Abstract
Wounding triggers organ regeneration in many plant species, and application of plant hormones, such as auxin and cytokinin, enhances their regenerative capacities in tissue culture. Recent studies have identified several key players mediating wound- and/or plant hormone-induced cellular reprogramming, but the global architecture of gene regulatory relationships underlying plant cellular reprogramming is still far from clear. In this study, we uncovered a gene regulatory network (GRN) associated with plant cellular reprogramming by using an enhanced yeast one-hybrid (eY1H) screen systematically to identify regulatory relationships between 252 transcription factors (TFs) and 48 promoters. Our network analyses suggest that wound- and/or hormone-invoked signals exhibit extensive cross-talk and regulate many common reprogramming-associated genes via multilayered regulatory cascades. Our data suggest that PLETHORA 3 (PLT3), ENHANCER OF SHOOT REGENERATION 1 (ESR1) and HEAT SHOCK FACTOR B 1 (HSFB1) act as critical nodes that have many overlapping targets and potentially connect upstream stimuli to downstream developmental decisions. Interestingly, a set of wound-inducible APETALA 2/ETHYLENE RESPONSE FACTORs (AP2/ERFs) appear to regulate these key genes, which, in turn, form feed-forward cascades that control downstream targets associated with callus formation and organ regeneration. In addition, we found another regulatory pathway, mediated by LATERAL ORGAN BOUNDARY/ASYMMETRIC LEAVES 2 (LOB/AS2) TFs, which probably plays a distinct but partially overlapping role alongside the AP2/ERFs in the putative gene regulatory cascades. Taken together, our findings provide the first global picture of the GRN governing plant cell reprogramming, which will serve as a valuable resource for future studies.
Keywords: Auxin, Callus formation, Cellular reprogramming, Cytokinin, Regeneration, Wound stress
Introduction
Multicellular organisms are often exposed to biotic and abiotic types of stress that cause severe wounding, leading to the partial or complete organ loss. Upon injury, many organisms activate internal mechanisms responsible for initiating wound repair and regeneration. Plant and animal species, however, possess highly varying regeneration capabilities. Arabidopsis roots, for instance, regenerate full apical meristems upon partial loss (Sena et al. 2009, Efroni et al. 2016). Similarly, zebrafish replace amputated fins by regenerating the organ; the new fin grows to be the same size as the original, lost one (Pfefferli and Jaźwińska 2015). Even more strikingly, in many plants, whole organisms can grow from small pieces of injured tissues, such as detached leaves and cuttings from shoot and roots (Ikeuchi et al. 2016). An important aspect of organ regeneration is the reactivation of cell proliferation at wound sites, which leads to the formation of a cell mass called callus, and subsequent establishment of shoot or root apical meristems (Ikeuchi et al. 2013). It is also possible for de novo organogenesis to occur without the formation of a macroscopic callus but, even in such cases, reactivation of the cell cycle is important for meristem formation and subsequent organogenesis. Another key cellular event in the regeneration of new organs and organisms is the acquisition of pluripotency, i.e. the competence to differentiate into multiple cell types (Kareem et al. 2015). Groups of cells in root explants, for example, can be reprogrammed so that they gain shoot identity during shoot regeneration (Atta et al. 2009).
How exogenous stimuli perturb intrinsic developmental signals, in turn leading to cell reprogramming and initiation of new tissue formation, is one of the central unanswered questions in regenerative biology. Recent studies have started to uncover important molecular crossroads between stress responses and organogenesis. A subclade of APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factors (TFs) named WOUND INDUCED DEDIFFERENTIATION 1–4 (WIND1–WIND4) are rapidly induced upon wounding and play key roles in callus formation at wound sites (Iwase et al. 2011). Plants overexpressing any one of the WIND genes develop callus that is competent for shoot and root regeneration as well as somatic embryogenesis (Iwase et al. 2011, Ikeuchi et al. 2013). Key to WIND1-triggered shoot regeneration is the ability of this TF to up-regulate directly ENHANCER OF SHOOT REGENERATION 1 (ESR1) (Iwase et al. 2017), which, together with its close homolog, ESR2, plays an important role in shoot development and regeneration (Banno et al. 2001, Ikeda et al. 2006, Chandler et al. 2007). Another wound-inducible AP2/ERF TF, known as ERF115, is required for the replenishment of root stem cells and it activates genes which encode peptide hormones, such as PHYTOSULFOKINE 5 (PSK5) (Heyman et al. 2013, Heyman et al. 2016). PSK peptides were originally identified as diffusible signals that promote cell proliferation in cell culture (Matsubayashi and Sakagami 1996), and later studies demonstrated that they facilitate tissue repair at wound sites (Amano et al. 2007). A DNA-binding with one zinc finger (Dof) TF, OBP1, may have similar physiological functions, as it is also wound inducible and activates cell proliferation through direct transcriptional up-regulation of cell cycle regulators, including CYCLIN D3; 3 (CYCD3; 3) (Skirycz et al. 2008).
Accumulating evidence suggests that wounding also modulates endogenous hormonal homeostasis to promote regeneration. Arabidopsis hypocotyls develop callus at wound sites, and this cellular response involves the transcriptional activation of genes, such as LONELY GUY (LOG); these genes encode enzymes involved in cytokinin biosynthesis (Ikeuchi et al. 2017). This causes cytokinin accumulation, and thus an increase in cytokinin response, leading to cell cycle re-entry and callus formation through the induction of CYCD3 expression. The de novo regeneration of roots from Arabidopsis leaf explants offers another model system to study wound-induced organ regeneration and, in this case, endogenous auxin plays a central role in promoting regeneration. Recent studies have shown that both basipetal transport of auxin to wound sites and YUCCA (YUC)-dependent de novo biosynthesis contribute to this type of root regeneration (Liu et al. 2014, Chen et al. 2016).
While it is clear that wound stress serves as a primary trigger for organ regeneration, it alone is often insufficient to provoke the entire suite of regenerative responses. Early studies in the 1950s already demonstrated that an exogenous supply of both auxin and cytokinin enhances regenerative capacities (Skoog and Miller 1957), and tissue culture techniques utilizing these plant hormones are now widely employed to propagate a wide range of plant species asexually. A two-step tissue culture method is usually used to induce shoot regeneration in Arabidopsis, in which explants are first incubated on auxin-rich callus-inducing medium (CIM) and subsequently on cytokinin-rich shoot-inducing medium (SIM) (Valvekens et al. 1988). Recent studies have shown that callus formation on CIM follows a developmental program also essential for that of both lateral root formation and adventitious root formation (Atta et al. 2009, Sugimoto et al. 2010, Liu et al. 2014). As reported for lateral root development, auxin in CIM probably activates AUXIN RESPONSE FACTORs (ARFs), such as ARF7 and ARF19, which in turn up-regulate LATERAL ORGAN BOUNDARY DOMAIN 16 (LBD16) and LBD29 (Okushima et al. 2007). In line with this idea, overexpression of LBD16, LBD17, LBD18 or LBD29 promotes callus formation in the wild type, as does LBD16 overexpression in arf7 arf19, a mutant which exhibits reduced callus formation on CIM (Fan et al. 2012). LBD18 and a close homolog, LBD33, are also known to activate the expression of E2Fa, a key cell cycle regulator (Berckmans et al. 2011), implying that one transcriptional module underlying auxin-induced callus formation is an ARF–LBD–E2Fa pathway. Recent studies identified another auxin-dependent mechanism for shoot regeneration in vitro, which is mediated by AP2/ERF TFs, PLETHORA 3 (PLT3), PLT5 and PLT7 (Kareem et al. 2015). Importantly, these PLTs are prerequisite for both the acquisition of pluripotency and the initiation of shoot meristem fate through the transcriptional up-regulation of PLT1 and PLT2 as well as the NAM, ATAF1, 2 and CUC2 (NAC) family TF-encoding genes CUP SHAPED COTYLEDON 1 (CUC1) and CUC2 (Kareem et al. 2015). Cytokinin promotes shoot regeneration through the up-regulation of WUSCHEL (WUS), and recent studies demonstrated that key cytokinin signaling components, namely type-B ARABIDOPSIS RESPONSE REGULATORSs (ARRs), directly bind the WUS promoter and regulate transcription of this gene (Meng et al. 2017, Zhang et al. 2017).
Systematic elucidation of a gene regulatory network (GRN) is a powerful approach used to gain a holistic picture of how transcriptional regulation controls specific biological events (Gaudinier and Brady 2016). Chromatin immunoprecipitation-sequencing (ChIP-seq) is one TF-based strategy to identify comprehensively DNA-binding sites in vivo. Performing ChIP-seq with the intent of constructing a GRN is, however, laborious, since a large number of transgenic plants expressing individual TFs must be generated. Another technique, termed DNA affinity purification sequencing (DAP-seq), can systematically identify TF–DNA interactions in vitro, and publicly available data (http://neomorph.salk.edu/dev/pages/shhuang/dap_web/pages/index.php) provide an excellent platform to search for TF–DNA interactions in silico (O’Malley et al. 2016). In contrast, yeast one-hybrid (Y1H) screening offers an alternative, promoter-based approach to identify potential TFs that bind to a collection of promoters of interest (Gaudinier and Brady 2016). Y1H has been applied to infer a GRN important for cellular differentiation (Taylor-Teeples et al. 2015) and defense response (Li et al. 2014), but similar systematic approaches have not been applied to investigate the GRN underlying plant cell reprogramming. In this study, we performed an enhanced Y1H (eY1H) screen (Gaudinier et al. 2011), using 559 TFs and 48 promoters associated with stress response, hormonal signaling and/or cellular reprogramming. We identified 1,162 TF–promoter interactions, many of which have not been reported in previous studies. Our data have thus revealed the most comprehensive GRN to date which governs cellular reprogramming in plants.
Results
eY1H assays identified 1,162 interactions between TFs and promoters
In order to uncover a GRN important for plant cell reprogramming during plant regeneration, we performed an eY1H screen with 48 bait promoters that are likely to be relevant for regeneration. Considering the rationale that regeneration occurs at the intersection of stress response, hormone signaling and organogenesis, we selected bait promoters based on the following functional annotations: ‘stress response’ [WIND1, WIND2, WIND3, WIND4, HEAT SHOCK FACTOR A1A (HSFA1A), HSFA2, HSFB1, HSFB2A, HSFB2B, WRKY48 and DUF313], ‘auxin’ (ARF5 and ARF19), ‘cytokinin’ (ARR1, ARR12, LOG1, LOG5 and LOG7), ‘cell proliferation’ [CYCD1; 1, CYCD3; 1, CYCD3; 3, E2Fa, KIP RELATED PROTEIN 2 (KRP2), KRP3, OBP1, PSK1, PSK2, PSK3, PSK4 and PSK5] and ‘development’ [CUC1, CUC2, ERF115, SCARECLOW LIKE 21 (SCL21), WUS, WOX3, WOX13, ESR1, ESR2, LBD18, LBD29, PLT3, PLT5, PLT7, LEAFY COTYLEDON 1 (LEC1), LEC2, HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE 2 (HSI2) and HSFB4/SCHIZORIZA (SCZ)]. To obtain promoters for each of the aforementioned set of genes, we then cloned 2 kb of upstream sequence (beginning from the start codon) into vectors harboring either LacZ or HIS3 for selection in yeast. We then chose a subset (558) of TFs from the collection described in Pruneda-Paz et al. (2014) as prey, against which to screen our 48 bait promoters. In addition, we cloned WIND4 and supplemented the prey TF collection with this gene, since it was not present in the library (Supplementary Table S1). We based our TF prey selection on the transcriptional profiles of genes present within the cDNA collection. In particular, we chose genes that are highly expressed in calli (Iwase et al. 2011), during shoot regeneration on SIM (Che et al. 2006) and during protoplast regeneration (Chupeau et al. 2013), as well as several that are responsive to wounding, auxin or cytokinin (TAIR; http://www.arabidopsis.org/). Given that these prey TFs include 15 genes which we also selected as bait promoters, we aimed to uncover multilayered putative regulatory relationships in our GRN.
Using a semi-automated eY1H screening system previously described by Gaudinier et al. (2011), we identified 1,162 interactions between 252 TFs and 48 promoters (Fig. 1; Supplementary Tables S2, S4). In order to test the reliability of these results, we first examined TF–promoter binding for HSFs, a group of TFs that are known to bind their downstream promoters via the heat shock elements (HSEs), 5'-nGAAnnTTCn-3' and 5'-nTTCnnGAAn-3' (Yoshida et al. 2011). We found that 13 out of 14 HSF targets identified in our eY1H screen harbor HSEs in their promoters (Supplementary Table S5). Further, included in this list are previously reported TF–promoter interactions, such as binding of HSFA1 TF to the promoter of HSFA2 (Yoshida et al. 2011), binding of the HSFB1 TF to the HSFA2 promoter (Ikeda et al. 2011) and binding of an HSFA2 TF to its own promoter (Liu et al. 2013) (Supplementary Table S4). These data thus suggest that our eY1H screening successfully reproduced previously described transcriptional relationships and our data can be used to identify novel TF–promoter interactions.
Fig. 1.
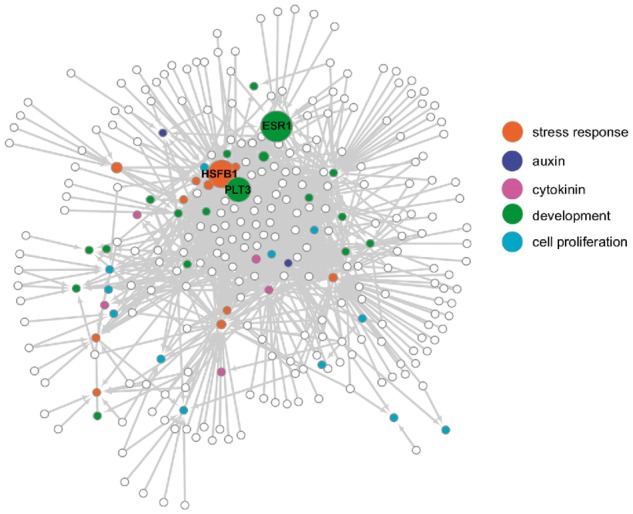
A graphic summary of the GRN for plant cellular reprogramming identified by eY1H assays. An eY1H screening identified 1,162 interactions (arrows) connecting 286 nodes (circles). Open circles represent prey TFs and filled circles represent bait promoters, with each color indicating a specific functional annotation. The size of circles indicates the betweenness centrality for each node, and three nodes, PLT3, ESR1 and HSFB1, that display the highest betweenness centrality are labeled. Prey TFs with a small number of targets are located on the periphery of the network, while those harboring a larger number of targets are located in the center. Bait promoters are often located between the periphery and the center of the network.
PLT3, ESR1 and HSFB1 may act as critical nodes in the reprogramming GRN
In order to reveal network motifs and features that are statistically overrepresented within our GRN, we first ran the NetworkAnalyzer Cytoscape plugin and examined TF interactions for each bait promoter (Assenov et al. 2008). This analysis revealed that each bait promoter has between 1 and 83 incoming TF interactions, with the median number of interactions being 13.5 (Supplementary Table S3). Interestingly, we found that promoters of closely related homologs generally have a comparable number of interactions with TFs (Supplementary Table S3), implying that our interaction data probably reflect genuine biological features. Among the PSK1–PSK5 promoters we tested, for instance, all promoters, with the exception of PSK5, have only one or a few TF interactions, and three promoters, PSK1, PSK2 and PSK3, are all bound by a NAC family TF encoded by At3g12910 (Supplementary Table S3). In contrast, among the six HSF promoters we tested, five have >40 TF interactions and HSFB4/SCZ shares most of its upstream TFs with other HSFs (Supplementary Tables S3, S4). We are aware that generally speaking, Y1H data ought to contain some false-negative or false-positive results and they do not identify, for instance, interactions regulated by TFs acting as heterodimers or in higher order transcriptional complexes. Nevertheless, our data provide the first global TF–promoter relationships that can be further validated and functionally tested.
We next analyzed the number of interactions for each TF and found that the distribution of the number of interactions fits a power-law distribution (y = 60.742x−1.179, R2 = 0.762; Supplementary Table S3; Supplementary Fig. S1). This statistical feature suggests that the obtained GRN has a property of being a scale-free network, which has often been reported for other biological networks (Barabási and Oltvai 2004). Given that the impact of nodes can be evaluated by the ‘betweenness centrality’, a measure for the shortest paths within the network passing through specific nodes (Freeman 1978), we calculated this parameter in our GRN (Supplementary Table S6). This analysis revealed that ESR1, HSFB1 and PLT3 have, by far, the highest betweenness centrality values (0.0120 for ESR1, 0.0101 for HSFB1 and 0.0087 for PLT3) among the nodes in our GRN, strongly suggesting that they act as critical nodes (Fig. 1; Supplementary Table S6). Interestingly, we detected that ESR1 and PLT3 have >20 incoming and 10 outgoing interactions, while HSFB1 has 73 incoming but only two outgoing interactions (Supplementary Table S6), implying that the mode of action of these TFs might be different in the GRN.
PLT3- and LOB/AS2-mediated pathways engage in cross-talk and may regulate reprogramming genes
To reveal key regulatory modules within our GRN more comprehensively, we also performed a power graph compression analysis (Ahnert 2013, Ahnert 2014). This compression reduces the GRN into power nodes consisting of groups of highly connected TFs that bind to a common set of targets through a so-called power edge. In our GRN, we identified 34 power edges, together representing 75 TFs interacting with 35 promoters (Supplementary Table S7). Within these power nodes, AP2/ERF, ABI3/VP1, HB, LOB/AS2 and NAC TFs are represented more substantially than other classes of TFs, as they are present in >10 of the 34 power nodes, suggesting that they are the most critical TF families within the cellular reprogramming GRN (Supplementary Table S8). To elucidate how these classes of TFs co-occur in our power nodes, we applied a multiple correspondence analysis (MCA) and found two groups of co-occurring TFs, namely, LOB/AS2–HB and AP2/ERF–ABI3/VP1 (Supplementary Fig. S2, dimension 2). Our MCA also showed that a NAC class of TFs exhibit similar co-occurrence with both of these groups (Supplementary Fig. S2, dimension 2).
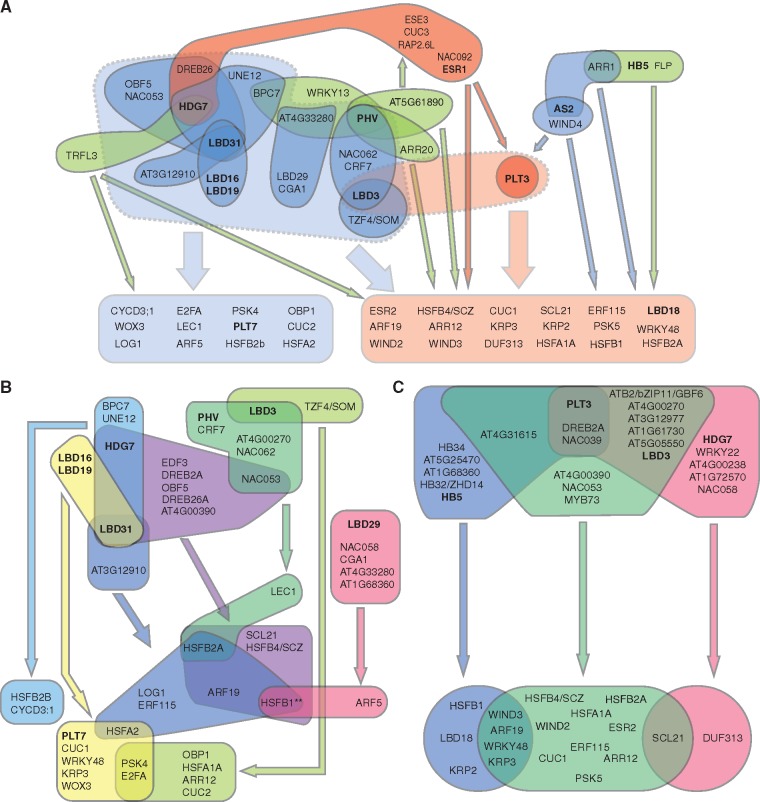
In order to visualize network characteristics driven by these groups of TF classes, we drew a simplified network depicting the power nodes containing these TFs (Fig. 2). Given our prediction that PLT3 and ESR1 are likely to be critical nodes (Fig. 1), we selected them as representative members of the AP2/ERF–ABI/VP1 group, while we kept all nine LOB/AS2–HB TFs for further analysis. This analysis confirmed that PLT3-containing power nodes and LOB/AS2-containing power nodes may constitute two regulatory modules within the network, which appear to target distinct sets of downstream promoters (Fig. 2A). Interestingly, ESR1 is represented in a single power node that acts upstream of PLT3, but the ESR1 power node also binds many promoters that are targeted by the PLT3 power nodes (Fig. 2A). Similarly, WIND4 and AS2 appear to bind both PLT3 and many of its targets (Fig. 2A), suggesting that PLT3-mediated feed-forward transcriptional regulation is a key feature of the PLT3 power nodes. The LOB/AS2 power nodes bind a set of 12 promoters that are not regulated by the PLT3 power nodes (Fig. 2A). On the other hand, 11 out of 18 targets of the PLT3 power nodes are also targeted by at least one of the LOB/AS2 power nodes (Fig. 2B, C), suggesting possible cross-talk between these two regulatory modules. We found that LBD3 is located at the boundary of LOB/AS2 and PLT3 power nodes, thus it might function as a regulator of interactions between both regulatory modules (Fig. 2A). Other candidate regulators that might also act at the boundary between these two regulatory modules include HDG7, NAC053 and NAC058 TFs, which are represented in both regulatory modules (Fig. 2B, C).
Fig. 2.
A power graph of the cellular reprogramming GRN. (A) A simplified representation of a subset of 34 power edges identified in the GRN for plant cell reprogramming using the power graph compression. All power nodes containing members of the LOB/AS2 TFs (colored in blue), HB TFs (colored in green), PLT3 and ESR1 (colored in red), representative of the AP2/ERF TFs, are shown, since they best explain the group structure of the power nodes based on the MCA. For simplicity, three PLT3 power nodes are represented as a single node, and all accompanying TFs, except for HB TFs, in the PLT3 power nodes that overlap with the LOB/AS2 power nodes are not shown. The target promoters are divided into two groups, depending on whether they interact with LOB/AS2 power nodes (colored in light blue) or primarily with PLT3 power nodes (colored in light red). Power edges between the nodes and these two promoter classes are represented by thin arrows, with colors corresponding to their regulatory nodes. Dotted, shaded collections of power nodes indicate the two main regulatory modules, the LOB/AS2-regulatory module (marked in light blue) and the PLT3-regulatory module (marked in light red), and thick arrows indicate their interaction with the promoters. See also Supplementary Table S7 for the full list of represented and non-represented genes. (B) (C) Detailed power edges within the LOB/AS2 (B) and PLT3 (C) regulatory modules, with the different power nodes and their power edges marked by the same colors. TFs belonging to the LOB/AS2 or HB TF classes and PLT3 and ESR1 are shown in bold.
Wound-induced and/or CIM-induced TFs form a heavily overlapping GRN which regulates reprogramming genes
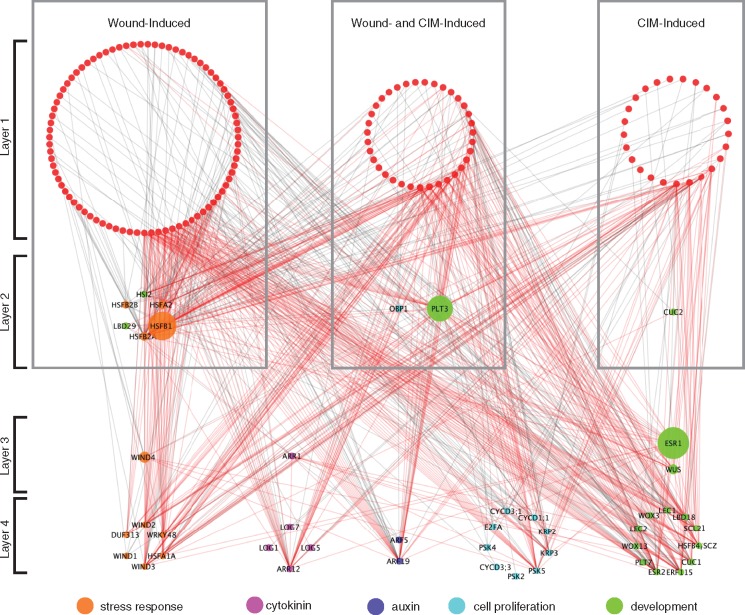
Recent studies have started to uncover some important links among key regulators of regeneration (Ikeuchi et al. 2016), but how wounding stimuli and hormonal pathways converge to control cellular reprogramming is still not known. Having generated the first comprehensive GRN for plant cell reprogramming, we sought to address this question by extracting a subnetwork that includes prey TFs which are highly expressed either after wounding or on CIM. Based on our gene expression profiling during wound-induced callus formation (Ikeuchi et al. 2017), we selected 127 TFs for which expression is induced within 24 h after wounding (Supplementary Table S2). We also selected TFs that are highly expressed upon CIM incubation (Che et al. 2006). In total, we found that 63 of the TFs in our GRN are CIM inducible, of which 37 are also included in our set of wound-induced TFs (Supplementary Table S2). Using the 153 TFs and 45 promoters bound by these TFs, we generated a subnetwork with 188 nodes connected by 707 TF–promoter interactions (Fig. 3). To infer a global trend of how wound stress and hormonal cues trigger transcriptional responses, we compared the number of TF–promoter interactions for the three classes of TFs, i.e. those induced only by wounding, only by CIM or by both wounding and CIM, within this subnetwork. To our surprise, we found that these three classes of TFs all bind, without obvious functional preferences, to promoters associated with stress response, cytokinin, auxin, cell proliferation and development (Fig. 3; Supplementary Table S9). We also detected extensive feed-forward regulatory loops (Fig. 3), and our statistical analysis showed that the number of feed-forward modules is significantly more than expected for a random scale-free network generated by the Watts and Strogatz model (483 vs. 92; P-value <0.02). These data suggest that inducible signals underlying plant cell reprogramming may be transmitted by a robust, multilayered transcriptional network. Taken together, these analyses revealed that the transcriptional cascades underlying wound- or CIM-induced cellular reprogramming are heavily interwoven and target a wide range of promoters associated with both stress response and developmental regulation.
Fig. 3.
A gene regulatory subnetwork highlights how wound-induced and/or CIM-induced TFs bind the promoters of downstream target genes. Wound-induced TFs were selected based on their up-regulation within 24 h after wounding as reported in Ikeuchi et al. (2017). CIM-induced TFs were defined as those significantly up-regulated in explants incubated on CIM for 7 d (FDR <0.01 and FC >0) (Che et al. 2006). These TFs were further layered based on their degrees of interactions within the subnetwork. TFs in layer 1 are induced by wounding and/or CIM and they have only outgoing interactions, i.e. binding to target promoters. TFs in layer 2 are also induced by wounding and/or CIM, and they engage in both incoming, i.e. promoter binding by upstream TFs, and outgoing interactions. TFs in layer 3 are not significantly induced by wounding and/or CIM, but they have both incoming and outgoing interactions. Genes in layer 4 represents bait promoters, thus they have only incoming interactions. Genes in layer 1 are colored in red, and those in layers 2, 3 and 4 are colored based on their functional annotations. Node size represents the betweenness centrality of the TFs in the network. Red edges represent TF–promoter interactions that are components of feed-forward loops.
A GRN for wound-induced cellular reprogramming
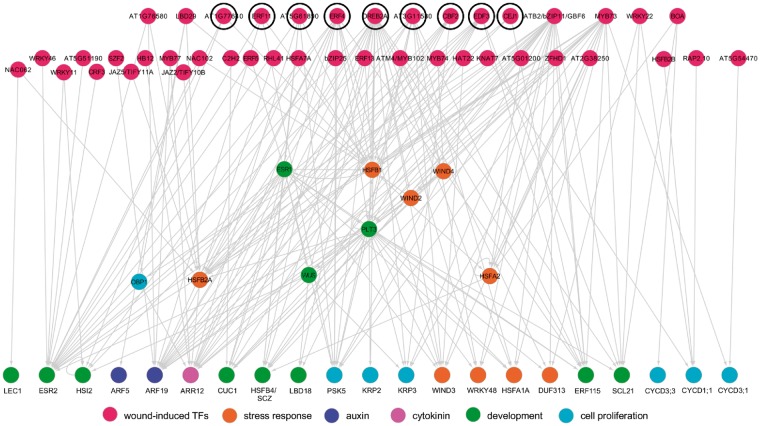
To investigate how early wound-responsive genes trigger cellular reprogramming more specifically, we next selected prey TFs for which gene expression is induced within 1 h after wounding (Ikeuchi et al. 2017) and examined their downstream transcriptional subnetwork. For the selected 42 fast wound-induced TFs, our eY1H identified 208 TF–promoter interactions with 30 promoters. Interestingly, we found that nine out of 15 TFs with more than two targets belong to the AP2/ERF family that include previously described key regulators of wound-induced cellular reprogramming, such as WINDs, ERF115 and PLT3/5/7 (Fig. 4). Importantly, many of these AP2/ERF TFs have not been characterized previously or are only known to function in other stress responses. DEHYDRATION RESPONSIVE ELEMENT BINDING PROTEIN 2 A (DREB2A), for example, is so far best described for its role in the regulation of drought and heat stress (Sakuma et al. 2006), and we confirmed previously reported binding of DREB2A to HSFB1 and HSFB2A promoters (Fig. 4). Our eY1H data further revealed DREB2A binding to the promoters of PLT3, PSK5, KRP2, KRP3, LBD18, WIND2 and WIND3, suggesting that DREB2A is also involved in wound-induced cellular reprogramming. We further noticed that some of these nine AP2/ERFs bind promoters of other AP2/ERF TFs and form multilayered regulatory cascades (Fig. 4). AT5G61890, for instance, binds the ESR1 promoter, and ESR1, in addition, binds the promoter of its closely related homolog ESR2 (Fig. 4). Similarly, EDF3, ERF4 and DREB2A all bind the PLT3 promoter, and PLT3, in turn, binds the promoter of ESR2 and ERF115 (Fig. 4). We should also mention that some of the wound-induced TFs, e.g. BOA and RAP2.10, directly bind promoters of core cell cycle regulators, such as CYCD1; 1, CYCD3; 1 and CYCD3; 3, implying that early wound signaling also regulates cell cycle progression in a direct manner.
Fig. 4.
A subnetwork highlighting how wound-induced TFs bind the promoters of downstream target genes. Wound-induced TFs, marked by red circles, directly or indirectly regulate downstream targets associated with cellular reprogramming. TFs interacting with more than two targets are positioned in the top tier and, among them, AP2/ERF family members are denoted by black circles. Downstream bait promoters are colored according to their functional annotations.
In order to examine whether the detected TF–promoter interactions reflect the actual transcriptional regulation under physiological conditions, we compared our eY1H results with the gene expression profiling data after wounding (Ikeuchi et al. 2017, Iwase et al. 2017). In support of the regulatory relationships inferred from our Y1H data, AT5G61890 is first induced within 1 h after wounding and its target, ESR1, is in turn induced after 3 h (Fig. 4; Supplementary Fig. S3; Iwase et al. 2017). These early transcriptional activations also precede the induction of further downstream target genes such as PLT3 and WIND3 (Fig. 4; Supplementary Fig. S3). Our transcriptome data, in addition, show that the expression of HSFB1 and HSFA2 is rapidly induced within 1 h upon wounding, followed by the induction of their common target HSFB4/SCZ after 6 h (Fig. 4; Supplementary Fig. S3). These data thus suggest that the TF–promoter interactions identified in this study are relevant for wound-induced transcriptional activation and should help identify novel regulators in wound-induced cellular reprogramming. HSFB4/SCZ, for instance, is so far only known to play roles in cell fate determination (Pernas et al. 2010, ten Hove et al. 2010), but our Y1H data suggest that it could also be involved in stress-induced cellular reprogramming downstream of HSFs.
A GRN for auxin-mediated cellular reprogramming
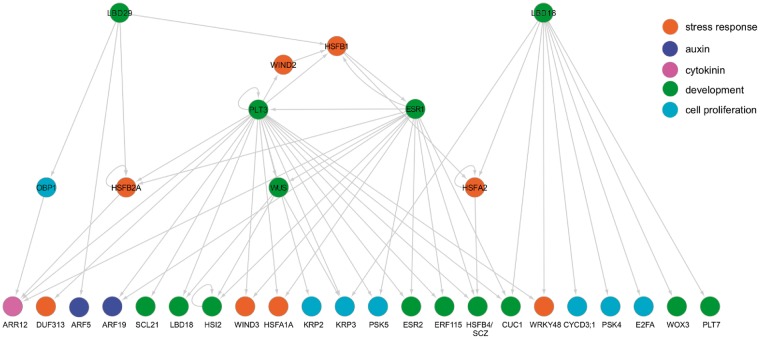
In order to gain further molecular insights into how auxin promotes cellular reprogramming, we selected three key regulators of this process that are highly represented in our GRN; LBD16, LBD29 and PLT3 (Fig. 2; Fan et al. 2012, Kareem et al. 2015), and examined their downstream subnetworks. As shown in Fig. 5, our eY1H data demonstrated that PLT3 forms a regulatory feedback loop together with ESR1 and HSFB1, each of which has some unique, but partially overlapping, incoming and outgoing interactions. Our data further support an autoactivation of PLT3, which was previously suggested by transcriptional induction of PLT3 upon its post-translational activation in PLT3–glucocorticoid receptor (GR) plants (Santuari et al. 2016). Similarly, our data show that PLT3 directly binds the promoters of HSFB2A, WIND3, PSK5 and HSFB4/SCZ, which are all misexpressed shortly after PLT3–GR activation (Santuari et al. 2016). Interestingly, PLT3 and ESR1 share 10 direct targets and they include important regulators of stress response (WIND3 and HSFA1A), auxin (ARF19), cytokinin (ARR12), development (ERF115, ESR2, LBD18, WUS and CUC1) and cell proliferation (PSK5) (Fig. 5), strongly suggesting that PLT3 and ESR1 together form a robust, overlapping transcriptional network. We have previously shown that ESR1 is required for the expression of WUS and ESR2 on SIM (Iwase et al. 2017). Our eY1H data confirm these results and further suggest that ESR1 regulates WUS and ESR2 expression both directly and indirectly through PLT3 (Fig. 5). It is also interesting to note that PLT3 and ESR1 form several feed-forward loops through WUS which regulate downstream genes. PLT3, for instance, appears to regulate LBD18 and KRP3 directly and also indirectly through WUS (Fig. 5).
Fig. 5.
A subnetwork highlighting how auxin-mediated pathways may regulate plant cell reprogramming. PLT3, ESR1 and HSFB1 appear to form a feedback loop which regulates a partially overlapping set of genes involved in development (ESR2, CUC1 and WUS) and cell proliferation (KRP2, KRP3 and PSK5). LBD29 and LBD16 appear to regulate a largely distinct set of downstream targets involved in cell proliferation. Colored circles indicate functional annotations of prey TFs and bait promoters.
As suggested by our power graph analysis (Fig. 2), direct targets of the PLT3–ESR1–HSFB1 pathway only overlap slightly with those targeted by LBD16 and LBD29 (Fig. 5). Of note, our data suggest that LBD16 directly targets several genes associated with cell proliferation, including E2FA, CYCD3; 1, KRP3 and PSK4 (Fig. 5). A previous study by Berckmans et al. (2011) has shown that a close homolog of LBD16, LBD18, directly regulates E2Fa expression. Our data thus further substantiate the role of LBDs in the control of cell cycle progression. We also noticed that CUC1 is among the few common targets of the LBD16/29- and PLT3–ESR1–HSFB1-mediated pathways identified in our GRN (Fig. 5). These data confirm previous reports demonstrating direct regulation of CUC1 expression by ESR1 and PLT3 (Matsuo et al. 2009, Kareem et al. 2015), strongly suggesting that CUC1 integrates upstream signals from both PLT3–ESR1–HSFB1 and LBD16/29 pathways and, in turn, controls cellular reprogramming.
A GRN for cytokinin-mediated cellular reprogramming
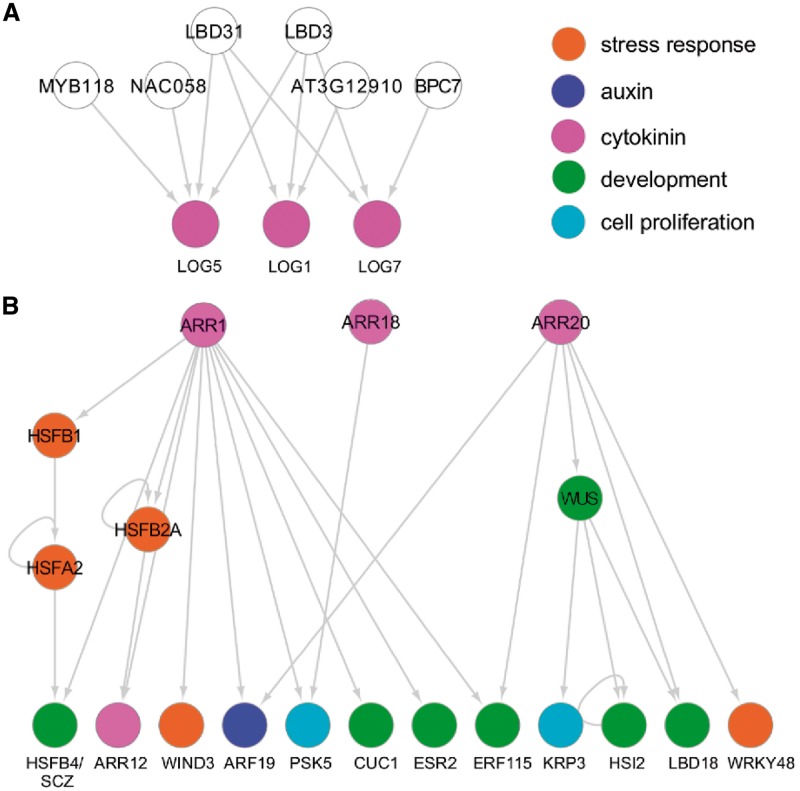
It is becoming increasingly clear that activation of both cytokinin biosynthesis and signaling plays key roles in plant cell reprogramming (Ikeuchi et al. 2013, Ikeuchi et al. 2017). We have recently shown that the transcriptional activation of LOG1, LOG5 and LOG7, which encode enzymes involved in cytokinin biosynthesis, is a core process which drives callus formation at wound sites (Ikeuchi et al. 2017). We thus searched for potential upstream regulators of the cytokinin pathway and identified six TFs that bind these types of promoters (Fig. 6A). Interestingly, LBD3 and LBD31 bind to the promoters of LOG1, LOG5 and LOG7 (Fig. 6A), raising the possibility that they influence cytokinin biosynthesis by up-regulating these genes.
Fig. 6.
Subnetworks highlighting how cytokinin-mediated pathways may regulate plant cell reprogramming. (A) Six TFs in our GRN directly bind the promoters of three LOG genes involved in cytokinin biosynthesis, potentially regulating their expression directly. Among them, both LBD3 and LBD31 bind the promoters of LOG1, LOG5 and LOG7. (B) Key regulators of cytokinin signaling, ARR1, ARR18 and ARR20, potentially regulate genes associated with stress response (HSF genes), development (WUS, CUC1 and ESR2), cell proliferation (PSK5 and KRP3) and auxin (ARF19). Colored circles indicate functional annotations of prey TFs and bait promoters.
To explore how cytokinin signaling promotes cellular reprogramming, we next screened for downstream targets of the type-B ARRs that were included in our set of prey TFs, ARR1, ARR12, ARR14, ARR18 and ARR20, and found 15 promoters that are bound by ARR1, ARR18 and/or ARR20 (Fig. 6B). Importantly, 12 out of these 15 promoters possess at least one of the previously described cytokinin response motifs 5'-(A/G)GAT(T/C)TT-3' or 5'-AAAGAT(T/C)TT-3' (Ramireddy et al. 2013; Supplementary Table S10), implying that these ARRs bind target promoters via these motifs. As might be expected, targets of these ARRs include those involved in stress response (HSFB1, HSFB2A, HSFA2, WIND3 and WRKY48), auxin (ARF19), cytokinin (ARR12), development (WUS, HSFB4/SCZ, CUC1, ESR2, ERF115, HSI2 and LBD18) and cell proliferation (PSK5 and KRP3) (Fig. 6B), suggesting that type-B ARR-mediated cytokinin signaling directly regulates genes associated with various aspects of cellular reprogramming. Previous studies have shown that type-B ARRs, including ARR1, ARR10 and ARR12, directly regulate WUS expression (Meng et al. 2017, Wang et al. 2017, Zhang et al. 2017, Zubo et al. 2017). Our data did not detect binding of ARR1 and ARR12 to the WUS promoter, but we did find that another type-B ARR, ARR20, binds the WUS promoter (Fig. 6B). Our data, in addition, showed that ARR1 binds the promoters of CUC1 and ESR2, other key regulators of shoot regeneration (Daimon et al. 2003, Ikeda et al. 2006) (Fig. 6B), suggesting that ARR-mediated cytokinin signaling directs shoot regeneration through multiple pathways. Our data also highlight the cross-talk between cytokinin and auxin signaling, as we found that both ARR1 and ARR20 directly bind the promoter of ARF19 (Fig. 6B). One unexpected link uncovered in our eY1H data set is the cross-talk between type-B ARRs and HSFs since we detected ARR1 binding to the promoters of HSFB1, HSFB2A and HSFB4/SCZ, as well as HSFB2A binding to the ARR12 promoter (Fig. 6B). These interactions together form several intersecting feed-forward loops (Fig. 6B), which potentially serve as integrators of stress response and cytokinin signaling.
Discussion
Identification of core regulatory pathways in a cellular reprogramming GRN
In this study we mapped the first GRN for cellular reprogramming in plants, based on large-scale eY1H analysis, and provided comprehensive molecular insights into how wound stress and hormonal cues may integrate and collectively regulate a large collection of genes implicated in stress response and developmental decisions. Further functional validations of new TF–promoter interactions found in this study should help identify novel regulators of cellular reprogramming and unveil how they shape the heavily interwoven transcriptional cascades. Our network analysis suggests that PLT3, ESR1 and HSFB1 form a feedback loop and act as the most critical nodes in our GRN, probably integrating upstream signaling to promote coherent downstream developmental transitions (Figs. 3, 5). Our data show that the PLT3 promoter is bound by many wound- and/or CIM-induced TFs (Fig. 3; Supplementary Table S4). This is in line with previously reported observations that the expression of PLT3 is strongly induced both by wounding and by incubation on CIM (Kareem et al. 2015, Ikeuchi et al. 2017). In addition, previous genetic evidence suggest that PLT3 is involved in both wound-induced callus formation and CIM/SIM-induced shoot regeneration (Kareem et al. 2015, Ikeuchi et al. 2017). As expected, our data indicate that PLT3 has many downstream targets associated with stress response, cytokinin, auxin, cell proliferation and development (Fig. 2; Supplementary Table S4). Similarly, our network analysis revealed that ESR1 is under the control of multiple inputs from both wound- and CIM-induced TFs (Fig. 3; Supplementary Table S4). This is also in agreement with our previous observation that ESR1 is up-regulated by wounding and its expression is further enhanced by exogenous application of both auxin and cytokinin (Iwase et al. 2017). Furthermore, our Y1H data include 14 downstream targets of ESR1 that also range from genes involved in stress response to those associated with developmental regulation (Fig. 2; Supplementary Tables S2, S4). Functional roles of HSFB1 in stress-induced cellular reprogramming have not been reported before, but it probably participates in this process, since many wound- and/or CIM-induced TFs bind the HSFB1 promoter, and HSFB1, in turn, binds the ESR1 promoter as part of the PLT3–ESR1–HSFB1 regulatory loop (Figs. 3, 5).
Our power graph analysis, in addition, highlighted the LOB/AS2-mediated pathway, which appears to play distinct but partially overlapping roles with the PLT3–ESR1–HSFB1 pathway in our GRN (Fig. 2). TFs that appear to act in the LOB/AS2-mediated pathway include LBD16 and LBD29, which have been implicated in the regulation of auxin-dependent establishment of root meristems (Okushima et al. 2007), as well as many TFs with unknown functions (Fig. 2). Our Y1H data identified a large set of downstream genes that are bound by these TFs and, importantly, uncovered several putative molecular links, such as LBD3, HDG7, NAC053 and NAC058, which might function in both PLT3–ESR1–HSFB1- and LOB/AS2-mediated pathways (Fig. 2). Further functional characterization of these regulators should advance our understanding of how the reprogramming GRN responds to inductive cues and collectively promotes organ regeneration through these core regulatory pathways. We should note that many of these regulators such as PLT3, ESR1, LBD16 and LBD29 also play important roles in normal organogenesis (Kirch et al. 2003, Okushima et al. 2007, Prasad et al. 2011) and they might bind their downstream targets identified in our Y1H in either context. Further studies are therefore needed to examine these regulatory relationships under different physiological conditions and reveal context-dependent specificity of TF–promoter binding.
Roles of AP2/ERFs in wound-induced cellular reprogramming
AP2/ERFs form a large family of TFs consisting of 147 members in Arabidopsis (Nakano et al. 2006). Some subfamily members, such as DREBs, are known to mediate various stress responses, while others, such as AP2 and AINTEGUMENTA, are more associated with developmental roles in the context of organ growth and differentiation (Jofuku et al. 1994, Mizukami and Fischer 2000). Importantly, previous studies have also identified several AP2/ERF TFs, including WINDs and ERF115, as important regulators of cellular reprogramming (Iwase et al. 2011, Heyman et al. 2013). A previous study has shown that ERF115 transcriptionally regulates WIND1 and PLT3 (Heyman et al. 2016) but interactions between ERF115 and these or other tested promoters were not detected in our eY1H experiments. Given that organ regeneration is the process of external stimuli perturbing endogenous developmental programs, it is plausible that there are many more AP2/ERF TFs which will be identified as key players in this process. Our eY1H analysis identified nine AP2/ERF TFs that are rapidly induced by wounding and bind promoters of many genes relevant to cellular reprogramming (Fig. 4; Supplementary Tables S2, S4). A previous study by Yang et al. (2005) has shown that ERF4 is induced by ethylene, jasmonate and ABA, and, in turn, it modulates cellular sensitivity to ABA. ABA is inhibitory for callus formation at wound sites (Ikeuchi et al. 2017), thus it would be interesting to test whether ERF4 is involved in ABA-dependent regulation of callus formation. Our eY1H data, in addition, suggest that some AP2/ERFs regulate the expression of other AP2/ERFs in the reprogramming GRN, thus forming multilayered cascades within AP2/ERF TFs (Fig. 4). There is precedent for this in other biological contexts, implying that this is one of the key features of GRNs. Three basic helix–loop–helix TFs, for instance, sequentially regulate stomatal differentiation (Pillitteri et al. 2007), and an interwoven cascade of HSF TFs mediates the heat shock response (Ohama et al. 2017). Further functional annotation of AP2/ERFs identified in this study should reveal the physiological relevance of AP2/ERF-mediated transcriptional cascades in stress-induced cellular reprogramming.
Roles of LBDs in the regulation of cytokinin biosynthesis
De novo biosynthesis of cytokinin promotes wound-induced callus formation, but we have recently shown that WIND1 is not required for this physiological response, since accumulation of endogenous cytokinin after wounding is not defective in WIND1–SRDX plants (Ikeuchi et al. 2017). In this study, we have identified LBD3 and LBD31 as putative upstream regulators of three wound-induced cytokinin biosynthesis genes, LOG1, LOG5 and LOG7 (Fig. 6). Intriguingly, however, the expression of LBD3 and LBD31 themselves is not wound inducible(Ikeuchi et al. 2017); thus, they might be activated, for example through post-translational mechanisms, in order to induce LOG expression upon wounding. It is also possible that other LBD homologs that are induced by wounding, such as LBD16 and LBD29 (Ikeuchi et al. 2017), also bind these promoters in planta and induce LOG genes, although our eY1H assay did not detect such TF–promoter interactions. It is worth noting that the expression of LBD3 is induced by the elevation of endogenous cytokinin levels (Wang et al. 2015). Therefore, LBD3 might act as a part of a feedback loop which enhances cytokinin-mediated signaling pathways.
Materials and Methods
Promoter cloning
The 2 kb promoter sequence upstream of the start codon was amplified by PCR using Prime star max (TAKARA) and subjected to A-tailing by Ex Taq polymerase (TAKARA). Amplicons were then introduced into either the pENTR 5'-TOPO (Thermo Fisher Scientific) or pDONRG P4-P1r (Oshima et al. 2011) vector. Promoter sequences were then subcloned into pMW2 and pMW3 (Addgene) by LR reactions (Thermo Fisher Scientific) and transformed into Escherichia coli (DH5α). After verifying the border sequence, purified plasmids were transformed into yeast (YM4271) as described in Gaudinier et al. (2017). A set of primers used for cloning and sequencing in this study are listed in Supplementary Table S11.
eY1H screening
The 48 bait promoters were screened against the 559 TFs as described previously (Gaudinier et al. 2011, Gaudinier et al. 2017). The 558 transcription factors were obtained from the Arabidopsis TF collection at the proteomics core facility of UC Davis (http://proteomics.ucdavis.edu/yeast-one-hybrid-services-brady-lab/). To supplement this library, WIND4 cDNA was cloned into the pDONR221 vector and recombined into the pDESTAD-2 μ vector by an LR reaction (Thermo Fisher Scientific). The recombined plasmid was transformed into yeast cells (Yα1867) as described in Gaudinier et al. (2017). Yeast cells containing a bait vector were mated with yeast cells containing a prey vector. Mated yeast cells were spotted at two independent foci for experimental replication. Mating and transferring of yeast cells were assisted by an automated system using ROTOR HDA (Singer Instruments). Diploid yeast cells were then subjected to the X-gal assay and 3-amino-1, 2, 4-triazole (3-AT) resistance assay. For the X-gal assay, plates were photographed at the earliest time point when any colonies turned blue, with the plates checked at 30 min and at 1, 2, 3, 4, 7 and 24 h. If no colony turned blue, plates were photographed at 7 and 24 h. For the 3-AT resistance assay, plates were kept in the dark at room temperature and observed every 2 d up to 10 d. Plates were photographed at 10 d and the 3-AT resistance was scored as ‘positive’ when both duplicate yeast colonies showed normal growth. The TF–promoter binding was regarded as ‘positive’ if either the X-gal assay or 3-AT resistance assay was positive compared with the pDESTAD-2 μ plasmid control with no TF.
Visualization of network graphs and power graph compression analysis
The network graphs were visualized by Cytoscape version 3.5.1 (http://www.cytoscape.org/) as previously described in Shannon et al. (2003). The power graph compression analysis was performed as previously described in Ahnert (2013) and Taylor-Teeples et al. (2015).
Supplementary Data
Supplementary data are available at PCP online.
Funding
This work was supported the Ministry of Education, Culture, Sports, Science and Technology (MEXT) [grant No. 15H05961 to K.S. and a MEXT fellowhip to D.C.]; the Japan Society for the Promotion of Science (JSPS) [grant Nos.17K15146 and 17J40121 to M.I., 16J07464 to M.S., 17K07461 to A.I. and 17H03704 to K.S., and a fellowhip to M.I., M.S. and D.F.]; RIKEN [an incentive research project grant 201701100428 to B.R. and a Special Postdoctoral Researcher Program to M.I.]; and the Naito Foundation [support to M.I.].
Disclosures
The authors have no conflicts of interest to declare.
Supplementary Material
Acknowledgments
We thank Mariko Mouri and Chika Ikeda for their technical assistance.
Glossary
Abbreviations
- AP2/ERF
APETALA2/ETHYLENE RESPONSE FACTOR
- ARR
ARABIDOPSIS RESPONSE REGULATOR
- ARF
AUXIN RESPONSE FACTOR
- 3-AT
3-amino-1, 2, 4-triazole
- CDK
CYCLIN DEPENDENT KINASE
- CIM
callus-inducing medium
- ChIP
chromatin immunoprecipitation
- CUC
CUP SHAPED COTYLEDON
- CYC
cyclin
- DAP
DNA affinity purification
- DREB
DEHYDRATION RESPONSIVE ELEMENT BINDING PROTEIN
- ESR
ENHANCER OF SHOOT REGENERATION
- eY1H
enhanced yeast one-hybrid
- GR
glucocorticoid receptor
- GRN
gene regulatory network
- HSE
heat shock element
- HSF
HEAT SHOCK FACTOR
- HSI
HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE
- KRP
KIP-RELATED PROTEIN
- LBD
LATERAL ORGAN BOUNDARY DOMAIN
- LEC
LEAFY COTYLEDON
- LOB/AS2
LATERAL ORGAN BOUNDARY/ASYMMETRIC LEAVES2
- LOG
LONELY GUY
- MCA
multiple correspondence analysis
- NAC
NAM, ATAF1, 2 and CUC2
- PSK
PHYTOSULFOKINE
- PLT
PLETHORA
- SCL
SCARECROW-LIKE
- SCZ
SCHIZORIZA
- SIM
shoot-inducing medium
- TF
transcription factor
- WIND
WOUND INDUCED DEDIFFERENTIATION
- WOX
WUSCHEL-RELATED HOMEOBOX
- WUS
WUSCHEL
- YUC
YUCCA
References
- Ahnert S.E. (2013) Power graph compression reveals dominant relationships in genetic transcription networks. Mol. Biosyst. 9: 2681–2685. [DOI] [PubMed] [Google Scholar]
- Ahnert S.E. (2014) Generalised power graph compression reveals dominant relationship patterns in complex networks. Sci. Rep. 4: 4385–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y. (2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenov Y., Ramírez F., Schelhorn S.E., Lengauer T., Albrecht M. (2008) Computing topological parameters of biological networks. Bioinformatics 24: 282–284. [DOI] [PubMed] [Google Scholar]
- Atta R., Laurens L., Boucheron-Dubuisson E., Guivarc’h A., Carnero E., Giraudat-Pautot V., et al. (2009) Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57: 626–644. [DOI] [PubMed] [Google Scholar]
- Banno H., Ikeda Y., Niu Q.W., Chua N.-H. (2001) Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13: 2609–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A.L., Oltvai Z.N. (2004) Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5: 101–113. [DOI] [PubMed] [Google Scholar]
- Berckmans B., Vassileva V., Schmid S.P., Maes S., Parizot B., Naramoto S., et al. (2011) Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23: 3671–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J.W., Cole M., Flier A., Grewe B., Werr W. (2007) The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134: 1653–1662. [DOI] [PubMed] [Google Scholar]
- Che P., Lall S., Nettleton D., Howell S.H. (2006) Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 141: 620–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Tong J., Xiao L., Ruan Y., Liu J., Zeng M., et al. (2016) YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 67: 4273–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupeau M.-C., Granier F., Pichon O., Renou J.-P., Gaudin V., Chupeau Y. (2013) Characterization of the early events leading to totipotency in an Arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell 25: 2444–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon Y., Takabe K., Tasaka M. (2003) The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 44: 113–121. [DOI] [PubMed] [Google Scholar]
- Efroni I., Mello A., Nawy T., Ip P.-L., Rahni R., DelRose N., et al. (2016) Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M., Xu C., Xu K., Hu Y. (2012) LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 22: 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L.C. (1978) Centrality in social networks: conceptual clarification. Social Networks 1: 215–239. [Google Scholar]
- Gaudinier A., Brady S.M. (2016) Mapping transcriptional networks in plants: data-driven discovery of novel biological mechanisms. Annu. Rev. Plant Biol. 67: 575–594. [DOI] [PubMed] [Google Scholar]
- Gaudinier A., Tang M., Bågman A.M., Brady S.M. (2017) Identification of protein–DNA interactions using enhanced yeast one-hybrid assays and a semiautomated approach. Methods Mol. Biol. 1610: 187–215. [DOI] [PubMed] [Google Scholar]
- Gaudinier A., Zhang L., Reece-Hoyes J.S., Taylor-Teeples M., Pu L., Liu Z., et al. (2011) Enhanced Y1H assays for Arabidopsis. Nat. Methods 8: 1053–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J., Cools T., Canher B., Shavialenka S., Traas J., Vercauteren I., et al. (2016) The heterodimeric transcription factor complex ERF115–PAT1 grants regeneration competence. Nat. Plants 2: 16165–16167. [DOI] [PubMed] [Google Scholar]
- Heyman J., Cools T., Vandenbussche F., Heyndrickx K.S., Van Leene J., Vercauteren I., et al. (2013) ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863. [DOI] [PubMed] [Google Scholar]
- Ikeda Y., Banno H., Niu Q.W., Howell S.H., Chua N.-H. (2006) The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 47: 1443–1456. [DOI] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. (2011) Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157: 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Sugimoto K., Iwase A. (2013) Plant callus: mechanisms of induction and repression. Plant Cell. 25: 3159–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Iwase A., Rymen B., Lambolez A., Kojima M., Takebayashi Y., et al. (2017) Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 175: 1158–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K. (2016) Plant regeneration: cellular origins and molecular mechanisms. Development 143: 1442–1451. [DOI] [PubMed] [Google Scholar]
- Iwase A., Harashima H., Ikeuchi M., Rymen B., Ohnuma M., Komaki S., et al. (2017) WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 29: 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A., Mitsuda N., Koyama T., Hiratsu K., Kojima M., Arai T., et al. (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 21: 508–514. [DOI] [PubMed] [Google Scholar]
- Jofuku K.D., den Boer B.G., Van Montagu M., Okamuro J.K. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareem A., Durgaprasad K., Sugimoto K., Du Y., Pulianmackal A.J., Trivedi Z.B., et al. (2015) PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25: 1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch T., Simon R., Grünewald M., Werr W. (2003) The DORNRÖSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem ccll fate and lateral organ development. Plant Cell 15: 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Gaudinier A., Tang M., Taylor-Teeples M., Nham N.T., Ghaffari C., et al. (2014) Promoter-based integration in plant defense regulation. Plant Physiol. 166: 1803–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sheng L., Xu Y., Li J., Yang Z., Huang H., et al. (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sun N., Liu M., Liu J., Du B., Wang Z., et al. (2013) An autoregulatory loop controlling Arabidopsis HsfA2 expression: role of heat shock-induced alternative splicing. Plant Physiol. 162: 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. (1996) Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 93: 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N., Mase H., Makino M., Takahashi H., Banno H. (2009) Identification of ENHANCER OF SHOOT REGENERATION 1-upregulated genes during in vitro shoot regeneration. Plant Biotechnol. 26: 385–393. [Google Scholar]
- Meng W.J., Cheng Z.J., Sang Y.L., Zhang M.M., Rong X.F., Wang Z.W., et al. (2017) Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y., Fischer R.L. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97: 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N., Sato H., Shinozaki K., Yamaguchi-Shinozaki K. (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22: 53–65. [DOI] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., et al. (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y., Mitsuda N., Nakata M., Nakagawa T., Nagaya S., Kato K., et al. (2011) Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol. 28: 201–210. [Google Scholar]
- Pernas M., Ryan E., Dolan L. (2010) SCHIZORIZA controls tissue system complexity in plants. Curr. Biol. 20: 818–823. [DOI] [PubMed] [Google Scholar]
- Pfefferli C., Jaźwińska A. (2015) The art of fin regeneration in zebrafish. Regeneration 2: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007) Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505. [DOI] [PubMed] [Google Scholar]
- Prasad K., Grigg S.P., Barkoulas M., Yadav R.K., Sanchez-Perez G.F., Pinon V., et al. (2011) Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr. Biol. 21: 1123–1128. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Nagel D.H., Kang S.E., Bonaldi K., Doherty C.J., et al. (2014) A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Rep. 8: 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramireddy E., Brenner W.G., Pfeifer A., Heyl A., Schmülling T. (2013) In planta analysis of a cis-regulatory cytokinin response motif in Arabidopsis and identification of a novel enhancer sequence. Plant Cell Physiol. 54: 1079–1092. [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 103: 18822–18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuari L., Sanchez-Perez G.F., Luijten M., Rutjens B., Terpstra I., Berke L., et al. (2016) The PLETHORA gene regulatory network guides growth and cell differentiation in arabidopsis roots. Plant Cell 28: 2937–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena G., Wang X., Liu H.-Y., Hofhuis H., Birnbaum K.D. (2009) Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D.. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., Radziejwoski A., Busch W., Hannah M.A., Czeszejko J., Kwaśniewski M., et al. (2008) The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J. 56: 779–792. [DOI] [PubMed] [Google Scholar]
- Skoog F., Miller C.O. (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 54: 118–130. [PubMed] [Google Scholar]
- Sugimoto K., Jiao Y., Meyerowitz E.M. (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18: 463–471. [DOI] [PubMed] [Google Scholar]
- Taylor-Teeples M., Lin L., de Lucas M., Turco G., Toal T.W., Gaudinier A., et al. (2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 29: 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove C.A., Willemsen V., de Vries W.J., van Dijken A., Scheres B., Heidstra R. (2010) SCHIZORIZA encodes a nuclear factor regulating asymmetry of stem cell divisions in the Arabidopsis root. Curr. Biol. 20: 452–457. [DOI] [PubMed] [Google Scholar]
- Valvekens D., Montagu M.V., Van Lijsebettens M. (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85: 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian C., Zhang C., Shi B., Cao X., Zhang T.Q., et al. (2017) Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29: 1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shen W., Chan Z., Wu Y. (2015) Endogenous cytokinin overproduction modulates ROS homeostasis and decreases salt stress resistance in Arabidopsis thaliana. Front. Plant Sci. 6: 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tian L., Latoszek-Green M., Brown D., Wu K. (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58: 585–596. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Ohama N., Nakajima J., Kidokoro S., Mizoi J., Nakashima K., et al. (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 286: 321–332. [DOI] [PubMed] [Google Scholar]
- Zhang T.-Q., Lian H., Zhou C.M., Xu L., Jiao Y., Wang J.W. (2017) A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo Y.O., Blakley I.C., Yamburenko M.V., Worthen J.M., Street I.H., Franco-Zorrilla J.M., et al. (2017) Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: E5995–E6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.