-3 [DOI] [PubMed] [Google Scholar]
- 79.Lawrence H.R., Martin M.P., Luo Y., Pireddu R., Yang H., Gevariya H. et al. (2012) Development of o-chlorophenyl substituted pyrimidines as exceptionally potent aurora kinase inhibitors. J. Med. Chem. 55, 7392–7416 10.1021/jm300334d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coumar M.S., Leou J.-S., Shukla P., Wu J.-S., Dixit A.K., Lin W.-H. et al. (2009) Structure-based drug design of novel Aurora kinase A inhibitors: structural basis for potency and specificity. J. Med. Chem. 52, 1050–1062 10.1021/jm801270e [DOI] [PubMed] [Google Scholar]
- 81.Coumar M.S., Tsai M.-T., Chu C.-Y., Uang B.-J., Lin W.-H., Chang C.-Y. et al. (2010) Identification, SAR studies, and X-ray co-crystallographic analysis of a novel furanopyrimidine aurora kinase A inhibitor. ChemMedChem 5, 255–267 10.1002/cmdc.200900339 [DOI] [PubMed] [Google Scholar]
- 82.Wu J.M., Chen C.T., Coumar M.S., Lin W.H., Chen Z.J., Hsu J.T. et al. (2013) Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules. Proc. Natl Acad. Sci. U.S.A. 110, E1779–E1787 10.1073/pnas.1220523110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burgess S.G., Oleksy A., Cavazza T., Richards M.W., Vernos I., Matthews D. et al. (2016) Allosteric inhibition of Aurora-A kinase by a synthetic vNAR domain. Open Biol. 6, 160089 10.1098/rsob.160089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levinson N.M., Kuchment O., Shen K., Young M.A., Koldobskiy M., Karplus M. et al. (2006) A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 4, e144 10.1371/journal.pbio.0040144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McClendon C.L., Kornev A.P., Gilson M.K. and Taylor S.S. (2014) Dynamic architecture of a protein kinase. Proc. Natl Acad. Sci. U.S.A. 111, E4623–E4631 10.1073/pnas.1418402111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson L.N., Noble M.E.M. and Owen D.J. (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85, 149–158 10.1016/S0092-8674(00)81092-2 [DOI] [PubMed] [Google Scholar]
- 87.Gilburt J.A.H., Sarkar H., Sheldrake P., Blagg J., Ying L. and Dodson C.A. (2017) Dynamic equilibrium of the Aurora A kinase activation loop revealed by single-molecule spectroscopy. Angew. Chem. Int. Ed. Engl. 56, 11409–11414 10.1002/anie.201704654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ruff E.F., Muretta J.M., Thompson A.R., Lake E.W., Cyphers S., Albanese S.K. et al. (2018) A dynamic mechanism for allosteric activation of Aurora kinase A by activation loop phosphorylation. eLife 7, e32766 10.7554/eLife.32766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith K.J., Carter P.S., Bridges A., Horrocks P., Lewis C., Pettman G. et al. (2004) The structure of MSK1 reveals a novel autoinhibitory conformation for a dual kinase protein. Structure 12, 1067–1077 10.1016/j.str.2004.02.040 [DOI] [PubMed] [Google Scholar]
- 90.De Bondt H.L., Rosenblatt J., Jancarik J., Jones H.D., Morgan D.O. and Kim S.H. (1993) Crystal structure of cyclin-dependent kinase 2. Nature 363, 595–602 10.1038/363595a0 [DOI] [PubMed] [Google Scholar]
- 91.Xu W., Harrison S.C. and Eck M.J. (1997) Three-dimensional structure of the tyrosine kinase c-Src. Nature 385, 595–602 10.1038/385595a0 [DOI] [PubMed] [Google Scholar]
- 92.Burgess S.G., Mukherjee M., Sabir S., Joseph N., Gutiérrez-Caballero C., Richards M.W. et al. (2018) Mitotic spindle association of TACC3 requires Aurora-A-dependent stabilization of a cryptic α-helix. EMBO J. 37, e97902 10.15252/embj.201797902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levinson N.M., Seeliger M.A., Cole P.A. and Kuriyan J. (2008) Structural basis for the recognition of c-Src by its inactivator Csk. Cell 134, 124–134 10.1016/j.cell.2008.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Novotny M. and Kleywegt G.J. (2005) A survey of left-handed helices in protein structures. J. Mol. Biol. 347, 231–241 10.1016/j.jmb.2005.01.037 [DOI] [PubMed] [Google Scholar]
- 95.Eisenberg D. and McLachlan A.D. (1986) Solvation energy in protein folding and binding. Nature 319, 199–203 10.1038/319199a0 [DOI] [PubMed] [Google Scholar]
- 96.Nozaki Y. and Tanford C. (1971) The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J. Biol. Chem. 246, 2211–2217 PMID: [PubMed] [Google Scholar]
- 97.Janeček M., Rossmann M., Sharma P., Emery A., Huggins D.J., Stockwell S.R. et al. (2016) Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2. Sci. Rep. 6, 28528 10.1038/srep28528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McIntyre P.J., Collins P.M., Vrzal L., Birchall K., Arnold L.H., Mpamhanga C. et al. (2017) Characterization of three druggable hot-spots in the Aurora-A/TPX2 interaction using biochemical, biophysical, and fragment-based approaches. ACS Chem. Biol. 12, 2906–2914 10.1021/acschembio.7b00537 [DOI] [PubMed] [Google Scholar]
- 99.Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.-P. et al. (2012) A landscape of driver mutations in melanoma. Cell 150, 251–263 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gold H.L., Wengrod J., de Miera E.V.-S., Wang D., Fleming N., Sikkema L. et al. (2014) PP6C hotspot mutations in melanoma display sensitivity to Aurora kinase inhibition. Mol. Cancer Res. 12, 433–439 10.1158/1541-7786.MCR-13-0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brodeur G.M., Seeger R.C., Schwab M., Varmus H.E. and Bishop J.M. (1984) Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224, 1121–1124 10.1126/science.6719137 [DOI] [PubMed] [Google Scholar]
- 102.Brockmann M., Poon E., Berry T., Carstensen A., Deubzer H.E., Rycak L. et al. (2013) Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell 24, 75–89 10.1016/j.ccr.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gustafson W.C., Meyerowitz J.G., Nekritz E.A., Chen J., Benes C., Charron E. et al. (2014) Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 26, 414–427 10.1016/j.ccr.2014.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Richards M.W., Burgess S.G., Poon E., Carstensen A., Eilers M., Chesler L. et al. (2016) Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proc. Natl Acad. Sci. U.S.A. 113, 13726–13731 10.1073/pnas.1610626113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DuBois S.G., Marachelian A., Fox E., Kudgus R.A., Reid J.M., Groshen S. et al. (2016) Phase I study of the Aurora A kinase inhibitor alisertib in combination with irinotecan and temozolomide for patients With relapsed or refractory neuroblastoma: A NANT (new approaches to neuroblastoma therapy) trial. J. Clin. Oncol. 34, 1368–1375 10.1200/JCO.2015.65.4889 [DOI] [PMC free article] [PubMed] [Google Scholar]