Abstract
Guidelines recommend patients be informed of their incidental results (IR) when undergoing genomic sequencing (GS), yet there are limited tools to support patients’ decisions about learning IR. The aim of this study is to develop and test the usability of a decision aid (DA) to guide patients’ selection of IR, and to describe patients’ preferences for learning IR following use of the DA. We developed and evaluated a DA using an iterative, mixed-methods process consisting of (1) prototype development, (2) feasibility testing, (3) cognitive interviews, (4) design and programming, and (5) usability testing. We created an interactive online DA called the Genomics ADvISER, a genomics decision AiD about Incidental SEquencing Results. The Genomics ADvISER begins with an educational whiteboard video, and then engages users in a values clarification exercise, knowledge quiz and final choice step, based on a ‘binning’ framework. Participants found the DA acceptable and intuitive to use. They were enthusiastic towards GS and IR; all selected multiple categories of IR. The Genomics ADvISER is a new patient-centered tool to support the clinical delivery of incidental GS results. The Genomics ADvISER fills critical care gaps, given the health care system’s limited genomics expertise and capacity to convey the large volume of IR and their myriad of implications.
Introduction
Guidelines established by the American College of Medical Genetics and Genomics (ACMG) recommend that a set of ‘medically actionable’ incidental results (IR) be sought out and offered to patients undergoing genomic sequencing (GS), with patient consent [1]. In contrast, recommendations from the European Society of Human Genetics recommend that steps be taken to avoid the discovery of IR in sequence analysis, and that variants lacking in clinical utility should not be analyzed or reported. However, literature indicates that the majority of patients are interested in learning both medically actionable and non-medically actionable IR [2]. While a majority of patients choose to learn IR, variation exists in patient preferences [2], and patients value having a choice about which IR they receive [3], indicating a need for a tool to help personalized decision-making.
There has been some discussion as to whether “incidental” or “secondary” is the preferable term to use to refer to this class of result [4]. “Secondary” generally indicates that findings are explicitly sought (e.g., 59 medically actionable genes selected by the ACMG), whereas “incidental” implies that they were discovered unintentionally [5]. European recommendations offer the term “unsolicited findings” for pathogenic results that are discovered through GS and not related to the reason for testing [6]. Some argue that the term “incidental” could make these findings seem less important or relevant than other types of findings [7], and some research suggests that the term “additional findings” may in fact be preferred by individuals receiving sequencing [4], though the latter term has not been widely used in literature. In America, “secondary” has come to be the preferred term following a Presidential Commission on secondary findings [8], and the most recent recommendations from the ACMG [1]. However, in these contexts, “secondary” is used to refer specifically to pre-determined, medically actionable and deliberately sought out findings, not other types of results (e.g., non-actionable results, carrier status). The categories of results in our decision aid reflect both “secondary” findings which would be deliberately sought out (e.g., ACMG medically actionable genes) and findings that would be inadvertently discovered (e.g., carrier status, pharmacogenomics variants, neurological disease risks, etc.). Therefore, for the purpose of this study, the term “incidental result” was used. This terminology also reflects a wider policy debate about the return of incidental results in other areas of medicine such as radiology [9].
The Institute of Medicine and the ACMG suggest that clinicians engage in shared decision-making with patients when considering IR [10, 11]. To participate in shared decision-making for GS, patients need to fully understand the relative benefits and harms of GS and thousands of possible IR. The standard care of pre-test genetic counseling, which consists of hours of counseling per test/disease, is highly costly and infeasible in usual clinical practice, given the thousands of possible results [12, 13]. As GS use increases, the demand for genetic counselors will exceed their availability [14], and there is limited genomics knowledge among other health care providers [15]. Novel approaches to patient education and decision support are needed to address the counseling burden, limited resources, and genomics expertise [14].
Decision aids (DAs) can help meet this genomics health services challenge because they improve decision quality, satisfaction [16], promote decisions congruent with patients’ values, reduce decisional conflict [16] and enhance patient-provider communication [17]. DAs improve patient knowledge, risk perception, and agreement between their values and their choice, compared to standard of care [17], though the evidence is mixed on whether DAs reduce the time clinicians spend with patients [12]. Electronic DAs have been shown to improve patients’ knowledge acquisition compared to paper-based educational materials [18] or genetic counseling alone [12]. Users’ knowledge also improves when electronic DAs are used in conjunction with genetic counseling [12]. Electronic DAs are thus effective clinical tools to help counsel patients on the thousands of incidental findings available, and to empower patients’ shared decision-making for GS.
Despite the long-standing practice of medical genetics, there are relatively few decision support tools for genetic testing and very few that have been rigorously evaluated [19]. Even fewer decision support tools exist on IR from GS; existing tools target pediatric contexts [13], focus on GS education-only [14] or on the return of results [20]; they do not cover all possible IR with decision support to simulate the genetic counseling pre-test experience, limiting their use and applicability in clinical care. Our primary aim was to develop and conduct usability testing on a DA to facilitate shared decision-making for the selection of IR from GS. Our secondary objective was to explore the usability of a framework for “binning” IR.
Materials and methods
Phase 1: Development of prototype DA
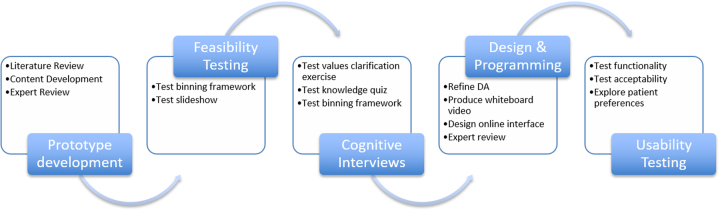
There were five phases in the development of the DA (Fig. 1). We developed a DA prototype using the Ottawa Decision Support Framework (ODSF) [21], which is an evidence-based framework designed to support health decisions that require effort to deliberate about a substantial amount of information. The ODSF offers guidance on how to provide decisional support, which should involve identifying patients’ decisional needs, making the decision clear, educating patients about the decision and possible outcomes, clarifying patients’ values, and monitoring decision making progress [22]. A key component of the ODSF is the emphasis on evaluating patients’ knowledge of the decision, their options and the benefits and harms, and providing them with this knowledge in different ways (e.g., expressing probabilities numerically and graphically) to facilitate comprehension [22]. Comprehension enables realistic expectations regarding the decision, risks, benefits and possible outcomes [22]. In addition to patient education, the ODSF emphasizes other critical aspects of decision making such as supporting patients in identifying their personal values related to the decision [22]. The framework has been validated and has been shown to be effective for improving patients’ knowledge and supporting informed, value-congruent decision making [23, 24].
Fig. 1.
The stages of decision aid (DA) development and testing
The ODSF is congruent with patients’ need to understand the myriad of IR available through GS and their implications. Consistent with the ODSF, our DA prototype comprised of four parts: (1) an educational, narrated slideshow, (2) a knowledge questionnaire, (3) a values clarification exercise, and (4) a final choice step. Given the multitude of choices of possible IR, we “binned” IR into five categories of medical actionability and disease characteristics based on Berg et al.’s “Binning” framework [25]. Bin 1 contained medically actionable and pharmacogenetic variants (e.g., Breast and ovarian cancer, Aortic aneurysm, Long QT syndrome, etc.—ACMG genes [1]); Bin 2a contained common disease susceptibility single nucleotide polymorphisms (e.g., coronary artery disease, type 2 diabetes, etc.); Bin 2b contained Mendelian disease variants (e.g., retinitis pigmentosa, neuropathy, etc.); Bin 2c contained variants associated with early-onset neurological disease (e.g., early-onset Alzheimer’s disease, early-onset Parkinson’s disease, etc.); Bin 3 contained carrier status (e.g., phenylketonuria, cystic fibrosis, sickle cell disease, etc.). Bin 4 contained variants of uncertain significance (VUS), which are generally not available [25] (Supplemental Appendix).
The DA prototype was reviewed by experts in clinical genetics, oncology, decision support science, knowledge translation, genomics literacy, genomics education, and health psychology. We refined the DA based on their feedback. We used the International Patient Decision Aids Standards (IPDAS) quality assessment framework [26] to evaluate our DA prototype.
Phase 2: Feasibility testing of DA prototype
Participants
A purposive sample [27] was recruited from the Clinical Genetics Service (CGS) at Memorial Sloan Kettering Cancer Center (MSK) in New York, NY between 2012–2013, comprising of cancer patients who had undergone genetic testing for a cancer predisposition.
Data collection
Participants worked through the DA prototype and were interviewed about the organization of bins and clarity of the slideshow. Ethical approval was obtained from the Institutional Review Board (IRB) of MSK.
Phase 3: Cognitive interviews of the DA prototype
Participants
A convenience sample of students were recruited via email from the CGS at MSK between 2013–2014 to participate in cognitive interviews [28].
Data collection
Participants used the DA prototype and completed values and knowledge questionnaires. These activities were followed by interview questions related to those questionnaires, as well as questions assessing the participants’ general understanding of the bins. Interviews and questionnaire responses were recorded. In addition, demographic data were collected on each participant. We summarized the problems that participants encountered and their suggested solutions. Ethical approval was obtained from the IRB of MSK.
Phase 4: Design and programming of the online DA
Based on feedback from the cognitive interviews, we refined the knowledge questionnaire and values clarification exercise, and converted the educational slideshow into a professional whiteboard video. The content of the video script was based on the slideshow from the DA prototype, converted to lay language with visual illustrations of the key concepts. The video script was reviewed by a panel of experts in medical genetics, genetic counseling, decision science, health psychology, knowledge translation, genomics literacy, genomics education, and health services research. The DA was modified based on their feedback. In addition, we evaluated the reading level of the video script and DA content using the Flesch–Kincaid readability test.
In partnership with a design company, we converted the sections of the DA into an online interface, based on design principles for navigability and user experience [29].
Phase 5: Usability testing
The final version of the DA underwent usability testing through an iterative process [30], where feedback from participants in each cycle informed updates and modifications to the DA; the updated DA was then tested in the subsequent cycle. Usability testing is an established technique with the objective to systematically test the navigability and content comprehension of a tool prior to its distribution [31], and is commonly used in the development of DAs for health decisions [13, 32, 33]. Usability testing does not aim to validate tools for clinical use. The goals of our usability testing were to determine whether participants were able to use the DA, to assess the DA’s acceptability, and to elicit feedback on both the tool and binning framework. Usability testing sessions were carried out in-person or via WebEx. Ethical approval was obtained from the IRB of St. Michael’s Hospital (SMH).
Participants & sampling
We recruited breast and colon cancer patients from clinics at SMH in Toronto, Ontario between April and July 2016. Participants were eligible to participate if they were current patients at the clinic, were over 18 years of age, had some experience using a computer, were able to speak and read English and were visiting the clinic for a follow-up appointment. Patients under 18 years of age, in advanced stage cancer (stage 4) or unable to use a computer were excluded. As is typical of usability testing, our sample was not intended to be representative of all individuals who may receive GS.
They were selected as a population of surrogate decision-makers reflective of those eligible for GS for the purpose of usability testing [34]. Patients making real decisions for GS were not selected as, as decision tools should be evaluated before they are used in actual clinical decisions. Prior to implementing the DA for clinical decisions, it must be tested among patients making hypothetical decisions. We targeted a convenience sample of fifteen patients, reflecting an appropriate sample size for usability studies [35–37], as 80% of usability issues can be identified through 5–8 participants [28].
Usability
During usability testing sessions, participants used the DA to make a hypothetical decision regarding receipt of IR, while the study coordinator (MC) observed and took notes. To elicit feedback on the DA’s functionality, participants were asked to “think aloud” while they used the DA, verbalizing their actions and decision-making process [30]. Participants were asked to reflect on the DA’s clarity, navigability and face validity using standard prompts from the ODSF [21].
Immediately following use of the DA, semi-structured interviews were conducted by MC about participants experience using the DA, navigability, the volume and presentation of information, their likes and dislikes, and suggestions for improvement. Participants were also asked to consider their preferences for clinical use of the DA: as a stand-alone decision tool or used in conjunction with a genetic counselor.
Acceptability
To assess the acceptability of the DA, participants completed a validated questionnaire [38], modified to apply to the assessment of a DA for GS. The questionnaire contained questions about elements of the DA including presentation, length, amount and clarity of information, and whether the DA was sufficient to make a decision. Participants were also asked to provide written feedback on what they liked about the DA and suggestions for improvement. Participants provided demographic information including gender, age, race/ethnicity, country of birth, employment, annual household income and education.
Perceptions of the binning framework
We qualitatively explored participants’ perceptions of the binning framework and preferences for results using a semi-structured interview guide. Interviews probed participants’ motivations for selecting specific categories and their perceived uses for IR. All sessions were audio-recorded and transcribed verbatim.
Analysis
Quantitative data
We used descriptive statistics to describe participants’ demographic characteristics, summarize acceptability results and summarize participants’ category choices for IR using Microsoft Excel.
Qualitative data
Qualitative data from the “think aloud” processes and cognitive interviews were analyzed using content analysis [39]. In keeping with qualitative methodology, analysis occurred concurrently with data collection [40], which informed subsequent iterations of the interview guide and modifications to the DA. Interviews were coded independently by MC, CM, LC, and SC, and differences were compared and reconciled. Descriptive and thematic codes were applied to the data. Initial codes were derived from themes explored in the interview guide; constant comparison allowed novel codes to emerge from the data. A codebook was developed following the initial coding structure and was expanded and adapted based on emergent codes.
Results
Stage 1: Development of the DA prototype
The DA prototype was developed based on literature by a multi-disciplinary team consisting of health services researchers, medical geneticists, oncologists, and genetic counselors. Consistent with other decision support tools [21, 41], our DA prototype comprised of four parts. It began with an educational slideshow that describes GS and the categories of IR offered for return, followed by a values clarification exercise that reviewed the benefits and harms of each option. Next, a knowledge questionnaire reviewed concepts and options of IR. The final part was a choice step in which a menu of choices was presented that outlined the categories of IR from which participants could choose.
Our DA met criteria from the IPDAS quality criteria checklist, including development via a systematic review process, engagement of patients and practitioners, review by external experts, presenting risks and benefits of each option, engaging patients in reflection on which risks and benefits matter most to them, providing a step-by-step way to make a decision, engagement of health professionals not involved in producing the DA in the development process, and providing adequate information on options [26].
Stage 2: Feasibility testing of the DA prototype
Participants
Of the six participants involved in the feasibility testing, two were clinical patients with a personal or family history of cancer from the CGS at MSK. All participants were female, ages 25–87, two of whom had children.
Perceptions of the binning framework
Overall the binning framework helped participants manage the volume of information, though some categories were less clear to participants than others. For example, Bin 2b was difficult to understand because it consisted of a heterogeneous group of variants (carrier, diagnostic results and progressive diseases, per Berg et al’s original framework [25]). These results pointed to the need to re-conceptualize Bin 2.
Stage 3: Cognitive interviews
Participants
Our convenience sample included 9 participants ages 19–26. Six participants were female; all had completed some postsecondary education.
Views of the DA
Cognitive interviews identified several components of the DA that were challenging for participants. In the educational slideshow, some key terms caused confusion for participants, including incidental results, actionable results, and healthy carriers. Many participants did not recall key concepts from the slideshow, which led to difficulty making choices in the subsequent components of the DA.
Views of the Bins
Participants’ understanding of bins was often tied to their familiarity with the disease example presented in the bin, indicating the need to provide several example diseases per bin. Participants also expressed confusion about Bin 4 (VUS) since results from that bin would not be made available. Finally, participants struggled with bin organization, for example being unsure as to whether bins 2a, 2b, and 2c could be selected individually, and what their relationship was beyond lack of medical actionability.
Stage 4: Development of the online version of the DA
Refinements of the DA
Based on the results of the cognitive interviews, the slideshow, knowledge and values clarification exercises as well as choice step were modified. The bins were renumbered as Categories 1–5 rather than the original 1, 2a, 2b, 2c, 3. The original Bin 4 containing VUS was omitted from the online DA, as these results were never intended to be returned (the bin had been added only to explain the fact that uncertain results are also generated from GS but would not be provided). In addition, more disease examples were added to the explanation of each category. The reading level of the video and DA were found to be at grade level 8.5 using the Flesch–Kincaid readability test.
Stage 5: Usability testing
Participants
Participants were predominately white/European (10/15), females (9/15), over 50 years old (11/15) and had high levels of education (n = 13/15 completed postsecondary education) (Table 1).
Table 1.
Demographic characteristics of participants in cognitive interviews (Stage 3) and usability testing (Stage 5)
| Cognitive interviews (n = 9) | Usability testing (n = 15) | |
|---|---|---|
| Age | ||
| 19–29 | 9/9 | 0/15 |
| 30–39 | 0/9 | 2/15 |
| 40–49 | 0/9 | 2/15 |
| >50 | 0/9 | 11/15 |
| Gender | ||
| Female | 6/9 | 8/15 |
| Education (highest level completed) | ||
| High school | 6/9 | 2/15 |
| College/Trade school | 0/9 | 5/15 |
| Bachelor’s degree | 3/9 | 4/15 |
| Post-graduate degree | 0/9 | 4/15 |
| Employment status | ||
| Full time | 0/9 | 5/15 |
| Part time | 9/9 | 1/15 |
| Disability | 0/9 | 4/15 |
| Retired | 0/9 | 4/15 |
| Income | ||
| <$80,000 | – | 9/15 |
| ≥$80,000 | – | 6/15 |
| Country of birth | ||
| Canada | – | 12/15 |
| Outside Canada | – | 3/15 |
| Ethnicity | ||
| Asian—East | – | 1/15 |
| Asian—South | – | 1/15 |
| Black—Caribbean | – | 1/15 |
| White—European | – | 10/15 |
| Other | – | 1/15 |
| Personal history of cancer | ||
| Breast | – | 8/15 |
| Colon | – | 6/15 |
| Breast & colon | – | 1/15 |
| Family history of cancer | ||
| Breast | 4/9 | 1/15 |
| Colon | 0/9 | 2/15 |
| Other/Multiple | 2/9 | 3/15 |
| None | 3/9 | 9/15 |
| Personal experience with genetic testing | ||
| Yes | 1/9 | 6/15 |
Refinements of the DA
Three iterative rounds of usability testing with five participants in each round allowed us to identify and resolve usability issues throughout the testing process.
Categories
A major challenge for participants was differentiating between the categories of IR. Initially, categories were identified by number and color alone. Participants were able to recall attributes of categories but struggled to assign the corresponding number to those attributes. Based on participant feedback, a short descriptor of each category was added after the number throughout the DA, such as “Category 1: Medically Actionable,” prior to the final round of usability testing. This allowed users to accurately recall and assign attributes to each category; none of the last five users testing the revised version had difficulty with category recall. By the last round of testing, participants could describe the categories well and liked their organization.
Knowledge test
Participants in the initial two rounds of usability testing struggled on the knowledge test, largely due to category recall as described above, and because some questions overlapped. New questions were written after the first two rounds of testing, which focused on the category content and general information about GS. The last five users had no issues with the new test questions; one user had one incorrect answer, but this was seen as normal variation.
General refinements
Other revisions made to the DA included clarifying transitions in the DA, adding a summary table of risks and benefits, changing disease examples to more familiar diseases, simplifying the values clarification exercise and adding the ability for users to review sections of the video based on the results of their knowledge test—before they make their final decision.
Usability
Interviews demonstrated strong face validity and content comprehension. Participants found the DA easy to navigate and intuitive to use. Overall, the participants reacted positively to the video. Participants found the video to be effective for learning and retaining the large volume of information about GS and IR. Many participants stated that while the DA was sufficient to make a decision, if they were to use it in a real clinical scenario they would prefer to also have the opportunity to consult a professional to ask questions, seek reassurance in their decision, and have a personal interaction. On average, it took participants 20 min to use the DA.
Acceptability
All participants responded favorably to the DA, with the majority rating it as highly acceptable. Most patients found the length and amount of information “just right” (13/15), clear (13/15) and balanced (14/15). (Table 2). All participants reported that Genomics ADvISER was sufficient for them to reach a decision; that the DA was easy to use and would recommend it (Table 2).
Table 2.
Acceptability results from usability testing (Stage 5)
| The way general information was presented | |
| Excellent | 13/15 |
| Good | 2/15 |
| Fair/Poor | 0/15 |
| The way disease risk categories were presented | |
| Excellent | 8/15 |
| Good | 5/15 |
| Fair | 1/15 |
| Poor | 0/15 |
| The way risks and benefits were presented | |
| Excellent | 9/15 |
| Good | 5/15 |
| Fair | 1/15 |
| Poor | 0/15 |
| The way questions and answers were presented | |
| Excellent | 8/15 |
| Good | 5/15 |
| Fair | 2/15 |
| Poor | 0/15 |
| Length of presentation | |
| Just right | 13/15 |
| Too long | 2/15 |
| Too short | 0/15 |
| Amount of information | |
| Just right | 12/15 |
| Too much | 3/15 |
| Too little | 0/15 |
| I found the DA | |
| Slanted toward certain categories | 14/15 |
| Balanced | 1/15 |
| Clarity of Information | |
| Everything clear | 5/15 |
| Most things clear | 8/15 |
| Balanced | 1/15 |
| Some unclear | 1/15 |
| Many unclear | 0/15 |
| Enough information to make decision | |
| Yes | 15/15 |
| Recommend DA to others | |
| Definitely recommend | 9/15 |
| Probably recommend | 6/15 |
| Probably/Definitely not recommend | 0/15 |
Perceptions of the binning framework
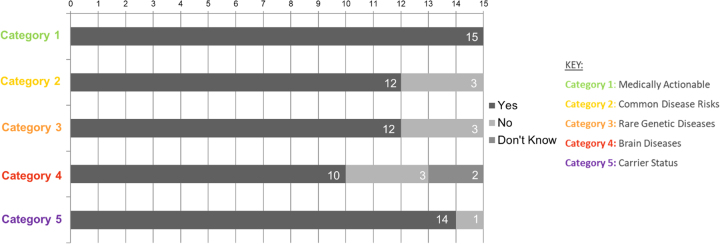
Participants were able to use the binning framework to hypothetically deliberate over which categories of IR they would select. A major theme that emerged from qualitative analysis was patients’ enthusiasm for GS and IR; indeed, all participants selected to receive some IR (Table 3, Fig. 2). The motivations for learning IR included being able to engage in disease prevention, plan for the future, share disease risks with family members, as well as a values-based stance on the perceived importance of the information. These motivations varied across categories and were modulated by past personal or family health experiences, and age or stage of life of the participant. For example, Category 1 was seen as “very important” (P13) and was selected by all participants, primarily because of the possibility to prevent or treat disease based on results. Participants’ perceptions of results from Category 2—small changes in genetic risk that could be mitigated by lifestyle changes—varied. Some saw this as valuable information that would motivate risk-reducing behaviors, whereas others dismissed its value since “diet and exercise, well that’s something that everybody should be focusing on anyway” (P8). Some participants perceived Category 3 (rare Mendelian disorders) as unclear, largely due to the unfamiliar nature of diseases. Nonetheless, many still selected it to inform relatives or to plan for the future, citing the ability to be “financially prepared” (P10) or “not living in a three-story walk-up if I am going to have a spinal disease” (P10), or to “read about [the disease] in case something new comes up” (P8). Category 4 (early-onset brain diseases) was well understood but perceived as “scary” (P5). Participants who did not choose Category 4 worried these results would cause excessive anxiety and rumination (it would “eat away at me” [P13]) due to the severity and untreatable nature of the diseases. Participants who selected Category 4 did so because “you can plan for it,” (P1) and “to inform your family because they are going to be in some ways more impacted than you are” (P5). Many participants perceived value in learning Category 5 (carrier) results as the information “could affect your decision for having children, and then of course letting your family and relatives know” (P1). Some participants were particularly anxious about non-preventable diseases or the possibility that insurance companies “won’t give you a policy or will give you a ridiculous price” (P2).
Table 3.
Participants’ quotes regarding the categories of IR from usability testing (Stage 5)
| Category | Illustrative quote | Illustrative quote | Illustrative quote |
|---|---|---|---|
| Category 1: Medically actionable | I think the first category […] there was no pausing on that one, […] I would want to know that I […] had that gene, and what […] could be done, because if there is surgery, or there [are] things that can be done ahead of time to deal with it, that’s the type of person I would be.’ (P15) | “ 'How important is it for you to learn about disease risks that you can reduce by taking medications, screening tests or having surgery?' I am going to say very important for that.” (P13) | “You know, none of the speaking changed my decision on how I would react to [categories] one and two, I would want them no matter what.” (P15) |
| Category 2: Common disease risks | “ 'How important is it for you to learn about your risk for common diseases you can reduce with a healthy diet and exercise?' Everybody knows healthy diet and exercise can already do that […]. So […] not very important for me.” (P13) | “Category two is the category which will tell me if I am at risk for common diseases, so yes, definitely” (P1) | “I still want to know if I am going to have diabetes, obviously I am not going to have that extra donut tonight. I mean, if I am going to have something. I still think even if you say, I can’t, interventions are not available, I still think there are things you can do.” (P10) |
| Category 3: Rare genetic diseases | “Even though they are not preventable I want to know, what I am in for. If something is in my future I can sort of plan ahead of time, so I will be comfortable.” (Pt 010) | “[…] These are not going to be diseases that are treatable or preventable. But it would mean that I could read about them in case something new comes up.” (P8) | “You know, […] if there is nothing I can do about it, I don’t really want to to know about it.' (P13) |
| Category 4: Brain diseases | “[…] Moving on to category four, oh gosh, this is even harder because it deals with brain diseases. […] I guess what makes it important is you can plan for it.” (P1) | “I don’t know, I just don’t see why I really need that information. I’d rather just live my life, you know, just continue on the way I am […] Eventually I’ll get over it but it’s still going to eat away at me, like it’s going to sit there in the back of my brain and if there is nothing I can do about it I just, I don’t want to know about it.” (P13). | “So finding out is a kind of, how do you plan for having Alzheimer’s? And then, maybe you don’t have [it]. And I think that’s one of the more anxiety [provoking], especially for somebody who is middle age or approaching middle age.” (P9) |
| Category 5: Carrier status | “…Okay, so I am moving to category five. Okay. So this would be very important because you I would be I carrier. So I would say, yeah of course it could affect your decision for having children, and then of course letting your family and relatives know if that’s important, even though [it] wouldn’t be easy to do.” (P1) | “Then you can prepare your children for it because I have a son […] I would want to let him know everything possible that, again, could make his life the best that it could be.” (P15) | “It is not applicable to me today. But I’d still like to know myself, even for my own personal reasons, I would say somewhat important. I want to know because if I have it my brother may have it still, or family [members]. I think it is important that we all share this information, whatever it is.” (P10) |
Fig. 2.
Participants’ selections of categories of incidental results (IR) from usability testing (Stage 5)
Final version of DA: Genomics ADvISER
At the end of this iterative process we produced a patient-centered, web-based tool, the Genomics ADvISER (www.genomicsadviser.com), in which audio, visual, text, and graphic components are integrated to aid comprehension and enhance user experience. Genomics ADvISER consists of four stages (Fig. 3): Learn, Explore, Prepare, and Decide.
Fig. 3.
The steps of the final decision aid (DA), The Genomics ADvISER. Adapted with permission from Shickh et al. [49]
In Learn, users watch a 10-min whiteboard video, which reviews GS and introduces the categories of IR. The categories of IR are presented throughout the DA as Category 1: Medically actionable, Category 2: Common disease risks, Category 3: Rare genetic diseases, Category 4: Brain diseases and Category 5: Carrier status. Following the video, users review the risks and benefits of the categories of IR. Users are then presented with a visual summary of the genetic risk (high, moderate, low) associated with results in each category, and the medical actionability of results in each category.
In Explore, users answer a series of values questions that allow them to review which risks and benefits of each category are important to them. After exploring their values, users indicate which categories they are thinking about selecting and view a summary of their preferences.
In Prepare users are quizzed on the key features of each category of IR. Next, they have the option to review the section of the video that describes each results category. For any question they answered incorrectly in the quiz, the DA suggests which part of the video they should review.
Lastly, the DA identifies users’ decision-making needs using the SURE test [42] and, depending on their answers, provides feedback about where they may need support. In the final choice step, Decide, users can view the video again if desired, and then select the categories they would like to receive. Their choice selections are then presented back to them on a summary page.
Discussion
We have created an innovative, patient-centered tool to support the clinical deployment of GS and mobilize patients’ informed decisions for receiving IR. The Genomics ADvISER is one of the first tools for use in the context of pre-test GS covering all categories of IR. The creation of this tool is timely; as GS is poised to become a part of clinical care [43, 44], the generation of IR will only increase. Returning IR to patients shifts the nature of the patient’s relationship with healthcare professionals by increasing the patient’s need to understand a large volume of choices and make informed decisions. The limited genomics expertise coupled with the volume and complexity of potential results makes pre-counseling each patient infeasible and beyond the capacity of our healthcare system, indicating a need for decision support tools. Our results suggest that Genomics ADvISER is effective at supporting informed decision-making; users with little or no prior knowledge of GS engaged in thoughtful and in-depth conversations about the different categories of IR and their utility. Participants were able to deliberate over the benefits and risks associated with learning IR and frame these risks and benefits in the context of their own stage of life, family dynamics and medical history. All users indicated that the Genomics ADvISER provided enough information for them to make a decision about which IR to receive. If used as an adjunct to standard counseling, the Genomics ADvISER could equip patients for more informed clinical conversations, alleviating some of the burden of information delivery and decision support from providers when engaging patients in informed consent or pre-test counseling. The Genomics ADvISER has the potential to improve the quality of patients’ decisions and ultimately save clinicians’ counseling time and health care costs.
We gained insight into the ways that patients’ values of IR may converge and also conflict with those of practitioners. First, medically actionable results seemed to be most highly valued among participants, as that was the only category selected by all participants. This is higher than would be expected from the literature, which suggests that most but not all participants elect to learn medically actionable IR [2]. However, this may be a product of our small sample size or may be due to the fact that real-life choices regarding genetic tests can be more conservative than hypothetical interest [45]. Nevertheless, participants’ interest in these results concurs with ACMG guidance recommending the return of only medically actionable IR [1]. However, participants were also interested in learning a wider range of results, as all chose to receive more IR than those considered medically actionable. This is consistent with an emerging literature which demonstrates interest in non-medically actionable results and the value that patients place in participating in the decision of what results to learn [2, 3]. The divergence between patient preferences and guidelines may cause gatekeeping challenges for providers if they are positioned to offer only medically actionable IR, and variations in access to information for patients.
Participants’ perceptions of utility also appeared to diverge from clinical expectations. Participants placed an overriding value in knowledge and information, particularly genetic information. Participants’ motivations to learn IR reflected a general enthusiasm towards GS and IR but also notions of personal utility [46], since they cited actions they could take in response to IR that included changing future plans, preparing for the impact of disease and informing family members. An understanding of participants’ values beyond clinical utility could guide clinicians’ pre-test conversations with patients undergoing GS and support shared-decision making [46]. Indeed, participants traded-off perceived clinical or lifestyle benefits and informing life-plans with the potential for distress when deliberating their choices of IR. Taken together, these results support the value-sensitive nature of these decisions, and more importantly, the need for decision support tools such as the Genomics ADvISER.
Consistent with others [47], we found that benefit to relatives was a prominent motivation for selection across all categories of IR. Enthusiasm to share results with family members coupled with the fact that patients will likely receive multiple IR could lead to a significant demand for testing among relatives, and possible misuse if misunderstandings exist. For example, if participants relay this information to the side of the family that does not carry the variant, unnecessary anxiety could ensue. This has been found to occur in other contexts [48]. Research is needed to assess such potential consequences of returning IR.
Our study had several limitations. While our sample size was sufficient for usability testing, it consisted of mainly older, highly educated females of European descent. We tested our DA in a population of participants making a hypothetical decision. This tool needs to be evaluated among patients making real decisions about IR and in more diverse populations. While others also found that participants using DAs prefer to speak with a practitioner [13], it is unclear if the Genomics ADvISER can be used as adjunct to, or replacement of, some genetic counseling functions. It is also unclear at this point whether the Genomics ADvISER would save clinicians’ time, While we did measure the time it took participants to compete the Genomics ADvISER (average 20 min) our study design did not have a comparison group, therefore time saved remains to be evaluated. We are evaluating the Genomics ADvISER compared to genetic counseling alone as part of a randomized controlled trial to address these questions and validate the tool [49].
Despite these limitations, we created a novel, patient-centered decision tool to support patients’ decision-making for IR from GS, which through usability testing was found to be acceptable to patients, effective for delivering a substantial amount of information and sufficient for them to make an informed hypothetical decision. Once clinically validated, this tool could mobilize informed consent and support the clinical delivery of GS and ultimately, save health care costs.
Electronic supplementary material
Acknowledgements
This research was supported by the Canadian Institutes of Health Research (CIHR) and the University of Toronto McLaughlin Centre. JGH, KO, and MER were supported by NCI P30 CA008748. We would like to thank Rohini Rau Murthy, Marina Corinnes, Melissa Salerno, Shari Gitlin, and Chelsea LeBlang for their contributions to the cognitive interviews. We are grateful to the following people for their feedback on the DA, video script and animation: Raymond Kim, Melyssa Aronson, June Carroll, Christian Storm, Jina Saikia, and Brian Lila. We thank Helios Design Labs for their work designing and programing the DA and our study participants for their time and valuable input.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0144-0) contains supplementary material, which is available to authorized users.
References
- 1.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SFv2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–55. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 2.Fiallos K, Applegate C, Mathews DJH, Bollinger J, Bergner AL, James CA. Choices for return of primary and secondary genomic research results of 790 members of families with Mendelian disease. Eur J Hum Genet. 2017;25:530–7. doi: 10.1038/ejhg.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clift KE, Halverson CME, Fiksdal AS, Kumbamu A, Sharp RR, McCormick JB. Patients’ views on incidental findings from clinical exome sequencing. Appl Transl Genom. 2015;4:38–43. doi: 10.1016/j.atg.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan N, Amendola LM, O’Daniel JM, et al. Is “incidental finding” the best term?: a study of patients’ preferences. Genet Med. 2017;19:176–81. doi: 10.1038/gim.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vears DF, Senecal K, Clarke AJ, et al. Points to consider for laboratories reporting results from diagnostic genomic sequencing. Eur J Hum Genet. 2018;26:36–43. doi: 10.1038/s41431-017-0043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van El CG, Cornel MC, Borry P, et al. Whole-genome sequencing in health care: recommendations of the European Society of Human Genetics. Eur J Hum Genet. 2013;21:580–4. doi: 10.1038/ejhg.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roche MI, Berg JS. Incidental findings with genomic testing: implications for genetic counseling practice. Curr Genet Med Rep. 2015;3:166–76. doi: 10.1007/s40142-015-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner C. Anticipate and communicate: ethical management of incidental and secondary findings in the clinical, research, and direct-to-consumer contexts (December 2013 report of the Presidential Commission for the Study of Bioethical Issues) Am J Epidemiol. 2014;180:562–4. doi: 10.1093/aje/kwu217. [DOI] [PubMed] [Google Scholar]
- 9.Smith-Bindman R. Use of advanced imaging tests and the not-so-incidental harms of incidental findings. JAMA Intern Med. 2018;178:227–8. doi: 10.1001/jamainternmed.2017.7557. [DOI] [PubMed] [Google Scholar]
- 10.Institute of Medicine. Evolution of translational omics: lessons learned and the path forward. Washington, DC: The National Academies Press; 2012. [PubMed] [Google Scholar]
- 11.American College of Medical Genetics and Genomics. Incidental findings in clinical genomics: a clarification. Genet Med. 2013;15:664-6. [DOI] [PubMed]
- 12.Birch PH. Interactive e-counselling for genetics pre-test decisions: where are we now? Clin Gen. 2015;87:209–17. doi: 10.1111/cge.12430. [DOI] [PubMed] [Google Scholar]
- 13.Birch P, Adam S, Bansback N, et al. DECIDE: a decision support tool to facilitate parents’ choices regarding genome-wide sequencing. J Genet Couns. 2016;25:1298–308. doi: 10.1007/s10897-016-9971-8. [DOI] [PubMed] [Google Scholar]
- 14.Sanderson SC, Suckiel SA, Zweig M, Bottinger EP, Jabs EW, Richardson LD. Development and preliminary evaluation of an online educational video about whole-genome sequencing for research participants, patients, and the general public. Genet Med. 2016;18:501–12. doi: 10.1038/gim.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonter K, Desjardins C, Currier N, Pun J, Ashbury FD. Personalised medicine in Canada: a survey of adoption and practice in oncology, cardiology and family medicine. BMJ Open. 2011;1:e000110. doi: 10.1136/bmjopen-2011-000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stacey D, Légaré F, Col NF et al. Decision aids for people facing health treatment or screening decisions. Cochrane Database Syst Rev. 2014;28:CD001431. 10.1002/14651858.CD001431.pub4. [DOI] [PubMed]
- 17.Stacey D, Légaré F, Lewis K et al. Decision aids for people facing health treatment or screening decisions. Cochrane Database Syst Rev. 2017. [DOI] [PMC free article] [PubMed]
- 18.Kuppermann M, Norton ME, Gates E, et al. Computerized prenatal genetic testing decision-assisting tool: a randomized controlled trial. Obstet Gynecol. 2009;113:53–63. doi: 10.1097/AOG.0b013e31818e7ec4. [DOI] [PubMed] [Google Scholar]
- 19.Légaré F, Robitaille H, Gane C, Hébert J, Labrecque M, Rousseau F. Improving decision making about genetic testing in the clinic: an overview of effective knowledge translation interventions. PLoS ONE. 2016;11:e0150123. doi: 10.1371/journal.pone.0150123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabor HK, Jamal SM, Yu JH, et al. My46: a web-based tool for self-guided management of genomic test results in research and clinical settings. Genet Med. 2017;19:467–75. doi: 10.1038/gim.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor A. Ottawa decision support framework to address decisional conflict. Ottawa Health Research Institute; Ottawa, ON, Canada 2006. http://www.ohri.ca/decisionaid/
- 22.O’Connor A, Stacey D, Boland L. Ottawa decision support tutorial. Ottawa Hospital Research Institute; Ottawa, ON, Canada 2015. http://www.ohri.ca/decisionaid/
- 23.Stacey D, O’Connor A, Legare F, et al. Ottawa decision support framework: update, gaps, and research priorities. Ottawa Hospital Research Institute; Ottawa, ON, Canada 2010. http://www.ohri.ca/decisionaid/
- 24.O’Connor AM, Tugwell P, Wells GA, et al. A decision aid for women considering hormone therapy after menopause: decision support framework and evaluation. Patient Educ Couns. 1998;33:267–79. doi: 10.1016/S0738-3991(98)00026-3. [DOI] [PubMed] [Google Scholar]
- 25.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med. 2011;13:499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 26.Elwyn G, O’Connor A, Stacey D, et al. Developing a quality criteria framework for patient decision aids: online international Delphi consensus process. BMJ. 2006;333:417. doi: 10.1136/bmj.38926.629329.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coyne IT. Sampling in qualitative research. Purposeful and theoretical sampling; merging or clear boundaries? J Adv Nurs. 1997;26:623–30. doi: 10.1046/j.1365-2648.1997.t01-25-00999.x. [DOI] [PubMed] [Google Scholar]
- 28.Kushniruk AW, Patel VL. Cognitive and usability engineering methods for the evaluation of clinical information systems. J Biomed Inform. 2004;37:56–76. doi: 10.1016/j.jbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Google Design. Google, 2014. Accessed on October 2015. https://design.google/ (2017).
- 30.Kushniruk AW, Patel VL, Cimino JJ. Usability testing in medical informatics: cognitive approaches to evaluation of information systems and user interfaces. Proc AMIA Annu Fall Symp. 1997:218–22. [PMC free article] [PubMed]
- 31.What and Why of Usability. U.S. Department of Health and Human Services, 2013. Accessed on October 2015. https://www.usability.gov/ (2016).
- 32.Berry DL, Halpenny B, Bosco JLF, Bruyere J, Jr., Sanda MG. Usability evaluation and adaptation of the e-health personal patient profile-prostate decision aid for Spanish-speaking Latino men. BMC Med Inform Decis Mak. 2015;15:56. doi: 10.1186/s12911-015-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis MA, Paquin RS, Roche MI, et al. Supporting parental decisions about genomic sequencing for newborn screening: the NC NEXUS decision aid. Pediatrics. 2016;137:S16–23. doi: 10.1542/peds.2015-3731E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldman-Stewart D, Brundage MD, Van Manen L. A decision aid for men with early stage prostate cancer: theoretical basis and a test by surrogate patients. Health Expect. 2001;4:221–34. doi: 10.1046/j.1369-6513.2001.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neri PM, Pollard SE, Volk LA, et al. Usability of a novel clinician interface for genetic results. J Biomed Inform. 2012;45:950–7. doi: 10.1016/j.jbi.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devine EB, Lee CJ, Overby CL, et al. Usability evaluation of pharmacogenomics clinical decision support aids and clinical knowledge resources in a computerized provider order entry system: a mixed methods approach. Int J Med Inform. 2014;83:473–83. doi: 10.1016/j.ijmedinf.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grim K, Rosenberg D, Svedberg P, Schon UK. Development and usability testing of a web-based decision support for users and health professionals in psychiatric services. Psychiatr Rehabil J. 2017;40:293–302. doi: 10.1037/prj0000278. [DOI] [PubMed] [Google Scholar]
- 38.User Manual—Acceptability. The Ottawa Hospital Research Institute, 1996. https://decisionaid.ohri.ca/docs/develop/user_manuals/UM_acceptability.pdf (2016).
- 39.Hsieh HF, Shannon SE. Three approaches to qualitative content analysis. Qual Health Res. 2005;15:1277–88. doi: 10.1177/1049732305276687. [DOI] [PubMed] [Google Scholar]
- 40.Charmaz KC. Constructing grounded theory: a practical guide through qualitative analysis. London: SAGE Publications Ltd.; 2006. [Google Scholar]
- 41.Metcalfe K, Poll A, O’Connor A, et al. Development and testing of a decision aid for breast cancer prevention for women with a BRCA1 or BRCA2 mutation. Clin Gen. 2007;72:208–17. doi: 10.1111/j.1399-0004.2007.00859.x. [DOI] [PubMed] [Google Scholar]
- 42.Légaré F, Kearing S, Clay K, et al. Are you SURE?: Assessing patient decisional conflict with a 4-item screening test. Can Fam Physician. 2010;56:e308–e14. [PMC free article] [PubMed] [Google Scholar]
- 43.Bombard Y. Translating personalized genomic medicine into clinical practice: evidence, values, and health policy. Genome. 2015;58:491–7. doi: 10.1139/gen-2015-0145. [DOI] [PubMed] [Google Scholar]
- 44.Iglesias A, Anyane-Yeboa K, Wynn J, et al. The usefulness of whole-exome sequencing in routine clinical practice. Genet Med. 2014;16:922–31. doi: 10.1038/gim.2014.58. [DOI] [PubMed] [Google Scholar]
- 45.Sanderson SC, O’Neill SC, Bastian LA, Bepler G, McBride CM. What can interest tell us about uptake of genetic testing? Intention and behavior amongst smokers related to patients with lung cancer. Public Health Genom. 2010;13:116–24. doi: 10.1159/000226595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohler JN, Turbitt E, Biesecker BB. Personal utility in genomic testing: a systematic literature review. Eur J Hum Genet. 2017;25:662–8. doi: 10.1038/ejhg.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malek J, Slashinski MJ, Robinson JO, et al. Parental perspectives on whole-exome sequencing in pediatric cancer: a typology of perceived utility. JCO Precis Oncol. 2017;1:1–10. doi: 10.1200/PO.17.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bombard Y, Miller FA, Barg CJ, et al. A secondary benefit: the reproductive impact of carrier results from newborn screening for cystic fibrosis. Genet Med. 2017;19:403–11. doi: 10.1038/gim.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shickh S, Clausen M, Mighton C et al. Evaluation of a decision aid for incidental genomic results: protocol for a mixed-methods randomized controlled trial. BMJ Open. 2018. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.