Abstract
Mutations in C2orf71 are causative for autosomal recessive retinitis pigmentosa and occasionally cone-rod dystrophy. We have recently discovered that the protein encoded by this gene is important for modulation of the ciliary membrane through the recruitment of an actin assembly module, and have therefore renamed the gene to PCARE (photoreceptor cilium actin regulator). Here, we report on the identification of two copies of the c2orf71/pcare gene in zebrafish, pcare1 and pcare2. To study the role of the gene most similar to human PCARE, pcare1, we have generated a stable pcare1 mutant zebrafish model (designated pcare1rmc100/rmc100) in which the coding sequence was disrupted using CRISPR/Cas9 technology. Retinas of both embryonic (5 dpf) and adult (6 mpf) pcare1rmc100/rmc100 zebrafish display a clear disorganization of photoreceptor outer segments, resembling the phenotype observed in Pcare−/− mice. Optokinetic response and visual motor response measurements indicated visual impairment in pcare1rmc100/rmc100 zebrafish larvae at 5 dpf. In addition, electroretinogram measurements showed decreased b-wave amplitudes in pcare1rmc100/rmc100 zebrafish as compared to age- and strain-matched wild-type larvae, indicating a defect in the transretinal current. Altogether, our data show that lack of pcare1 causes a retinal phenotype in zebrafish and indicate that the function of the PCARE gene is conserved across species.
Introduction
Retinitis pigmentosa (RP, MIM 268000) is an inherited retinal disease (IRD) characterized by progressive visual impairment due to the loss of rod photoreceptors. The prevalence of RP in the population ranges between 1:3,000 and 1:5,000, making it the most common form of IRD. In 2010, we and others reported that mutations in C2orf71 underlie non-syndromic autosomal recessive RP (arRP)1,2. Consecutive studies detected additional disease-causing missense and nonsense mutations along its entire coding sequence in cases with RP or cone-rod dystrophy (CRD)3–8. Of interest, C2orf71 mutations represent a frequent cause of arRP in the Swiss population9.
C2orf71 is almost exclusively expressed in the retina1 and codes for a ciliary protein that consists of 1,288 amino acids. In the photoreceptor cell, actin localizes at the base of the outer segments (OS), where OS disk neogenesis is initiated10. In C2orf71−/− mice, the OS are shortened and disorganized, and are marked by a mislocalization of proteins such as rhodopsin and cone opsins11. Recently, we have discovered that the protein encoded by C2orf71 recruits a ciliary actin dynamics module that modifies the ciliary membrane, and hypothesize that this principle may be the driver of photoreceptor OS disk morphogenesis (manuscript submitted). Hence, we have renamed the C2orf71 gene to PCARE which stands for photoreceptor cilium actin regulator. If PCARE plays part of a conserved actin-related function for the process of vision, we may expect a similar function for this gene in a non-mammalian organism.
The zebrafish, Danio rerio, is a tropical freshwater fish that is commonly used in IRD research12–14. Advantages of using zebrafish for such studies are mainly their short life cycle and the large number of offspring. In addition, their ex utero development allows for easy tracking of eye development15. Despite being genetically distant to humans, zebrafish eyes share some similarities in morphology and physiology to human eyes. Like humans, zebrafish are also diurnal animals, and their retinas present a similar layered structure16. A whole-genome duplication event that occurred during the divergence of the teleost fish17,18 made the present day zebrafish genome to contain many duplicated genes, of which over 64 corresponding to genes mutated in IRD, including pcare15.
Transient morpholino-based knockdown of one of the two copies of pcare in zebrafish, pcare1, resulted in shortening of photoreceptor outer segments and visual dysfunction2. However, over the recent years, the specificity of morpholino-based knockdown studies has been debated, mostly due to non-specific binding that may occur19. In addition, the transient nature of the knock-down does not allow to study long-term effects.
Here, we have generated a stable pcare1 zebrafish mutant using CRISPR/Cas9 technology to investigate the effect of the absence of pcare in a non-mammalian model, and demonstrate that, like in humans, the pcare gene is essential for retinal function.
Results
Pcare is duplicated in the zebrafish genome
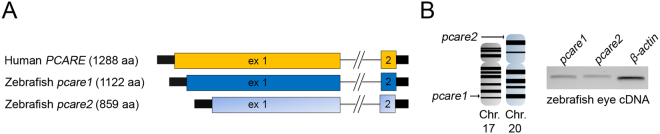
The zebrafish pcare paralogues, pcare1 and pcare2, were identified by bioinformatic analysis using the software programs BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Ensembl (https://www.ensembl.org/). Reciprocal BLAST searches revealed that both genes are true orthologues of the human PCARE gene. Bioinformatic analysis revealed that pcare1 is located on chromosome 17 and codes for a protein of 1122 amino acids, while pcare2 is found on chromosome 20 and encodes a protein of 859 amino acids (Fig. 1). A synteny between part of the zebrafish chromosome 17 and the human chromosome 2 on which PCARE is located, suggests that pcare1 is the true orthologue of the human counterpart. To study the expression of these genes, we performed RT-PCR on wild-type zebrafish eye cDNA at 5 days post fertilization (dpf). We observed that both genes are expressed in the zebrafish eye (Fig. 1B). Because of the higher similarity of pcare1 with the human PCARE protein sequence (32.1% of pcare1 vs 25.6% of pcare2, see Supplementary Figure S1) we decided to disrupt the pcare1 gene.
Figure 1.
Pcare is duplicated in the zebrafish genome. (A) Graphical representation of the human PCARE gene and the two orthologous zebrafish genes, pcare1 and pcare2. (B) Genomic location of the duplicated pcare genes in zebrafish. pcare1 is located in chromosome 17 and pcare2 is found in chromosome 20 (left). Expression analysis using zebrafish eyes cDNA of pcare1 and pcare2 (right). A full image of this gel can be found in the Supplementary Information file (Supplementary Figure S3).
Generation of a pcare1 mutant zebrafish line
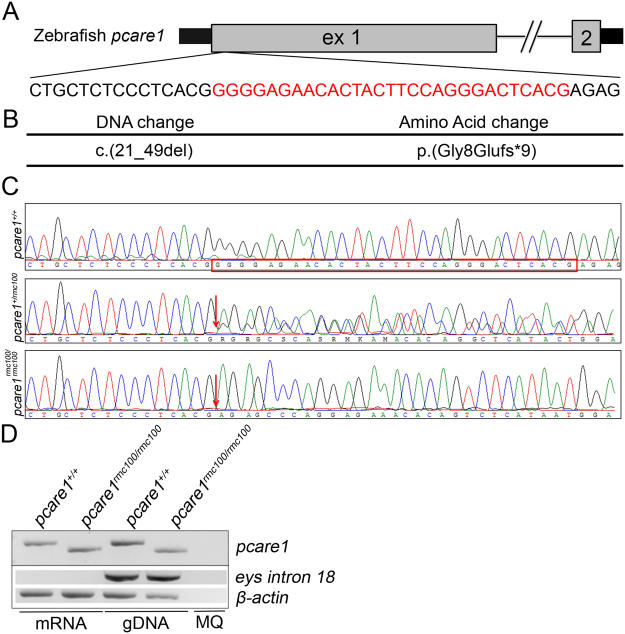
For the generation of pcare1 mutant zebrafish, a pcare1 sequence-specific guide RNA was designed, and co-injected with Cas9 mRNA into single cell zebrafish embryos. Mosaic fish containing indels around the target region were then out-crossed with wild-type fish to determine germline transmission and the exact genomic lesion that was introduced. We identified four adult F1 fish containing a heterozygous 29-basepair deletion in exon 1 of pcare1 (c. 21_49del) (Fig. 2A–C). Subsequently, heterozygous pcare1 carriers were mated to generate homozygous mutant fish, named pcare1rmc100/rmc100, and subsequently crossed for a number of generations. Homozygous mutant fish did not show any signs of gross morphological abnormalities. The 29-basepair deletion is predicted to result in premature termination of translation after amino acid 16 of pcare1 (p.Gly8Glufs*9), most likely resulting in a functional null allele. RT-PCR analysis showed that mutant mRNA is expressed in the pcare1 mutants, indicating that it does not, or at least not completely, undergo nonsense-mediated decay (Fig. 2D). Staining with different commercial and home-made antibodies targeting the pcare protein in zebrafish did not yield any specific signals, nor any differences between wild-type and pcare1rmc100/rmc100 mutant fish (data not shown). These results suggest that the antibodies targeting human PCARE do not recognize the zebrafish protein, most likely due to the low conservation between the human and zebrafish PCARE sequences (Supplementary Figure S1).
Figure 2.
Generation of pcare1rmc100/rmc100 zebrafish. (A) The 2 exons of zebrafish pcare1 gene are shown. In red, the sequence deleted by the CRISPR/Cas9 is indicated. (B) The detected DNA change (c.21_49del) leads to a 29 bp deletion causing a frame shift after aminoacid 8 of pcare1, predicted to result in a truncated protein (p.Gly8Glufs*9). (C) Sequence validation of the targeted region in pcare1+/+, pcare1+/rmc100 and pcare1rmc100/rmc100 zebrafish. The deleted 29 bp are marked in red in the pcare1+/+ sequence and absent from the homozygous sequence (red arrows indicate start of the deletion). (D) Upper panel: (RT-)PCR analysis of Pcare 1 in wild-type and Pcare1rmc100/rmc100 zebrafish, showing the presence of a shorter transcript in the mutant fish. Lower panels: analysis of eys (only present in genomic DNA) and β-actin (present in mRNA and genomic DNA) for comparison and as loading controls.
Pcare1rmc100/rmc100 zebrafish show aberrant photoreceptor morphology
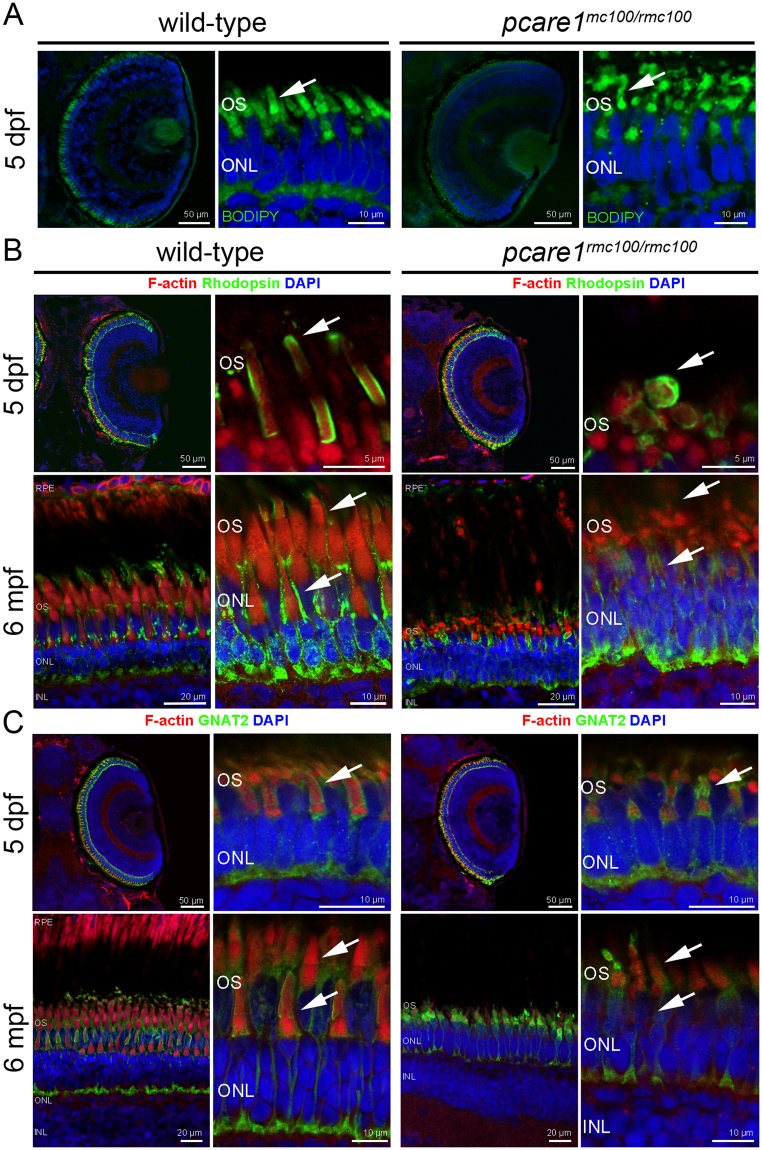
To analyze the morphology of the photoreceptor OS, pcare1rmc100/rmc100 and wild-type strain-matched embryos of 5 dpf were sectioned and stained with boron-dipyrromethene (BODIPY). We observed a pronounced dysmorphology of the outer segments in the pcare1rmc100/rmc100 zebrafish as compared to wild-type embryos (Fig. 3A). Since we recently discovered that the human PCARE protein is associated to actin, we also stained retinas, from embryonic (5 dpf) as well as adult (6 months post fertilization, mpf) fish, with an antibody against F-actin. The localization of actin further supported the disorganization of the OS (Fig. 3B,C) in the pcare1rmc100/rmc100 retinas. In addition, both the rod-specific protein rhodopsin and the cone-specific protein Gnat2 exhibited a different pattern in pcare1rmc100/rmc100 fish compared to wild-type fish, presumably affected by the change of morphology of the outer segments (Fig. 3B,C). Additionally, a reduction in the thickness of both outer and inner nuclear layers was observed in pcare1rmc100/rmc100 fish of 6 months of age (Supplementary Figure S2).
Figure 3.
Pcare1rmc100/rmc100 zebrafish show aberrant photoreceptor morphology. (A) Analysis of the morphology of the pcare1rmc100/rmc100 zebrafish larval retina (5 dpf) using boron-dipyrromethene (BODIPY) revealed disorganization of photoreceptor outer segments as compared to those of strain- and age-matched wild-type larvae (arrows). (B) Siblings of adult zebrafish without (wild-type) or containing (pcare1rmc100/rmc100) the 29 bp deletion in pcare1 were sectioned and stained with antibodies against F-actin (red), Rhodopsin (green) or, in (C) GNAT2 (green). Arrows indicate normal outer segments in control fish and dysmorphic outer segments in mutant fish. Nuclei were stained with DAPI.
Pcare1rmc100/rmc100 larvae are visually impaired
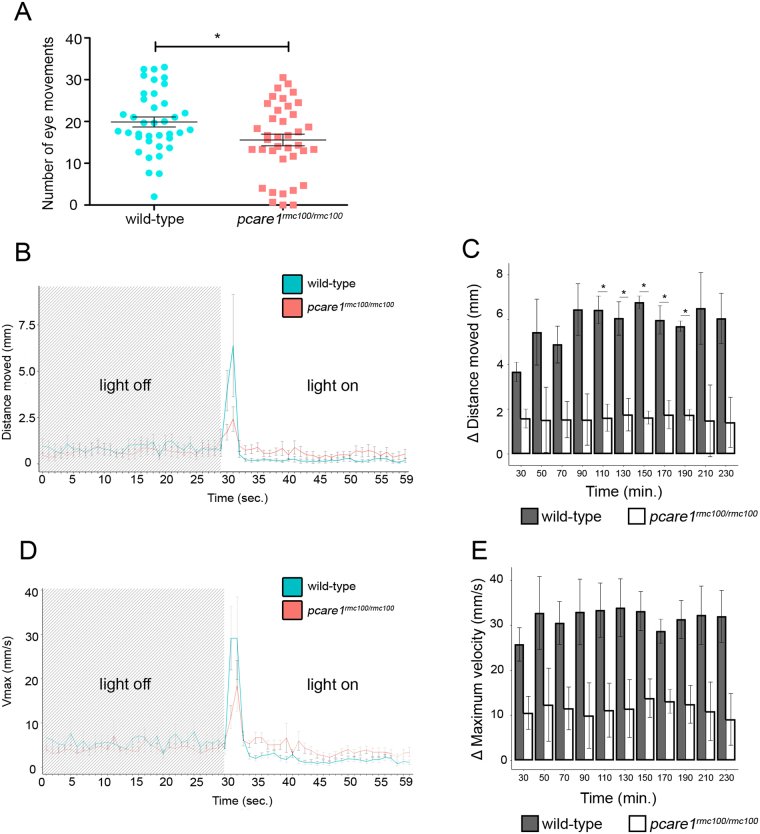
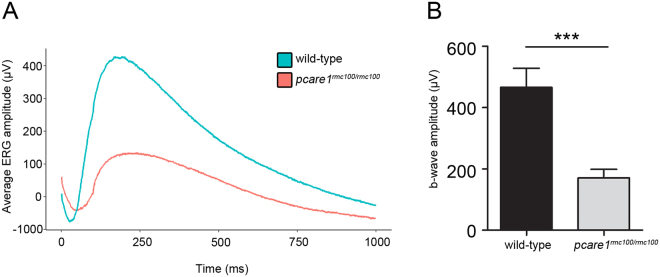
To study the response of the zebrafish to visual stimuli, we performed optokinetic response (OKR) and visual motor response (VMR) measurements in wild-type and pcare1rmc100/rmc100 larvae at 5 dpf. OKR analysis, that measures eye movements in a rotating drum, showed a significant decrease in the number of movements in pcare1rmc100/rmc100 larvae compared to wild types (Fig. 4A). Twenty-one percent of the mutant larvae responded with less than 5 eye movements, which can be considered as spontaneous movements, while 5% of the mutant larvae showed no response at all (Fig. 4A). For VMR measurements, we employed the DanioVision system that automatically measures the activity of animals following a series of dark-to-light transitions. Our analysis showed a major decrease of the distance moved after the dark-to-light transitions in pcare1rmc100/rmc100 fish compared to wild-type larvae (Fig. 4B,C). The velocity of the movements was also higher in the wild-type larvae (Fig. 4D,E), though it was not statistically significant. To determine the response of the outer retina to light stimuli, we performed ERG measurements in dark-adapted zebrafish larvae at 5 dpf. The b-wave response, which measures the action potentials of ON bipolar cells20, was drastically reduced in pcare1rmc100/rmc100 compared to age-matched control larvae (Fig. 5).
Figure 4.
Pcare1rmc100/rmc100 zebrafish are visually impaired. (A) Optokinetic response (OKR) measurements revealed a decrease in the number of eye movements of pcare1rmc100/rmc100 larvae (n = 38) compared to control larvae (n = 38). (B) Representative graph showing the distance moved (maximum velocity in (D)) of the larvae between light-off to light-on change in a one-minute interval. (C) Graphs showing the differences in the distance moved (maximum velocity in (E)) between wild-type and pcare1rmc100/rmc100 larvae in different time points and combining three independent experiments. Bars indicate the standard error of the mean. P-values are corrected for multiple testing using Benjamini-Hochberg method; the comparisons marked with an asterisk showed statistical significance (*)p-value < 0.05.
Figure 5.
Pcare1rmc100/rmc100 mutant zebrafish show reduced ERG responses. (A) The average ERG amplitude to light stimuli of wild-type larvae (n = 15) and pcare1 mutant larvae (n = 17) was measured. (B) Comparison of maximal b-wave amplitudes between wild-type and pcare1rmc100/rmc100 zebrafish larvae. The b-wave amplitude is significantly reduced in pcare1rmc100/rmc100 mutants compared to wild-type larvae; p-value = 0.0004. Bars indicate the standard error of the mean.
Discussion
In humans, mutations in the gene PCARE (formerly known as C2orf71) lead to phenotypes where either the rod photoreceptors, in case of arRP, or the cone photoreceptors, in arCRD, are primarily affected. The outer segments of photoreceptor cells are specialized sensory cilia composed of stacked membrane disks that contain all the necessary elements for phototransduction21. Very recently, we have found evidence that PCARE could be an important ciliary actin modulator postulated to drive outer segment disk morphogenesis (manuscript submitted). This study was done using a Pcare−/− mouse model and human hTERT RPE-1 cells. Here, we have generated a zebrafish pcare1 mutant (named pcare1rmc100/rmc100) in order to obtain a better understanding of the function of pcare in a non-mammalian vertebrate. Based on the data presented here, it seems that the function of PCARE is conserved, as the photoreceptor outer segments are disorganized in pcare1rmc100/rmc100 zebrafish.
The age of disease onset in patients with PCARE mutations ranges from the first to the sixth decade9. We observed that pcare1rmc100/rmc100 zebrafish show a worse response than wild-type zebrafish, both in a behavioral assay and after electrophysiological recordings; however, pcare1rmc100/rmc100 zebrafish are not completely blind. These observations correlate with the human phenotype, since patients do not become fully blind until a more advanced age. In mice, disruption of Pcare causes severe retinal dystrophy with an early onset, with flat ERG responses already at 8 weeks of age11. Behavioral analysis using DanioVision did not show statistically significant difference for velocity and some points of distance moved; however, we can observe a clearly different trend between wild-type and pcare1rmc100/rmc100 larvae. This observation correlates with the high variability in results using visual motor assays that other authors have reported22. Additionally, photoreceptive molecules in the zebrafish brain may mediate some of the behavioral responses to light23,24.
In our study, pcare1rmc100/rmc100 zebrafish ERG responses show a reduction in the b-wave amplitude as early as 5 dpf, similar to the phenotype observed in mice. Since rod photoreceptors are still not functional in zebrafish at 5 dpf 25,26, this indicates that cone photoreceptor cells and/or bipolar cells could be affected by depletion of pcare1, although we only measured b-wave responses that are derived from the bipolar cells. In addition, light detection through non-retinal tissues could be accounting for some of the responses.
Because the outer segments are not completely disrupted in pcare1rmc100/rmc100 zebrafish, we hypothesize that other proteins involved in outer segment disk formation may still be functional at their action site. Unlike for other vertebrates, the common ancestor from the teleost lineage underwent a whole-genome duplication event18. A consequence of this is the presence of 26,206 protein-coding genes, more than any other sequenced vertebrate species27. For some of the duplicated genes, their function has diverged since then28,29, while for others it has remained unchanged30. Interestingly, parts of zebrafish chromosome 17 and 20, where pcare1 and pcare2 are located, descent from the same pre-duplication in the ancestral chromosome. Although disruption of the pcare1 copy is sufficient to generate a strong retinal phenotype, there is still a possibility that pcare2 may be involved in a similar process within the photoreceptor cells. Thus, a more severe retinal defect could be expected if both gene copies are disrupted, and future studies will be needed to address the function of pcare2.
Alternatively, other proteins involved in photoreceptor outer segment formation or maintenance could be responsible for our observations. Examples of such proteins are the intraflagellar transport (IFT) protein complex and the motor kinesin-231, which are needed to maintain the extraordinary traffic of proteins along the connecting cilium. Zebrafish knockdown of IFT proteins causes severe retinal degeneration32. Ift57-deficient zebrafish mutants however are able to form a primitive photoreceptor connecting cilium, rod sacs and flattened outer segment disks, but the disks fail to acquire a proper length and subsequently the photoreceptors die32,33. Because of the similarities between the phenotypes of Ift57 mutants and pcare1rmc100/rmc100 zebrafish mutants, we hypothesize that a similar degenerative process of pcare1rmc100/rmc100 zebrafish outer segments might take place.
Contrary to mammals, where Müller glia cells rarely divide followed by retinal injury, retinal regeneration does occur in zebrafish34,35. When injury happens, Müller glia are able to reprogram into retinal stem cells36. We however observed a disorganization of the photoreceptor outer segments and degeneration of the outer nuclear layer in pcare1rmc100/rmc100 zebrafish, both at 5 dpf and at 6 mpf. Thus, the regeneration occurring in zebrafish is apparently not sufficient to halt the photoreceptor degeneration caused by absence of pcare1.
In summary, we have generated a pcare1-deficient zebrafish line using CRISPR/Cas9 technology. Analysis of the morphology and physiology of embryonic and adult pcare1rmc100/rmc100 zebrafish results in a clear retinal disease phenotype. We provide an additional excellent model organism to study retinal degeneration caused by PCARE mutations which will be useful to understand the disease mechanisms. Furthermore, our findings support the hypothesis of the involvement of PCARE in forming and/or maintaining photoreceptor outer segments and, thereby, proper vision.
Methods
Ethics statement
Animal experiments were conducted in accordance with the Dutch guidelines for the care and use of laboratory animals, with the approval of the local Animal Experimentation Committee (Dier Experimenten Commissie [DEC]) (Protocol #DEC 2016–0091).
Zebrafish maintenance
Experimental procedures were conducted in accordance with international and institutional guidelines. Wild-type adult Tupfel Long Fin (TLF) zebrafish were used. Zebrafish eggs were obtained from natural spawning of wild-type breeding fish. Larvae were maintained and raised by standard methods37. In brief, embryos were raised in E3 medium (5 mM NaCL, 0.17 mM KCL, 0.33 mM CaCl2, 0.33 mM MgSO4, supplemented with 0.1% methylene blue) in a 28 °C incubator with the same day/night cycle (14 h light / 10 h dark). The medium was changed on a daily basis and during the process curved and dead larvae were discarded. Functional and behavioral testing was carried out at 5 days post fertilization (dpf).
Target site selection and gRNA synthesis
Sites for targeted genome editing were selected using the online software ZiFiT Targeter version 4.2 (http://zifit.partners.org/ZiFiT/)38,39. Templates for gRNA transcription were generated by annealing gene-specific oligonucleotides containing the T7 (5′-TAATACGACTCACTATA-3′) promoter sequence, the 20-base target sequence without the PAM (5′-GGGGGAGAACACTACTTCCA-3′), and a complementary region to a constant oligonucleotide encoding the reverse complement of the tracrRNA tail. T4 DNA polymerase (New England Biolabs, Ipswich, Massachusetts, USA) was used to fill the ssDNA overhang and the template was purified using columns (Sigma, St. Louis, Missouri, USA). Transcription of the gRNAs was performed using the MEGAshortscript Kit (Ambion®, Thermo Fisher Scientific, Waltham, Massachusetts, USA).
Microinjections
Zebrafish embryos were collected and injected at single cell stage with 1 nl of a mix consisting of 4.5 µl gRNA (173,72 ng/ul), 2.5 µl Cas9 protein (2 µg/µl, PNA Bio Inc, Newbury Park, California, USA), 2 µl 1 M KCl and 1 µl 0.5% phenol red dye. To screen for genomic lesions, genomic DNA was extracted from a pool of 15 embryos at 2.5 dpf.
Genotyping PCR
After injections, genomic DNA was extracted from 2.5 dpf larvae or caudal fin tissue of adult zebrafish. Tissue was incubated in 25 µl (larvae) or 75 µl (fin tissue) lysis buffer (40 mM NaOH, 0.2 mM EDTA) for 20 min at 95 °C. The lysed samples were diluted 10 times after which 1 µl was incubated together with 0.5 µM of the forward and reverse primer, 100 µM dNTPs (Roche, Basel, Switzerland), 0.25 U Taq polymerase (Roche, Basel, Switzerland) and 10 × PCR buffer + 15 mM MgCl2 (Roche, Basel, Switzerland) in a total volume of 25 µl. Samples were denatured at 94 °C for 5 min followed by 35 cycles of amplification consisting of 30 seconds (sec) at 94 °C, 30 sec at 58 °C and 60 sec at 68 °C, followed by a final primer extension of 10 min at 72 °C. To screen for genomic lesions, PCR products were sequenced directly. Primer sequences are listed in Supplementary Table S1.
Establishment of a pcare1rmc100/rmc100 zebrafish line
Mosaic fish were out-crossed with wild-type TLF fish to determine germline transmission and the exact introduced genomic lesion. Heterozygous mutants were mated to generate homozygous pcare1 mutants, which were subsequently crossed for a number of generations. A pcare1rmc100/rmc100 zebrafish line was established containing a 29-basepair deletion at the start of exon 1 of pcare1 (c. 21_49del). This is predicted to lead to premature termination of translation after amino acid 16.
RT-PCR analysis
To analyze the expression of pcare1 and pcare2, we performed RT-PCR using cDNA extracted from wild-type zebrafish eyes. One microgram of total RNA was incubated with 1 µl iScript Reverse Transcriptase (1708891, Bio-Rad, Hercules, California, USA) and 1x reaction mix in a total volume of 20 µl nuclease free water. For the RT-reaction, the mixture was incubated for 5 min at 25 °C, 30 min at 42 °C and the reaction was stopped by heating at 85 °C for 5 min. PCR analysis was performed as described above. The primers used are listed in Supplementary Table S1.
Preparation of zebrafish sections
Larvae (5 dpf) were collected in an eppendorf tube and sedated with 2-phenoxyethanol (1:500). After one wash with PBS, embryos or adult eyes (6 mpf) were fixed in 4% PFA overnight, brought gradually to 100% MetOH and stored overnight at −20 °C. After the overnight incubation, samples were rehydrated (75% MetOH in 0.1% PBS-Tween20 (PBS-T), 50% MetOH in 0.1% PBS-Tween20, 25% MetOH in 0.1% PBS-Tween20 for 15 min each), followed by 15 min incubation in 0.1% PBS-Tween20. After this, 10% sucrose in 0.1% PBS-Tween20 was added, followed by an incubation step in 30% sucrose in 0.1% PBS-Tween20 for 1 h at room temperature (RT). Samples were embedded in O.C.T. compound (Tissue-Tek®, 25608-930, VWR, Radnor, Pennsylvania, USA) and snap frozen in isopentane cooled by liquid nitrogen. Cryosectioning was done following standard protocols (seven μm thickness along the lens/optic nerve axis).
Bodipy staining
To analyze outer segment morphology, cryostat sections of 5 dpf larvae were briefly washed with 1xPBS, permeabilized with 0.5% Triton X-100 in PBS, washed in PBS and incubated for 20 min with TR methyl ester (Bodipy, 1:5000, Life Technologies, Carlsbad, Califoria, USA) and DAPI (1:8000, ITK Diagnostics, Uithoorn, The Netherlands). Sections were washed in PBS, followed by a brief wash with MilliQ and mounted with ProLong(R) Gold antifade reagent (P36930, Life Technologies, Carlsbad, California, USA).
Immunohistochemistry
Slides were incubated for 2 min in 0.1% PBS-T. Antigen retrieval was done by autoclaving the slides in 10 mM sodium citrate for 1 min at 121 °C. After that, sections were washed three times in 0.1% PBS-T and blocked for 1 h in blocking solution (10% non-fat dry milk in 0.1% PBS-T). Primary antibody (Rhodopsin clone 4D2, mouse, 1:2000, Novus Biological NBP1-48334; GNAT2, rabbit, 1:500, MBL PM075) was incubated in blocking solution overnight at 4 °C. After primary antibody incubation, sections were washed in 0.1% PBS-T, followed by secondary antibody (Alexa Fluor 568® phalloidin, 1:100, Molecular Probes A-12380; Goat IgG, Alexa Fluor 488, donkey, 1:500, Molecular Probes A11055; Mouse IgG, Alexa Fluor 488, donkey, 1:500, Life Technologies A21202) incubation for 45 min in blocking solution. Afterwards, sections were washed in PBS-T and mounted with anti-fade Prolong(R) Gold antifade reagent (P36930, Life technologies, Carlsbad, California, USA). To analyze the thickness of ONL and INL in 6 mpf zebrafish, up to 10 different measurements were taken for each section. Two different sections were analyzed for one wild-type fish and one pcare1 mutant fish (Supplementary Figure S2).
Optokinetic response (OKR) assay
All measurements were conducted between 10 am and 5 pm. The OKR was measured as described previously40–42. In brief, zebrafish larvae were mounted in an upright position in 3% methylcellulose in a small Petri dish placed on a platform surrounded by a rotating drum. A pattern of alternating black and white vertical stripes was displayed on the drum interior. Larvae (5 dpf) were visualized through a stereomicroscope positioned over the drum. Eye movements were recorded while larvae were optically stimulated by the rotating stripes. Larvae were subjected to a protocol of a 30 sec counterclockwise rotation, a 10 sec rest, and a 30 sec clockwise rotation. Measurements were recorded blinded. Graphs were generated and statistical analysis was done with Graphpad Prism6 software (La Jolla, CA, USA).
Visual motor response (VMR) assay (DanioVision)
Locomotor activity in response to light-dark conditions was analyzed using the DanioVision system (Noldus B.V., Wageningen, The Netherlands). Larvae were transferred to a 48-wells plate filled with 200 µl E3 medium without methylene blue. Each run consisted of 24 mutant larvae and 24 age-matched control wild-type zebrafish larvae using a 48-wells plate. During the experiment, the temperature was constant at 28 °C using a heating/cooling system (Noldus B.V., Wageningen, The Netherlands). The protocol consisted of 20 min of acclimation (with lid of the system open; room light: 500–650 lux), closing of the lid followed by alternating periods of 10 min dark, 10 min bright light (about 3000 lux) and 10 min dark (in total 12 cycles).
Variables of interest were: distance moved (mm) for general activity under dark and light conditions; change in maximum velocity (mm∙s-1), to measure the change from dark to light (maximum velocity first 30 sec with light minus maximum velocity last 30 sec without light); change in distance moved (mm), to measure the change from light to dark (distance moved first 30 seconds without light minus distance moved from last 30 sec with light)43–45. Zebrafish tracks were analysed using the analysis software (EthioVision). First, the missed subjects (<10%) were filtered out. Next, the heat maps of 5-minute interval were plotted, to check for general activity. Fish not moving were removed from the analysis. The distance moved (DM (mm)) per unit of time (1 min, 30 sec or 1 sec intervals) for each animal and group was used as a measure for locomotor activity. In addition, the speed of movement (Vmax (mm/s)) was measured in the dark-to-light transition. The values of 1-second interval were used to study more precisely immediate effects of a sudden change from the dark to the light transition and from the light to dark transition. First, the distance moved was expressed by 30 sec/1 min intervals and compared between the different groups; Second, the more precise per 1 sec interval was used to analyze the direct effects of light/dark and dark/light transitions. EthioVision XT was used to quantify the experiments. Complementary to it, R programming language was used to generate plots, calculate mean values and SEM values, and perform statistical tests. The difference between wild-type and mutant animals was analyzed using two-tailed, unpaired Student’s t-test, and p-values were corrected for multiple testing using Benjamini-Hochberg method. The means vs. standard errors of the mean are shown. Exact p-values can be found in Supplementary Table S2.
Electroretinograms (ERG)
ERG measurements were performed on isolated larval eyes (5 dpf) as previously described46. Larvae were dark-adapted for a minimum of 30 min prior to the measurements and subsequently handled under dim red illumination. The isolated eye was positioned to face the light source. Under visual control via a standard microscope equipped with red illumination (Stemi 2000C, Zeiss, Oberkochen, Germany), the recording electrode with an opening of approximately 20 μm at the tip was placed against the center of the cornea. This electrode was filled with E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl, and 0.33 mM MgSO4), the same in which the embryos were raised and held. The electrode was moved with a micromanipulator (M330R, World Precision Instruments Inc., Sarasota, USA). A custom-made stimulator was invoked to provide light pulses of 100 ms duration, with a light intensity of 6000 lux. It uses a ZEISS XBO 75 W light source and a fast shutter (Uni-Blitz Model D122, Vincent Associates, Rochester, NY, USA) driven by a delay unit interfaced to the main ERG recording setup. Electronic signals were amplified 1000 times by a pre-amplifier (P55 A.C. Preamplifier, Astro-Med. Inc, Grass Technology) with a band pass between 0.1 and 100 Hz, digitized by DAQ Board NI PCI-6035E (National Instruments) via NI BNC-2090 accessories and displayed via self-developed NI Labview program47. All the experiments were performed at room temperature (22 °C). Statistical analysis was performed using GraphPad Prism6 (La Jolla, CA, USA), and graphs were generated in R (Boston, USA) or GraphPad Prism6. For statistical analysis of b-wave amplitude (Fig. 5B), two-tailed unpaired t-test with Welch’s correction was performed; p-value = 0,0004; t = 4.309, df = 19.
Electronic supplementary material
Acknowledgements
We thank Tom Spanings, Antoon van der Horst and Franziska Wagner for technical assistance, and Dyah Karjosukarso for assistance with OKR and DanioVision data analysis. This work was funded by the FP7-PEOPLE-2012-ITN programme EyeTN, Brussels, Belgium (grant agreement no.: 317472) to R.W.J.C.
Author Contributions
J.C.C.S., M.M., M.D., T.P. and L.K. performed experiments. E.v.W. and R.W.J.C. supervised research. J.C.C.S. prepared figures. J.C.C.S. and R.W.J.C. wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27928-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Collin RW, et al. Mutations in C2ORF71 cause autosomal-recessive retinitis pigmentosa. American journal of human genetics. 2010;86:783–788. doi: 10.1016/j.ajhg.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishimura DY, et al. Discovery and functional analysis of a retinitis pigmentosa gene, C2ORF71. Am J Hum Genet. 2010;86:686–695. doi: 10.1016/j.ajhg.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hebrard M, et al. Combining gene mapping and phenotype assessment for fast mutation finding in non-consanguineous autosomal recessive retinitis pigmentosa families. Eur J Hum Genet. 2011;19:1256–1263. doi: 10.1038/ejhg.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Y, et al. Mutations of 60 known causative genes in 157 families with retinitis pigmentosa based on exome sequencing. Human genetics. 2014;133:1255–1271. doi: 10.1007/s00439-014-1460-2. [DOI] [PubMed] [Google Scholar]
- 5.Bocquet B, et al. Homozygosity mapping in autosomal recessive retinitis pigmentosa families detects novel mutations. Molecular vision. 2013;19:2487–2500. [PMC free article] [PubMed] [Google Scholar]
- 6.Sergouniotis PI, et al. A survey of DNA variation of C2ORF71 in probands with progressive autosomal recessive retinal degeneration and controls. Invest Ophthalmol Vis Sci. 2011;52:1880–1886. doi: 10.1167/iovs.10-6043. [DOI] [PubMed] [Google Scholar]
- 7.Audo I, et al. Novel C2orf71 mutations account for approximately 1% of cases in a large French arRP cohort. Human mutation. 2011;32:E2091–2103. doi: 10.1002/humu.21460. [DOI] [PubMed] [Google Scholar]
- 8.Liu YP, et al. Putative digenic inheritance of heterozygous RP1L1 and C2orf71 null mutations in syndromic retinal dystrophy. Ophthalmic Genet. 2017;38:127–132. doi: 10.3109/13816810.2016.1151898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerth-Kahlert C, et al. C2orf71 Mutations as a Frequent Cause of Autosomal-Recessive Retinitis Pigmentosa: Clinical Analysis and Presentation of 8 Novel Mutations. Invest Ophthalmol Vis Sci. 2017;58:3840–3850. doi: 10.1167/iovs.17-21597. [DOI] [PubMed] [Google Scholar]
- 10.Chaitin MH, Schneider BG, Hall MO, Papermaster DS. Actin in the photoreceptor connecting cilium: immunocytochemical localization to the site of outer segment disk formation. J Cell Biol. 1984;99:239–247. doi: 10.1083/jcb.99.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kevany BM, Zhang N, Jastrzebska B, Palczewski K. Animals deficient in C2Orf71, an autosomal recessive retinitis pigmentosa-associated locus, develop severe early-onset retinal degeneration. Hum Mol Genet. 2015;24:2627–2640. doi: 10.1093/hmg/ddv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Z, et al. Ablation of EYS in zebrafish causes mislocalisation of outer segment proteins, F-actin disruption and cone-rod dystrophy. Sci Rep. 2017;7:46098. doi: 10.1038/srep46098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu M, et al. Eyes shut homolog is required for maintaining the ciliary pocket and survival of photoreceptors in zebrafish. Biol Open. 2016;5:1662–1673. doi: 10.1242/bio.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F, et al. Knockout of RP2 decreases GRK1 and rod transducin subunits and leads to photoreceptor degeneration in zebrafish. Hum Mol Genet. 2015;24:4648–4659. doi: 10.1093/hmg/ddv197. [DOI] [PubMed] [Google Scholar]
- 15.Slijkerman RW, et al. The pros and cons of vertebrate animal models for functional and therapeutic research on inherited retinal dystrophies. Prog Retin Eye Res. 2015;48:137–159. doi: 10.1016/j.preteyeres.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Bibliowicz J, Tittle RK, Gross JM. Toward a better understanding of human eye disease insights from the zebrafish, Danio rerio. Prog Mol Biol Transl Sci. 2011;100:287–330. doi: 10.1016/B978-0-12-384878-9.00007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/S0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 18.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 20.Dowling JE. What can a zebrafish see with only an off-pathway and other fish stories? J Ophthalmic Vis Res. 2012;7:97–99. [PMC free article] [PubMed] [Google Scholar]
- 21.May-Simera H, Nagel-Wolfrum K, Wolfrum U. Cilia - The sensory antennae in the eye. Prog Retin Eye Res. 2017;60:144–180. doi: 10.1016/j.preteyeres.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Emran, F., Rihel, J. & Dowling, J. E. A behavioral assay to measure responsiveness of zebrafish to changes in light intensities. J Vis Exp, 10.3791/923 (2008). [DOI] [PMC free article] [PubMed]
- 23.Fernandes AM, et al. Deep brain photoreceptors control light-seeking behavior in zebrafish larvae. Curr Biol. 2012;22:2042–2047. doi: 10.1016/j.cub.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies WI, et al. An extended family of novel vertebrate photopigments is widely expressed and displays a diversity of function. Genome Res. 2015;25:1666–1679. doi: 10.1101/gr.189886.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branchek T, Bremiller R. The development of photoreceptors in the zebrafish, Brachydanio rerio. I. Structure. J Comp Neurol. 1984;224:107–115. doi: 10.1002/cne.902240109. [DOI] [PubMed] [Google Scholar]
- 26.Morris AC, Fadool JM. Studying rod photoreceptor development in zebrafish. Physiol Behav. 2005;86:306–313. doi: 10.1016/j.physbeh.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins JE, White S, Searle SM, Stemple DL. Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res. 2012;22:2067–2078. doi: 10.1101/gr.137901.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collery R, et al. Duplication and divergence of zebrafish CRALBP genes uncovers novel role for RPE- and Muller-CRALBP in cone vision. Invest Ophthalmol Vis Sci. 2008;49:3812–3820. doi: 10.1167/iovs.08-1957. [DOI] [PubMed] [Google Scholar]
- 29.Seiler C, et al. Duplicated genes with split functions: independent roles of protocadherin15 orthologues in zebrafish hearing and vision. Development. 2005;132:615–623. doi: 10.1242/dev.01591. [DOI] [PubMed] [Google Scholar]
- 30.Morrow JM, et al. A second visual rhodopsin gene, rh1-2, is expressed in zebrafish photoreceptors and found in other ray-finned fishes. J Exp Biol. 2017;220:294–303. doi: 10.1242/jeb.145953. [DOI] [PubMed] [Google Scholar]
- 31.Trivedi D, Colin E, Louie CM, Williams DS. Live-cell imaging evidence for the ciliary transport of rod photoreceptor opsin by heterotrimeric kinesin-2. J Neurosci. 2012;32:10587–10593. doi: 10.1523/JNEUROSCI.0015-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sukumaran S, Perkins BD. Early defects in photoreceptor outer segment morphogenesis in zebrafishift57, ift88 and ift172 Intraflagellar Transport mutants. Vision Res. 2009;49:479–489. doi: 10.1016/j.visres.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krock BL, Perkins BD. The intraflagellar transport protein IFT57 is required for cilia maintenance and regulates IFT-particle-kinesin-II dissociation in vertebrate photoreceptors. J Cell Sci. 2008;121:1907–1915. doi: 10.1242/jcs.029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan J, Goldman D. Retina regeneration in zebrafish. Curr Opin Genet Dev. 2016;40:41–47. doi: 10.1016/j.gde.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bringmann A, et al. Cellular signaling and factors involved in Muller cell gliosis: neuroprotective and detrimental effects. Prog Retin Eye Res. 2009;28:423–451. doi: 10.1016/j.preteyeres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Goldman D. Muller glial cell reprogramming and retina regeneration. Nat Rev Neurosci. 2014;15:431–442. doi: 10.1038/nrn3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 38.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sander JD, et al. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips JB, et al. Harmonin (Ush1c) is required in zebrafish Muller glial cells for photoreceptor synaptic development and function. Dis Model Mech. 2011;4:786–800. doi: 10.1242/dmm.006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huber-Reggi SP, Mueller KP, Neuhauss SC. Analysis of optokinetic response in zebrafish by computer-based eye tracking. Methods in molecular biology. 2013;935:139–160. doi: 10.1007/978-1-62703-080-9_10. [DOI] [PubMed] [Google Scholar]
- 42.Hodel C, Neuhauss SC. Computer-based analysis of the optokinetic response in zebrafish larvae. CSH protocols. 2008;2008:pdbprot4961. doi: 10.1101/pdb.prot4961. [DOI] [PubMed] [Google Scholar]
- 43.de Esch C, et al. Locomotor activity assay in zebrafish larvae: influence of age, strain and ethanol. Neurotoxicology and teratology. 2012;34:425–433. doi: 10.1016/j.ntt.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Gao Y, et al. Computational classification of different wild-type zebrafish strains based on their variation in light-induced locomotor response. Comput Biol Med. 2016;69:1–9. doi: 10.1016/j.compbiomed.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, et al. Statistical Analysis of Zebrafish Locomotor Response. PLoS One. 2015;10:e0139521. doi: 10.1371/journal.pone.0139521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sirisi S, et al. Megalencephalic leukoencephalopathy with subcortical cysts protein 1 regulates glial surface localization of GLIALCAM from fish to humans. Human molecular genetics. 2014;23:5069–5086. doi: 10.1093/hmg/ddu231. [DOI] [PubMed] [Google Scholar]
- 47.Rinner O, Makhankov YV, Biehlmaier O, Neuhauss SC. Knockdown of cone-specific kinase GRK7 in larval zebrafish leads to impaired cone response recovery and delayed dark adaptation. Neuron. 2005;47:231–242. doi: 10.1016/j.neuron.2005.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.