Abstract
Microbiome composition has been associated to several inflammatory diseases, including asthma. There are few studies exploring the relationships of gut microbiota with airway obstruction pheonotypes in adult asthma, especially those living in the tropics. We sought to evaluate the relationships of gut microbiota with the airway obstruction and other variables of interest in asthmatic patients living in the tropics according to three phenotypes: No Airway Obstruction (NAO), Reversible Airway Obstruction (RAO) or Fixed Airway Obstruction (FAO). We found that Streptococcaceae:Streptococcus and Enterobacteriaceae:Escherichia-Shigella consistently discriminated asthmatic individuals suffering FAO from NAO or RAO, plus Veillonellaceae:Megasphaera when comparing FAO and RAO (p < 0.05; FDR < 0.05). In the FAO, the network showing the genus relations was less complex and interconnected. Several Rumminococcaceae, Lachnospiraceae and Clostridiales were enriched in patients with low specific IgE levels to mites and Ascaris. All patients shared a common exposure framework; control medication usage and smoking habit were uncommon and equally distributed between them. In conclusion, in this tropical asthmatic population, components of human gut microbiota are associated with the presence of a FAO phenotype and lower specific IgE response to mites and Ascaris.
Introduction
Asthma is a chronic inflammatory respiratory disease affecting about 10% of humans. Although genetic predisposition is determinant for disease development, environmental factors also have a role. Microbial colonization of body sites may be shaped by both, host genetics and environment (i.e. diet and geography). Microbiome composition has been associated to several inflammatory diseases, including asthma1–3. There are an increasing number of publications that shows a link between intestinal or airway microbial composition and the incidence or severity of asthma in animal models and humans4–9. Different mechanisms derived from microbiome-host interactions and metabolites could explain how a microorganism could impact disease presentation1,2,4,5. Moreover, restoration of gut microbiota, through the use of prebiotics or probiotics, has shown positive results in asthma-related clinical outcomes10. Heterogeneity at different levels, personal and geographical, makes it difficult to identify microorganisms or microbial networks associated with diseases11. In the case of asthma, it is also recognized that it has different disease phenotypes12. Further profiling and characterization of the microbiome associated with different asthma phenotypes is necessary for identifying novel microbiota-related mechanisms of disease.
Other authors have found association of airway obstruction phenotypes with different immune traits and disease severity13. Bronchodilator response (BDR) is a classical asthma phenotype that means reversibility of airway obstruction after the administration of a short-acting Beta 2 agonist, a common relief medication for asthma symptoms. Although its patophysiology and relationships with clinical outcomes are partially understood, it is recognized as complex trait involving interactions of different cell types and genetical determinants14. Reversible airway obstruction is considered as a criterium for asthma diagnosis and it has been associated with biomarkers of eosinophilic inflammation, atopy and bronchial hyperreactivity15–17. Cluster analysis has suggested that the BDR is an important component in defining asthma phenotypes18. On the other hand, lack of airway obstruction reversibility (also known as fixed airway obstruction) is another phenotype of asthma found in some asthmatic individuals19,20.
In general, there are few studies about microbiome differences in regard to asthma phenotypes, but less information exist about microbiome composition of tropical populations, especially from Latin America21,22. The aim of this study was to evaluate the relationships of gut microbiota with the airway obstruction and other variables of interest in adult asthmatics living in the tropics.
Methods
Design, location and study population
This study analyzes the gut microbiome of asthmatic subjects included in the Ascaris and Asthma Severity program; which assesses the relationship between asthma severity and Ascaris lumbricoides sensitization23. Patients live in Cartagena de Indias in the North Caribbean coast of Colombia, a tropical city (10° 23′ 59″ North, 75° 30′ 52″ West) with around 1 million inhabitants, most of them poor according to the govermental indexes24. Socioeconomic stratification in Colombia ranges from 1 to 6, and the majority of the city population, as well as the study participants, belonged to the lowest strata 1–3 and shared environmental conditions. The genetic background resulted from racial admixture between Native Americans, Spaniards, and an important proportion (37.9%) of African ancestry25,26. The study was approved by the Ethics Commitee of the University of Cartagena (Cartagena, Colombia), all experiments were performed in accordance with and following the Declaration of Helsinki Principles. Signed informed consent was obtained from patients or their parents. Stool sample for microbiota analysis was collected from 202 subjects of the program.
Eligibility criteria and enrollment procedures
Subjects attending five public primary health care centers and the University Hospital were screened for eligibility by physicians of the research staff between June 2010 and March 2011. These centers serve the lowest social strata in the city. Eligibility criteria were: subjects in the age range of 8 to 70 years who answered affirmatively to the question: Have you ever been diagnosed with asthma? Inclusion in the study depended on the confirmation of asthma diagnosis made by the physician. Control medication usage was defined as the regular use of oral (OCS) or inhaled corticosteroids (ICS) and evaluated by questionnaires. Patients with chronic obstructive pulmonary disease (COPD) or another chronic respiratory co-morbidity were excluded as well as those belonging to the highest socio-economical strata of the city (4 to 6).
Asthma diagnosis
Eligible subjects were further interviewed and asthma diagnosis was confirmed in those with at least two respiratory symptoms (cough, wheezing, dyspnea, and nocturnal cough/wheezing/dyspnea) or a history of recurrent asthma attacks. These questions were done by staff physicians following a validated questionnaire27,28.
Skin prick test
Skin prick test (SPT) was done in the forearm with a battery of allergen extracts (kindly supplied by Leti; Madrid, Spain) as previously described23. Atopy was defined as at least one positive SPT to any of the tested allergens.
Assessment of lung function and the airway obstruction phenotypes
Spirometry was performed with a Microlab spirometer (Carefusion Corporation, San Diego, USA) following the American Thoracic Society recommendations29. Height and weight were measured; patients were instructed to avoid use of short-acting bronchodilators for at least 12 hours before testing. The best forced expiratory volume in one second (FEV1) was selected and was taken as an objective indicator of basal airway obstruction; bronchodilator response was defined as >12% improvement in the predicted basal FEV1 levels after 200 µg of salbutamol inhalation. According to spirometry results, subjects were classified into 3 different phenotypes: No Airway Obstruction (NAO: Symptoms + FEV1 ≥ 80% predicted; n = 66), Reversible Airway Obstruction (RAO: Symptoms + FEV1 < 80% + Bronchodilator Response; n = 74), Fixed Airway Obstruction (FAO: Symptoms + FEV1 < 80% predicted + Lack of Bronchodilator Response; n = 42).
Quantification of total and specific IgE
In the tropics, helminthiases are common and house dust mites (HDM) exposure is perennial. We previously showed that Ascaris and HDM sensitization is associated with indicators of asthma severity23; therefore, we evaluated the relationship of specific IgE levels with microbiota composition. Briefly, blood samples were taken by venipuncture using anticoagulant-free tubes to obtain serum for antibody determinations. Serum total IgE and specific IgE levels against B. tropicalis, D. pteronyssinus and A. lumbricoides were determined by ImmunoCap system (Phadia100, Thermo, Sweden).
Stool DNA extraction
Participants were asked to bring a recently collected stool sample in a hermetic recipient provided by the research team. Samples kept refrigerated less than 12 hours between collection and frozing at −20 °C until processing. Samples were thawed for the first time and DNA extraction was performed using Qiamp DNA stool minikit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA concentration was quantified using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, USA). Eluted DNA was stored at −80 °C until amplification reaction and V4 region sequencing.
Amplification and sequencing of variable 4 (V4) region of 16S rRNA gene
Genomic bacterial DNA extraction methods were optimized to maximize the yield of bacterial DNA while keeping background amplification to a minimum. 16S rRNA gene sequencing methods were adapted from the methods developed for the NIH-Human Microbiome30,31. Briefly, the 16S rDNA V4 region was amplified by PCR and sequenced in the MiSeq platform (Illumina, San Diego, USA) using the 2 × 250 bp paired-end protocol yielding pair-end reads that overlap almost completely. The primers used for amplification contain adapters for MiSeq sequencing and dual-index barcodes so that the PCR products may be pooled and sequenced directly32, targeting at least 10,000 reads per sample.
Our standard pipeline for processing and analyzing the 16S rRNA gene data incorporates phylogenetic and alignment-based approaches to maximize data resolution. The read pairs were demultiplexed based on the unique molecular barcodes, and reads were merged using USEARCH v7.0.100133, allowing zero mismatches and a minimum overlap of 50 bases. Merged reads were trimmed at first base with Q5. In addition, a quality filter as applied to the resulting merged reads and reads containing above 0.05 expected errors were discarded.
16S rRNA gene sequences were assigned into OTUs or phylotypes at a similarity cutoff value of 97% using the UPARSE algorithm. OTUs were then mapped to an optimized version of the SILVA Database34,35 containing only the 16S v4 region to determine taxonomies. Abundances were recovered by mapping the demultiplexed reads to the UPARSE OTUs. A custom script constructed an OTU table from the output files generated in the previous two steps, which was then used to calculate alpha-diversity, beta-diversity36, and provide taxonomic summaries that were leveraged for all subsequent analyses discussed below.
Parasitological examination
Parasitological analyses were done using 0.85% saline solution and lugol staining; counting helminth eggs were done by the Kato Katz method using a commercial kit (Copro Kit, C&M Medical, Campinas, Brazil). The presence of eggs from geohelminths or parasite visualization was considered diagnostics of active infection.
Statistical and network analysis
In socio-demographic analyses, differences between proportions were analyzed by Pearson chi-squared test (or Fisher exact test when appropriate). Total and specific IgE values were not normally distributed and they were therefore reported as the median value and its interquartile range, except total IgE (geometric mean). Kruskal-Wallis (KW) test was used for comparison of continuous variables among three groups. Post hoc analyses were also performed.
Analysis of microbiota datasets presented several technical challenges. First, OTU abundances are compositional and not independent, thus conventional comparison or correlation between OTU abundances using traditional statistical methods can lead to spurius results. Secondly, variables in an OTU table outnumber the tested individual samples, thus inference of OTU-OTU association networks is underpowered37. Analyses were done using R scripts and to reduce false positive, only OTUs with at least 1 count in 50% of samples were included in the analysis. The compositional nature of the data, characterized by non-negative counts whose weight is relative to the other components of a given sample, restricts the analysis of the data to a simplex space. In order to carry the data to a normal space, a Centered Log Ratio Transformation (CLR) of Aitchinson was applied, this transformation was done using the function logratio.transfo of the mixOmics package38. Since this is a logarithmic transformation, it requires a pre-conditioning of the data to replace the zeros, this substitution was performed with a multiplicative Bayesian replacement strategy using the cmultRepl function of the zCompositions package39.
Alpha diversity was calculated using Chao 1 and Shannon indexes using the Phyloseq package40. sPLS-DA used an approach that asks and identifies which features (OTUs) separates patients according to airway obstruction phenotypes based on a discriminant analysis of the partial least square metric. To select the most predictive/discriminative OTUs classifying the samples according to the airway obstruction phenotypes or specific IgE levels, Sparce Partial Least Square Linear Dyscriminant Analysis (sPLS-DA) was done using the package Mixomics including 10 variables in the first component38. The Percentile 25th and 75th were calculated to select the extremes of the distribution of specific IgE levels to Ascaris, B. tropicalis and D. pteronyssinus in order to discriminate between high IgE responders (HR, levels ≥ percentile 75th to the highest) and low IgE responders (LR, levels ≤ percentile 25th). Since percentile 25th values were 0,2, 0,2 and 0,1 kUL for Ascaris, B. tropicalis and D. pteronyssinus – specific IgE, respectively, all patients included in LR were not sensitized to that specific source. Percentile 75th values were 3,1, 23,7 and 13,03 for Ascaris, B. tropicalis and D. pteronyssinus – specific IgE, respectively. Thus, those in HR group were sensitized individuals with the the highest responses.
As individuals living in the tropics are exposed to Ascaris infection and this could alterate the gut microbiota, sPLS-DA was also done using Ascaris infection status as a binary category. Iterative cross validation method allowed us to select the most stable OTUs in the model of classification; wich means those OTUs always indicating differences in the individuals belonging to the category of interest after iterative comparison with the individuals of the other category. To avoid a type 1 error after the selection of the most consistent OTUs (Stability Index = 1), row counts were later compared between phenotypes of inteterest using the Metagenomeseq package normalizing the data with Cumulative Sum Scaling by applying the cumNorm function and performing the analysis with a zero-inflated log-normal model by applying the fitFeatureModel, this package employ the Wilcoxon signed-rank test and posterior correction for multiple comparisons with the Benjamini-Hochberg method41. Due to significant differences in age among phenotypes, the influence of age as a continuous variable on OTU counts was evaluated the MaAsLin package42.
To predict OTUs networks’ interactions within each asthma group, undirected conditional independence network graphs were constructed using Sparce and Compositionally Robust Inference of Microbial Ecological Networks (SPIEC-EASI) statistical method37, which used the Meinshausen-Buhlmann strategy as the graph estimator. Finally, differentially enriched OTUs, detected by sPLS-DA, were localized into the predicted interaction network to find nodes and clusters of interest differentiating the groups.
Results
Characteristics of the study population
Twenty out of 202 sequenced samples were excluded from the analysis because OTU counts were too low (n = 3) or due to lack of spirometry data23. The V4 region of the 16S rRNA was amplified and sequenced in all of them obtaining at least 10.000 sequencing reads per sample (see Supplementary Fig. S1,2). Sociodemographic and clinical features of the study population are shown in Table 1. Overall, we included asthmatic patients sharing an environmental exposure framework. NAO patients were younger than those with obstruction. Specific IgE levels to D. pteronyssinus were different among phenotypes, being higher in RAO patients than those with FAO.
Table 1.
Socio-demographic characteristics of the study population.
| Variables | All patients (n = 182) | FAO (n = 42) | RAO (n = 74) | NAO (n = 66) | p-value |
|---|---|---|---|---|---|
| Gender (male) | 23.6 | 21.4 | 18.9 | 30.3 | 0.26 |
| Age (mean ± SD) | 33.7 ± 17.3 | 36.5 ± 17.5 | 39.4 ± 16.9 | 25.4 ± 14.3 | <0.0001 |
| Socio-economical strata | 0.32 | ||||
| 1 | 63.18 | 64.28 | 68.91 | 56.06 | |
| 2 | 29.12 | 28.57 | 21.62 | 37.87 | |
| 3 | 7.69 | 7.14 | 9.45 | 6.06 | |
| Smoking habit | 2.74 | 2.38 | 4.05 | 1.51 | 0.64 |
| Co-habitation with a smoker | 25.27 | 33.33 | 25.67 | 19.69 | 0.26 |
| Predicted baseline FEV1 (%) | 71.87 ± 21.47 | 66.03 ± 12.29 | 56.63 ± 18.79 | 92.50 ± 8.67 | <0.0001 |
| Predicted baseline FEV1/FVC (%) | 86.57 ± 15.44 | 88.04 ± 14.56 | 77.14 ± 15.41 | 96.5 ± 7.65 | <0.0001 |
| Episodes of severe dyspnea | 66.48 | 69.04 | 71.62 | 59.09 | 0.27 |
| Nocturnal awakenings | 87.36 | 79.57 | 93.24 | 86.36 | 0.06 |
| >4 ER visits int the last year | 26.37 | 23.80 | 35.13 | 18.18 | 0.07 |
| Hospitalizations in the last year | 13.73 | 23.80 | 13.51 | 7.57 | 0.06 |
| Bronchodilator responsiveness | 12.63 ± 19.40 | 2.45 ± 6.90 | 27.95 ± 21.92 | 1.93 ± 5.29 | <0.0001 |
| Use of oral corticosteroids | 30.81 | 26.31 | 35.71 | 28.12 | 0.50 |
| Use of inhaled corticosteroids | 30.81 | 26.31 | 35.71 | 28.12 | 0.5 |
| Current Ascaris infection | 4.39 | 4.76 | 5.40 | 3.03 | 0.81 |
| Antecedent of worm expulsion | 60.43 | 66.66 | 62.16 | 54.54 | 0.43 |
| Allergic rinitis | 84.61 | 88.69 | 83.78 | 83.33 | 0.80 |
| Atopy (>1 positive SPT) | 83.51 | 78.5 | 86.4 | 83.3 | 0.54 |
| Sensitization to B. tropicalis | 64.20 | 59.52 | 69.53 | 61.53 | 0.46 |
| Sensitization to D. Pteronyssinus | 69.88 | 64.28 | 76.81 | 66.15 | 0.27 |
| Total IgE‡† | 654.6695 ± 602.8 | 525.70 ± 979.54 | 826.76 ± 1767.43 | 543.78 ± 637.61 | 0.09 |
| Specific IgE to B. Tropicalis† | 19.52 ± 33.54 | 20.31 ± 30.39 | 19.42 ± 33.60 | 19.12 ± 35.79 | 0.6 |
| Specific IgE to D. Pteronyssinus † | 19.12 ± 36.61 | 11.02 ± 24.51 | 27.08 ± 44.70 | 15.36 ± 31.32 | 0.01 |
| Specific IgE to A. lumbricoides† | 3.82 ± 2.745 | 2.82 ± 6.51 | 4.29 ± 8.4 | 3.93 ± 9.41 | 0.07 |
†Comparison by Krukal-Wallis test.
Frequency rates (%) for categorical variables are shown.
‡Geometric mean (standard deviation of mean) are reported.
Gut microbial richness and diversity was similar among airway obstruction phenotypes
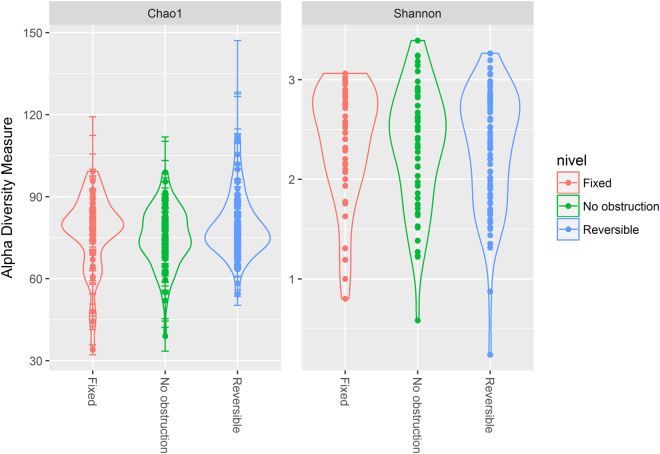
Microbial richness and diversity were evaluated by means of the Chao 1 and Shannon indexes, respectively. Based on the OTUs distribution, the average value of Chao 1 was not different between phenotypes, showing the following mean values: 76.8 ± SD 13.91, 79.1 ± SD 11.25 and 75 ± SD 11.02 for FAO, RAO and NAO phenotype, respectively (KW Test; p = 0.25; Fig. 1A). Mean Shannon index values were neither different among groups: 2.37 ± SD 0.58, 2.31 ± SD 0.59 and 2.32 ± SD 0.59 in asthmatics with FAO, RAO and NAO, respectively (KW test; p = 0.79; Fig. 1B).
Figure 1.
Comparison of alpha diversity among different airway obstruction phenotypes using Chao 1 (A) and Shannon diversity indexes (B). Violin plot includes the median, 95% CI, IQR, and density plot where the width of the differently colored lines indicate frequency.
Streptococcaceae:Streptococcus and Enterobacteriaceae:Escherichia-Shigella consistently discriminated and were enriched in patients with the FAO phenotype
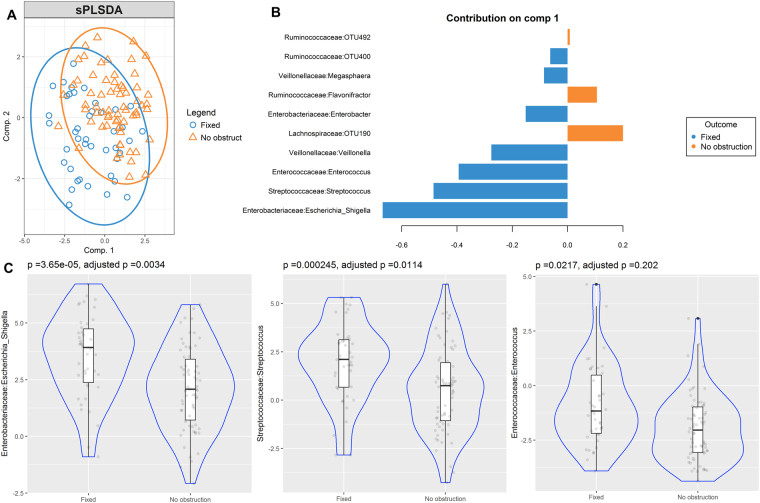
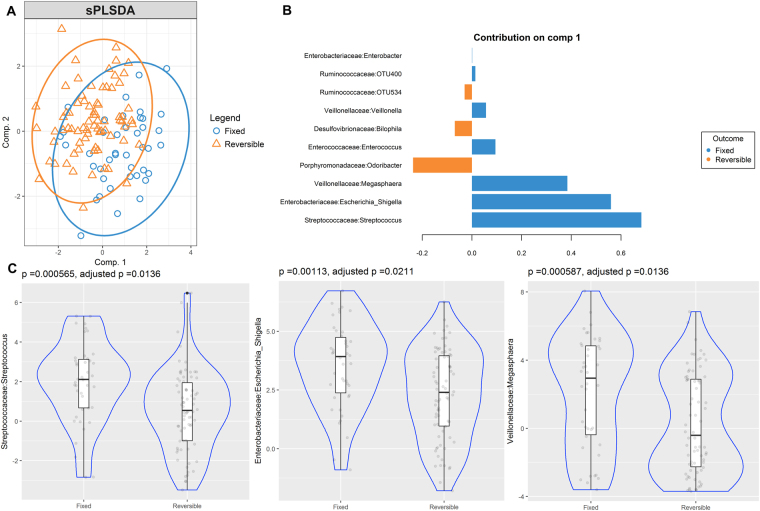
sPLS-DA analyses were used to identify OTUs associated with airway obstruction phenotypes. As observed in the bi-plot, microbial composition showed a clear differentiation between patients with FAO and NAO (Fig. 2A). Several differentially abundant OTUs in FAO and NAO stool samples that contributed to their separation in component 1 were identified (Fig. 2B). Enterobacteriaceae:Escherichia-Shigella, Streptococcaceae:Streptococcus and Enterococcacea:enterococcus were enriched in FAO patients and accurately discriminated them from those with NAO. These bacteria were also the most consistent OTUs (Stability index = 1), being present in 90.9%, 98.8% and 56.7% of samples, respectively. After correction by multiple comparisons, only Enterococcus:Escherichia-Shigella and Streptococcaceae:Streptococcus showed significant enrichment (Fig. 3C and Supplementary Table S1). Microbial composition of patients with FAO was also clearly discriminated from those with RAO. Streptococcaceae:Streptococcus and Enterococcus:Escherichia-Shigella plus Veillonellaceae:megasphaera were significantly enriched in FAO and were the most consistent taxa (Stability index = 1) and (Fig. 3A,B). Furthermore, after correction by multiple comparisons, all of them kept significantly enriched (Fig. 3C and Supplementary Table S1). In addition, none of the above was significantly associated with age and remained discriminant after removing patients younger than 18 years in sensitivity analysis with sPLS-DA (data not shown). The sPLS-DA analysis showed that in females the same OTUs remained discriminant, but in males only Enterobacteriaceae:Escherichia-Shigella remained discriminant (see Supplementary Figs S3,4). However, as we found no differences on male proportion between phenotypes (Table 1), it seems that gender distribution is not a confounding variable. Comparison of the two other phenotypes (NAO vs RAO) did not let to significantly enriched taxa after adjustment (see Supplementary Fig. S5 and Table S1).
Figure 2.
Partial least squares discriminant analysis of gut microbiome composition between patients with fixed airway obstruction versus no obstruction sPLS-DA plot based on the relative abundance of bacterial taxa of gut microbiota from patients with fixed airway obstruction (blue circle) or without obstruction (orange triangle) and their 95% confidence ellipses (A). Contribution plot indicating genera contributing to component 1 of the sPLS-DA plot that discriminate these phenotypes (B). The abundance of the most consistent OTUs was compared using Metagenomeseq and presented on a violin plot, which includes the median, 95% CI, IQR, and density plot where the width of the blue lines indicate frequency (C).
Figure 3.
Partial least squares discriminant analysis of gut microbiome composition between patients with fixed versus reversible obstruction. sPLS-DA plot based on the relative abundance of bacterial taxa of gut microbiota from patients with fixed airway obstruction (blue circle) or reversible airway obstruction (orange triangle) and their 95% confidence ellipses (A). Contribution plot indicating genera contributing to component 1 of the sPLS-DA plot that discriminate these phenotypes (B). The abundance of the most consistent OTUs was compared using Metagenomeseq and presented on a violin plot, which includes the median, 95% CI, IQR, and density plot where the width of the blue lines indicate frequency (C).
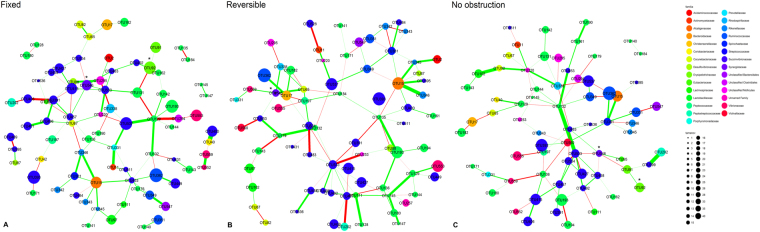
FAO phenotype harbour a distinctive network of OTUs
The networks were dominated by the Rumminococcaceae and Lachnospiraceae families and were clustered by taxonomic families in the network graph. Airway obstruction phenotype groups showed a similar number of OTU nodes in the networks (λ index: 0.56 for the three groups) (Fig. 4A–C). However, the networks in FAO group were less complex (see Supplementary Fig. S6A) and interconnected as showed by a lower sparsity index (SI: 0.55) when compared with NAO (SI: 0.75) and RAO phenotypes (SI: 0.79); however, the strengths of edges were greater (see Supplementary Fig. S6B). Also, this network highlighted the enrichment of Streptococcaceae:Streptococcus (OTU546) and Enterococcus:Escherichia-Shigella (OTU92) and their difference in the connectivity compared to those networks derived from individuals in the NAO and RAO or NAO phenotypes (Fig. 4A–C and Supplementary Table S3).
Figure 4.
MB Networks from patients with fixed (A), reversible (B) and no airway obstruction (C) using SPIEC-EASI method. Overall, networks display a differential association pattern between OTUs according to airway obstruction phenotypes. Consistently discriminative OTUs identified in sPLS-DA as differentially enriched are marked with asterisks inside the graph.
Several Rumminococcaceae, Lachnospiraceae and Clostridiales were enriched in patients with lower levels of IgE to Ascaris and HDM
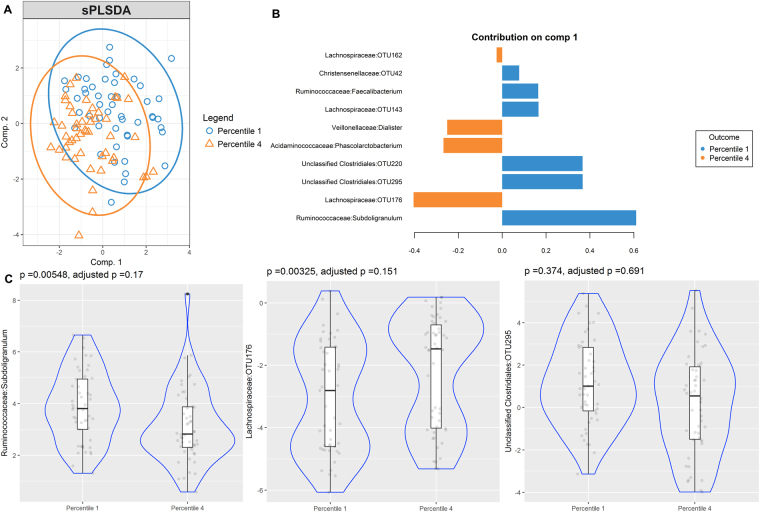
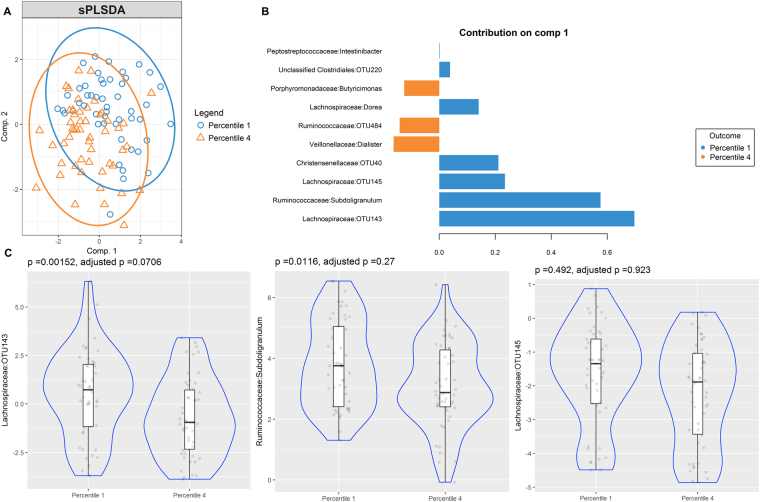
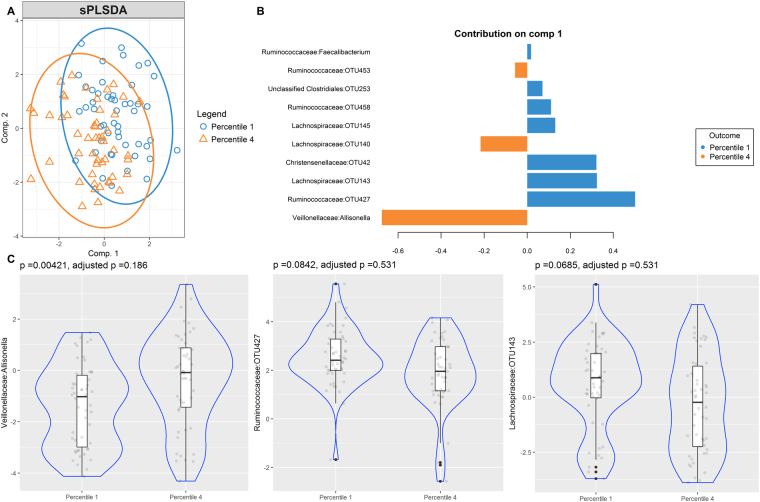
Although cases of current Ascaris infection were scarce (n = 8), our analysis showed two groups according to infection status and various differentially enriched OTUs however it did not reach statistical significance after correction by multiple comparisons (see Supplementary Fig. S7). Gut microbiome of high and low specific IgE responders to Ascaris and HDM species, B. tropicalis and D. pteronyssinus, was also compared. The sPLS-DA bi-plots show discrimination of OTUs between low and high IgE responders to Ascaris (Fig. 5A), B. tropicalis (Fig. 6A) and D. pteronyssinus (Fig. 7A). Several OTUs supported those differences and were differentially enriched, contributing to the separation in the first component (Figs 5B, 6B and 7B). However, none of them kept significantly associated after adjustment by multiple comparisons (Figs 5C, 6C and 7C).
Figure 5.
Partial least squares discriminant analysis of gut microbiome composition between low and high IgE responders to Ascaris. sPLS-DA plot based on the relative abundance of bacterial taxa of gut microbiota from low (blue circle, percentile 1) and high IgE responders (orange triangle, percentile 4) and their 95% confidence ellipses (A). Contribution plot indicating genera contributing to component 1 of the sPLS-DA plot that discriminate first and fourth sIgE percentiles (B). The abundance of the most consistent OTUs was compared using Metagenomeseq and presented on a violin plot, which includes the median, 95% CI, IQR, and density plot where the width of the blue lines indicate frequency (C).
Figure 6.
Partial least squares discriminant analysis of gut microbiome composition between low and high IgE responders to B. tropicalis. sPLS-DA plot based on the relative abundance of bacterial taxa of gut microbiota from low (blue circle, percentile 1) and high IgE responders (orange triangle, percentile 4) and their 95% confidence ellipses (A). Contribution plot indicating genera contributing to component 1 of the sPLS-DA plot that discriminate first and fourth sIgE percentiles (B). The abundance of the most consistent OTUs was compared using Metagenomeseq and presented on a violin plot, which includes the median, 95% CI, IQR, and density plot where the width of the blue lines indicate frequency (C).
Figure 7.
Partial least squares discriminant analysis of gut microbiome composition between low and high IgE responders to D. pteronyssinus. sPLS-DA plot based on the relative abundance of bacterial taxa of gut microbiota from low (blue circle, percentile 1) and high IgE responders (orange triangle, percentile 4) and their 95% confidence ellipses (A). Contribution plot indicating genera contributing to component 1 of the sPLS-DA plot that discriminate first and fourth sIgE percentiles (B). The abundance of the most consistent OTUs was compared using Metagenomeseq and presented on a violin plot, which includes the median, 95% CI, IQR, and density plot where the width of the blue lines indicate frequency (C).
Discussion
In this study, we compared gut microbiota composition among different airway obstruction phenotypes in asthmatic patients, representative of a socio-economically deprived population living in a tropical city. Although gut microbial richness and diversity was similar among phenotypes, there were differentially enriched microorganisms, being Streptococcaceae:Streptococcus and Enterococcus:Escherichia-Shigella more abundant in patients with FAO. In addition, a distinctive interconnected network enriched with those OTUs was characteristic.
A previous report by Hevia et al. comparing the gut microbiota composition between asthmatics and a healthy control group showed that patients with long term asthma harbour lower levels of Bifidobacterium adolescentis in the gut, but they did not found microbiota clustering according to asthmatic status, suggesting to some extent that globally gut microbiota in asthmatics is similar to healthy control subjects, except for the abundance of particular commensals43. Also, Hua et al. reported that the gut microbiota does not differ when compared between asthmatics and healthy controls according to α or β-diversity, but the identification of asthmatic category was not specific as included other lung disease44. The present study is the first evaluating associations between gut microbiota and the airway obstruction in a population of adult asthmatic individuals living in a tropical region. We sougth to explore differences in the gut microbiota composition explaining the airway obstruction phenotype in a population sharing the environmental exposure framework, looking for particular OTUs and their networks. In our study we found a consistent association between Streptococcaceae:Streptococcus and Enterococcus:Escherichia-Shigella with the FAO, and also Veillonellaceae:Megasphaera when comparing the FAO with RAO phenotype. Interestingly, a recent report by Arrieta et al. found an association between various Streptococcus and Veillonella species in the gut and the occurrence of wheeze in children from a low income tropical population at three months of age22 and at least for Streptococcus spp, associations for their increased abundance in the airway and wheeze45,46 or a severe asthma phenotype have been reported before8. Also, it has been shown that various species of Streptococcus and Veillonella populate the small intestine47 and appeared to block IL-12p70 production, while augmenting IL-8, IL-6, IL-10, and TNF-a responses by dendritic cells in in-vitro assays48, indeed these cytokines have been linked to airway obstruction and severe asthma phenotype49–53.
Although there is not a consensus definition for the FAO phenotype, it have been recognized as an asthma phenotype differentiated from COPD54–58 and explained by the progressive decline in FEV1 as a result of airway remodelling asscociated with allergic airway inflammation, allergen exposure and elevated IgE levels throughout life, in agreement with this is the fact that NAO patients were younger than the patients in the other phenotypes. As showed in other cohorts of patients19,20, in our study the FAO phenotype was not necessarily associated with more asthma symptoms, but a slightly increased prevalence of hospitalizations in the last year that deserves some mention. In contrast, in the RAO phenotype there was a stronger sIgE response to D. pteronyssinus and a tendency to more symptoms as described in other studies17,59,60, but a lower prevalence of hospitalizations, possibly those two different phenotypes of airway obstruction reflects two different clinical patterns of asthma. Smoking habit and exposure to cigarrete smoke can be aggravating factors, but they are not the only necessary condition for fixed airway obstruction outcome to occur in long lasting asthmatics61. As smoking habit and exposure to cigarrete smoke were uncommon and equally distributed across airway obstruction phenotypes in our population, it unlikely that those exposures explained the FAO phenotype. Instead, specific gut microbiota components appear as risks factors associated with the FAO phenotype, and this finding deserves more investigation employing more refined methodologies such as microbial whole genome sequencing to disentangle possible genetic pathways explaining the association.
In the other hand, NAO individuals showed an enrichment of various Ruminococcaceae and Lachnospiraceae when compared with the RAO or FAO phenotypes, but they were not significant after adjustment for multiple comparisons. Also, those belonging to the lowest sIgE responders to Ascaris and B. tropicalis showed enrichment of members from those two families plus various Clostridiales members. Various members of those families cooperatively metabolize the diet fiber and produce anti-inflammatory metabolites in the colon62 that decrease sIgE levels against D. pteronyssinus extract in a mice model of mite sensitization feeded with a high fiber diet63. On the other hand, gut microbiota signalling through TLR-5 and MyD88 dependent pathways regulates IgG and IgE levels against bacterial and non-bacterial antigens in mice64–66, also signalling through various pattern recognition receptor such as TLR-2 and TLR-4 has been associated with IgE mediated eccema67 and food allergy in humans68, suggesting that microbiota produced metabolites or specific structural components could stimulate innate immunity and influence systemic antibody levels. Although, after correction by multiple comparisons the abundance of commensal associated with protection was not significant, we speculate that protection against airway obstruction and sIgE levels could be accomplished by the combined presence of all those commensal as showed by sPLS-DA, but we can not assure causality or a specific mechanism.
As the gut comprises an extensive area populated with lymphoid organs and is highly vascularized and is proximal to the lung, the migration of dendritic, or another proinflammatory immune cells and cytokines induced by the microbiota to the lung is possible69, indeed previous findings in another inflammatory diseases such as arthritis and cerebrovascular diseases supports this possibility70–72. However, because of the cross sectional design of the study and the lack of an experimental animal model evaluating the effects of those commensals on allergic asthma, we cannot assure causality between the found taxa and airway obstruction phenotypes.
In our analysis we used sPLS-DA and network analysis assuming data to be compositional and sparse; in consequence CLR transformation was done, avoiding spurius association between some taxa and airway obstruction phenotypes. In undirected network construction, we used the SPIEC-EASI algorithm to robustly predict OTU-OTU interrelationships and construct a characteristic network for each phenotype. This algorithm begins with a neighborhood selection of commensals and selects those that better characterize the network (represented by the λ index) and calculate inverse covariance between them based on the concept of conditional independence to construct edge between nodes. The construction of edges is based on the abundance of commensals in the metadata for each patient belonging to a particular phenotype, remaining only with those conections that robustly reproduce in all patients after iteration and showing their number and weights represented by sticks connecting the nodes. Our results shows that OTUs clustered with another family related OTUs, as was described by Kurtz et al.37 for the analysis of fecal samples of healthy individuals belonging to the AGP. The fact that those OTUs discriminating between phenotypes of airway obstruction in SPLS-LDA (Streptococcaceae:Streptococcus and Enterococcus:Escherichia-Shigella) were also enriched in undirected network graphs supports our findings, as these two independent methods using compositinal data highlighted the importance of the same commensals and their networks.
Several limitations of this study should be mentioned. Although we only explored the association between gut microbiota composition and various clinical phenotypes of airway obstruction in asthma, the use of biological markers would have contributed to better characterize the severity of airway inflammation and its relationship with microbiome configuration. Although differences in age distribution among asthma phenotypes could introduce bias in the results, its relevance is low according to our sensitivity and age-OTU abundance association analyses. Also, dietary information and weight associated variables were not obtained. Corticosteroid use might also influence gut microbiome; however, in this study it was scarce and equally distributed between patients.
In summary, we found that Streptococcaceae:Streptococcus and Enterococcus:Escherichia-Shigella were consistently enriched in asthmatic individuals suffering a fixed airway obstruction in a socio-economically deprived population with poor controlled asthma sharing the environmental framework.
Electronic supplementary material
Acknowledgements
We thank all patients that voluntarily participated in this study and the staff of the E.S.E. Cartagena de Indias, by their administrative support. This research was supported by a grant (Contract 602–2009) from the Colombian Government (Departamento Administrativo de Ciencia y Tecnología e Innovación, Colciencias), FITP (Fundación para Promoción de la Investigación y la Tecnología, Grant 201521), University of Cartagena and Fundemeb.
Author Contributions
L.C. conceived the investigation, designed the epidemiological study, supervised the general aspects of the work, analyzed the data and wrote the manuscript. E.B. conceived the investigation, designed the epidemiological study, contributed with the clinical work, analyzed data and wrote the manuscript. J.Z. participated in study design, organized the clinical work, managed databases, and contributed to manuscript writing. E.Z. analyzed the data and revised the manuscript. H.S.V. analyzed the data and revised the manuscript. N.A. performed the 16S v4 region sequencing, OTUs table generation and revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27964-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thorburn AN, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 2.Fujimura KE, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stokholm J, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. 2018;9:141. doi: 10.1038/s41467-017-02573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gollwitzer ES, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 2014;20:642–647. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- 5.Arrieta MC, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 6.Stiemsma LT, et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin Sci (Lond) 2016;130:2199–2207. doi: 10.1042/CS20160349. [DOI] [PubMed] [Google Scholar]
- 7.Turturice BA, et al. Atopic asthmatic immune phenotypes associated with airway microbiota and airway obstruction. PLoS One. 2017;12:e0184566. doi: 10.1371/journal.pone.0184566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, et al. Airway Microbiota in Severe Asthma and Relationship to Asthma Severity and Phenotypes. PLoS One. 2016;11:e0152724. doi: 10.1371/journal.pone.0152724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang YJ, et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol. 2015;136:874–884. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SC, Yang YH, Chuang SY, Huang SY, Pan WH. Reduced medication use and improved pulmonary function with supplements containing vegetable and fruit concentrate, fish oil and probiotics in asthmatic school children: a randomised controlled trial. Br J Nutr. 2013;110:145–155. doi: 10.1017/S0007114512004692. [DOI] [PubMed] [Google Scholar]
- 11.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agache I, Akdis C, Jutel M, Virchow JC. Untangling asthma phenotypes and endotypes. Allergy. 2012;67:835–846. doi: 10.1111/j.1398-9995.2012.02832.x. [DOI] [PubMed] [Google Scholar]
- 13.Lotvall J, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127:355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 14.Himes BE, et al. Genome-wide association analysis in asthma subjects identifies SPATS2L as a novel bronchodilator response gene. PLoS Genet. 2012;8:e1002824. doi: 10.1371/journal.pgen.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar R, et al. Bronchodilator responses in Chinese children from asthma index families and the general population. J Allergy Clin Immunol. 2006;117:1257–1263. doi: 10.1016/j.jaci.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 16.Puckett JL, et al. An elevated bronchodilator response predicts large airway inflammation in mild asthma. Pediatr Pulmonol. 2010;45:174–181. doi: 10.1002/ppul.21172. [DOI] [PubMed] [Google Scholar]
- 17.Sharma S, et al. Clinical predictors and outcomes of consistent bronchodilator response in the childhood asthma management program. J Allergy Clin Immunol. 2008;122:921–928 e924. doi: 10.1016/j.jaci.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore WC, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2009;181:315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yii AC, Tan GL, Tan KL, Lapperre TS, Koh MS. Fixed airways obstruction among patients with severe asthma: findings from the Singapore General Hospital-Severe Asthma Phenotype Study. BMC Pulm Med. 2014;14:191. doi: 10.1186/1471-2466-14-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tashkin DP, Chipps BE, Trudo F, Zangrilli JG. Fixed airflow obstruction in asthma: a descriptive study of patient profiles and effect on treatment responses. J Asthma. 2014;51:603–609. doi: 10.3109/02770903.2014.895012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escobar JS, Klotz B, Valdes BE, Agudelo GM. The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014;14:311. doi: 10.1186/s12866-014-0311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arrieta, M. C. et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol (2017). [DOI] [PMC free article] [PubMed]
- 23.Buendia E, Zakzuk J, Mercado D, Alvarez A, Caraballo L. The IgE response to Ascaris molecular components is associated with clinical indicators of asthma severity. World Allergy. Organ J. 2015;8:8. doi: 10.1186/s40413-015-0058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rueda, E. Will today’s poor be tomorrow’s poor? Determinants of poverty and magnitude of the vulnerability in Cartagena de Indias (2009).
- 25.Vergara C, et al. African ancestry is associated with risk of asthma and high total serum IgE in a population from the Caribbean Coast of Colombia. Hum Genet. 2009;125:565–579. doi: 10.1007/s00439-009-0649-2. [DOI] [PubMed] [Google Scholar]
- 26.Mathias RA, et al. A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat Commun. 2016;7:12522. doi: 10.1038/ncomms12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennis RJ, et al. Prevalence of asthma and other allergic conditions in Colombia 2009-2010: a cross-sectional study. BMC Pulm Med. 2012;12:17. doi: 10.1186/1471-2466-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dennis R, et al. Asthma and other allergic conditions in Colombia: a study in 6 cities. Ann Allergy Asthma Immunol. 2004;93:568–574. doi: 10.1016/S1081-1206(10)61265-3. [DOI] [PubMed] [Google Scholar]
- 29.Miller MR, et al. Standardisation of spirometry. Eur Respir J. 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 30.Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium HMP. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 35.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurtz ZD, et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11:e1004226. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Cao KA, et al. MixMC: A Multivariate Statistical Framework to Gain Insight into Microbial Communities. PLoS One. 2016;11:e0160169. doi: 10.1371/journal.pone.0160169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Javier Palarea-Albaladejo JAM-F. zCompositions- R package for multivariate imputation of left-censored data under a compositional approach. Chemometrics and Intelligent Laboratory Systems. 2015;143:85–96. doi: 10.1016/j.chemolab.2015.02.019. [DOI] [Google Scholar]
- 40.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hevia A, et al. Allergic Patients with Long-Term Asthma Display Low Levels of Bifidobacterium adolescentis. PLoS One. 2016;11:e0147809. doi: 10.1371/journal.pone.0147809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hua X, Goedert JJ, Pu A, Yu G, Shi J. Allergy associations with the adult fecal microbiota: Analysis of the American Gut Project. EBioMedicine. 2016;3:172–179. doi: 10.1016/j.ebiom.2015.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bisgaard H, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 46.Schwerk N, Brinkmann F, Soudah B, Kabesch M, Hansen G. Wheeze in preschool age is associated with pulmonary bacterial infection and resolves after antibiotic therapy. PLoS One. 2011;6:e27913. doi: 10.1371/journal.pone.0027913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Bogert B, et al. Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol Ecol. 2013;85:376–388. doi: 10.1111/1574-6941.12127. [DOI] [PubMed] [Google Scholar]
- 48.van den Bogert B, Meijerink M, Zoetendal EG, Wells JM, Kleerebezem M. Immunomodulatory properties of Streptococcus and Veillonella isolates from the human small intestine microbiota. PLoS One. 2014;9:e114277. doi: 10.1371/journal.pone.0114277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ordonez CL, Shaughnessy TE, Matthay MA, Fahy JV. Increased neutrophil numbers and IL-8 levels in airway secretions in acute severe asthma: Clinical and biologic significance. Am J Respir Crit Care Med. 2000;161:1185–1190. doi: 10.1164/ajrccm.161.4.9812061. [DOI] [PubMed] [Google Scholar]
- 50.Yalcin AD, Bisgin A, Gorczynski RM. IL-8, IL-10, TGF-beta, and GCSF levels were increased in severe persistent allergic asthma patients with the anti-IgE treatment. Mediators Inflamm. 2013;2012:720976. doi: 10.1155/2012/720976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silvestri M, et al. High serum levels of tumour necrosis factor-alpha and interleukin-8 in severe asthma: markers of systemic inflammation? Clin Exp Allergy. 2006;36:1373–1381. doi: 10.1111/j.1365-2222.2006.02502.x. [DOI] [PubMed] [Google Scholar]
- 52.Choi IW, et al. TNF-alpha induces the late-phase airway hyperresponsiveness and airway inflammation through cytosolic phospholipase A(2) activation. J Allergy Clin Immunol. 2005;116:537–543. doi: 10.1016/j.jaci.2005.05.034. [DOI] [PubMed] [Google Scholar]
- 53.Peters MC, et al. Plasma interleukin-6 concentrations, metabolic dysfunction, and asthma severity: a cross-sectional analysis of two cohorts. Lancet Respir Med. 2016;4:574–584. doi: 10.1016/S2213-2600(16)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dales RE, Spitzer WO, Tousignant P, Schechter M, Suissa S. Clinical interpretation of airway response to a bronchodilator. Epidemiologic considerations. Am Rev Respir Dis. 1988;138:317–320. doi: 10.1164/ajrccm/138.2.317. [DOI] [PubMed] [Google Scholar]
- 55.Lange P. Persistent airway obstruction in asthma. Am J Respir Crit Care Med. 2013;187:1–2. doi: 10.1164/rccm.201211-1966ED. [DOI] [PubMed] [Google Scholar]
- 56.Bumbacea D, et al. Parameters associated with persistent airflow obstruction in chronic severe asthma. Eur Respir J. 2004;24:122–128. doi: 10.1183/09031936.04.00077803. [DOI] [PubMed] [Google Scholar]
- 57.Hudon C, Turcotte H, Laviolette M, Carrier G, Boulet LP. Characteristics of bronchial asthma with incomplete reversibility of airflow obstruction. Ann Allergy Asthma Immunol. 1997;78:195–202. doi: 10.1016/S1081-1206(10)63387-X. [DOI] [PubMed] [Google Scholar]
- 58.Fabbri LM, et al. Differences in airway inflammation in patients with fixed airflow obstruction due to asthma or chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2003;167:418–424. doi: 10.1164/rccm.200203-183OC. [DOI] [PubMed] [Google Scholar]
- 59.Heffler E, et al. Bronchodilator response as a marker of poor asthma control. Respir Med. 2016;112:45–50. doi: 10.1016/j.rmed.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 60.Galant SP, et al. The relationship of the bronchodilator response phenotype to poor asthma control in children with normal spirometry. J Pediatr. 2011;158:953–959 e951. doi: 10.1016/j.jpeds.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sears MR, et al. A longitudinal, population-based, cohort study of childhood asthma followed to adulthood. N Engl J Med. 2003;349:1414–1422. doi: 10.1056/NEJMoa022363. [DOI] [PubMed] [Google Scholar]
- 62.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 63.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 64.Zeng MY, et al. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh JZ, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hill DA, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.West CE, et al. Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin Exp Allergy. 2015;45:1419–1429. doi: 10.1111/cea.12566. [DOI] [PubMed] [Google Scholar]
- 68.Ling Z, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schroeder BO, Backhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079–1089. doi: 10.1038/nm.4185. [DOI] [PubMed] [Google Scholar]
- 70.Teng F, et al. Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity. 2016;44:875–888. doi: 10.1016/j.immuni.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maeda Y, et al. Dysbiosis Contributes to Arthritis Development via Activation of Autoreactive T Cells in the Intestine. Arthritis Rheumatol. 2016;68:2646–2661. doi: 10.1002/art.39783. [DOI] [PubMed] [Google Scholar]
- 72.Benakis, C. et al. Commensal microbiota affects ischemic stroke outcome by regulating intestinal gammadelta T cells. Nat Med (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.