Abstract
Context
Previous genome-wide association studies have shown that single-nucleotide polymorphism (SNP) rs2439302 in chromosome 8p12 is significantly associated with papillary thyroid carcinoma (PTC) risk and dysregulated NRG1 expression. The underlying mechanisms remain to be discovered.
Objective
To evaluate the expression of NRG1 isoforms, candidate functional variants, and potential genes downstream of NRG1 in thyroid tissue.
Methods
Quantitative reverse transcription polymerase chain reaction was applied for gene expression analysis. SNaPshot assay, haplotype, and computer analyses were performed to evaluate candidate functional variants. Other functional assays [chromatin immunoprecipitation (ChIP) assay, luciferase assay, small interfering RNA knockdown, and RNA sequencing] were performed.
Results
Three NRG1 isoforms (NM_004495, NM_013958, and NM_001160008) tested were highly expressed in thyroid tissue. The expression levels of the three isoforms were significantly correlated with the genotypes of rs2439302. A DNA block of ~32 kb containing the risk G allele of rs2439302 was revealed, harboring multiple candidate functional variants. ChIP assay for active chromatin markers indicated at least nine regions in the DNA block showing strong H3Kme1 and H3K27Ac signals in thyroid tissue. Luciferase reporter assays revealed differential allelic activities associated with seven SNPs. Knocking down NRG1 in primary thyroid cells revealed downstream or interacting genes related to NRG1.
Conclusions
Our data suggest a role for transcriptional regulation of NRG1 in the predisposition to PTC.
The thyroid cancer risk [G] allele of rs2439302 belongs to a haplotype whose SNPs regulate NRG1 expression followed by transcriptional regulation of NRG1-targeting or -interacting genes.
Thyroid cancer, the most common endocrine malignancy, represents ~1% of newly diagnosed cancers. A recent study revealed that the overall incidence of thyroid cancer in the United States increased 3% annually from 1974 to 2013; this may be a true increase, or it may be due to overdiagnosis of papillary microcarcinoma (1, 2). It is estimated that 56,870 people in the United States will be diagnosed with thyroid cancer in 2017 (3). The majority of thyroid cancers are nonmedullary thyroid carcinoma. Papillary thyroid carcinoma (PTC) is the main form of nonmedullary thyroid carcinoma, representing ~80% of all thyroid cancers. Although the etiology of PTC is not well characterized, it is clearly influenced by both genetic and environmental factors. On one hand, radiation exposure of the thyroid during childhood is the best-defined environmental factor associated with PTC. On the other hand, PTC shows a high degree of heritability. Genetic predisposition plays a major role, as evidenced by case control studies (4, 5). The genetic factors can probably be attributed to many low-penetrance DNA variants in the human genome (6–8).
The genetic predisposition to PTC has been widely studied, and advances have been made through genome-wide association studies (GWAS). Previously, we identified a single-nucleotide polymorphism (SNP), rs2439302, in chromosome 8p12 that was associated with PTC (9). The association of rs2439302 and thyroid cancer risk has been independently replicated by subsequent association studies in different populations (7, 10–12). Recently we performed meta-GWAS analyses and confirmed SNP rs2439302 as a PTC risk variant (13). In this analysis, SNP rs2466076 in the 8p12 locus was found with the lowest P value [odds ratio (OR) 1.32, P = 1.5 × 10−17]. These two SNPs are in high linkage disequilibrium (LD), with r2 = 0.94. Both SNPs are located in introns of the neuregulin 1 (NRG1) gene.
The NRG1 gene encodes a membrane glycoprotein that mediates cell–cell signaling and plays a critical role in the growth and development of multiple organ systems (14). NRG1 is produced in multiple isoforms by alternative splicing and usage of distinct promoters, which allows it to perform a wide variety of functions. Additionally, NRG1 has been demonstrated to be involved in carcinoma development and metastasis (15–17).
It was recently reported that the risk allele [G] of rs2439302 was associated with decreased expression of NRG1 in thyroid tissue (11). This observation conflicts with the GTEx data, in which the [G] allele is correlated with increased expression of NRG1 in thyroid tissue (https://www.gtexportal.org/) (18). In another study the risk [G] allele of SNP rs2439302 was associated with lower expression levels of NRG1 in blood, which is consistent with the GTEx data (9, 18). In an effort to elucidate the functional basis of the underlying disease risk, we performed gene expression and correlation analysis between the genotypes of rs2439302 and the isoform abundance of NRG1. We evaluated multiple candidate functional variants in a ~32-kb DNA block harboring the GWAS SNPs (rs2439302 and rs2466076) by performing haplotype analysis and functional assays. We attempted to explore the downstream and interacting genes by small interfering RNA (siRNA) knockdown of NRG1 in primary thyroid cells followed by RNA sequencing (RNA-seq) gene expression analysis.
Materials and Methods
The studies were approved by the Institutional Review Board at The Ohio State University (OSU), and all subjects gave written informed consent before participation.
Thyroid tissue samples and cell lines
Fresh frozen unaffected thyroid tissue samples (n = 163) were obtained from patients with PTC during surgery. Blood samples (1359 PTC cases/1605 controls) were collected at OSU as part of ongoing studies. The clinical information is provided in Supplemental Table 1. The thyroid cancer cell lines (BCPAP, KTC1, TPC1, and SW1736) were incubated in RPMI1640 medium supplemented with 10% fetal bovine serum at 37°C in humidified air with 5% CO2.
Quantitative reverse transcription polymerase chain reaction
Quantitative reverse transcription polymerase chain reaction (RT-PCR) was performed with an ABI PRISM 7700 DNA Sequence Detection System (Applied Biosystems). TaqMan assays with custom probes (Life Technologies) were applied for measuring three NRG1 isoforms (NM_004495, NM_013958, and NM_001160008). The primer and probe sequences are provided in Supplemental Table 2. These primers were designed to amplify specific transcripts based on the unique exon structure of each isoform. A predesigned TaqMan Assay (Hs00247620_m1, Life Technologies) was used for detecting multiple NRG1 isoforms. A GAPDH TaqMan kit was used as an internal control, and the equation 2−ΔCt, where ΔCt = Ct(GENE) − Ct(GAPDH), was used to calculate the relative expression levels.
SNP genotyping
Blood genomic DNA was used for genotyping SNP rs2439302 with a SNaPshot assay as described (19). The polymerase chain reaction (PCR) primer and extension primer sequences are provided in Supplemental Table 2. The PCR assays were performed according to a standard protocol as follows: 5 minutes at 94°C; followed by 30 cycles of 30 seconds at 94°C, 30 seconds at 58°C, and 30 seconds at 72°C; followed by a final extension of 10 minutes at 72°C. An ABI 3730 DNA Analyzer was used for allele analysis.
Expression quantitative trait loci, LD, and haplotype analyses
The NRG1 expression quantitative trait loci data were obtained from the Genotype and Tissue Expression Project (https://www.gtexportal.org/). LD analysis was performed with the genotypes from the 1000 Genomes Project (phase 3, EUR) in Haploview 4.2 software (20). Haplotype analysis was performed in an Ohio cohort of 1359 cases and 1605 controls with 27 selected SNPs. The genotypes were obtained with the use of SNP arrays followed by computer imputation performed in deCODE (13). We used the SHAPEIT program to estimate haplotype frequencies (21). For each haplotype, counts were compared between cases and controls. The ORs and P values were calculated by comparing the given haplotype with the rest of the haplotypes. Fisher exact test and univariate analysis were applied.
Quantitative chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was performed as previously described, with minor modifications (22). Briefly, protein-DNA cross-linking was performed by incubating cells with formaldehyde at a final concentration of 1% for 10 minutes at room temperature. After sonication, chromatin was immunoprecipitated with specific antibodies at 4°C overnight. The antibodies against H3K4me1 (ab8895), H3K4me3 (ab8580), and H3K27Ac (ab4729) were from Abcam. The immune complexes were then eluted from the magnetic beads, and the crosslinks were reversed by incubating at 65°C overnight. The DNA was purified with QIAquick PCR purification kit and used as the template in quantitative PCRs. Primers were designed to yield amplicons ranging in length from 100 to 250 bp, spanning the tested SNP regions (Supplemental Table 2).
Constructs, transient transfections, and luciferase reporter gene assays
DNA fragments containing the wild type or risk allele in the selected regions were PCR amplified and cloned into the PGL4.10-E4TATA vector. This vector contains a 50-bp minimal E4 TATA promoter sequence. The constructs were validated by Sanger sequencing. The primer sequences for the PCR reactions are provided in Supplemental Table 2. Cotransfection of KTC1 or HeLa cells was performed with empty vector or the enhancer constructs and a Renilla luciferase reporter plasmid. Cells were harvested after 24 hours, and lysates were used for luciferase assays.
Human primary thyroid cell culture and nucleofection
Human primary thyroid cells were cultured in customized 6H medium as previously described (23). Fresh nontumorous thyroid tissue samples were obtained from patients with PTC. The tissue was digested with 1% trypsin (Life Technologies) and 0.35% collagenase 4 (Worthington Biochemical) solution for 90 minutes at 37°C. Thyroid primary cells were seeded to a density of 105 to 106 cells per well on six-well plates. After being cultured for 5 days, cells were electroporated by a Nucleofector® II device (Amaxa) with 75 pmol siRNA in 100 μL of Basic Nucleofector Medium (VPI-1005; Lonza). Human NRG1 siRNA (ON-TARGET plus SMART pool, L-004608-02-0005) and negative control siRNA (ON-TARGET plus Nontargeting pool, D-001810-10-05) were purchased from Dharmacon. After the nucleofection step, cells were cultured for 24 hours and then harvested for further analysis.
RNA-seq sample preparation and differential gene expression analysis
RNA-seq samples were prepared and analyzed as previously described (24). Briefly, the total RNA samples were extracted by TRIzol reagent (Invitrogen) and then treated by DNase-1 (Ambion). Illumina TruSeq Stranded Total RNA Sample Prep Kit with Ribo-Zero Gold (catalog #RS-122-2201) was used to transform RNA into complementary DNA after removing ribosomal RNA and mitochondrial RNA. Then, 75-bp paired-end sequencing was performed with the Illumina HiSEquation 2500 system. The RNA-seq data have been submitted to the GEO database (accession #GSE100322).
Statistical analysis
Associations between quantitative PCR expression estimates and genotypes were analyzed with generalized linear models. RNA-seq gene expression data from NRG1 siRNA knockdown experiments were normalized and log2 transformed before comparative analysis. Quantitative RT-PCR expression data were analyzed by applying nonparametric Kruskal–Wallis analysis of variance tests and Wilcoxon rank sum tests. All P values reported are two-sided. Network, functional, and canonical pathway analyses of differentially expressed gene expression data were performed in Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems Inc.).
Results
Expression of NRG1 isoforms and correlation between rs2439302 and NRG1 expression
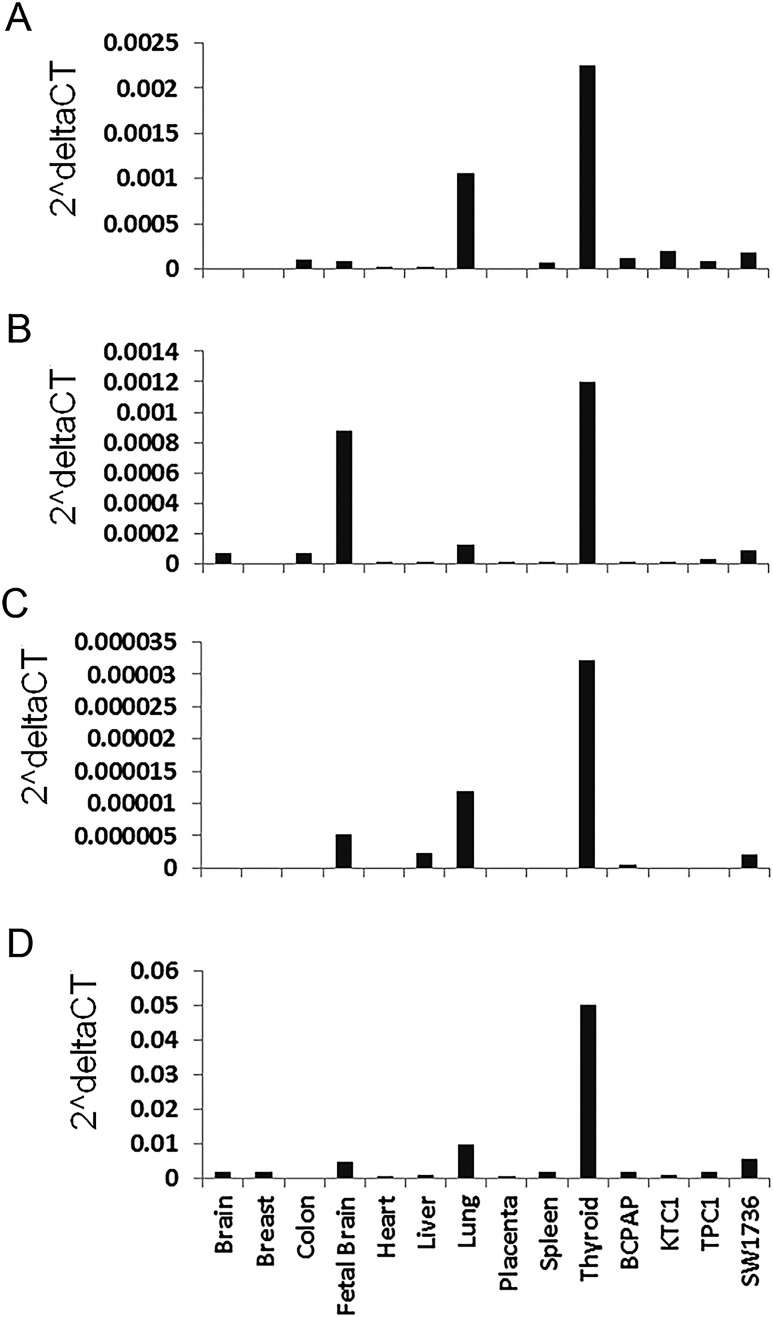
The NRG1 gene encodes multiple splicing isoforms. To analyze NRG1 expression in human tissues and thyroid cancer cell lines, we selected three NRG1 isoforms (NM_004495, NM_013958, and NM_001160008) by using the RNA-seq data from The Cancer Genome Atlas and our previous study (24, 25). We performed quantitative RT-PCR to measure the isoforms by using probe-specific, custom-designed TaqMan assays. We also used a predesigned TaqMan assay for measuring multiple NRG1 isoforms (designated as multiNRG1 in this study). The NRG1 isoforms and gene structures are shown in Supplemental Fig. 1A. A schematic representation of the three isoforms and the corresponding probes and primers used in the TaqMan assays are shown in Supplemental Fig. 1B. These three isoforms and multiNRG1 are abundantly expressed in thyroid, moderately expressed in fetal brain and lung, and marginally expressed in other tissues tested, including breast, colon, heart, placenta, and spleen and four thyroid cancer cell lines (BCPAP, KTC1, TPC1, and SW1736) (Fig. 1). The three isoforms share the exons 1 to 5 but differ in other exons. Their features are listed in Supplemental Table 3.
Figure 1.
Gene expression analysis of NRG1 isoforms in human tissues and thyroid cancer cell lines. Probe-specific TaqMan quantitative RT-PCR assays were performed. (A) NM_004495. (B) NM_013958. (C) NM_001160008. (D) MultiNRG1 isoforms. The depicted expression level was normalized with GAPDH used as an internal control. The multiNRG1 in (D) was measured with a predesigned TaqMan assay kit #Hs00247620_m1 (http://www.thermofisher.com).
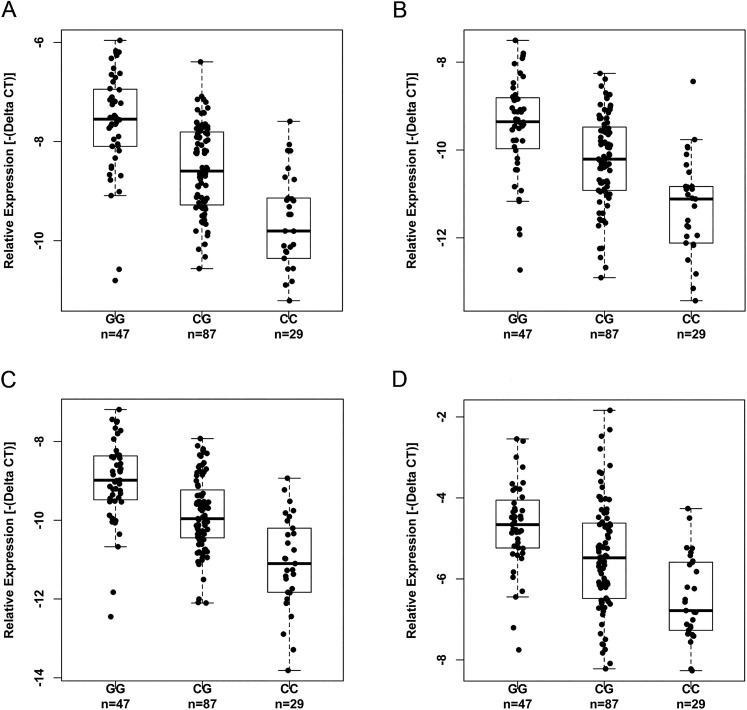
To test whether the DNA variant rs2439302 has any effect on the expression of the NRG1 isoforms, we measured the expression levels of the three isoforms and the multiNRG1 in unaffected thyroid tissue samples of an Ohio cohort (n = 163), genotyped rs2439302, and performed correlation analysis between the genotypes and the expression levels of NRG1 isoforms. We observed a significant correlation between the risk [G] allele and increased expression of all the three NRG1 isoforms and the multiNRG1 transcript levels when we compared them with the wild type [C] allele, with P < 0.0001 (Fig. 2). Our observation is consistent with the GTEx data (Supplemental Fig. 2).
Figure 2.
Correlation between the genotypes of rs2439302 and expression of NRG1 isoforms in unaffected thyroid tissue samples. (A) NM_004495, P = 2.81E-13. (B) NM_013958, P = 1.30E-09. (C) NM_001160008, P = 7.14E-14. (D) MultiNRG1, P = 1.17E-06. The P values were obtained by multivariate analysis and adjusted for age, sex, and chronic lymphocytic thyroiditis. CC, CG, GG, genotypes of rs2439302.
Haplotype analysis of SNPs in the NRG1 region and the identification of candidate functional SNPs
In the NRG1 genomic region a cluster of SNPs is significantly associated with the expression levels of NRG1 in thyroid tissue in the GTEx data, including the GWAS SNPs rs2439302 and rs2466076. We designated this region as a “focused region” with genomic coordinates of chromosome 8: 32181948 to 32469369 (Supplemental Fig. 3A). To evaluate the role of these SNPs in PTC predisposition, we performed an association analysis in an Ohio cohort of 1359 cases and 1605 controls. The regional association plot indicated that the cluster of SNPs are significantly associated with PTC (Supplemental Fig. 3B).
We next performed LD analyses in the “focused region” in an attempt to narrow down candidate functional SNPs. Haploview LD analysis revealed an LD block of ~32 kb around rs2439302 (Supplemental Fig. 3C). Most of the LD block is located in intron 1 of the three NRG1 isoforms we tested; it also spans the common exon 1 and a small region immediately upstream of exon 1. This LD block is contained in a gene regulatory hotspot, as shown by the H3K27Ac signals in ENCODE data (Supplemental Fig. 4). In this DNA block there are 46 SNPs significantly associated with PTC risk and NRG1 expression; all these SNPs are in introns of the NRG1 gene. To establish whether these SNPs have a functional role in transcriptional regulation, we studied the RegulomeDB database, which contains experimental data sets from ENCODE and other sources in addition to computational predictions of regulatory potential. These combinatory analyses revealed 27 SNPs to represent candidate functional and causal variants in the NRG1 locus (Supplemental Table 4).
We assessed the association of the 27 SNP haplotypes with PTC risk in the Ohio cohort described earlier. The estimated haplotypes with frequencies ≥0.01 in cases or controls are listed in Table 1. Notably, one haplotype (Hap1) containing the risk [G] allele of rs2439302 and the risk [G] allele of rs2466076 is significantly associated with increased PTC risk, with an allelic OR of 1.32, and accounts for ~16.2% of the cases. One haplotype (Hap2) appears as a protective one, with an OR of 0.65, accounting for 4.5% of the cases. These analyses suggest that multiple SNPs in this ~32-kb block are likely to be functional and contribute to the increased PTC risk.
Table 1.
Haplotype Analysis of the ~32-kb LD Block
| Haplotype | Controls (n) | Cases (n) | Frequency in Control | Frequency in Case | OR | P a | rs3802160 | rs3802158 | rs36213229 | rs7820838 | rs7834206 | rs73234136 | rs113350646 | rs4733128 | rs4733129 | rs4733130 | rs12548687 | rs7835688 | rs17646763 | rs2439312 | rs9642727 | rs17646936 | rs17719705 | rs7825175 | rs13258892 | rs35233333 | rs2439304 | rs2439303 | rs2439302 | rs2466077 | rs2466076 | rs2466075 | rs71512640 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap 1 | 409 | 439 | 0.127 | 0.162 | 1.32 | 1.96E-04 | G | T | T | C | A | C | A | T | C | C | G | C | C | G | C | A | T | A | C | T | A | T | G | G | G | A | G |
| Hap 2 | 216 | 121 | 0.067 | 0.045 | 0.65 | 1.57E-04 | A | C | G | T | C | T | G | C | T | T | A | G | T | A | A | G | A | G | T | C | G | C | C | T | T | G | A |
| Hap 3 | 145 | 88 | 0.045 | 0.032 | 0.71 | 1.29E-02 | A | C | G | C | C | T | G | C | T | T | A | G | T | G | A | G | A | G | T | C | G | C | C | T | T | G | A |
| Hap 4 | 47 | 22 | 0.015 | 0.008 | 0.55 | 2.07E-02 | A | C | G | C | C | T | G | C | T | T | A | G | T | G | A | A | A | G | T | T | A | C | C | T | T | G | G |
| Hap 5 | 69 | 83 | 0.021 | 0.031 | 1.43 | 3.18E-02 | G | T | T | C | A | C | G | T | C | C | G | C | C | G | C | A | T | G | C | T | A | T | G | G | G | G | G |
| Hap 6 | 524 | 499 | 0.163 | 0.184 | 1.15 | 4.18E-02 | G | T | T | C | A | C | G | T | C | C | G | C | C | G | C | A | T | G | C | T | A | T | G | G | G | A | G |
| Hap 7 | 43 | 22 | 0.013 | 0.008 | 0.60 | 5.99E-02 | A | C | G | T | C | T | G | C | T | T | A | G | T | A | A | A | A | G | C | T | G | C | C | T | T | G | A |
| Hap 8 | 96 | 65 | 0.030 | 0.024 | 0.79 | 1.73E-01 | A | C | G | T | C | T | G | C | T | T | A | G | T | A | A | G | A | G | T | C | G | C | C | T | T | G | G |
| Hap 9 | 387 | 298 | 0.121 | 0.110 | 0.90 | 1.92E-01 | A | C | G | C | C | T | G | C | T | T | A | G | T | G | A | A | A | G | C | T | G | C | C | T | T | G | G |
| Hap 10 | 83 | 84 | 0.026 | 0.031 | 1.20 | 2.70E-01 | A | C | G | C | C | T | G | C | T | T | A | G | T | G | A | A | A | G | C | T | G | C | C | T | T | A | G |
| Hap 11 | 81 | 79 | 0.025 | 0.029 | 1.16 | 3.77E-01 | G | T | T | C | A | C | A | T | C | C | G | C | C | G | C | A | T | A | C | T | A | T | G | T | T | G | G |
| Hap 12 | 145 | 110 | 0.045 | 0.040 | 0.89 | 4.04E-01 | A | C | G | C | C | T | G | C | T | T | A | G | T | G | A | G | A | G | T | C | G | C | C | T | T | G | G |
| Hap 13 | 43 | 32 | 0.013 | 0.012 | 0.88 | 6.41E-01 | G | T | T | C | A | C | A | T | C | C | G | C | C | G | C | A | T | A | C | T | A | T | G | G | G | G | G |
| Hap 14 | 77 | 70 | 0.024 | 0.026 | 1.08 | 6.76E-01 | A | C | G | C | C | T | G | C | T | T | A | G | T | G | A | A | A | G | C | T | G | C | C | T | T | G | A |
| Hap 15 | 84 | 73 | 0.026 | 0.027 | 1.03 | 8.71E-01 | G | T | T | C | A | C | G | T | C | C | G | C | C | G | C | A | T | G | C | T | A | T | G | T | T | G | G |
| Hap 16 | 103 | 88 | 0.032 | 0.032 | 1.01 | 1.00E+00 | A | C | G | C | C | T | G | C | T | T | A | G | T | G | A | A | A | G | C | T | G | C | C | G | G | A | G |
| Hap 17 | 99 | 83 | 0.031 | 0.031 | 0.99 | 1.00E+00 | A | C | G | T | C | T | G | C | T | T | A | G | T | A | A | G | A | G | T | C | G | C | C | T | T | A | G |
Haplotypes with a frequency of ≥0.01 are included in the table.
Fisher exact test.
Enrichment of active chromatin markers in the NRG1 locus in thyroid tissue
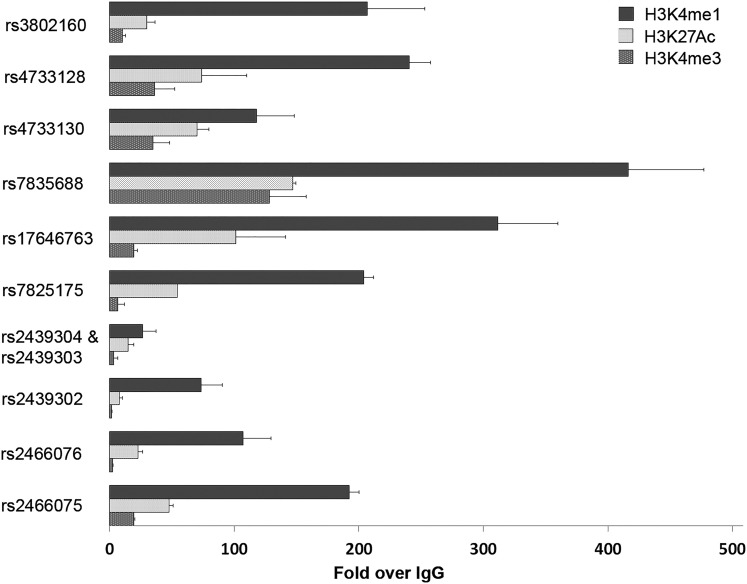
Because the active chromatin markers can predict enhancer or promoter activity at many loci, we performed site-directed ChIP assays for H3K4me1, H3K4me3, and H3K27Ac with thyroid tissue samples. We tested areas corresponding to 11 selected SNPs in the ~32-kb LD block (Fig. 3 and Supplemental Fig. 4). ChIP assay results revealed significant enrichment of H3K4me1 and H3K27Ac in all of the SNP areas tested. The enrichment of H3K4me3 was significant in several areas, such as rs4733128, rs4733130, and rs7835688, but overall the enrichment of H3K4me3 was much lower than that of H3K4me1. The increased H3K4me1/H3K4me3 ratios suggests that this DNA block region should be classified as an enhancer or superenhancer in thyroid tissue (26).
Figure 3.
ChIP assays of active chromatin markers H3K4me1, H3K27Ac, and H3K4me3 in the ~32-kb LD block as shown in Supplemental Fig. 4. Two fresh frozen thyroid tissue samples were used for the ChIP assays. The relative abundance of the three markers was normalized to immunoglobulin G control. H3K4me1 and H3K27Ac showed significant binding in all the tested regions, with P < 0.005, Student t test.
Allelic transcriptional luciferase activities in the ~32-kb LD block
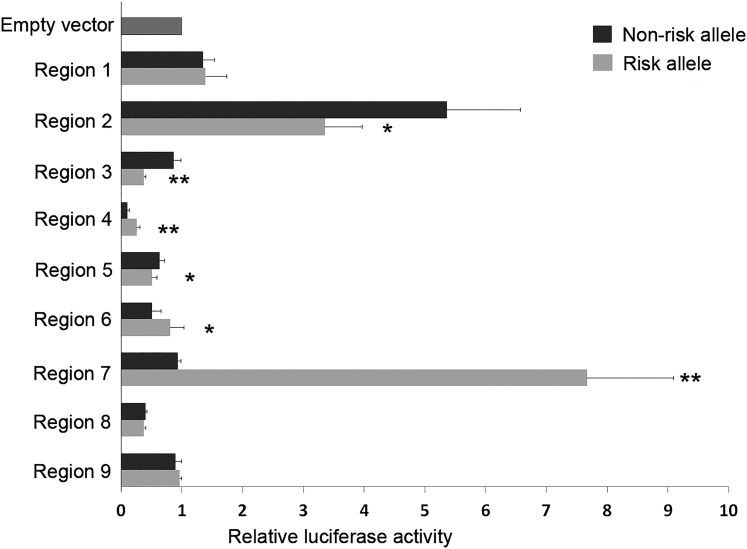
To test the potential functions of the 11 SNPs in the LD block, we performed enhancer luciferase assays. We hypothesized that these SNPs may be functional SNPs leading to a detectable allele-specific functional effect on transcription. To test this hypothesis, we amplified DNA fragments (~200 bp to 1.3 kb) in nine regions corresponding to the 11 SNPs (Supplemental Fig. 4). The DNA fragments carrying either risk or wild type alleles were cloned into luciferase reporter constructs, driven by a minimal promoter vector. Enhancer activities of these fragments were determined by transient transfection and luciferase assays in the BCPAP cell line (Fig. 4). When comparing with an empty vector control, we observed significant constitutive enhancer activities in two regions, corresponding to SNPs rs4733128 (region 2) and rs2439304 and rs2439303 (region 7). The rest of the regions showed slightly increased or decreased luciferase activities. Nevertheless, we observed significant differential allelic luciferase activity between the risk allele and the wild type allele in six regions (regions 2 to 7), with P < 0.001. The risk alleles of SNPs rs7835688 in region 4, rs7825175 in region 6, and rs2439304 and rs2439303 in region 7 showed increased transcription, whereas the risk alleles of rs4733128 in region 2, rs4733130 in region 3, and rs17646763 in region 5 showed reduced transcription. We performed the same experiments by using Hela cells and observed similar differential allelic luciferase activities in the six regions, although the patterns of luciferase activities could differ between BCPAP and HeLa cells (Supplemental Fig. 5). These six regions harbor seven SNPs, suggesting that at least these seven SNPs could alter the expression of firefly luciferase.
Figure 4.
Differential allelic luciferase activities associated with multiple SNPs in the NRG1 locus. DNA fragments containing the risk or wild type alleles in each region (regions 1 to 9) were cloned into a luciferase enhancer reporter vector with a minimal promoter. The 11 regions and the corresponding 11 SNPs in the ~32-kb LD block are shown in Supplemental Fig. 4. The luciferase assay was performed in at least three experiments. The luciferase activities were normalized with the empty vector control. *P < 0.05; **P < 0.01, Student t test.
Taken together, the results of the ChIP experiments and luciferase reporter assays are consistent with the hypothesis that the NRG1 locus harbors intronic regulatory elements, and multiple SNPs appear to be functioning by affecting the transcriptional activities.
NRG1 is involved in the regulation and interaction of coding and noncoding genes in thyroid primary cells
In an attempt to explore the downstream targets and signaling pathways of NRG1 in thyroid, we performed knockdown of NRG1 with siRNAs in human thyroid primary cells (23). We transiently transfected primary cultures of thyroid cells with siRNA oligos targeting multiple NRG1 isoforms (Supplemental Fig. 1). Knockdown of NRG1 resulted in low-level gene expression changes consistently shown in three separate thyroid primary cultures and determined by RNA deep sequencing. There were 141 dysregulated genes with fold changes ≥1.25 (upregulated, n = 71; downregulated, n = 70; P < 0.05), with the NRG1 gene being the top downregulated one. There were 27 dysregulated genes with fold changes ≥1.3 (upregulated, n = 6; downregulated, n = 21) (Table 2). Notably, among the downregulated genes, the majority (18 out of 21, 85.7%) are categorized as noncoding RNAs, pseudogenes, antisense, or uncharacterized transcripts. Biological functional analysis of coding genes via IPA software indicated that the top three categories of the differentially expressed genes belong to neurologic disease, psychological disorder, and cancer (Supplemental Table 5). The major molecular functions are protein synthesis, cell growth and proliferation, and cell death and survival (Supplemental Table 6).
Table 2.
Differentially Expressed Genes After siRNA Knocking Down NRG1
| Gene ID | Gene Name | Gene Type | Direction | Fold Changes a | P |
|---|---|---|---|---|---|
| ENSG00000157168.17 | NRG1 | Protein_coding | Down | 1.64 | 3.10E-11 |
| ENSG00000266718.1 | RP11-466A19.1 | Antisense | Down | 1.36 | 0.004 |
| ENSG00000223522.1 | AC093690.1 | Antisense | Down | 1.36 | 0.006 |
| ENSG00000253636.1 | RP11-531A24.5 | Antisense | Down | 1.36 | 0.006 |
| ENSG00000165181.15 | C9orf84 | Protein_coding | Down | 1.33 | 0.007 |
| ENSG00000250290.1 | CTC-820M8.1 | Processed_pseudogene | Down | 1.33 | 0.010 |
| ENSG00000254263.1 | RP11-473O4.4 | LncRNA | Down | 1.33 | 0.010 |
| ENSG00000232295.6 | RP11-154D6.1 | Processed_transcript | Down | 1.32 | 0.005 |
| ENSG00000259167.2 | NMNAT1P1 | Processed_pseudogene | Down | 1.32 | 0.001 |
| ENSG00000116690.10 | PRG4 | Protein_coding | Down | 1.31 | 0.014 |
| ENSG00000182021.8 | RP11-381O7.3 | LncRNA | Down | 1.31 | 0.010 |
| ENSG00000271705.1 | RPL17P3 | Unprocessed_pseudogene | Down | 1.31 | 0.007 |
| ENSG00000256399.1 | RP11-274J7.3 | Processed_pseudogene | Down | 1.30 | 0.014 |
| ENSG00000178440.5 | LINC00843 | LncRNA | Down | 1.30 | 0.009 |
| ENSG00000230638.4 | RP11-486B10.4 | Processed_pseudogene | Down | 1.30 | 0.014 |
| ENSG00000200816.1 | SNORA38 | snoRNA | Down | 1.30 | 0.014 |
| ENSG00000228817.4 | BACH1-IT2 | LncRNA | Down | 1.30 | 0.017 |
| ENSG00000268758.4 | EMR4P | Transcribed_unprocessed_pseudogene | Down | 1.30 | 0.012 |
| ENSG00000233478.1 | RP1-187B23.1 | Antisense | Down | 1.30 | 0.008 |
| ENSG00000235253.1 | AC010240.2 | Processed_pseudogene | Down | 1.30 | 0.015 |
| ENSG00000233844.1 | KCNQ5-IT1 | Sense_intronic | Down | 1.30 | 0.014 |
| ENSG00000234664.1 | HMGN2P5 | Processed_pseudogene | Up | 1.38 | 0.003 |
| ENSG00000126453.8 | BCL2L12 | Protein_coding | Up | 1.34 | 0.006 |
| ENSG00000250588.5 | IQCJ-SCHIP1 | Protein_coding | Up | 1.34 | 0.005 |
| ENSG00000137331.11 | IER3 | Protein_coding | Up | 1.32 | 0.001 |
| ENSG00000198576.3 | ARC | Protein_coding | Up | 1.32 | 0.010 |
| ENSG00000007520.3 | TSR3 | Protein_coding | Up | 1.31 | 0.001 |
NRG1 vs control, three repeated experiments.
Discussion
Our understanding of NRG1 regulation and its involvement in PTC is challenged by the complexity of the gene, which gives rise to an extraordinary variety of isoforms. These isoforms differ significantly in their structures and expression in a tissue-specific manner. The three NRG1 isoforms we tested are categorized as NRG1 type 1; they are also named HRG-β-3, HRG-β-2b, and HRG-γ, respectively. Our data indicate that at least these three NRG1 isoforms show high expression in thyroid tissue compared with other tissues we tested. Interestingly, much lower NRG1 expression was observed in four thyroid cancer cell lines (BAPAP, KTC1, TPC1, and SW1736), implying aberrant regulation of NRG1 in these cell lines. Our study showed that the risk [G] allele of rs2439302 was significantly associated with increased expression of the three NRG1 isoforms and multiNRG1 in unaffected thyroid tissue samples. It is believed that members of the NRG1 family of growth factors bind to and activate the ErbB-3 and ErbB-4 receptors (14, 27). The isoform HRG-β2b is a transmembrane prototype NRG1, which could become a soluble secreted ligand for ErbBs. However, the other two isoforms (HRG-β-3 and HRG-γ) lack the transmembrane and cytoplasmic domains, are presumably retained intracellularly, and are considered cytosolic NRG1 isoforms. Their potential functional implications are poorly understood, possibly independent of the ErbB receptor system (28–30). Our data suggest that multiple isoforms with higher NRG1 expression in thyroid tissue contribute to increased PTC risk.
In this study we fine-mapped the NRG1 genomic region and identified a ~32-kb LD block containing rs2439302 and rs2466076, which was located mostly in the first intron of the three NRG1 isoforms we tested. We performed ChIP assays in this LD block on thyroid tissue samples and observed strong H3K4me1 and H3K27Ac activities in areas corresponding to 11 SNPs, whereas H3K4me3 activities were low or much lower. The specific histone modifications of H3K4me1 and H3K27Ac are often the readout of active enhancers, whereas the H3K4me3 marker usually corresponds to proximal promoters (26, 31–33). Our observations suggest a cluster of multiple enhancer elements or a superenhancer in thyroid in this locus (34). Although our data favor enhancer involvement, we could not exclude a promoter-type effect. It is possible that the same sequences encode both enhancer and promoter activities (35). Nevertheless, our data suggest that transcriptional regulation underlies the mechanisms of NRG1 involvement in thyroid cancer.
We and others previously observed that multiple enhancer variants within a given locus typically target the same gene and that multiple enhancer variants cooperatively contribute to altered expression of their target gene (36). In this case, luciferase experiments detected variable luciferase enhancer activities in different regions within the LD block. We focused on comparing the effects of the risk and wild type alleles, respectively, in each region and observed seven SNPs with significantly differential allelic luciferase activities, implying their being functional variants in the thyroid. The GWAS SNPs rs2439302 and rs2466076, though involved in the risk haplotypes, are seemingly not functional. The impact of the risk alleles on transcriptional regulation seems conflicting in that some of the risk alleles showed increased luciferase activities, whereas other risk alleles showed reduced transcription. It might be explained by the fact that the complex transcriptional regulatory mechanisms could not be completely represented by the cloned short DNA fragments. The effects of the NRG1 intronic variants we describe here are probably combinatorial in dictating the gene expression of NRG1 and in conferring susceptibility to PTC.
Studies of the mechanisms by which NRG1 influences unaffected thyroid and thyroid cancer are limited in the literature. We used siRNA to knock down NRG1 in human primary thyroid cells and revealed a subset of differentially expressed genes. Notably, most of the downregulated genes belong to the noncoding RNA gene category. IPA analysis of the differentially expressed coding genes indicated that genes with biological functions in neurologic diseases, psychological disorders, and cancer were enriched; these genes could be involved in cell growth and proliferation and cell death and survival. These signaling molecules that were targeted by or interacting with NRG1 appear to function in a thyroid-specific manner. Additional work is needed to decipher the role of these genes and molecules in thyroid function and thyroid cancer.
In conclusion, our study is an initial effort to functionally annotate the GWAS SNP rs2439302 in 8p12 as a factor contributing to PTC risk. The risk allele [G] of rs2439302 correlates with high expression of multiple NRG1 isoforms in unaffected thyroid tissue. Transcriptional regulation of NRG1, mediated by allele-specific enhancer activities, is probably an important mechanism predisposing to thyroid cancer.
Supplementary Material
Acknowledgments
We thank Jan Lockman and Barbara Fersch for administrative help, the OSU Comprehensive Cancer Center Nucleic Acid Shared Resource for RNA-seq and SNP genotyping, the tissue bank, and the OSU supercomputer center. We thank Julius Gudmundsson at deCODE for performing SNP genotyping and imputation for the association study.
Financial Support: This work was supported by National Cancer Institute Grants P30CA16058 and P50CA168505.
Disclosure Summary: The authors have nothing to disclose.
Abbreviations:
- ChIP
chromatin immunoprecipitation
- GWAS
genome-wide association studies
- IPA
Ingenuity Pathway Analysis
- LD
linkage disequilibrium
- OR
odds ratio
- OSU
The Ohio State University
- PCR
polymerase chain reaction
- PTC
papillary thyroid carcinoma
- RNA-seq
RNA sequencing
- RT-PCR
reverse transcription polymerase chain reaction
- siRNA
small interfering RNA
- SNP
single-nucleotide polymorphism.
References
- 1. Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. JAMA. 2017;317(13):1338–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. La Vecchia C, Negri E. Thyroid cancer: the thyroid cancer epidemic: overdiagnosis or a real increase? Nat Rev Endocrinol. 2017;13(6):318–319. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 4. Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst. 1994;86(21):1600–1608. [DOI] [PubMed] [Google Scholar]
- 5. Giordano TJ, Beaudenon-Huibregtse S, Shinde R, Langfield L, Vinco M, Laosinchai-Wolf W, Labourier E. Molecular testing for oncogenic gene mutations in thyroid lesions: a case–control validation study in 413 postsurgical specimens. Hum Pathol. 2014;45(7):1339–1347. [DOI] [PubMed] [Google Scholar]
- 6. Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He H, Blondal T, Geller F, Jakobsdottir M, Magnusdottir DN, Matthiasdottir S, Stacey SN, Skarphedinsson OB, Helgadottir H, Li W, Nagy R, Aguillo E, Faure E, Prats E, Saez B, Martinez M, Eyjolfsson GI, Bjornsdottir US, Holm H, Kristjansson K, Frigge ML, Kristvinsson H, Gulcher JR, Jonsson T, Rafnar T, Hjartarsson H, Mayordomo JI, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Kong A, Stefansson K. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41(4):460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liyanarachchi S, Wojcicka A, Li W, Czetwertynska M, Stachlewska E, Nagy R, Hoag K, Wen B, Ploski R, Ringel MD, Kozłowicz-Gudzinska I, Gierlikowski W, Jazdzewski K, He H, de la Chapelle A. Cumulative risk impact of five genetic variants associated with papillary thyroid carcinoma. Thyroid. 2013;23(12):1532–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Landa I, Robledo M. Association studies in thyroid cancer susceptibility: are we on the right track? J Mol Endocrinol. 2011;47(1):R43–R58. [DOI] [PubMed] [Google Scholar]
- 9. Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Masson G, He H, Jonasdottir A, Sigurdsson A, Stacey SN, Johannsdottir H, Helgadottir HT, Li W, Nagy R, Ringel MD, Kloos RT, de Visser MCH, Plantinga TS, den Heijer M, Aguillo E, Panadero A, Prats E, Garcia-Castaño A, De Juan A, Rivera F, Walters GB, Bjarnason H, Tryggvadottir L, Eyjolfsson GI, Bjornsdottir US, Holm H, Olafsson I, Kristjansson K, Kristvinsson H, Magnusson OT, Thorleifsson G, Gulcher JR, Kong A, Kiemeney LA, Jonsson T, Hjartarson H, Mayordomo JI, Netea-Maier RT, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Rafnar T, Stefansson K. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet. 2012;44(3):319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y-L, Feng S-H, Guo S-C, Wei W-J, Li D-S, Wang Y, Wang X, Wang Z-Y, Ma Y-Y, Jin L, Ji Q-H, Wang J-C. Confirmation of papillary thyroid cancer susceptibility loci identified by genome-wide association studies of chromosomes 14q13, 9q22, 2q35 and 8p12 in a Chinese population. J Med Genet. 2013;50(10):689–695. [DOI] [PubMed] [Google Scholar]
- 11. Rogounovitch TI, Bychkov A, Takahashi M, Mitsutake N, Nakashima M, Nikitski AV, Hayashi T, Hirokawa M, Ishigaki K, Shigematsu K, Bogdanova T, Matsuse M, Nishihara E, Minami S, Yamanouchi K, Ito M, Kawaguchi T, Kondo H, Takamura N, Ito Y, Miyauchi A, Matsuda F, Yamashita S, Saenko VA. The common genetic variant rs944289 on chromosome 14q13.3 associates with risk of both malignant and benign thyroid tumors in the Japanese population. Thyroid. 2015;25(3):333–340. [DOI] [PubMed] [Google Scholar]
- 12. Son H-Y, Hwangbo Y, Yoo S-K, Im SW, Yang SD, Kwak SJ, Park MS, Kwak SH, Cho SW, Ryu JS, Kim J, Jung YS, Kim TH, Kim SJ, Lee KE, Park DJ, Cho NH, Sung J, Seo JS, Lee EK, Park YJ, Kim JI. Genome-wide association and expression quantitative trait loci studies identify multiple susceptibility loci for thyroid cancer. Nat Commun. 2017;8:15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gudmundsson J, Thorleifsson G, Sigurdsson JK, Stefansdottir L, Jonasson JG, Gudjonsson SA, Gudbjartsson DF, Masson G, Johannsdottir H, Halldorsson GH, Stacey SN, Helgason H, Sulem P, Senter L, He H, Liyanarachchi S, Ringel MD, Aguillo E, Panadero A, Prats E, Garcia-Castaño A, De Juan A, Rivera F, Xu L, Kiemeney LA, Eyjolfsson GI, Sigurdardottir O, Olafsson I, Kristvinsson H, Netea-Maier RT, Jonsson T, Mayordomo JI, Plantinga TS, Hjartarson H, Hrafnkelsson J, Sturgis EM, Thorsteinsdottir U, Rafnar T, de la Chapelle A, Stefansson K. A genome-wide association study yields five novel thyroid cancer risk loci. Nat Commun. 2017;8:14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Talmage DA. Mechanisms of neuregulin action. Novartis Found Symp. 2008;289:74–84, discussion 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ocaña A, Díez-González L, Esparís-Ogando A, Montero JC, Amir E, Pandiella A. Neuregulin expression in solid tumors: prognostic value and predictive role to anti-HER3 therapies. Oncotarget. 2016;7(29):45042–45051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones FE, Jerry DJ, Guarino BC, Andrews GC, Stern DF. Heregulin induces in vivo proliferation and differentiation of mammary epithelium into secretory lobuloalveoli. Cell Growth Differ. 1996;7(8):1031–1038. [PubMed] [Google Scholar]
- 17. Le X-F, McWatters A, Wiener J, Wu J-Y, Mills GB, Bast RC Jr. Anti-HER2 antibody and heregulin suppress growth of HER2-overexpressing human breast cancer cells through different mechanisms. Clin Cancer Res. 2000;6(1):260–270. [PubMed] [Google Scholar]
- 18. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, Foster B, Moser M, Karasik E, Gillard B, Ramsey K, Sullivan S, Bridge J, Magazine H, Syron J, Fleming J, Siminoff L, Traino H, Mosavel M, Barker L, Jewell S, Rohrer D, Maxim D, Filkins D, Harbach P, Cortadillo E, Berghuis B, Turner L, Hudson E, Feenstra K, Sobin L, Robb J, Branton P, Korzeniewski G, Shive C, Tabor D, Qi L, Groch K, Nampally S, Buia S, Zimmerman A, Smith A, Burges R, Robinson K, Valentino K, Bradbury D, Cosentino M, Diaz-Mayoral N, Kennedy M, Engel T, Williams P, Erickson K, Ardlie K, Winckler W, Getz G, DeLuca D, MacArthur D, Kellis M, Thomson A, Young T, Gelfand E, Donovan M, Meng Y, Grant G, Mash D, Marcus Y, Basile M, Liu J, Zhu J, Tu Z, Cox NJ, Nicolae DL, Gamazon ER, Im HK, Konkashbaev A, Pritchard J, Stevens M, Flutre T, Wen X, Dermitzakis ET, Lappalainen T, Guigo R, Monlong J, Sammeth M, Koller D, Battle A, Mostafavi S, McCarthy M, Rivas M, Maller J, Rusyn I, Nobel A, Wright F, Shabalin A, Feolo M, Sharopova N, Sturcke A, Paschal J, Anderson JM, Wilder EL, Derr LK, Green ED, Struewing JP, Temple G, Volpi S, Boyer JT, Thomson EJ, Guyer MS, Ng C, Abdallah A, Colantuoni D, Insel TR, Koester SE, Little AR, Bender PK, Lehner T, Yao Y, Compton CC, Vaught JB, Sawyer S, Lockhart NC, Demchok J, Moore HF; GTEx Consortium . The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He H, Nagy R, Liyanarachchi S, Jiao H, Li W, Suster S, Kere J, de la Chapelle A. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res. 2009;69(2):625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. [DOI] [PubMed] [Google Scholar]
- 21. Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–181. [DOI] [PubMed] [Google Scholar]
- 22. He H, Li W, Wu D, Nagy R, Liyanarachchi S, Akagi K, Jendrzejewski J, Jiao H, Hoag K, Wen B, Srinivas M, Waidyaratne G, Wang R, Wojcicka A, Lattimer IR, Stachlewska E, Czetwertynska M, Dlugosinska J, Gierlikowski W, Ploski R, Krawczyk M, Jazdzewski K, Kere J, Symer DE, Jin V, Wang Q, de la Chapelle A. Ultra-rare mutation in long-range enhancer predisposes to thyroid carcinoma with high penetrance [published correction appears in PLoS ONE. 2013;8(9):10] PLoS One. 2013;8(5):e61920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y, Li W, Phay JE, Shen R, Pellegata NS, Saji M, Ringel MD, de la Chapelle A, He H. Primary cell culture systems for human thyroid studies. Thyroid. 2016;26(8):1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liyanarachchi S, Li W, Yan P, Bundschuh R, Brock P, Senter L, Ringel MD, de la Chapelle A, He H. Genome-wide expression screening discloses long noncoding RNAs involved in thyroid carcinogenesis. J Clin Endocrinol Metab. 2016;101(11):4005–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Britsch S. The neuregulin-I/ErbB signaling system in development and disease. Adv Anat Embryol Cell Biol. 2007;190:1–65. [PubMed] [Google Scholar]
- 28. Breuleux M. Role of heregulin in human cancer. Cell Mol Life Sci. 2007;64(18):2358–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284(1):14–30. [DOI] [PubMed] [Google Scholar]
- 30. Fluge O, Akslen LA, Haugen DRF, Varhaug JE, Lillehaug JR. Expression of heregulins and associations with the ErbB family of tyrosine kinase receptors in papillary thyroid carcinomas. Int J Cancer. 2000;87(6):763–770. [DOI] [PubMed] [Google Scholar]
- 31. Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type–specific gene expression. Nature. 2009;459(7243):108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Corradin O, Scacheri PC. Enhancer variants: evaluating functions in common disease. Genome Med. 2014;6(10):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nguyen TA, Jones RD, Snavely AR, Pfenning AR, Kirchner R, Hemberg M, Gray JM. High-throughput functional comparison of promoter and enhancer activities. Genome Res. 2016;26(8):1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal.lari R, Lupien M, Markowitz S, Scacheri PC. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.