Abstract
High-throughput sequencing (HTS) of human genome coding regions allows the simultaneous screen of a large number of genes, significantly improving the diagnosis of non-syndromic intellectual disabilities (ID). HTS studies permit the redefinition of the phenotypical spectrum of known disease-causing genes, escaping the clinical inclusion bias of gene-by-gene Sanger sequencing. We studied a cohort of 903 patients with ID not reminiscent of a well-known syndrome, using an ID-targeted HTS of several hundred genes and found de novo heterozygous variants in TCF4 (transcription factor 4) in eight novel patients. Piecing together the patients from this study and those from previous large-scale unbiased HTS studies, we estimated the rate of individuals with ID carrying a disease-causing TCF4 mutation to 0.7%. So far, TCF4 molecular abnormalities were known to cause a syndromic form of ID, Pitt–Hopkins syndrome (PTHS), which combines severe ID, developmental delay, absence of speech, behavioral and ventilation disorders, and a distinctive facial gestalt. Therefore, we reevaluated ten patients carrying a pathogenic or likely pathogenic variant in TCF4 (eight patients included in this study and two from our previous ID-HTS study) for PTHS criteria defined by Whalen and Marangi. A posteriori, five patients had a score highly evocative of PTHS, three were possibly consistent with this diagnosis, and two had a score below the defined PTHS threshold. In conclusion, these results highlight TCF4 as a frequent cause of moderate to profound ID and broaden the clinical spectrum associated to TCF4 mutations to nonspecific ID.
Introduction
Since a few years, the development of new high throughput sequencing technologies (HTS) permitted the study of either a large number of genes or the entire exome/genome in patients with non-syndromic ID. These allowed the identification of disease-causing variants in genes involved in syndromic forms of intellectual disability (ID) in patients whom clinical manifestations were not typical of the corresponding disorders. In our previous targeted sequencing (TES) study performed in 106 individuals with unexplained ID using a panel of 217-ID genes, only four genes were found to be mutated in more than one family. Three mutations were identified in MECP2 (MIM *300005, involved in Rett syndrome #312750), two de novo point mutations in two girls and one maternally inherited complex rearrangement in exon 4 of the gene in one boy removing 60 amino acids inherited from his mother (speech delay) [1]. Two disease-causing variants were identified in another X-linked gene, KDM5C (MIM *314690), and in two autosomal genes DYRK1A (MIM *600855) and TCF4 (MIM *602272). TCF4 (transcription factor 4) is located in 18q21, and encodes a class I basic helix-loop-helix transcription factor binding to E-boxes on DNA after dimerization, which is involved in cell signaling, cell survival and neurodevelopment [5]. So far, TCF4 is the single gene involved in Pitt-Hopkins Syndrome (PTHS, MIM #610954) [2–4], a rare, well-characterized, neurodevelopmental disorder usually presenting with severe intellectual disability associated with distinctive facial features, various neurological and behavioral impairment and gastro-intestinal dysfunction, hypotonia, ataxia, breathing abnormalities, and seizures [6]. This provided a rationale for TCF4 Sanger-sequencing in patients with syndromic ID after ruling out differential diagnoses by PTHS clinical scores [6, 7]. Since implementation of HTS in ID screening, we and others have suggested TCF4 implication in isolated ID [1, 8–10].
To assess the frequency of TCF4 molecular abnormalities in non-syndromic ID patients, we studied 903 novel patients with mild to severe ID and reviewed the previous published targeted, exome or genome sequencing studies [1, 11–15]. To better delineate the phenotype related to TCF4 mutations we re-analyzed a posteriori the phenotype of all the patients carrying a pathogenic or likely pathogenic variant in this gene (as defined by the American College of Medical Genetics and Genomics), but for whom PTHS diagnostic was not clinically suspected.
Materials and methods
Patients
DNA samples (from peripheral blood or saliva) of the 903 patients were referred to the laboratory of genetic diagnosis. Patients presented with non-specific intellectual disability and no major congenital anomalies. The cohort includes patients with mild ID or ID of unknown severity (around 25%), moderate (around 40%), or severe to profound (around 35%) ID, based on clinician’s appreciations. The most current causes of cognitive impairment were dismissed by fragile-X test, array-CGH, and metabolic explorations (in 90% of patients or more). Among the more recurrent tests, UBE3A (MIM *601623) sequencing or methylation analysis were performed in <20% of the patients, and MECP2, ARX (MIM *300382) or DMPK (MIM *605377) in around 12%. Clinical data were recorded before inclusion following a standardized clinical questionnaire highlighting prenatal history, developmental milestones, neurological, and behavioral disorders. ID severity was assessed by medical geneticists upon clinical evaluation and was not a discriminating inclusion criterion. However, the cohort was enriched in severe and moderate forms of ID compared to the distribution in ID population. After obtaining the molecular diagnosis, the patient was reevaluated by the clinical geneticist. All the clinical data were re-collected, with a specific attention to PTHS clinical signs. This study was approved by the local Ethics Committee of the Strasbourg University Hospital (Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale - CCPPRB). For all patients, a written informed consent for genetic testing was obtained from their legal representative.
Targeted genes and capture design
DNA samples were extracted from peripheral blood or saliva. HTS targeted libraries were prepared, as previously described [1] with individual in-solution SureSelect capture reaction for each DNA sample (custom design for genes known to be involved in ID, Agilent, Santa Clara, California, USA). Capture experiments were performed using probes corresponding to a panel of 275 (in 207 patients), 451 (in 66 patients) or 456 (in 630 patients) ID genes. Paired-end sequencing (2 × 101-bp) was performed on an Illumina HiSeq 2500, multiplexing in average 32 samples per sequencing lane. Read mapping, variant calling and annotation were performed, as previously described [1]. Detected variants, short indels and single nucleotide variants (SNVs), were annotated and ranked by VaRank software [16].
Sanger sequencing confirmation
TCF4 pathogenic or likely pathogenic variants identified by HTS were confirmed in patients and the de novo status was checked in their parents by Sanger sequencing. Pedigree (parents-child) concordance was confirmed by checking the segregation of several highly polymorphic microsatellite markers (PowerPlex 16 HS System, Promega, Madison, WI, USA) or frequent variants (when TES was also performed for parental DNA). We reported the variants identified in TCF4 in a specific database (https://databases.lovd.nl/shared/genes/TCF4).
PTHS clinical scoring
To facilitate the clinical diagnosis of PTHS two scoring tests have been developed in 2012. The first one, established by Whalen et al., was based on the scoring of the following criteria: facial gestalt (8 points), severe motor delay (2 points), absent language (2 points), stereotypic movements (2 points), hyperventilation (1 point), anxiety (1 point), hypotonia (1 point), smiling appearance (1 point), ataxic gait (1 point), and strabismus (1 point). This score was validated in patients evocative of PTHS with (n = 33) or without (n = 100) pathogenic variant identified in TCF4. A threshold of 15/20 was considered as a good indicator of TCF4. A score between 10 and 15 could also be suggestive of this diagnosis, especially for young patients [6]. The second scoring, established by Marangi et al. scored the following symptoms: typical/partial facial features (4 points/2 points), moderate/severe intellectual disability (2 points), poor/absent language (1 point/2 point), normal growth parameters at birth (1 point), microcephaly (1 point), epilepsy/EEG abnormalities (1 point), ataxic gait (1 point), hyperventilation (1 point), constipation (1 point), brain MRI abnormalities (1 point) and strabismus or ophthalmologic abnormalities (1 point) [7]. These criteria were evaluated in patients evocative of PTHS with (n = 18) or without (n = 60) pathogenic variants in TCF4 and a score above 10/16 was recommended for a molecular study of TCF4. Whalen and Marangi’s scores were calculated after a clinical reexamination (a posteriori after obtaining the molecular diagnosis) for the patients described in this paper plus the two we previously described [1].
Results
Pathogenic or likely pathogenic TCF4 variants in undiagnosed ID patients
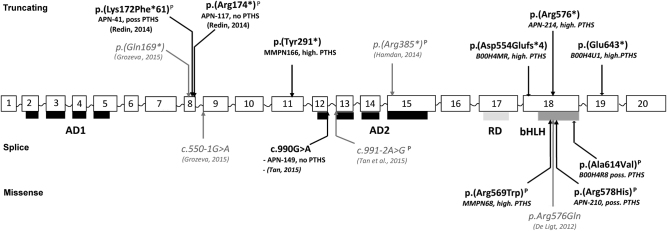
Through HTS targeted sequencing of several hundred of ID genes in 903 patients with undiagnosed ID, we identified eight pathogenic or likely pathogenic TCF4 variants among which four were novel (Table 1, Fig. 1). All these variants occurred de novo, were not reported in ExAC general population database and affected amino acids included in all the isoforms of the gene. Named here according to the NM_001083962.1, we identified four nonsense or frameshift variants c.873C>A p.(Tyr291*), c.1662del p.(Asp554Glufs*4), c.1726C>T p.(Arg576*) and c.1927G>T p.(Glu643*), three missense variants affecting conserved amino acid located in the bHLH domain of the protein and predicted to be damaging by in silico tools (SIFT, Polyphen2): c.1705C>T p.(Arg569Trp), c.1733G>A p.(Arg578His) and c.1841C>T p.(Ala614Val), and one silent variant altering the last nucleotide of exon 12 (according to NG_011716) and predicted to modify the donor splice site (c.990G>A, p.?). In addition, two variants affecting only one alternative isoform (NM_001243231.1: c.7G>T p.(Glu3*) and c.2T>C, p.(Met1?)) have been identified, both inherited from an unaffected parent and were therefore classified as likely benign.
Table 1.
Pathogenic or likely pathogenic variants in TCF4 identified in patients with intellectual disability (ID) by large-scale sequencing approaches
| Reference | Variant nomenclature | Inheritance | dbSNP | Previously reported in Clinvar | Previously described in PTHS screening | TCF4-LOVD Individual ID | SIFT | PP2 | Individual | Gender | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| This report | c.873C>A | p.(Tyr291*) | de novo | NA | NA | no | 000100 | NA | NA | MMPN166 | F |
| This report Tan et al. [15] |
c.990G>A | p.? | de novo, de novo | rs587784469 | RCV000147730.1 (Likely. pathogenic) | no | 000098 | NA | NA | - APN149 -Patient 6 |
M F |
| This report | c.1662del | p.(Asp554Glufs*4) | de novo | NA | NA | no | 000101 | NA | NA | B00H4MR | F |
| This report | c.1705C>T | p.(Arg569Trp) | de novo | NA | NA | yes [6] | 000026 | Deleterious | Prob. Damaging | MMPN68 | M |
| This report | c.1726C>T | p.(Arg576*) | de novo | NA | NA | no | 000102 | NA | NA | APN214 | M |
| This report | c.1733G>A | p.(Arg578His) | de novo | rs121909123 | RCV000079458.4|RCV000189738.1 (Pathogenic) | yes [22] | 000029 | Deleterious | Poss. Damaging | APN210 | F |
| This report | c.1841C>T | p.(Ala614Val) | de novo | NA | NA | yes [32] | 000021 | Deleterious | Prob. Damaging | B00H4R8 | F |
| This report | c.1927G>T | p.(Glu643*) | de novo | NA | NA | no | 000103 | NA | NA | B00H4U1 | F |
| Redin et al. [1] | c.514_517del | p.(Lys172Phefs*61) | de novo | rs398123561 | RCV000079461.4 (Pathogenic) | yes [6] | 000047 | NA | NA | APN41 | M |
| Redin et al. [1] | c.520C>T | p.(Arg174*) | de novo | NA | RCV000224478.1 (Pathogenic) | yes [6] | 000023 | NA | NA | APN117 | F |
| Grozeva et al. [12] | c.505C>T | p.(Gln169*) | NA | NA | NA | no | 000104 | NA | NA | 5410771 | M |
| Grozeva et al. [12] | c.550–1G>A g.52946888 C>T |
p.? | NA | NA | NA | no | 000105 | NA | NA | 5411380 | M |
| Tan et al. [15] | c.991–2A>G g.52927260 T>C |
p.? | NA | rs587784470 | RCV000147731.1 (Pathogenic) | no | 000106 | NA | NA | Patient 5 | F |
| Hamdan et al. [13] | c.1153C>T | p.(Arg385*) | de novo | rs121909122 | RCV000007797.4 (Pathogenic) | yes [4, 33] | 000003 | NA | NA | Case 045.400 | M |
| De Ligt et al. [11] | c.1727G>A | p.(Arg576Gln) | de novo | NA | RCV000431775.1 (Pathogenic) | yes [33] | 000027 | Deleterious | Poss. Damaging | Trio 15 | F |
All the c. positions were given according to NM_001083962.1 isoform (and NG_011716.2 isoform for intronic variants). PP2 Polyphen2 SIFT scores. Variants are classified following recommendations from the American College of Medical Genetics and Genomics
Fig. 1.
Schematic representation of disease-causing variants identified in TCF4. AD1, AD2 transactivation domains; RD repressor domain; bHLH DNA-binding domain. In bold: variants identified by TES in our cohort. Patient number is indicated as well as the severity of his PTHS phenotype: no not evocative of PTHS, poss. possibly evocative of PTHS, high. highly evocative of PTHS), in italic: variants identified in other HTS studies P: variants previously described in PTHS patients
TCF4 mutation rate is of 0.7% (16/2239) in individuals with undiagnosed ID
Piecing together the 8 patients out of the 903 of this study with the two out of 106 patients that we have previously reported [1], the frequency of TCF4 disease-causing variants is of 1% (10/1009) in our cohort of individuals with ID undiagnosed by a geneticist. Furthermore, we reviewed data from other large scale studies, including TES of ID genes [12, 15], and WES performed in patients with non-specific ID [11, 13, 14] and calculate the TCF4 mutation rate in patients with non-syndromic ID (Tables 1 and 2). Altogether with our results, 16 individuals with pathogenic or likely pathogenic TCF4 variants were identified during the large-scale sequencing studies performed in 2230 patients with nonspecific ID, providing a TCF4 mutation rate of 0.7% (Table 2, Fig. 1).
Table 2.
Pathogenic or likely pathogenic variants identified in TCF4 during targeted sequencing (TES), whole exome sequencing (WES) or whole genome sequencing (WGS) in patients with intellectual disability (ID)
| Cohort | Reference | Approach | Number of patients | TCF4 mutations |
|---|---|---|---|---|
| ID (mild to severe) | this study | TES (275–456 genes) | 903 | 8 |
| ID (mild to severe) | Redin et al. [1] | TES (217 genes) | 106 | 2 |
| ID (moderate to severe) | Grozeva et al. [12] | TES (575 genes) | 986 | 2 |
| ID | Tan et al. [15] | TES (90 genes) | 52 | 2 |
| ID (severe) | Rauch et al. [14] | WES | 51 | (1*) |
| ID (moderate to severe) | de Ligt et al. [11] | WES | 100 | 1 |
| ID (moderate to severe) | Hamdan et al. [13] | WES | 41 | 1 |
| Total | 2239 | 16 (0.7%) |
* de novo missense variant predicted to be benign, not included in the statistics
TCF4 mutations can cause ID poorly suggestive of PTHS
A posteriori clinical reevaluation was performed for the 10 patients (eight novel patients included in this study and two from our previous ID-HTS study) carrying a TCF4 disease-causing variant (Table 3, Fig. 2). All probands, except MMPN166, were born from unrelated healthy parents, with irrelevant family history. According to Whalen and Marangi scores, five patients (MMPN166, MMPN68, APN-214, B00H4MR, and B00H4U1) had features reminiscent of PTHS (>12/20 Whalen’s and 10/16 Marangi’s score), three individuals (B00H4R8, APN-210, and APN-41) were slightly evocative of PTHS (only one of the scores was upper to the threshold) and two patients (APN-149 and APN-117) were not consistent with PTHS (both scoring were below the threshold). To widely asses the phenotype of patients with a TCF4 pathogenic variant identified through TES or WES, we further evaluate the phenotype of the patients reported by other groups [11–13, 15] (Table 4). Clinical data were available for four out of the six reported patients. The phenotype could be evocative of a PTHS for three of the patients, but not in the last one who had only a mild ID. Taken together, in nearly half of the patients (6/13) studied by HTS and carrying a disease-causing TCF4 variant, clinical features were poorly or not evocative of PTHS.
Table 3.
A posteriori reevaluation of PTHS clinical signs in seven patients carrying a pathogenic mutation in TCF4
| Patient MMPN166 | Patient APN149 | Patient B00H4MR | Patient MMPN68 | Patient APN214 | Patient APN210 | Patient B00H4R8 | Patient B00H4U1 | Patient (Redin et al., 2014) APN41 | Patient (Redin et al., 2014) APN117 | |
|---|---|---|---|---|---|---|---|---|---|---|
| TCF4 variant | c.873 C>A, p.(Tyr291*) | ac.990 G > A, p.? | c.1662del, p.(Asp554Glufs4*) | ac.1705C>T, p.(Arg569Trp) | c.1726C>T, p.(Arg576) | ac.1733G>A, p.(Arg578His) | ac.1841C>T p.(Ala614Val) | c.1927G>T p.(Glu643*) | ac.514_517del p.(Lys172Phefs*61) | ac.520 C>T, p.(Arg174*) |
| Other variants/CNV | dup 22q11 | no | no | no | no | no | no | no | no | no |
| Patient | Female | Male | Female | Male | Male | Female | Female | Female | Male | Female |
| Age of inclusion | 4 y-o | 5 y-o | 4 y-o | 5 y-o | 18 y-o | 6 y-o | 7 y-o | 2 y-o | 3 y-o | 10 y-o |
| Age of reexamination | 6 y-o | 9 y-o | 5y 8 m-o | 8 y-o | 20 y-o | 9 y-o | 7 y-o | 4 y-o | 6 y-o | 13 y-o |
| Classical PTHS symptoms | ||||||||||
| Facial gestalt | typical | mild | typical | mild | typical | mild | not typicalb | typical | mild | mild |
| Growth (statural/ponderal) | M/M | M/M | −1/−2 | M/ + 5 SD | −3.5 SD/M | M/ + 3 SD | −3 SD/M | M/M | −2SD/−2SD | M/M |
| Head circumference | −1 DS | M | −2.5 | M | M | +0,5 SD | +1.5 SD | M | M | −1.8 SD |
| Cognitive impairment | profound | moderate | severe | profound | profound | moderate | severe | severe | severe | moderate |
| Walking | absent | 16 mo | NA | 5 y | 3 y | 25 mo | 5 y | absent | 3 y | 28 mo |
| Absent speech | yes | yes | no (few words) | yes | yes | yes | yes | yes | yes | no |
| Hyperventilation/apneas | no | no | no | no | yes | yes | no | yes | no | no |
| Happy appearance | yes | yes | no | no | yes | yes | NA | no | yes | yes |
| Sleep disturbance | no | no | yes | no | yes | yes | no | no | no | yes |
| Behavior problems | no | yes (Self-aggress.) | no | no | yes (Self-aggress.) | no | yes, severe | no? | yes (poor interactions) | no |
| Stereotypic behavior | yes | no | yes | yes | yes | no | yes | yes | yes | no |
| Seizures | yes (3–6 mo) | no | no | yes | yes | no | no | no | yes | no |
| Hypotonia | yes | no | yes | yes | yes | no | no | yes | yes | yes |
| Ataxic gait | no | no | yes | yes (mild) | yes | no | instable | no | yes | yes |
| Ophthalmologic anomalies | yes strabismus | no | yes strabismus | yes | yes strabismus, astigmatism | yes Duane anomaly | No (ptosis) | yes strabismus hyperopia | no | no |
| Constipation | no | yes | yes | yes | yes | yes | no | no | yes | yes |
| Gastro-esophageal reflux | no | yes | no | no | yes | no | NA | yes (infancy) | yes | no |
| Fetal pads | no | no | yes | yes | no | no | no | no | yes | yes |
| Cerebral MRI | normal | – | abormal (1) | abnormal | abnormal | normal | abnormal (2) | normal | normal | normal |
| Other signs | no | no | cervical syringomyelia | no | cryptorchidism, abolition of osteo-tendinous reflexes | headaches | no | heterotaxy, bifid uvula, long thumbs, labia minora hypoplasia | chronic otitis | no |
| Whalen’s score (>12 or 15/20) | 16 | 7 | 15 | 15 | 17 | 13 | 13 | 15 | 12 | 9 |
| Marangi’s score (>10/16) | 12 | 7 | 13 | 14 | 13 | 10 | 10 | 12 | 12 | 7 |
| Conclusion PTHS | highly | no | highly | highly | highly | possibly | possibly | highly | possibly | no |
PTHS Pitt-Hopkins syndrome, y-o year-old; mo: months, SD standard deviation, M value in normal range, MRI Magnetic resonance imaging
a mutation previously described in databases or literature
b Rubinstein-Taybi facial features suggested. MRI anomalies 1: posterior atrophy of corpus callosum; 2: Hypersignal of the subcortical white matter in temporal lobes. The cDNA nomenclature given according to NM_001083962.1 isoform for all the variants.
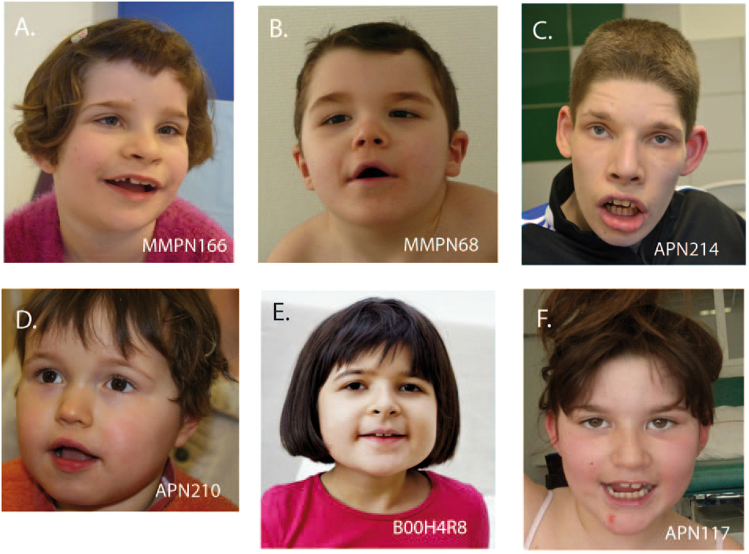
Fig. 2.
Pictures of Patients carrying de novo heterozygous disease-causing variants in TCF4. a Patient MMPN166, b Patient MMPN68, c Patient APN214, d Patient APN210, e Patient B00H4R8, and f Patient APN117
Table 4.
Summary of clinical information available for patients with TCF4 mutations identified by other large-scale sequencing studies (form supplementary information of De ligt et al. [11]; Hamdan et al. [13], Tan et al. [15], Grozeva et al. [12])
| Reference | Patient identification | TCF4 pathogenic variant | Clinical description | Previous genetic investigations |
|---|---|---|---|---|
| De Ligt et al. [11] | Trio 15 | de novo c.1727G>A, p.(Arg576Gln) | Female, 4 y-o: Moderate ID, feeding problems, recurrent otitis. Sitting: 22 months, walking 3,5 y-o; no speech. Mild dysmorphic features (epicanthic folds, a broad nasal tip and prominent ears). Hypotonia. Ataxic walking pattern. MRI: mildly enlarged ventricles without structural anomalies | CGH-array, 15q methylation, MECP2, EHMT1, UBE3A sequencing |
| Hamdan et al. [13] | Case 1045.400 | de novo c.1153 C>T, p.(Arg385*) | Male, 6 y-o: Severe ID. no stand nor walk without support, no speech. Hypotonia. Hypersalivation. No breathing problem. Had seizure once. Minor dysmorphy (wide mouth, bilateral single palmar creases, bilateral clinodactyly and overlapping 2nd toes). MRI: increased T2 and FLAIR signal in the periventricular regions, thin corpus callosum, myelination delay. | CGH-array, FMR1 CGG expansion testing, MECP2 sequencing, 15q methylation |
| Tan et al. [15] | Patient 5 | c.991–2 A>G, g.52927260 T>C | Female, 5 y-o: Global developmental delay, height and size: 10th percentile, minor facial dysmorphy. Myopia. MRI: tiny pineal cyst and peritrigonal white matter intensity | Karyotype, CGH-array, ZEB2 sequencing |
| Tan et al. [15] | Patient 6 | de novo c.990 G>A | Female, 7 y-o: Global developmental delay, facial dysmorphy (thick overfolded helix of the ear, wide mouth, coarse facial features, flat philtrum, bulbous nose). Ataxia. Strabismus. MRI: normal | CGH-array, FMR1 CGG expansion testing, SNRPN methylation, 17q deletion, MECP2 sequencing |
| Grozeva et al. [12] | UK10K_FINDWGA5411380 | c.550–1 G>A, g.52946888 C>T | Male: NA | NA |
| Grozeva et al. [12] | UK10K_FINDWGA5410771 | c.505 C>T, p.(Gln169*) | Male: NA | NA |
MRI: Magnetic resonance imaging; y-o: year-old; NA: non available. The variations are given according to hg19/GRC37 for the genomic nomenclature and the RefSeq transcript NM_001083962.1 for the cDNA nomenclature
Discussion
Targeted or whole exome HTS used in routine diagnosis have demonstrated their efficiency in the diagnosis of isolated ID [1, 11, 14]. Unexpected rates of pathogenic variants in genes implicated in syndromic cognitive impairment were found with these clinically unbiased approaches. We studied 903 patients with undiagnosed ID by targeted HTS of ID known genes, and identified eight novel patients carrying a pathogenic or likely pathogenic variant in TCF4. We also analyzed data from previous HTS studies, and found eight additional patients carrying a disease-causing variant in TCF4, including two patients reported by our group [1]. Taken together, we count 16 patients carrying a TCF4 disase-causing variant (of which 15 distinct variants) among 2239 ID patients and we obtained a TCF4 mutation rate of 0.7% in non-specific ID (Table 2). This mutation rate is close to those of the most frequent causes of ID such as FMR1 expansions [17, 18] or ARID1B mutations [19] in Fragile-X and Coffin-Siris syndromes. Otherwise, TCF4 mutation rate gets down to 0.3% (13/4293) in studies including patients with developmental disorders in which ID is not a mandatory sign, such as the Deciphering Developmental Disorder (DDD) project [20]. Indeed, a very recent study reported ID in 100% (47/47) of patients carrying a disease-causing variant in TCF4, collected though a web-based database [21]. However, in the DDD data, TCF4 still appears in the top-twenty of the most frequently mutated genes in with developmental disorders.
The patients included in our TES study were referred by a geneticist after several biological, radiological and molecular tests which did not allow a diagnosis. The a posteriori analysis of the clinical features of the ten patients carrying a TCF4 disease-causing variant showed that PTHS could have been suspected in five patients. However, even if the diagnosis would have been possible in three additional cases, by using Whalen and Marangi clinical scores, facial gestalt of those patients was not typical of PTHS. Furthermore, for two patients, Whalen and Marangi clinical scores were low and PTHS could not have been suspected clinically. Indeed, in absence of distinctive signs of PTHS, such as a typical facial gestalt (4/10) (Fig. 2) or hyperventilation (3/10) which can appear later in childhood [21, 22], the clinical diagnosis remains challenging, especially for the patients with moderate ID. In contrast, absence of speech (8/10), noticeable delay in walking (after 3 years of age, if acquired) (8/10), seizures (4/10), behavior problems (self-aggressiveness, poor social interactions) (4/10), smiling appearance (6/10), strabismus (4/10) and constipation (7/10) were observed in our patients, but were not sufficiently discriminatory signs of PTHS, as they can be also found in non-specific ID. This study suggests that even if some were a posteriori evocative of PTHS, other ones presenting nonspecific ID and only few PTHS features could not be diagnosed clinically showing that phenotypic spectrum associated to a TCF4 disease-causing variant is wider than we used to think.
The main differential diagnoses described for PTHS are Angelman and Rett syndromes [23]. Consistent with that, previous genetic tests performed in the patients, before identification of a disease-causing variant in TCF4, were UBE3A methylation testing or point mutation screening (64% of the patients), and MECP2 sequencing (36%). A third known differential diagnosis, the Mowat–Wilson syndrome, was suspected in one patient. This later syndrome is associated with cardiac and urogenital malformations and Hirschsprung disease, which are features more discriminative for clinical diagnosis. Surprisingly, a Steinert syndrome was suspected in four patients, maybe due to hypotonia observed in those patients. Analysis of the 17p11.2 deletion (Smith–Magenis syndrome) and of ARX coding sequences were also performed in two patients. Taken together, these explorations assess the difficulty to evoke clinically PTHS when the patient only presents with severe delay of psychomotor acquisitions with mild dysmorphic features.
Most of the disease-causing TCF4 variants previously associated to PTHS are truncating mutations localized between the exons 7 and 18 and are probably responsible of haploinsufficiency. Missense variants mainly concern the bHLH domain of the protein including the arginine residues 578 and 580, spots of recurrent mutations [6]. In in vitro functional studies, Sepp et al. highlighted the variation in expression, patterning, dimerization and DNA binding of different TCF4 mutants comparing to WT proteins, suggesting that disease-causing variants can have various functional effects ranging from selective heterodimerization defects to complete lack of DNA binding or possible dominant-negative effects [24]. These authors suggested that the variety of variations could explain the phenotypic variability. Other authors suggested that seizures are more often associated to missense than truncating variants [25] but this was not confirmed afterwards [6]. It is tempting to speculate that some milder phenotype might be explained by variants having a less severe effect, but no clear correlation between the type of variation (missense, truncating) or its location and the phenotype was reported so far [6]. Actually, in the patients reported here, no correlation between the PTHS score and the type or the location of the variant was found. Some of the patients, as for instance patient APN117, had a milder PTHS score while carrying disease-causing variants previously described in classical PTHS cases (Fig. 2). Finally, the c.990 G > A variant, predicted to affect the exon 12 splice donor site, was identified in two patients poorly evocative of PTHS (patient APN149 and Patient 6 reported by Tan et al., 2014). In this specific case, the presence of normal splicing in a part of transcripts might explain the milder phenotype of these patients. Due to the large number of TCF4 transcripts and to the tissue-variability, splicing effects are difficult to assess. Furthermore, the threshold of TCF4 normal transcript level sufficient to avoid a pathogenic effect is not known since several cases of typical PTHS with varying levels of mosaicism have been reported [26–29]. Interruptions of the TCF4 gene can also result in a broader phenotype than usually described, as suggested by Kalscheuer et al. in 2008 after reporting the case of a girl with mild ID, minor facial gestalt and a balanced 18;20 translocation disrupting TCF4 in exon 4 [9]. More recently, Schluth-Bolard et al. reported a case of a girl with severe developmental delay and microcephaly who was carrier of an apparently balanced translocation between chromosomes 1 and 18, which was disrupting TCF4 in intron 6 [30]. Similar complex chromosomal translocations have been reported in familial cases of mild ID with an autosomal dominant transmission pattern, without any feature of PTHS [10, 31]. Both breakpoints were located before exon 8. More than a dozen of transcripts isoforms are described for TCF4. Functional RNA studies carried on fibroblasts showed, as expected, a decrease of the long isoforms of TCF4 (affected by the breakpoint) in the patients while the short isoforms encoding nuclear TCF4 were upregulated [31]. The authors suggested that the persistence of the expression of TCF4 short isoforms may rescue part of PTHS phenotype. In our study, there is no correlation between the number of isoforms affected by the different disease-causing variations and the severity of the phenotype, suggesting that additional mechanisms than a rescue with short isoforms are responsible for the clinical variability. Finally, genetic background may also play a role and influence the severity of clinical manifestations caused by a disease-causing variant in TCF4. It is interesting to note that Patient MMPN166, one the most severely affected patient, also carries an inherited 22q11.21 duplication which segregates with various neurological signs in her family. The hypothesis of a second genetic hit should be considered to account for the phenotypic difference of patients carrying a disease-causing variant in TCF4.
The growing number of HTS realized in routine in patients with ID may allow to provide more data about the prevalence of disease-causing variants in TCF4 in patients with cognitive impairment and to assess its related phenotype in an unbiased manner. Our study extended the clinical spectrum associated to TCF4 mutation from PTHS to nonspecific intellectual disability. The high prevalence (0.7%) of disease-causing variants in TCF4 found in large cohorts of patients suffering from intellectual disability proves that the borders of PTHS are less stringent than we used to consider. This gene should therefore be included in all HTS panels used for diagnosis of unspecific ID. The use of “Pitt-Hopkins syndrome” when reporting a disease-causing variant in TCF4 in a patient with a low PTHS clinical score should also be discussed.
Web resources
The URLs for online tools and data presented herein are:
OMIM: http://www.omim/org/
UCSC: http://genome.ucsc.edu/
dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP/
Mutation Nomenclature: http://www.hgvs.org/mutnomen/recs.html
Exome Variant Server, NHLBI Exome Sequencing Project (ESP): http://evs.gs.washington.edu/EVS/
ExAC Browser (Beta) | Exome Aggregation Consortium: http://exac.broadinstitute.org/
Integrative Genomics Viewer (IGV): http://www.broadinstitute.org/igv/
These variants were submitted to Clinvar: http://www.ncbi.nlm.nih.gov/clinvar/
Electronic supplementary material
Acknowledgements
We thank the families for their participation to the study. We also thank the Fondation Jerome Lejeune and the Agence de la Biomédecine for their financial support. They want also to thank all the people from the Strasbourg Hospital molecular diagnostic lab, from the IGBMC and CNG/CEA sequencing platform from UMR_S 1112 (Bernard Jost, Stéphanie Le Gras, Mathieu Jung, Céline Keime, Jean Muller, Véronique Geoffroy) and from the Mondor Hospital molecular diagnostic lab (Thierry Gaillon) for their technical and bioinformatical supports.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Amélie Piton, Phone: +33369551652, Email: piton@igbmc.fr.
Irina Giurgea, Phone: +330144735295, Email: irina.giurgea@inserm.fr.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0096-4) contains supplementary material, which is available to authorized users.
References
- 1.Redin C, Gerard B, Lauer J, et al. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet. 2014;51:724–36. doi: 10.1136/jmedgenet-2014-102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amiel J, Rio M, de Pontual L, et al. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am J Hum Genet. 2007;80:988–93. doi: 10.1086/515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brockschmidt A, Todt U, Ryu S, et al. Severe mental retardation with breathing abnormalities (Pitt-Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum Mol Genet. 2007;16:1488–94. doi: 10.1093/hmg/ddm099. [DOI] [PubMed] [Google Scholar]
- 4.Zweier C, Peippo MM, Hoyer J, et al. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome) Am J Hum Genet. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–905. doi: 10.1128/MCB.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whalen S, Heron D, Gaillon T, et al. Novel comprehensive diagnostic strategy in Pitt-Hopkins syndrome: clinical score and further delineation of the TCF4 mutational spectrum. Hum Mutat. 2012;33:64–72. doi: 10.1002/humu.21639. [DOI] [PubMed] [Google Scholar]
- 7.Marangi G, Ricciardi S, Orteschi D, et al. Proposal of a clinical score for the molecular test for Pitt-Hopkins syndrome. Am J Med Genet A. 2012;158A:1604–11. doi: 10.1002/ajmg.a.35419. [DOI] [PubMed] [Google Scholar]
- 8.Hamdan FF, Daoud H, Patry L, et al. Parent-child exome sequencing identifies a de novo truncating mutation in TCF4 in non-syndromic intellectual disability. Clin Genet. 2013;83:198–200. doi: 10.1111/j.1399-0004.2012.01890.x. [DOI] [PubMed] [Google Scholar]
- 9.Kalscheuer VM, Feenstra I, Van Ravenswaaij-Arts CM, et al. Disruption of the TCF4 gene in a girl with mental retardation but without the classical Pitt-Hopkins syndrome. Am J Med Genet A. 2008;146A:2053–9. doi: 10.1002/ajmg.a.32419. [DOI] [PubMed] [Google Scholar]
- 10.Kharbanda M, Kannike K, Lampe A, Berg J, Timmusk T, Sepp M. Partial deletion of TCF4 in three generation family with non-syndromic intellectual disability, without features of Pitt-Hopkins syndrome. Eur J Med Genet. 2016;59:310–4. doi: 10.1016/j.ejmg.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 11.de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 12.Grozeva D, Carss K, Spasic-Boskovic O, et al. Targeted next-generation sequencing analysis of 1000 individuals with intellectual disability. Hum Mutat. 2015;36:1197–204. doi: 10.1002/humu.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamdan FF, Srour M, Capo-Chichi JM, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 15.Tan CA, Topper S, Del Gaudio D et al. Characterization of patients referred for non-specific intellectual disability testing: the importance of autosomal genes for diagnosis. Clin Genet. 2016;89:478–483. [DOI] [PubMed]
- 16.Geoffroy V, Pizot C, Redin C, et al. VaRank: a simple and powerful tool for ranking genetic variants. PeerJ. 2015;3:e796. doi: 10.7717/peerj.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biancalana V, Beldjord C, Taillandier A, et al. Five years of molecular diagnosis of Fragile X syndrome (1997–2001): a collaborative study reporting 95% of the activity in France. Am J Med Genet A. 2004;129A:218–24. doi: 10.1002/ajmg.a.30237. [DOI] [PubMed] [Google Scholar]
- 18.Hagerman PJ. The fragile X prevalence paradox. J Med Genet. 2008;45:498–9. doi: 10.1136/jmg.2008.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyer J, Ekici AB, Endele S, et al. Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am J Hum Genet. 2012;90:565–72. doi: 10.1016/j.ajhg.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Winter CF, Baas M, Bijlsma EK, van Heukelingen J, Routledge S, Hennekam RC. Phenotype and natural history in 101 individuals with Pitt-Hopkins syndrome through an internet questionnaire system. Orphanet J Rare Dis. 2016;11:37. doi: 10.1186/s13023-016-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zweier C, Sticht H, Bijlsma EK, et al. Further delineation of Pitt-Hopkins syndrome: phenotypic and genotypic description of 16 novel patients. J Med Genet. 2008;45:738–44. doi: 10.1136/jmg.2008.060129. [DOI] [PubMed] [Google Scholar]
- 23.Marangi G, Zollino M. Pitt-Hopkins syndrome and differential diagnosis: a molecular and clinical challenge. J Pediatr Genet. 2015;4:168–76. doi: 10.1055/s-0035-1564570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sepp M, Pruunsild P, Timmusk T. Pitt-Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum Mol Genet. 2012;21:2873–88. doi: 10.1093/hmg/dds112. [DOI] [PubMed] [Google Scholar]
- 25.Rosenfeld JA, Leppig K, Ballif BC, et al. Genotype-phenotype analysis of TCF4 mutations causing Pitt-Hopkins syndrome shows increased seizure activity with missense mutations. Genet Med. 2009;11:797–805. doi: 10.1097/GIM.0b013e3181bd38a9. [DOI] [PubMed] [Google Scholar]
- 26.Giurgea I, Missirian C, Cacciagli P, et al. TCF4 deletions in Pitt-Hopkins Syndrome. Hum Mutat. 2008;29:E242–51. doi: 10.1002/humu.20859. [DOI] [PubMed] [Google Scholar]
- 27.Kousoulidou L, Tanteles G, Moutafi M, Sismani C, Patsalis PC, Anastasiadou V. 263.4 kb deletion within the TCF4 gene consistent with Pitt-Hopkins syndrome, inherited from a mosaic parent with normal phenotype. Eur J Med Genet. 2013;56:314–8. doi: 10.1016/j.ejmg.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Rossi M, Labalme A, Cordier MP, et al. Mosaic 18q21.2 deletions including the TCF4 gene: a clinical report. Am J Med Genet A. 2012;158A:3174–81. doi: 10.1002/ajmg.a.35588. [DOI] [PubMed] [Google Scholar]
- 29.Steinbusch CV, van Roozendaal KE, Tserpelis D, et al. Somatic mosaicism in a mother of two children with Pitt-Hopkins syndrome. Clin Genet. 2013;83:73–77. doi: 10.1111/j.1399-0004.2012.01857.x. [DOI] [PubMed] [Google Scholar]
- 30.Schluth-Bolard C, Labalme A, Cordier MP, et al. Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. J Med Genet. 2013;50:144–50. doi: 10.1136/jmedgenet-2012-101351. [DOI] [PubMed] [Google Scholar]
- 31.Maduro V, Pusey BN, Cherukuri PF, et al. Complex translocation disrupting TCF4 and altering TCF4 isoform expression segregates as mild autosomal dominant intellectual disability. Orphanet J Rare Dis. 2016;11:62. doi: 10.1186/s13023-016-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Pontual L, Mathieu Y, Golzio C, et al. Mutational, functional, and expression studies of the TCF4 gene in Pitt-Hopkins syndrome. Hum Mutat. 2009;30:669–76. doi: 10.1002/humu.20935. [DOI] [PubMed] [Google Scholar]
- 33.Peippo MM, Simola KO, Valanne LK, et al. Pitt-Hopkins syndrome in two patients and further definition of the phenotype. Clin Dysmorphol. 2006;15:47–54. doi: 10.1097/01.mcd.0000184973.14775.32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.