Abstract
The description of comammox Nitrospira spp., performing complete ammonia-to-nitrate oxidation, and their co-occurrence with canonical β-proteobacterial ammonia oxidizing bacteria (β-AOB) in the environment, calls into question the metabolic potential of comammox Nitrospira and the evolutionary history of their ammonia oxidation pathway. We report four new comammox Nitrospira genomes, constituting two novel species, and the first comparative genomic analysis on comammox Nitrospira. Unlike canonical Nitrospira, comammox Nitrospira genomes lack genes for assimilatory nitrite reduction, suggesting that they have lost the potential to use external nitrite nitrogen sources. By contrast, compared to canonical Nitrospira, comammox Nitrospira harbor a higher diversity of urea transporters and copper homeostasis genes and lack cyanate hydratase genes. Additionally, the two comammox clades differ in their ammonium uptake systems. Contrary to β-AOB, comammox Nitrospira genomes have single copies of the two central ammonia oxidation pathway operons. Similar to ammonia oxidizing archaea and some oligotrophic AOB strains, they lack genes involved in nitric oxide reduction. Furthermore, comammox Nitrospira genomes encode genes that might allow efficient growth at low oxygen concentrations. Regarding the evolutionary history of comammox Nitrospira, our analyses indicate that several genes belonging to the ammonia oxidation pathway could have been laterally transferred from β-AOB to comammox Nitrospira. We postulate that the absence of comammox genes in other sublineage II Nitrospira genomes is the result of subsequent loss.
Introduction
Nitrification, the biological oxidation of ammonia to nitrate, is an essential process in terrestrial and aquatic environments as well as in water quality engineering applications. For the last century, nitrification was assumed a two-step process executed by two complementary functional groups, ammonia-oxidizing prokaryotes (AOP) and nitrite-oxidizing bacteria (NOB). Recently, several groups have shown that single microorganisms belonging to the genus Nitrospira can carry out the complete oxidation of ammonia to nitrate, a process abbreviated comammox [1, 2]. Nitrospira spp., long-recognized as nitrite-oxidizers, are widespread in both natural and engineered ecosystems associated with nitrogen cycling [3, 4]. The Nitrospira genus is extremely diverse and comprises at least six lineages, which frequently coexist [3, 5]. Comammox Nitrospira genomes described to date belong to Nitrospira lineage II, and comprise two clades (clade A and B) based on the phylogeny of their ammonia monooxygenases [1, 2]. Both clades were detected in samples retrieved from a groundwater well and in a groundwater-treating rapid sand filter, together with conventional Nitrospira spp. [1, 6]. The diversity of Nitrospira spp. points towards ecological niche-partitioning; nitrite and dissolved oxygen concentrations are potential niche determinants for Nitrospira lineages I and II [7–9]. The recently described metabolic versatility of some Nitrospira spp., which includes formate, hydrogen, urea and cyanate metabolisms [10–12] may also contribute to their coexistence. Nevertheless, little is known about differences in the functional potential of the two comammox Nitrospira clades and niche-partitioning between comammox Nitrospira and AOP. Only the hypothesis that comammox Nitrospira could outcompete canonical AOB in surface-attached and substrate-limited environments has been proposed [13]. The evolutionary history of ammonia oxidation in Nitrospira is unknown. Based on their ammonia monooxygenase (AMO) and hydroxylamine dehydrogenase (HAO) sequences, β-proteobacterial ammonia oxidizing bacteria (β-AOB) are most similar to comammox Nitrospira [1, 2, 6, 14]. Either AMO and HAO encoding genes are ancestral in Nitrospira spp. or were acquired by horizontal gene transfer from AOP, even though the high degree of phylogenetic divergence of these proteins between comammox Nitrospira and AOP makes a recent acquisition improbable [2].
To examine the functional potential which might allow niche-partitioning among comammox Nitrospira and between comammox Nitrospira and AOP, and to unravel the evolutionary history of ammonia oxidation in Nitrospira, we performed differential coverage genome binning on a Nitrospira composite population genome recovered from a metagenome in which comammox Nitrospira, canonical AOB and ammonia oxidizing archaea (AOA) co-occurred [3, 6]. Comparative genome analysis was conducted with the recovered genomes and high-quality published Nitrospira genomes. The evolutionary history of genes involved in ammonia oxidation in comammox Nitrospira was also explored with a special focus on understanding the potential role of horizontal gene transfer, gene duplication, and gene loss. For this purpose, we executed pairwise protein dissimilarity comparison between and within comammox Nitrospira clades as well as with other AOB, explored the genomic arrangement in the relevant pathways, and performed probabilistic and parsimony-based reconciliation analysis where trees for individual genes were compared to a clock-like species-tree constructed from ribosomal proteins. Our study revealed comammox-, comammox clade- and canonical Nitrospira-specific features related to nitrogen assimilation, electron donor versatility and substrate-limitation tolerance. Our analysis suggests that ammonia-oxidation related genes were transferred from β-proteobacterial ammonia oxidizers to comammox Nitrospira.
Materials and methods
Recovery of individual genomes
As described in Palomo et al. [6], DNA was extracted from triplicate 0.5 g samples taken from three adjacent (ca. 0.5 m apart; and two different depths from each) locations in a singular rapid sand filter (RSF) and subject to shotgun sequencing to describe the microbial community involved in groundwater purification. The work presented here focuses on the metagenomes from which a Nitrospira composite population genome (CG24) was recovered [6]. In order to separate the composite population genome into individual genomes, a strategy based on subsample assembly [15] followed by differential coverage binning was applied [16] (further details in Supplementary Information and Supplementary Figure S1). The quality of recovered genomes was evaluated using coverage plots from mmgenome [17], hidden Markov models (HMMs) for 107 essential single-copy genes, and CheckM [18] (further details in Supplementary Information). The selected draft genomes were refined using Anvi’o [19] to remove contigs with inconsistent coverage. Assembly quality was improved by alignment against related complete or draft genomes using the Multi-Draft based Scaffolder (MeDuSa) [20] and gaps were closed with GapFiller v.1.10 [21]. All raw sequence data and genome sequences have been deposited at NCBI under the project PRJNA384587, with sequence accession for the raw sequence data: SRR5739198—SRR5739203; and for the draft genomes: GCA_002869925.1, GCA_002869845.1, GCA_002869885.1, GCA_002869855.1 and GCA_002869895.1 (GenBank assembly accessions).
Comparative genome analysis
High quality published Nitrospira genomes (more than 90% complete and with less than 5% redundancy) were also included in the comparative genomic analysis. Gene calling on the recovered genome bins and published genomes was performed using Prodigal v.2.62 [22]. Annotation was conducted in RAST [23] and protein functional assignments of genes of interest were confirmed using KEGG and blastp. Pangenomic analysis was executed using the meta-pangenomic workflow of Anvi’o [19] with default parameters with the exception --maxbit = 0.3 (further details in Supplementary Information). All comparative genome information was visualized using the program anvi-interactive in Anvi’o [19].
Sequence dissimilarity and gene synteny analysis
Pairwise amino acid dissimilarities (Supplementary Information) among the genomes for 18 housekeeping genes, 27 ammonia oxidation-related proteins and 14 syntenic ribosomal proteins (Supplementary Table S1) were calculated using Clustal Omega v.1.2.1 [24]. Gene arrangement of ammonia oxidation related genes was visualized using the R package genoPlotR [25].
Construction of clock-based species-tree and guest-tree sets
The software BEAST, version 2.4.4, was used to construct a dated species-tree using a relaxed log-normal clock model [26, 27]. The input data was a set of 14 ribosomal proteins assumed to be inherited mostly vertically (Supplementary Information and Supplementary Table S1). The timetree.org resource [28] together with the substitution rate of 16S rRNA [29] were used as a main starting point for finding calibration information for internal nodes (Supplementary Information).
For each investigated gene the protein sequences were aligned using the G-INS-i method in MAFFT [30]. The software MrBayes version 3.2.6 [31] was then used to reconstruct phylogenies (further details in Supplementary Information).
Reconciliation analysis
The analysis of event probabilities was done using the software JPrIME-DLTRS version 0.3.6 [32] (Supplementary Information). The clock-based tree mentioned above was used as the reference species-tree for all analyses (Supplementary Information). For each gene, the sample file containing 1500 midpoint-rooted trees constructed using MrBayes was used as a guest-tree set from which JPrIME-DLTRS could sample guest-tree topologies. MCMC was allowed to run for 4 million iterations with a thinning of 200, and running 3 parallel chains for each gene (Supplementary Information). To investigate which branches the transfer events most likely involved, we used the software ecceTERA version 1.2.4 [33] (Supplementary Information). The software SylvX version 1.3 [34] was used for visualizing reconciliations computed by ecceTERA.
Phylogenetic analysis
Phylogenetic trees for AmoA and HaoA were constructed using MrBayes. The substitution model for both data sets was LG with gamma-distributed rates (determined using jmodeltest), using 5 million iterations for 2 sets of 3 chains, with a thinning of 5000 and a burn-in of 25%. Convergence was checked using Tracer and by computing the potential scale reduction factor (ensuring that R was close to 1.0 for all parameters) and the average standard deviation of split frequencies (which was well below 0.01 in both cases).
Statistical analysis
All statistical tests were performed using R v3.3.1 [35]. Correlations between genomes phylogenetic distance (based on 14 concatenate ribosomal proteins) and sequence divergence for housekeeping or the ammonia-oxidation related proteins were determined using a linear regression model. Significances between two slopes were calculated using ANOVA test. Significances between two means of sequence dissimilarities for either housekeeping proteins or ammonia-oxidation related proteins were evaluated using Welch Two Sample t-test.
Results and Discussion
Genome extraction from the metagenome
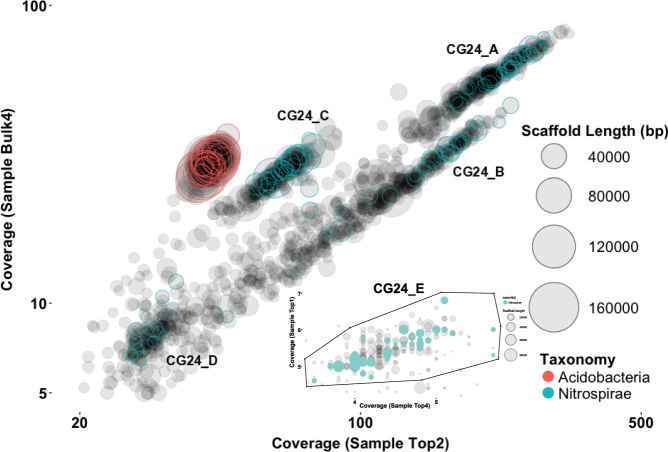
Metagenomes from adjacent locations in a RSF were used for individual genome extraction [6]. Five Nitrospira genomes (CG24_A, CG24_B, CG24_C, CG24_D, CG24_E) were recovered from the metagenomes using differential coverage binning [16] (Fig. 1). The characteristics of these and other genomes used in this study are shown in Table 1. The size of the comammox genomes assembled here range from 3.0–3.6 Mb, with a completeness of 95–100%. Based on phylogenetic analysis of 14 syntenic ribosomal proteins, all genomes belong to Nitrospira sublineage II (Supplementary Figure S2). Genes required for complete ammonia oxidation (amo and hao operons) were detected in four of the genomes. Based on amoA phylogeny CG24_A, CG24_C and CG24_E belong to comammox clade B and CG24_B to comammox clade A (Supplementary Figure S3). CG24_A was most abundant at an average relative abundance of 18.6% (of all metagenome sequence reads) followed by CG24_B (8.6%), CG24_C (4.6%), CG24_D (2.4%, the only canonical NOB Nitrospira recovered) and CG24_E (1.1%). (Supplementary Figure S4).
Fig. 1.
Differential coverage plot of two metagenomes obtained from Islevbro waterworks at different sampling depth. Scaffolds are displayed as circles, scaled by length and colored according to phylum-level taxonomic affiliation. Only scaffolds >4 kbp are shown. A second differential coverage plot showing the CG24_E bin extraction (enclosed by the polygon) is presented
Table 1.
Characteristics of examined genomes
| Bin ID or reference | CG24_A | CG24_B | CG24_C | CG24_D | CG24_E | A2_bin | AAU_ MBR1 | AAU_ MBR2 | Ca N. inopinata | Ca N. nitrificans | Ca N. nitrosa | OLB3 | GWW3 | GWW4 | N. mosco -viensis | Ca N defluvii |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | 3,555,106 | 3,236,455 | 3,000,613 | 3,392,746 | 3,477,523 | 4,111,999 | 3,525,021 | 4,296,753 | 3,295,117 | 4,117,083 | 4,422,398 | 3,754,263 | 3,501,770 | 2,981,542 | 4,589,485 | 4,317,083 |
| GC Content | 55.7 | 55.3 | 56.1 | 57.8 | 55.8 | 55.1 | 55.1 | 59.1 | 59.2 | 56.6 | 54.8 | 60.4 | 55.8 | 55.9 | 62.0 | 59.0 |
| N50 (bp) | 137,244 | 240,795 | 172,703 | 66,498 | 113,824 | 150,736 | 30,005 | 73,098 | 3,295,117 | 174,194 | 727,365 | 121,772 | 184,662 | 32,587 | 4,589,485 | 4,317,083 |
| Number of scaffolds | 58 | 36 | 28 | 135 | 63 | 61 | 225 | 172 | 1 | 36 | 15 | 79 | 38 | 167 | 1 | 1 |
| Number of coding sequences | 3697 | 3437 | 3044 | 3388 | 3628 | 4316 | 3607 | 4129 | 3308 | 4162 | 4380 | 3734 | 3433 | 3077 | 4572 | 4132 |
| Number of tRNAs | 42 | 35 | 44 | 39 | 36 | 42 | 39 | 49 | 49 | 46 | 49 | 40 | 43 | 37 | 50 | 49 |
| Completeness (107 HMM) | 95 | 95 | 97 | 97 | 100 | 93 | 90 | 99 | 100 | 100 | 100 | 94 | 98 | 94 | 100 | 100 |
| Redundancy (107 HMM) | 2.9 | 2.0 | 0.0 | 1.0 | 0.9 | 1.0 | 2.1 | 1.0 | 2.8 | 0.9 | 0.9 | 2.0 | 1.0 | 1.0 | 2.8 | 0.9 |
| Completeness (CheckM) | 88 | 92 | 92 | 92 | 94 | 88 | 86 | 97 | 97 | 97 | 97 | 91 | 92 | 84 | 96 | 98 |
| Redundancy (CheckM) | 5.1 | 6.9 | 2.7 | 2.7 | 3.2 | 2.8 | 2.7 | 5.5 | 4.8 | 2.7 | 2.3 | 4.3 | 3.2 | 1.8 | 6.6 | 2.3 |
| Comammox affiliation | clade B | clade A | clade B | No | clade B | clade A | clade A | No | clade A | clade A | clade A | No | clade B | clade B | No | No |
| Nitrospira Lineage | II | II | II | II | II | II | II | I | II | II | II | I | II | II | II | I |
| Reference | This study | This study | This study | This study | This study | [14] | [1] | [1] | [1] | [2] | [2] | [82] | [1] | [1] | [11] | [51] |
*Minimum sequence length in which 50% of the assembled bases occur
Comparative genomics
The newly recovered genomes were compared with each other and publicly-available, high-quality Nitrospira genomes, including in total 11 comammox and five canonical Nitrospira (Table 1). Based on average amino acid identity (AAI) analysis, the 16 studied Nitrospira genomes constitute 11 different species, seven of them comammox Nitrospira (at species level cut-off of 85% AAI [36]. CG24_A, CG24_D and CG24_E are divergent enough from each other and previously published genomes to be separate species (Supplementary Figure S5).
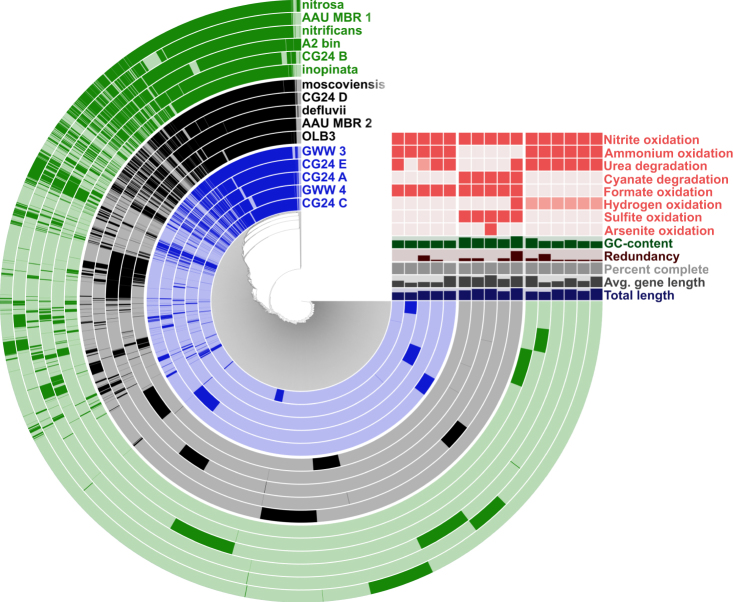
Comparison of the 16 Nitrospira genomes was based on SEED subsystems (further details in Supplementary Information and Supplementary Figure S6) and pangenomic analysis (Fig. 2). The 59,744 coding sequences (CDS) of the 16 Nitrospira genomes clustered into 12,337 protein clusters (PCs), with a core Nitrospira genome consisting of 1382 PCs. The core genome includes genes for the nitrite oxidation pathway, the reductive tricarboxylic acid cycle for CO2 fixation (rTCA), the gluconeogenesis, the pentose phosphate, and the oxidative TCA cycle. Chlorite dismutase and copper-containing nitrite reductase (nirK) are also present in the core genome (Fig. 3). 35 comammox-specific PCs were identified; 16 and 3 of these PCs had highest sequence similarity to homologs in β-AOB and methane oxidizers, respectively (Supplementary Table S2). In addition, we detected 57 and 52 comammox clade A and clade B-specific PCs, respectively (Supplementary Table S2 and Fig. 3). The specific metabolic characteristics inferred from the annotations of genes with known homologs, and identified with the pangenomic analysis are described below.
Fig. 2.
Communality and uniqueness in the Nitrospira pangenome as derived from the clustering of 16 genomes based on 12,337 protein clusters (PCs). Each radial layer represents a genome, and each bar in a layer represents the occurrence of a PC (dark presence, light absence). Clade A and Clade B comammox Nitrospira genomes are denoted in green and blue, respectively
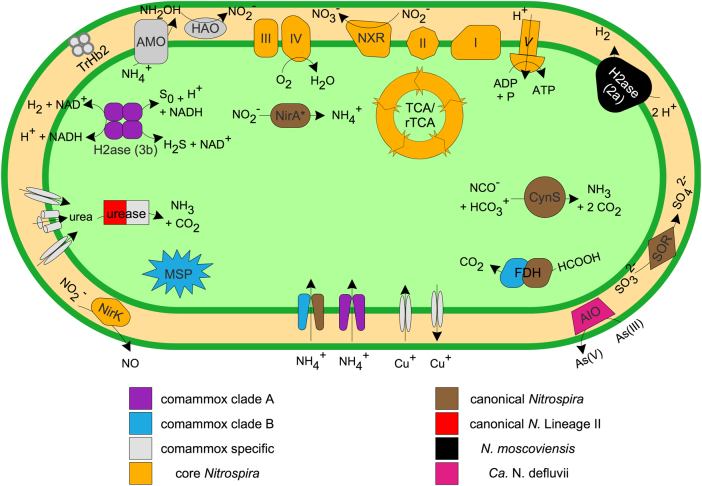
Fig. 3.
Cartoon of core and specific key metabolic features in the Nitrospira pangenome, as predicted from genome annotation. AIO arsenite oxidase, CynS cyanate hydratase, FDH formate dehydrogenase, H2ase hydrogenase, MSP methionine salvage pathway, SOR sulfite dehydrogenase, TrHb2 2/2 hemoglobin group 2. Enzyme complexes of the electron transport chain are labeled by Roman numerals. * N. moscoviensis possesses an octaheme nitrite reductase (ONR) putatively involved in nitrite reduction to ammonia
Nitrogen metabolism
All the recovered genomes encoded the nitrite oxidoreductase (NXR), consistent with other Nitrospira spp. The copy number of NXR in the investigated genomes varied from one to two in most of the comammox genomes (except Ca. N. nitrificans, which has four copies), to five in N. moscoviensis (Supplementary Table S2). The ammonia oxidation pathway (AMO structural genes amoCAB and the putative AMO subunits amoEDD2, as well as genes for HaoAB and the associated cytochromes CycAB) is present in four of the newly recovered genomes (CG24_A, CG24_B, CG24_C and CG24_E).
Regarding nitrogen uptake, clade B and canonical Nitrospira genomes encode MEP-type ammonia transporters, which are also found in AOA [37]. Clade A genomes encode an ammonia transporter with high homology (>70% amino acid similarity) to Rh-type transporters found in most of β-AOB (Supplementary Table S3). Experimental analysis in Nitrosomonas europea showed that its Rh-type transporter proteins have ammonium affinity in millimolar range and high uptake capacity [38], and similar results were observed for Rh-type ammonia transporter in other organisms [39]. In contrast, investigated MEP-type ammonium transporters have higher affinity (micromolar range) and lower uptake capacity [40, 41]. Whether these observations are also valid for Nitrospira spp. awaits experimental confirmation. Besides the uptake of exogenous ammonia by Nitrospira, ammonia can be produced through urea degradation as most of the comammox genomes harbor urease genes. This enzyme, which is functional in N. moscoviensis [11], is not detected in genomes of Nitrospira sublineage I. The comammox genomes contain a diversity of urea transporters. In addition to the high affinity urea ABC transporters (urtABCDE) present in some Nitrospira spp. such as N. lenta [11], comammox genomes also harbor two additional urea transporters: an outer-membrane porin (fmdC) involved in uptake of short-chain amides and urea at extremely low concentrations [42], and a urea carboxylase-related transporter (uctT). These extra urea transporters present in comammox Nitrospira might confer a competitive advantage in urea uptake with respect to other Nitrospira in environments with low and/or fluctuating urea concentrations. On the other hand, cyanate hydratase genes (cynS) are only detected in canonical Nitrospira. This may confer a benefit over comammox Nitrospira in environments with very low ammonium concentrations and the presence of cyanate coupled with reciprocal feeding with ammonia oxidizers [12]. All genomes except Ca. N. inopinata encode a NO2−/NO3− transporter gene (narK). Furthermore, canonical Nitrospira genomes contain additional transporters for the uptake of nitrite: lineage I Nitrospira and CG24_D genomes harbor a nitrite transporter (nirC) and a formate/nitrite family transporter, while N. moscoviensis and CG24_D contain a nitrite-nitrate ABC transporter (nrtABC). The comammox genomes lack an assimilatory nitrite reductase (nirA) or octaheme nitrite reductase (ONR) (which has been proposed to be associated with nitrite reduction to ammonia in N. moscoviensis [11]), which would prevent comammox Nitrospira growth in the presence of nitrite as sole N source, as observed in Ca. N. inopinata [1].
Alternative electron donors
Some Nitrospira can utilize substrates beyond nitrite as electron donors including formate and hydrogen [7, 10, 11, 43]. The genes coding for formate dehydrogenase (fdh) are present in canonical Nitrospira and comammox clade B but were not detected in comammox clade A. This could provide an opportunity for niche-differentiation particularly in oxic–anoxic transition zones where formate can be found as a product of fermentation. With respect to hydrogen oxidation, the group 2a [NiFe] hydrogenase and accessory proteins involved in aerobic hydrogen oxidation in N. moscoviensis [10] are absent from the other Nitrospira genomes. However, all comammox clade A genomes encode a complete group 3b [NiFe]-hydrogenase. Although this bidirectional hydrogenase (sulfhydrogenase) is extensively distributed across the bacterial domain [44], little is known about its actual role (further details in Supplementary Information). Comammox genomes do not contain genes for a periplasmic sulfite:cytochrome c oxidoreductase (sulfite dehydrogenase, sorAB), which is characteristic of canonical Nitrospira. These genomes also possess the genetic inventory for sulfur assimilation, suggesting that this sulfite dehydrogenase might be involved in sulfite oxidation, as suggested for another NOB (Nitrospina gracilis) [45]. Lastly, a complete arsenite oxidase was exclusively found in Ca. N. defluvii. Although all examined genomes harbor arsenic resistance genes, only Ca. N. defluvii has the genetic potential for energy conservation during arsenite oxidation (Supplementary Information).
Energy conservation and transduction
All Nitrospira genomes harbor two homologous sets of genes encoding the complex I of the electron transport chain (NADH:ubiquinone oxidoreductase, NUO). One of these NUO lacks the subunits involved in NADH binding (NuoE and NuoF). This feature has been observed in other prokaryotes [46]; in Nitrospira spp. this incomplete complex I may be used for reverse electron transport from quinol to low-potential ferredoxin, as proposed in Nitrospina gracilis [45], thereby providing reduced ferredoxin required for CO2 fixation in the rTCA cycle. Additionally, CG24_A, Ca. N. nitrosa, AAU_MBR1, A2_bin and N. moscoviensis possess a third complete homologous set of genes coding for complex I, which is divergent from the other two, and is closely related to the γ-proteobacterial clade E type. This type of complex I has also been found in other distantly related organisms, including Nitrosospira multiformis and some Bacteroidetes spp., suggesting acquisition by horizontal gene transfer [47]. Although the exact function of this additional complex I is not known, differential expression of distinct complex I isozymes has been observed in other microorganisms, pointing towards physiological versatility [48, 49]. Also, differences with respect to complex II, the succinate:quinone oxidoreductase (SQR), were noted across genomes. SQR connects the TCA cycle to the quinone pool. This complex is classified in five types based on the number of membrane-bound domains and haem content [50]. Nitrospira genomes do not all contain the same SQR type. Clade B, CG24_D, and genomes belonging to Nitrospira lineage I contain a SQR type E, whereas clade A genomes and Nitrospira moscoviensis possess a SQR type B. Differences were also detected for the cytochrome bc complex (complex III) which is involved in the transfer of electrons from quinol to a c-type cytochrome. The canonical Nitrospira genomes contain at least two copies of complex III (three in N. moscoviensis and OLB3), while the comammox genomes harbor just one copy of this complex. With respect to cytochrome oxidases (complex IV), the studied genomes harbor a CDS similar to the the uncharacterized but functionally transcribed ‘cyt bd-like oxidase’ (NIDE0901) of Ca. N. defluvii which was proposed to be a terminal oxidase [51]. In addition to this putative terminal oxidase, canonical Nitrospira (except CG24_D), comammox clade A genomes (excluding Ca. N. nitrosa) and the clade B GWW3 bin contain the cydAB genes encoding a heterodimeric cyt. bd quinol oxidase. The reaction catalyzed by this respiratory oxidase is not coupled to proton pumping [52], and may be important when electrons derive from low potential electron donors.
Carbon metabolism
Comammox Nitrospira, like other sequenced Nitrospira genomes [51], encode genes for glycolysis, gluconeogenesis, the pentose phosphate pathway and the TCA cycle. Moreover, clade B comammox genomes and AAU_MBR2 carry an acetate permease gene (actP). Nitrospira genomes have the potential to degrade catechol and protocatechuate (Supplementary Table S2). Although these degradation pathways are only partially complete in Nitrospira genomes, comammox Nitrospira possess an intradiol ring-cleavage dioxygenase, which could transform catecholate derivatives to TCA cycle intermediates [55]. Additionally, comammox clade B and N. moscoviensis harbor a 4-oxalocrotonate tautomerase, essential in the conversion pathway of various aromatic compounds such as catechol to intermediates for the TCA cycle [54]. The phaZ gene which codes for polyhydroxybutyrate (PHB) depolymerase involved in PHB degradation was exclusively found in Nitrospira comammox genomes. Genes related to PHB synthesis are not found in the studied genomes, hence whether the PBH polymerase is a relic or has any functional role remains unknown. Overall, the presence of enzymes and pathways involved in different carbon sources degradation suggests that comammox Nitrospira have the potential to grow mixotrophically as reported for other Nitrospira spp. [43, 55, 56].
Stress response, resistance and defense
Genes encoding catalase (cat) or superoxide dismutase (sod), providing protection against reactive oxygen species (ROS), are absent from the closely related comammox genomes Ca. N. nitrosa and AAU_MBR1 as well as from Ca. N. defluvii. On the other hand, CG24_A and N. moscoviensis harbor diverse genes related to ROS protection including one catalase and one peroxidase (two in both cases for N. moscoviensis), and two dissimilar superoxide dismutases. The remaining genomes encode for either catalases or superoxide dismutases and all genomes harbor the putative ROS defense mechanisms predicted for Ca. N. defluvii (consisting of cyt. c peroxidases, thioredoxin-dependent peroxiredoxins, manganeses, bacterioferritin and carotenoids) [51]. Contrary to canonical Nitrospira, the comammox genomes contain a 2/2 hemoglobin type II (TrHb2), which has been associated with oxidative stress resistance and oxygen scavenging [57, 58]. Thus, the distinct capacity to deal with oxidative stress within Nitrospira spp. may enable these organisms to coexist by occupying different microniches in environments with oxygen gradients such as biofilms and flocs. Most of the genomes encode two proteins homologous to RsbUV, related to environmental stress response, and comammox clade A genomes possess a fusion protein distinct from other Nitrospira spp. that is homologous to RsbUVW. N. moscoviensis also encodes all the genes for the stressosome (rsbRSTX), a complex that controls several signaling pathways in response to diverse environmental stresses [59] (further details in Supplementary Information). Hence, based on these genomes characteristics, comammox clade A and especially canonical N. moscoviensis might be better adapted to changing environmental conditions than other Nitrospira. On another note, genes associated with low-resource environments were encountered in some of the Nitrospira genomes. Comammox Nitrospira may have higher requirements for copper than canonical Nitrospira as ammonia monooxygenase contain Cu as cofactor. Different from canonical Nitrospira, the comammox genomes contain Cu homeostasis genes (copCD and copAB) with high sequence similarities to homologs in β-AOB. These proteins confer higher Cu2+ tolerance or increased Cu2+ uptake [60, 61] and may allow comammox Nitrospira to survive in conditions of low Cu2+ availability. Canonical Nitrospira genomes harbor the genes for the cytochrome c biogenesis system II. In contrast, comammox genomes contain the genes for the cytochrome c biogenesis system I. While system II requires less energy for cytochrome synthesis, system I is advantageous in iron-limited environments as this system has hemes with higher iron affinity [62]. As nitrifiers have a high iron requirement for their [FeS] cluster- and heme-containing enzymes, this could provide an important competitive advantage for comammox Nitrospira in iron-limited environments [63]. Comammox Nitrospira clade B genomes contain the methionine salvage pathway which was not detected in clade A or canonical Nitrospira genomes (Fig. 3 and Supplementary Table S2). This pathway is involved in the recycling of sulfur-containing metabolites to methionine and is upregulated under sulfur-limiting conditions [64, 65]. Thus, comammox clade B could better persist during periods of sulfur depletion. Genes related to arsenic and mercury resistance were found in all studied genomes. Especially striking were the cases of comammox CG24_B and AAU_MBR1 which contain phylogenetically distinct chromate, arsenic and mercury resistance proteins in a putative integrative conjugative element (Supplementary Information).
Comparison between comammoxNitrospiraand ammonia oxidizing microorganisms
Comammox Nitrospira generally harbor a single copy of AMO and HAO genes (Ca.N. nitrosa contains two amoA genes and all comammox genomes harbor extra amoC besides the one present in the amo operon). A similar scenario is observed for γ-proteobacterial AOB (γ-AOB) and AOA. In contrast, β-AOB genomes are characterized by the presence of two or three copies of amo and hao gene clusters. Differences are also detected for genes related to NOx metabolism. Copiotrophic AOB genomes generally contain cytochrome c nitric oxide reductase (cNOR), heme-copper nitric oxide reductase (sNOR), nitrosocyanin, and cytochrome P460. In the case of oligotrophic AOB genomes, they lack sNOR and in some cases nitrosocyanin or cNOR (Supplementary Table S3). On the other hand, comammox Nitrospira genomes do not have the four mentioned proteins related to NOx metabolism as also observed for AOA genomes (Supplementary Table S3). This lack of NOx metabolism related proteins may be associated with low ammonium concentration environments where nitrosative stress would be minor. Nevertheless, the investigated comammox Nitrospira genomes contain two proteins putatively linked to resistance to nitrosative stress: a NO-responsive regulator (nnrS) and a 2/2 hemoglobin type I (TrHb1), that are not found in AOA. The different AOP groups also clearly differ in their carbon fixation pathways. AOB and AOA utilize the oxygen-tolerant Calvin–Benson–Bassham and hydroxypropionate–hydroxybutyrate cycles, respectively, while comammox Nitrospira possess the microaerophilic related rTCA pathway [66]. Furthermore, canonical AOP genomes encode the low-affinity aa3-type heme-copper oxidase (Nitrosomonas eutropha also contains a high-affinity cytochrome c oxidase cbb3). In contrast, comammox Nitrospira harbor cytochrome bd-like oxidases. Cytochrome bd oxidases have high affinity for O2 [67] and are expressed under O2-limited conditions in other microorganisms [68, 69], but it remains unknown whether this is also the case for the cyt bd-like oxidases of Nitrospira spp. Additionally, the 2/2 hemoglobin type II (TrHb2), associated with oxidative stress resistance and oxygen scavenging, which was detected in comammox Nitrospira, is not universal for AOB (only present in Nitrosospira spp.) and has not been detected in AOA. It is to be experimentally confirmed whether these low oxygen-tolerance features are also physiologically expressed in comammox Nitrospira and may provide an advantage for growth in microaerophilic environments. Another characteristic of comammox Nitrospira is their genetic potential to compete under phosphorous and copper limiting conditions. Comammox Nitrospira genomes contain an alkaline phosphatase (phoD), which has been detected to be highly expressed under phosphorus limitation and starvation in other microorganisms [70, 71]. This enzyme was not detected in AOA genomes and is not universal in AOB (putative homologs are present in Nitrosomonas sp. Is79A3, Nitrosococcus halophilus and Nitrosospira spp.). In relation to Cu homeostasis, while copCD genes are present in both AOB and comammox genomes, the genes copAB detected in comammox Nitrospira are not common for AOB as just few species harbor homologs of these genes (Nitrosomonas europaea, Nitrosomonas eutropha, Nitrosospira multiformis and Nitrosospira sp. Nv17). Although AOA seem to have a high Cu2+ demand due to their high presence of copper-containing proteins, their Cu2+ acquisition mechanisms are unknown [72]. These two aspects could be of great importance as both phosphorous and copper deficiency can impact nitrification in engineered environments [73–75]. Recently, it was described that N. inopinata, the only comammox pure culture to date, has a high affinity for ammonia similar to AOA and much higher than AOB cultures [76]. Taken together, comammox Nitrospira might be adapted to nutrient-limited conditions. Consistently, comammox Nitrospira have so far mainly been detected in substrate-limited environments as well as in low oxygen or microaerophilic environments [1, 2, 6, 77–81].
Horizontal gene transfer events shaped comammoxNitrospira
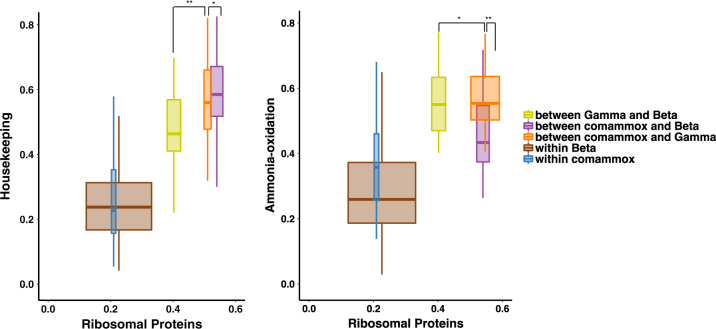
The AMO and HAO sequences of β-AOB are most closely related to those of comammox Nitrospira [1, 2, 6, 14]. In fact, the sequence similarity of amo and hao genes between β-AOB and comammox Nitrospira (ca. 60% amino acid sequence identity for amoA and ca. 66% for haoA) is higher than between β-AOB and γ-AOB (ca. 45% for amoA and ca. 53% for haoA), even though the latter two groups belong to the same phylum (Proteobacteria) and are only distantly related to the Nitrospirae phylum. Discrepancies between gene-trees and species-trees can be caused by horizontal gene transfer (HGT—in this case, for instance of amo and hao genes between comammox Nitrospira and β-AOB), or by gene duplication followed by differential loss (one copy lost in one group, the other copy lost in a second group). To explore whether HGT has played a role in the evolutionary history of comammox Nitrospira and β-AOB, we compared the sequence dissimilarity of proteins related to the ammonia oxidation pathway and 18 housekeeping proteins to that of a set of 14 ribosomal proteins (Supplementary Table S1). We included sequences from the 11 comammox genomes investigated in this study as well as from eight and two previously published β-AOB and γ-AOB genomes, respectively (Supplementary Table S4). Pairwise dissimilarity comparisons showed that housekeeping proteins are linearly related to the ribosomal proteins across essentially all genomes (Fig. 4 and Supplementary Figure S7) suggesting vertical inheritance. However, for most of the proteins associated with ammonia oxidation, the relationship is different with obvious discontinuities (Fig. 4 and Supplementary Figure S8). Generally, it was observed that the sequence dissimilarities between β-AOB and comammox genomes are smaller than expected, while the sequence dissimilarities are larger than expected between β-AOB and γ-AOB genomes (Fig. 4, Supplementary Figure S8, Supplementary Table S5 and Supplementary Information). Additionally, in some cases, the sequence dissimilarities between β-AOB and comammox genomes are as high as between comammox clade A and clade B (Supplementary Figure S8), although these two groups are closely related based on ribosomal proteins (Supplementary Figure S2).
Fig. 4.
Relationship between average phylogenetic distance of genomes and protein sequence divergence for housekeeping proteins (left) and for ammonia-oxidation related proteins (right) for comammox Nitrospira, β-AOB and γ-AOB genomes. Boxplot are colored according to the groups to which the compared genomes belong. The y-axis shows the pairwise protein dissimilarity (fraction of differing amino-acid sites) for a set of 18 housekeeping proteins or 27 ammonia-oxidation related proteins, while the x-axis shows the corresponding pairwise dissimilarity for a set of 14 ribosomal proteins. Asterisks(* and **) indicates non-significant (P > 0.01) and significant (P < 0.01) difference between means of sequence dissimilarities of two studied groups, respectively
This pairwise dissimilarity analysis indicates that the evolutionary history of the ammonia oxidation-related genes is complex, and might indicate the occurrence of horizontal transfer(s) between comammox Nitrospira and β-AOB.
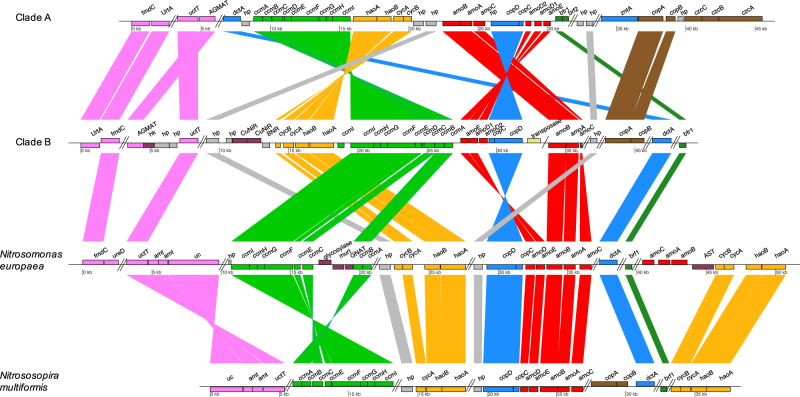
We, subsequently, analysed the genomic arrangement of ammonia oxidation-related genes for all comammox genomes and for representative β-AOB genomes (Fig. 5 and Supplementary Figure S9). For all comammox genomes except for Ca. N. inopinata, the AMO and HAO gene clusters as well as the cytochrome c-biosynthesis genes are situated in the same genomic region. This is not the case for AOB genomes. Additionally, comammox genomes have two copies of amoD, while AOB only have one copy. Among the comammox genomes, clade B genomes uniquely contain a duplication of one of the cytochrome c-biosynthesis proteins (CcmI). Clade A genomes have two duplicated hypothetical proteins-coding genes next to the hao operon, while comammox clade B and AOB genomes have only one copy (more details of genomic arrangement analysis in Supplementary Information). Shared features in the genomic regions encoding the ammonia oxidization pathways between the two comammox clades that are not shared with other ammonia oxidizers, suggests that the comammox clades have a common ancestor for this specific genetic capacity.
Fig. 5.
Schematic of the ammonia oxidation pathway genomic region as well as other AOB-related genomic features in comammox Nitrospira clade A (Ca. N. nitrificans), clade B (GWW3 bin) and selected ammonia-oxidizing bacteria (Nitrosospira multiformis and Nitrosomonas europaea). Homologous genes are connected by lines. Functions of the encoded proteins are represented by color. Parallel double lines designate a break in locus organization (other genes are in between the genes of interest). Single line designates a break probably due to contig fragmentation. Position of the blocks denotes the orientation of the coding strand (upper strands for forward; down strands for reverse)
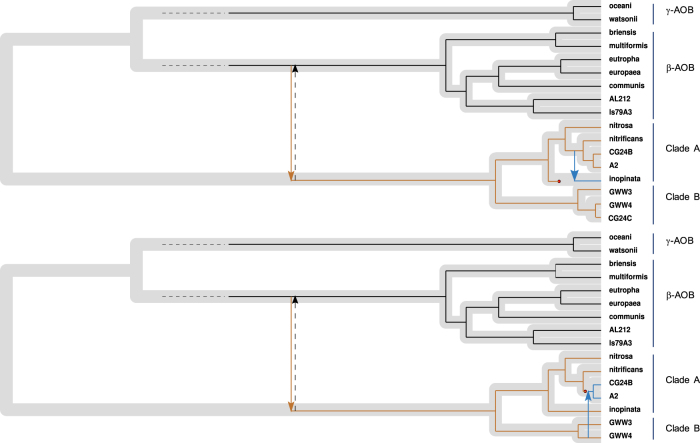
A reconciliation analysis was performed to stringently examine the possible occurrences of gene duplication(s), gene loss(es), and horizontal gene transfer(s). First, we constructed a clock-model based species-tree from the 14 ribosomal proteins (Supplementary Table S1), including the 16 examined Nitrospira genomes, Leptospirillum ferrooxidans (of the Nitrospirae phylum), eight and two publically available β-AOB and γ-AOB genomes, respectively, and two additional non-nitrifier β- and γ- proteobacterial genomes closely related to β-AOB and γ-AOB, respectively. (Supplementary Figure S10 and Supplementary Table S4). Gene trees were, subsequently, constructed for the ammonia oxidation-related genes under investigation (Supplementary Table S4). The probabilistic (Bayesian) analysis provided strong support for horizontal transfer of the majority of the investigated genes (Supplementary Table S6): for 12 genes, the posterior probability for at least one transfer event was between 95 and 100%, for other 10, the posterior probability was between 80 and 95%. For all investigated genes, we found very strong support (95–100%) for either a transfer or a duplication event.
The evolutionary history of amoA and haoA was further investigated. For amoA, the reconciliation model suggests a gene transfer event from β-AOB to an ancestor of comammox Nitrospira (Fig. 6). In addition, a transfer of amoA from the common ancestor of Ca. N. nitrosa and AAU_MBR_1 to Ca. N. inopinata was inferred. This transfer would explain the deviation in the tetranucleotide pattern of the amoCAB-containing region compared to the genome-wide signature identified in Ca. N. inopinata by Daims et al. [1]. Furthermore, unlike the other comammox genomes, Ca. N. inopinata does not have the amo operon located in the same genomic region as the hao operon and the genes for cytochrome c-biogenesis system II (Supplementary Figure S9).
Fig. 6.
Reconciliation of functional gene trees (based on amino acid sequences. Top. AmoA; bottom. HaoA) with species tree for comammox Nitrospira, β-AOB, and γ-AOB genomes. The species tree, based on 14 ribosomal proteins, is shown in gray with the gene-trees super-imposed on top in narrower black lines. Arrows and red dots denote transfer and loss events, respectively. The displayed tree is the most parsimonious tree. Dashed arrows indicate an alternative reconciliation scenario
The high dissimilarity between the amoA sequences of the otherwise closely related clade A and clade B comammox genomes (Supplementary Figure S10), may be explained by an adaptation of the comammox organisms to different niches. Based on the pairwise dissimilatory sequence analysis, clade B comammox genomes have a faster evolution rate of the ammonia oxidation-related proteins than clade A genomes (P < 0.0001; only for AmoA P < 0.0005), while the two clades do not have a different evolution rate for the housekeeping protein (P > 0.5).
Regarding haoA, the reconciliation model predicted two transfer events (Fig. 6). As for amoA, a transfer from β-AOB to an ancestor of comammox Nitrospira is suggested. Another transfer was inferred from clade B to the ancestor of the clade A genomes A2 and CG24_B. Consistent with this hypothesis, A2 and CG24_B contain genes coding for a copper-containing nitrite reductase next to the HAO cluster as also observed for clade B genomes (Supplementary Figure S10).
Besides the scenarios predicted from the reconciliation model, alternative scenarios cannot be ruled out due to the uncertainties associated with the limited number of sequenced Nitrospira and AOB genomes. An alternative scenario would be HGT of amoA and haoA from comammox Nitrospira to β-AOB (Fig. 6). In another possible scenario, both comammox Nitrospira and β-AOB would have acquired the ammonia-oxidation genes from an unknown third donor. This final scenario was tested in our reconciliation analysis by including the option of investigating transfers to and from unsampled species. The potential involvement of an unknown donor in the evolutionary history of amoA is not discarded, although these scenarios are less parsimonious than the one showed in Fig. 6. On the other hand, no evidence of HGT from an unsampled species was detected for haoA.
As comammox clade A and clade B do not form a monophyletic group within Nitrospira sublineage II (Supplementary Figure S2), loss events for both amoA and haoA are expected in some of the Nitrospira spp. that phylogenetically placed between these clades such as N. moscoviensis and CG24_D. One possible explanation for loss of ammonia oxidation capacity maybe the postulated trade-off between rate and yield of ATP production [13]. Under this hypothesis, shortening the nitrification pathway would lead to an increased specific growth rate which could be advantageous in some scenarios. Thus, the lack of the putatively acquired AMO and HAO-related genes in extant canonical Nitrospira spp. could be the result of selection for optimal pathway length.
The clock-based species-tree estimated the separation between clade A and clade B comammox genomes to have occurred approximately 300 ± 90 MYA, while the split between Nitrospira lineage I and II was dated at 375 ± 50 MYA (Supplementary Figure S10). Our model point towards an earlier transfer event for both amoA and haoA although it was not possible to date the HGT event with a high level of certainty. Hence, it remains unknown whether additional loss events would be required to explain the lack of comammox genes in Nitrospira genomes belonging to lineages other than lineage II.
Conclusions
In summary, our findings reveal diverse genetic capabilities of the two comammox clades, canonical Nitrospira and canonical ammonia oxidizing prokaryotes. The absence of NOx metabolism pathways in the studied comammox genomes, as observed in AOA and some oligotrophic AOB strains in comparison with copiotrophic AOB, and the diverse genetic capacity for tolerance of low micronutrient concentrations, together with the dominant detection of comammox Nitrospira in substrate-limited environments might indicate an oligotrophic lifestyle of comammox Nitrospira. In fact, an oligotrophic lifestyle has been experimentally demonstrated in N. inopinata [76], the only comammox isolated to date.
Additionally, we identified a high probability of transfer events between β-proteobacterial ammonia oxidizers and comammox Nitrospira for genes belonging to the ammonia oxidation pathway. Together, these results expand our knowledge of the ecology and evolution of the recently discovered comammox Nitrospira. Further discovery and characterization of new comammox Nitrospira, as well as canonical NOB and AOP genomes will help confirm these observations.
Electronic supplementary material
Acknowledgements
This research was financially supported by MERMAID (An initial training network funded by the People Program—Marie Skłodowska-Curie Actions- of the European Union’s Seventh Framework Program FP7/2007-2013/ under REA grant agreement n°607492), and a research grant (Expa-N, 13391) from VILLUM FONDEN.
Authors contributions
The study was conceived by AP, BFS and TSP. AP performed the genome binning and comparative genomic analyses. AGP conducted the evolutionary analysis supported by AP. AP led interpretation of the results supported by SJF, AD and AGP. AP drafted the manuscript helped by SJF, AGP, and BFS; all authors contributed to its revision, and approved the final submission.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Additional information:All raw sequence data and genome sequences have been deposited at NCBI under the project PRJNA384587, with sequence accession for the raw sequence data: SRR5739198—SRR5739203; and for the draft genomes: GCA_002869925.1, GCA_002869845.1, GCA_002869885.1, GCA_002869855.1 and GCA_002869895.1 (GenBank assembly accessions). The file containing the protein clusters sequences is available on figshare ((http://dx.doi.org/10.6084/m9.figshare.4924748).
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0083-3) contains supplementary material, which is available to authorized users.
References
- 1.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–9. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, et al. Complete nitrification by a single microorganism. Nature. 2015;528:555–9. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gülay A, Musovic S, Albrechtsen HJ, Al-Soud WA, Sørensen SJ, Smets BF. Ecological patterns, diversity and core taxa of microbial communities in groundwater-fed rapid gravity filters. ISME J. 2016;10:2209–22. doi: 10.1038/ismej.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Off S, Alawi M, Spieck E. Enrichment and physiological characterization of a novel nitrospira-like bacterium obtained from a marine sponge. Appl Environ Microbiol. 2010;76:4640–6. doi: 10.1128/AEM.00320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pester M, Maixner F, Berry D, Rattei T, Koch H, Lücker S, et al. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol. 2014;16:3055–71. doi: 10.1111/1462-2920.12300. [DOI] [PubMed] [Google Scholar]
- 6.Palomo A, Jane Fowler S, Gülay A, Rasmussen S, Sicheritz-Ponten T, Smets BF. Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. ISME J. 2016;10:2569–81. doi: 10.1038/ismej.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber-Dorninger C, Pester M, Kitzinger K, Savio DF, Loy A, Rattei T, et al. Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J. 2015;9:643–55. doi: 10.1038/ismej.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, et al. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol. 2006;8:1487–95. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- 9.Park HD, Noguera DR. Nitrospira community composition in nitrifying reactors operated with two different dissolved oxygen levels. J Microbiol Biotechnol. 2008;18:1470–4. [PubMed] [Google Scholar]
- 10.Koch H, Galushko A, Albertsen M, Schintlmeister A, Gruber-Dorninger C, Lucker S, et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science. 2014;345:1052–4. doi: 10.1126/science.1256985. [DOI] [PubMed] [Google Scholar]
- 11.Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci. 2015;112:11371–6. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palatinszky M, Herbold C, Jehmlich N, Pogoda M, Han P, von Bergen M, et al. Cyanate as an energy source for nitrifiers. Nature. 2015;524:105–8. doi: 10.1038/nature14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa E, Pérez J, Kreft JU. Why is metabolic labour divided in nitrification? Trends Microbiol. 2006;14:213–9. doi: 10.1016/j.tim.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Pinto AJ, Marcus DN, Ijaz Z, Bautista-de los Santos QM, Dick GJ, Raskin L. Metagenomic Evidence for the Presence of Comammox Nitrospira-Like Bacteria in a Drinking Water System. mSphere. 2015;1:e00054–15. doi: 10.1128/mSphere.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hug LA, Thomas BC, Sharon I, Brown CT, Sharma R, Hettich RL, et al. Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages. Environ Microbiol. 2016;18:159–73. doi: 10.1111/1462-2920.12930. [DOI] [PubMed] [Google Scholar]
- 16.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–8. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 17.Karst SM, Kirkegaard RH, Albertsen M. (2016). Mmgenome: a toolbox for reproducible genome extraction from metagenomes. bioRxiv 59121.
- 18.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eren AM, Esen OumlC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosi E, Donati B, Galardini M, Brunetti S, Sagot MF, Lió P, et al. MeDuSa: a multi-draft based scaffolder. Bioinformatics. 2015;31:2443–51. doi: 10.1093/bioinformatics/btv171. [DOI] [PubMed] [Google Scholar]
- 21.Boetzer M, Pirovano W, Zerbino D, Birney E, Simpson J, Wong K, et al. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer F, Paarmann D, D’Souza M, Olson R, Glass E, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guy L, Kultima JR, Andersson SGE, Quackenbush J. GenoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2011;27:2334–5. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:1–6. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:699–710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 29.Ochman H, Wilson AC. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 30.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 32.Sjöstrand J, Tofigh A, Daubin V, Arvestad L, Sennblad B, Lagergren J. A bayesian method for analyzing lateral gene transfer. Syst Biol. 2014;63:409–20. doi: 10.1093/sysbio/syu007. [DOI] [PubMed] [Google Scholar]
- 33.Jacox E, Chauve C, Szöllősi GJ, Ponty Y, Scornavacca C. ecceTERA: comprehensive gene tree-species tree reconciliation using parsimony. Bioinformatics. 2016;32:btw105. doi: 10.1093/bioinformatics/btw105. [DOI] [PubMed] [Google Scholar]
- 34.Chevenet F, Doyon JP, Scornavacca C, Jacox E, Jousselin E, Berry V. SylvX: a viewer for phylogenetic tree reconciliations. Bioinformatics. 2016;32:608–10. doi: 10.1093/bioinformatics/btv625. [DOI] [PubMed] [Google Scholar]
- 35.R Core Team (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. http://www.R-project.org/.
- 36.Luo C, Rodriguez-R LM, Konstantinidis KT. MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 2014;42:1–12. doi: 10.1093/nar/gkt1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Offre P, Kerou M, Spang A, Schleper C. Variability of the transporter gene complement in ammoniaoxidizing archaea. Trends Microbiol. 2014;22:665–75. doi: 10.1016/j.tim.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Weidinger K, Neuhäuser B, Gilch S, Ludewig U, Meyer O, Schmidt I. Functional and physiological evidence for a Rhesus-type ammonia transporter in Nitrosomonas europaea. FEMS Microbiol Lett. 2007;273:260–7. doi: 10.1111/j.1574-6968.2007.00805.x. [DOI] [PubMed] [Google Scholar]
- 39.Westhoff CM, Wylie DE. Transport characteristics of mammalian Rh and Rh glycoproteins expressed in heterologous systems. Transfus Clin Biol. 2006;13:132–8. doi: 10.1016/j.tracli.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Javelle A, Thomas G, Marini AM, Krämer R, Merrick M. In vivo functional characterization of the Escherichia coli ammonium channel AmtB: evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem J. 2005;390:215–22. doi: 10.1042/BJ20042094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter B, Kuspert M, Ansorge D, Kramer R, Burkovski A. Dissection of ammonium uptake systems in Corynebacterium glutamicum: mechanism of action and energetics of AmtA and AmtB. J Bacteriol. 2008;190:2611–4. doi: 10.1128/JB.01896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills J, Wyborn NR, Greenwood JA, Williams SG, Jones CW. An outer-membrane porin inducible by short-chain amides and urea in the methylotrophic bacterium Methylophilus methylotrophus. Microbiology. 1997;143:2373–9. doi: 10.1099/00221287-143-7-2373. [DOI] [PubMed] [Google Scholar]
- 43.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. In situ characterization of nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol. 2001;67:5273–84. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, et al. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2016;10:761–77. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lücker S, Nowka B, Rattei T, Spieck E, Daims H. The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol. 2013;4:1–19. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moparthi VK, Hägerhäll C. The evolution of respiratory chain complex i from a smaller last common ancestor consisting of 11 protein subunits. J Mol Evol. 2011;72:484–97. doi: 10.1007/s00239-011-9447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spero MA, Aylward FO, Currie CR, Donohue TJ. Phylogenomic analysis and predicted physiological role of the proton-translocating NADH:Quinone oxidoreductase (Complex I) across bacteria. MBio. 2015;6:e00389–15. doi: 10.1128/mBio.00389-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arai H, Jung HR, Kaplan S. Transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 2008;190:286–99. doi: 10.1128/JB.01375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spero MA, Brickner JR, Mollet JT, Pisithkul T, Amador-Noguez D, Donohue TJ. Different functions of phylogenetically distinct bacterial complex I isozymes. J Bacteriol. 2016;198:1268–80. doi: 10.1128/JB.01025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lancaster CR. Wolinella succinogenes quinol:fumarate reductase - 2.2-Å resolution crystal structure and the E-pathway hypothesis of coupled transmembrane proton and electron transfer. Biochim Biophys Acta. 2002;1565:215–31. doi: 10.1016/S0005-2736(02)00571-0. [DOI] [PubMed] [Google Scholar]
- 51.Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA. 2010;107:13479–84. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giuffrè A, Borisov VB, Arese M, Sarti P, Forte E. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim Biophys Acta. 2014;1837:1178–87. doi: 10.1016/j.bbabio.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 53.Zambonelli C and Roberts MF. Non-HKD phospholipase D enzymes: new players in phosphatidic acid signaling? Prog Nucleic Acid Res Mol Biol. 2005;79:133–181 [DOI] [PubMed]
- 54.Harayama S, Rekik M, Ngai KL, Ornston LN. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J Bacteriol. 1989;171:6251–8. doi: 10.1128/jb.171.11.6251-6258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spieck E, Hartwig C, McCormack I, Maixner F, Wagner M, Lipski A, et al. Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ Microbiol. 2006;8:405–15. doi: 10.1111/j.1462-2920.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 56.Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U. Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol. 1986;144:1–7. doi: 10.1007/BF00454947. [DOI] [Google Scholar]
- 57.Ouellet H, Ranguelova K, LaBarre M, Wittenberg JB, Wittenberg BA, Magliozzo RS, et al. Reaction of Mycobacterium tuberculosis truncated hemoglobin O with hydrogen peroxide: Evidence for peroxidatic activity and formation of protein-based radicals. J Biol Chem. 2007;282:7491–503. doi: 10.1074/jbc.M609155200. [DOI] [PubMed] [Google Scholar]
- 58.Torge R, Comandini A, Catacchio B, Bonamore A, Botta B, Boffi A. Peroxidase-like activity of Thermobifida fusca hemoglobin: the oxidation of dibenzylbutanolide. J Mol Catal B Enzym. 2009;61:303–8. doi: 10.1016/j.molcatb.2009.08.010. [DOI] [Google Scholar]
- 59.Marles-Wright J, Lewis RJ. The stressosome: molecular architecture of a signalling hub. Biochem Soc Trans. 2010;38:928–33. doi: 10.1042/BST0380928. [DOI] [PubMed] [Google Scholar]
- 60.Cha JS, Cooksey DA. Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon. Appl Environ Microbiol. 1993;59:1671–4. doi: 10.1128/aem.59.5.1671-1674.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph nitrosomonas europaea complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph nitrosomonas europaea †. J Bacteriol. 2003;185:2759–73. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richard-Fogal CL, Frawley ER, Feissner RE, Kranz RG. Heme concentration dependence and metalloporphyrin inhibition of the system I and II cytochrome c assembly pathways. J Bacteriol. 2007;189:455–63. doi: 10.1128/JB.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016;24:699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Albers E. Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5’-methylthioadenosine. IUBMB Life. 2009;61:1132–42. doi: 10.1002/iub.278. [DOI] [PubMed] [Google Scholar]
- 65.Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M. Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J. 2013;451:145–54. doi: 10.1042/BJ20121744. [DOI] [PubMed] [Google Scholar]
- 66.Lucker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci. 2010;107:13479–84. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belevich I, Borisov VB, Konstantinov AA, Verkhovsky MI. Oxygenated complex of cytochrome bd from Escherichia coli: stability and photolability. FEBS Lett. 2005;579:4567–70. doi: 10.1016/j.febslet.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 68.Le Laz S, Kpebe A, Bauzan M, Lignon S, Rousset M, Brugna M. A biochemical approach to study the role of the terminal oxidases in aerobic respiration in Shewanella oneidensis MR-1. PLoS One. 2014;9:e86343. doi: 10.1371/journal.pone.0086343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 2011;1807:1398–413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kageyama H, Tripathi K, Rai AK, Cha-Um S, Waditee-Sirisattha R, Takabe T. An alkaline phosphatase/phosphodiesterase, PhoD, induced by salt stress and secreted out of the cells of Aphanothece halophytica, a halotolerant cyanobacterium. Appl Environ Microbiol. 2011;77:5178–83. doi: 10.1128/AEM.00667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen YC, Hu YN, Shaw GC. Expressions of alkaline phosphatase genes during phosphate starvation are under positive influences of multiple cell wall hydrolase genes in Bacillus subtilis. J Gen Appl Microbiol. 2016;62:106–9. doi: 10.2323/jgam.62.106. [DOI] [PubMed] [Google Scholar]
- 72.Amin Sa, Moffett JW, Martens-Habbena W, Jacquot JE, Han Y, Devol A, et al. Copper requirements of the ammonia-oxidizing archaeon Nitrosopumilus maritimus SCM1 and implications for nitrification in the marine environment. Limnol Oceanogr. 2013;58:2037–45. doi: 10.4319/lo.2013.58.6.2037. [DOI] [Google Scholar]
- 73.De Vet WWJM, Van Loosdrecht MCM, Rietveld LC. Phosphorus limitation in nitrifying groundwater filters. Water Res. 2012;46:1061–9. doi: 10.1016/j.watres.2011.11.075. [DOI] [PubMed] [Google Scholar]
- 74.Scherrenberg SM, Menkveld HWH, Bechger M, Van Der Graaf JHJM. Phosphorus and nitrogen profile measurements to locate phosphorus limitation in a fixed bed filter. Water Sci Technol. 2009;60:2537–44. doi: 10.2166/wst.2009.606. [DOI] [PubMed] [Google Scholar]
- 75.Wagner FB, Nielsen PB, Boe-Hansen R, Albrechtsen HJ. Copper deficiency can limit nitrification in biological rapid sand filters for drinking water production. Water Res. 2016;95:280–8. doi: 10.1016/j.watres.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 76.Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature. 2017;549:269–72. doi: 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Camejo PY, Santo Domingo J, McMahon KD, Noguera DR. Genome-enabled insights into the ecophysiology of the Comammox Bacterium ‘ Candidatus Nitrospira nitrosa’. mSystems. 2017;2:e00059–17. doi: 10.1128/mSystems.00059-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao JF, Fan XY, Pan KL, Li HY, Sun LX. Diversity, abundance and activity of ammonia-oxidizing microorganisms in fine particulate matter. Sci Rep. 2016;6:38785. doi: 10.1038/srep38785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bartelme RP, McLellan SL, Newton RJ. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and Comammox Nitrospira. Front Microbiol. 2017;8:101. doi: 10.3389/fmicb.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pjevac P, Schauberger C, Poghosyan L, Herbold CW, van Kessel MAHJ, Daebeler A, et al. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of Comammox Nitrospira in the environment. Front Microbiol. 2017;8. e-pub ahead of print, 10.3389/fmicb.2017.01508. [DOI] [PMC free article] [PubMed]
- 81.Wang Y, Ma L, Mao Y, Jiang X, Xia Y, Yu K, et al. Comammox in drinking water systems. Water Res. 2017;116:332–41. doi: 10.1016/j.watres.2017.03.042. [DOI] [PubMed] [Google Scholar]
- 82.Speth DR, Guerrero-Cruz S, Dutilh BE, et al. Genome-based microbial ecology of anammox granules in a fullscale wastewater treatment system. Nat Commun. 2016;7:11172. doi: 10.1038/ncomms11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.