High-CO2 treatment (95% CO2, 1% O2) triggers deastringency in persimmon fruit via a cascade of interactions between several different classes of transcription factors leading to activation of genes required for anaerobic metabolism.
Keywords: Astringency removal, ERF, high CO2, hypoxia, MYB, persimmon fruit, transcriptional regulation
Abstract
Plant responses to anaerobic environments are regulated by ethylene-response factors (ERFs) in both vegetative and productive organs, but the roles of other transcription factors (TFs) in hypoxia responses are poorly understood. In this study, eight TFs (DkbHLH1, DkMYB9/10/11, DkRH2-1, DkGT3-1, DkAN1-1, DkHSF1) were shown to be strongly up-regulated by an artificial high-CO2 atmosphere (1% O2 and 95% CO2). Dual-luciferase assays indicated that some TFs were activators of previously characterized DkERFs, including DkMYB10 for the DkERF9 promoter, DkERF18/19 and DkMYB6 for the DkERF19 promoter, and DkERF21/22 for the DkERF10 promoter. Yeast one-hybrid and cis-element mutagenesis confirmed these physical interactions with one exception. The potential roles of these TFs in persimmon fruit deastringency were analysed by investigating their transient over-expression (TOX) in persimmon fruit discs, which indicated that DkMYB6TOX, DkMYB10TOX, DkERF18TOX, and DkERF19TOX were all effective in causing insolubilization of tannins, concomitantly with the up-regulation of the corresponding genes. These results indicated that multiple TFs of different classes are responsive to high-CO2/hypoxia in fruit tissues, and that a TF–TF regulatory cascade is involved in the hypoxia responses involving the Group VII DkERF10, and DkERFs and DkMYBs.
Introduction
Anoxia is a common abiotic stress for plants, usually caused by flooding (Yang et al., 2011). The response to anoxia involves a range of metabolic and morphological responses over different timescales, including a rapid induction of anaerobic metabolism (Kennedy et al., 1992; Voesenek and Bailey-Serres, 2015). Controlled-atmosphere storage in artificially reduced oxygen, usually supplemented with CO2, has been used for a long time to actively extend post-harvest storage and alleviate physiological disorders for various fruits (Ali et al., 2016; Bekele et al., 2016; Matityahu et al., 2016) and can induce anaerobic responses. A specific benefit for fruit quality conferred by a low-O2 environment has been reported for astringent-type persimmon (Diospyros kaki) incubated in an atmosphere of 1% O2 and 95% CO2 (Pesis and Ben-Arie, 1984; Taira et al., 1992, 2001; Min et al., 2012). The low-oxygen environment leads to acetaldehyde accumulation, which removes astringency in persimmon fruit by precipitation of soluble tannins (Taira et al., 2001; Salvador et al., 2007). Controlled atmospheres containing ethylene also promote deastringency, suggesting that ethylene signaling is involved (Ikegami et al., 2007; Min et al., 2012, 2014; Yin et al., 2012). Despite the fact that the commercial application of reduced oxygen for transportation and storage of fruit and some other plant products underpins a major industry and is important for food security, the underlying molecular mechanisms of fruit response to hypoxia are poorly understood.
In recent years, our knowledge of transcriptional regulatory mechanisms controlling hypoxia responses has been advanced significantly by the characterization of subfamily VII of the ethylene-response factors (ERF VII) (Sasidharan and Mustroph, 2011; Xie et al., 2016). In Arabidopsis, five ERF genes, namely hypoxia-responsive ERF1 (HRE1), HRE2, RAP2.2, RAP2.3, and RAP2.12, have been reported as the main plant oxygen-sensing regulators, and have been shown to control fermentation-related ADH and PDC genes (Hinz et al., 2010; Licausi et al., 2010; Yang et al., 2011; Bui et al., 2015; Papdi et al., 2015). This sensing system operates via the N-end rule pathway, which controls plant ERF hypoxia responses, via post-translational regulation (Gibbs et al., 2011; Licausi et al., 2011a). Involvement of ERFs in the regulation of hypoxia responses has also been reported in other plants, such as rice submergence tolerance-related Submergence 1 (Sub1; Xu et al., 2006), ERF VII in Rumex and Rorippa (van Veen et al., 2014), and ERF VII in apple fruit (Cukrov et al., 2016). Potential roles for ERFs in persimmon fruit responses to hypoxia have also been investigated. Eighteen DkERF genes were shown to be responsive to treatment with 95% CO2 (1% O2), but only DkERF9, 10, 19, and 22 were capable of trans-activation of the promoters of DkADH and DkPDC genes (Min et al., 2012, 2014). Moreover, of these four DkERF genes, only DkERF10, which has similarity to Arabidopsis HRE2, belongs to subfamily VII, indicating either that the hypoxia response is more complicated than revealed by investigations in Arabidopsis or that the ERF-VIIs may be regulated mainly at the post-translational level.
ERFs are one of the most comprehensively investigated transcription factor (TF) families with regards to involvement in plant hypoxia responses, although a few other hypoxia-related TFs have been reported, such as Arabidopsis AtMYB2 (Hoeren et al., 1998) and Heat shock factor A2 (HsfA2; Banti et al., 2010), wheat TaMYB1 (Lee et al., 2007), persimmon DkMYB6 (Fang et al., 2016) and DkTGA1 (Zhu et al., 2016). Omics-based analyses, however, have indicated many more TFs are responsive to hypoxia; for instance, at least 22 ERFs are regulated by anoxia in coleoptiles of rice (Lasanthi-Kudahettige et al., 2007), and additional differentially expressed TFs have been characterized from Arabidopsis roots, leaves, and seedlings (Branco-Price et al., 2005; Liu et al., 2005; Mustroph et al., 2009; Lee et al., 2011; Licausi et al., 2011b). These data indicated the involvement of a variety of TFs in hypoxia responses in plants, although whether and how they interact is unclear.
In the present research, using astringency loss as a reporter of the anaerobic response, we utilized RNA-seq data previously used for DkERF isolation (Min et al., 2012, 2014) and identified unigenes for TFs that were up-regulated by an artificial high-CO2 atmosphere (AHCA; 1% O2 and 95% CO2). Another treatment, AHNA (artificial high-N2 atmosphere; 99% N2 and 1% O2) was introduced to distinguish between responses to high CO2 and hypoxia. Both high-CO2-responsive and hypoxia-responsive TFs were selected for further analyses. Regulatory interactions of these TFs during hypoxia-triggered deastringency in persimmon fruit were investigated by dual-luciferase assays, yeast one-hybrid interactions, and promoter motif mutations. In the absence of a transformation system for persimmon, the functions of some potential regulators were analysed by transient over-expression in persimmon fruit discs.
Materials and methods
Plant material and treatments
Three astringent-type persimmon (Diospyros kaki) fruit were selected for this study, namely two Chinese cultivars, ‘Mopanshi’ and ‘Jingmianshi’, and one Japanese cultivar, ‘Tonewase’, all of which were collected from an orchard at Qingdao (Shandong, China) in 2014. Fruit without disease or signs of mechanical wounding were selected and divided into two batches: (1) the first batch was treated with AHCA (artificial high-CO2 atmosphere, 1% O2 and 95% CO2) in sealed in air-tight containers for 1 d to remove astringency, and (2) the second batch was sealed in similar containers containing air for 1 d, as a control.
In order to distinguish between the effects of high CO2 and low oxygen, AHNA (artificial high-N2 atmosphere, 99% N2 and 1% O2) treatments were performed using the cultivar ‘Gong cheng-shui shi’, which was obtained from a commercial orchard at Gongcheng (Guilin, China) in 2017. The fruit were divided into three batches: (1) the first batch was treated with AHCA in sealed air-tight containers for 1 d, (2) the second batch was treated with AHNA in similar containers, and (3) the third batch was sealed in containers containing air, as a control.
The treated fruit were transferred to storage in air at 20 °C. Fruit flesh from three replicate samples each of which consisted of four fruit were sampled for each treatment at all sampling points. The samples were frozen in liquid nitrogen and stored at –80 °C until further use.
Soluble condensed tannins
Soluble condensed tannins are the main source of astringency for persimmon fruit. Here, two different methods were selected to determine the content of soluble condensed tannins. The printing method was used for fruit flesh, according to Min et al., (2015). The whole fruit (1 d after picking, immediately after treatments) was cut into two parts and the cut surface was printed onto processed filter paper that had been soaked with 5% FeCl2 and then oven-dried at 60 °C. The content of soluble tannins was indicated by the intensity of the black color on the filter paper.
A more accurate measure of the content of soluble tannins from frozen samples was obtained with Folin–Ciocalteu reagent, with three biological replicates, according to the method described by Yin et al., (2012). The results were calculated using a standard curve of tannin acid equivalents.
Acetaldehyde and ethanol
Acetaldehyde and ethanol production were measured with a gas chromatograph (Agilent 6890N, USA), fitted with a FID column (HP-INNOWAX, 0.25 mm, 30 m, 0.25 μm, Agilent J&W, CA, USA), using the same parameters described previously by Min et al. (2012). In brief, 2 g frozen fruit flesh was ground in liquid nitrogen and added to 5 ml saturated NaCl solution. Then 3 ml of the mixture was transferred to 10-ml air-tight vials with crimp-top caps. The vials were placed in a water-bath at 60 °C for 1 h, after which 0.2 ml of head-space gas was removed for analysis. The injector, detector, and oven temperatures were set at 150, 160 and 100 °C, respectively. Sec-butyl alcohol (Sigma) was used as an internal control. The results were calculated using standard curves for acetaldehyde and ethanol. All measurements were conducted with three biological replicates.
RNA extraction and cDNA synthesis
Total RNA was extracted from frozen persimmon fruit flesh samples (2.0 g for each), using the method described by Chang et al. (1993). The total RNA was treated with a TURBO DNA-free kit (Ambion) to remove the genomic DNA. First-strand cDNA synthesis was initiated from 1.0 μg DNA-free RNA, using an iScriptTM cDNA Synthesis Kit (Bio-Rad). For each sampling point, three biological replicates were used for RNA extraction and the subsequent cDNA synthesis.
Gene isolation and sequence analysis
Using the same RNA-seq results described by Min et al. (2014), predicted TF-related hypoxia-responsive unigenes were isolated. The UTR regions of the transcripts were obtained using a SMART RACE cDNA amplification Kit (Clontech) and the primers are described in Supplementary Tables S1 and S2 at JXB online. The sequences of full-length TFs were confirmed and amplified with primers spanning the start and stop codons (Supplementary Table S3) and translated with the ExPASy software (http://web.expasy.org/translate). The newly isolated TFs were named after a BLAST analysis in Genbank and comparison with the reported TFs in persimmon.
Real-time PCR analysis
For real-time PCR, gene-specific oligonucleotide primers were designed (see Supplementary Table S4). The quality and specificity of each pair of primers were checked by melting curves and product resequencing. The housekeeping gene DkACT (Min et al., 2012) was chosen as the internal control and the 2–△△Ct method was used to calculate the relative expression levels of genes (Livak and Schmittgen, 2001). The expression at the time-point of fruit harvest (0 d) was set as 1 for each gene.
PCR reactions were performed on a CFX96TM Real-Time System (Bio-Rad). PCR reaction mixtures (20 μl) comprised 10 μl of SsoFastTM EvaGreen Supermix (Bio-Rad), 1 μl of each primer (10 μM), 2 μl diluted cDNA, and 6 μl DEPC-treated water. The PCR program was initiated with a preliminary step of 30 s at 95 °C, followed by 45 cycles of 95 °C for 5 s, 60 °C for 5 s, and completed with a melting-curve analysis program. For real-time PCR, three biological replicates were conducted for each gene at each sampling point of each treatment.
Dual-luciferase assay
The trans-activation by the TFs of genes related to deastringency was investigated by dual luciferase assays (Hellens et al., 2005). All constructs were electroporated into Agrobacterium tumefaciens GV3101. Full-length TFs were cloned into pGreen II 002962-SK vector (SK), using the primers described in Supplementary Table S5. The promoters of alcoholic fermentation-related genes (DkADH1 and DkPDC2) and deastringency-related ERFs (DkERF9, DkERF10, and DkERF19) were originally constructed by Min et al. (2012) and Fang et al. (2016), and were inserted into the pGreen II 0800-LUC vector.
The dual-luciferase assays were performed with Nicotiana benthamiana leaves, using the protocol described by Min et al. (2012, 2014). Agrobacterium carrying constructs were suspended in infiltration buffer (10 mM MES, 10 mM MgCl2. 150 mM acetosyringone, pH5.6) to an OD600 of approximately 0.75. TFs and promoters were combined at a ratio of 10:1 (v/v) and infiltrated into tobacco leaves by needleless syringes. Three days after infiltration, leaf discs were punched and assayed with dual-luciferase assay reagents (Promega). Dual-luciferase assays were performed with at least three independent experiments, with five biological replicates in each experiment.
Yeast one-hybrid assay
Yeast one-hybrid assays (Y1Hs) were performed in order to verify the gene–gene interactions, using the MatchmakerTM Gold Yeast One-Hybrid Library Screening System (Clontech, USA). The full-length DkMYB10 was subcloned into the pGADT7 AD vector and the promoter of DkERF9 was constructed into the pAbAi vector according to the ClonExpress II One-Step Cloning Kit (Vazyme, Nanjing) (primers are listed in Supplementary Table S6). Auto-activation and the interaction analyses were conducted according to the manufacturer’s protocol.
Site-directed mutagenesis of gene promoters
Due to auto-activation of the promoters of DkERF10 and DkERF19 in yeast (see Supplementary Fig. S7), site-directed mutagenesis was performed for the DkERF9, DkERF10, and DkERF19 promoters to eliminate the predicted binding sites for ERF and MYB TF (see Results). Motif mutations were carried out using the Fast Mutagenesis System (Transgene, Beijing) (primers are listed in Supplementary Table S7). Trans-activation effects of TFs on mutated promoters were further analysed by dual-luciferase assays.
Transient overexpression in persimmon fruit discs
In order to further verify the potential roles of TFs in persimmon fruit deastringency, transient overexpression (TOX) was performed with persimmon fruit discs. Discs of 1 cm diameter and 0.5 cm thickness were divided into five batches. The discs were incubated for 1 h with Agrobacterium carrying constructs in the same buffer used for the dual-luciferase assay. The discs were then transferred to filter papers (wetted by Murashige and Skoog medium) in tissue-culture dishes, and placed in an incubator at 25 °C for 3 d. All of the experiments (all genes and the empty vector) were performed with three biological replicates. At each sampling point (each day), the discs were dried on filter papers, frozen in liquid nitrogen and stored at –80 °C for further use.
Statistical analysis
The statistical significance of differences was determined using Student’s t-test by DPS2.05 (Zhejiang University, Hangzhou, China).
Results
Isolation and characterization of deastringency/hypoxia-responsive transcription factors from ‘Mopanshi’ persimmon fruit
From RNA-seq data from the ‘Mopanshi’ cultivar (Min et al., 2014), 13 full-length TFs were amplified by RACE and designated according to blast analysis as: DkbHLH1 and 2 (basic/helix-loop-helix, KY849612-3), DkMYB9, 10, 11, 12, and 13 (KY849603-7), DkRH2-1 and 2 (ring-H2 finger protein, KY849614-5), DkGT3-1 (trihelix transcription factor GT-3, KY849616), DkAN1-1 (zinc finger AN1 domain-containing protein, KY849617), DkHSF1 (heat shock factor, KY849619), and DkIAA1 (auxin-responsive protein, KY849618). These, together with previously reported TFs from persimmon (DkERFs, DkNACs, DkMYB6, DkTGA1) (Min et al., 2012, 2014, 2015; Fang et al., 2016; Zhu et al., 2016), were used to study transcriptional interactions in anaerobic persimmon fruit. A summary of AHCA-responsive transcription factors from persimmon fruit is given in Supplementary Table S8.
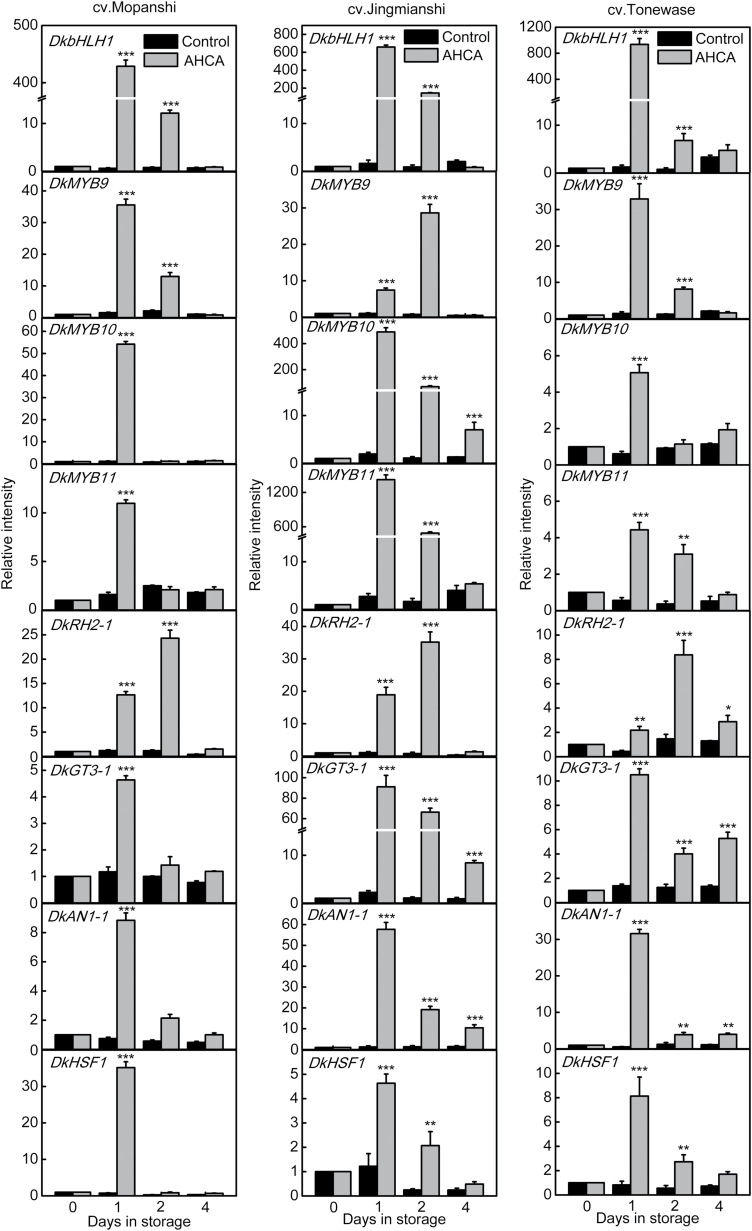
AHCA accelerated deastringency in persimmon fruit and triggered anaerobic fermentation, as indicated by bursts of acetaldehyde and ethanol production (see Supplementary Fig. S1). Expression of the 13 full-length TFs, which were predicted by RNA-seq, were analysed by real-time PCR. Eight genes were AHCA-responsive in ‘Mopanshi’, namely DkbHLH1, DkMYB9,10,11, DkRH2-1, DkGT3-1, DkAN1-1, and DkHSF1 (Fig. 1). Of these, DkbHLH1 showed the most striking response, increasing by about 429-fold after 1 d AHCA treatment, followed by DkMYB10, which increased by approximately 55-fold after 1 d. In ‘Jingmianshi’, DkMYB11 was the most responsive to high-CO2 treatment, increasing by about 1422-fold after 1 d, followed by DkbHLH1 and DkMYB10, with 658-fold and 489-fold increases, respectively. In ‘Tonewase’ only DkbHLH1 expression was very strongly responsive to AHCA treatment, increasing by about 935-fold after 1d. In contrast, the expression of the other five of the 13 TFs showed limited responses to AHCA treatment at 1 d (Supplementary Fig. S2), at which time the content of soluble tannins had almost reached its lowest level Supplementary Fig. S1). Thus, the subsequent responses (from 2 d onwards) of these genes were probably not related to deastringency.
Fig. 1.
Expression of TFs in response to treatment with artificial high-CO2 atmosphere (AHCA, 95% CO2 and 1% O2, 1 d) in persimmon fruit cultivars ‘Mopanshi’, ‘Jingmianshi’, and ‘Tonewase’ at 20 °C. For relative mRNA abundance of the genes, the values at day 0 were set as 1. Values are means (+SE) from three biological replicates (*P<0.05, **P<0.01, ***P<0.001).
In addition, a comparison was made between AHCA and AHNA using the cultivar ‘Gong cheng-shui shi’ (see Supplementary Fig. S3). Among the eight AHCA-responsive TFs, five (DkbHLH1, DkMYB9, DkMYB11, DkRH2-1, and DkHSF1) were responsive to both AHCA and AHNA, and thus can be termed as hypoxia-responsive; the expression of the other three TFs, DkGT3-1, DkAN1-1, and DkMYB10, remained constant in response to AHNA, and thus these genes were responsive to high CO2 (Supplementary Fig. S4).
Effect of high-CO2/hypoxia-responsive TFs on DkADH and DkPDC promoters
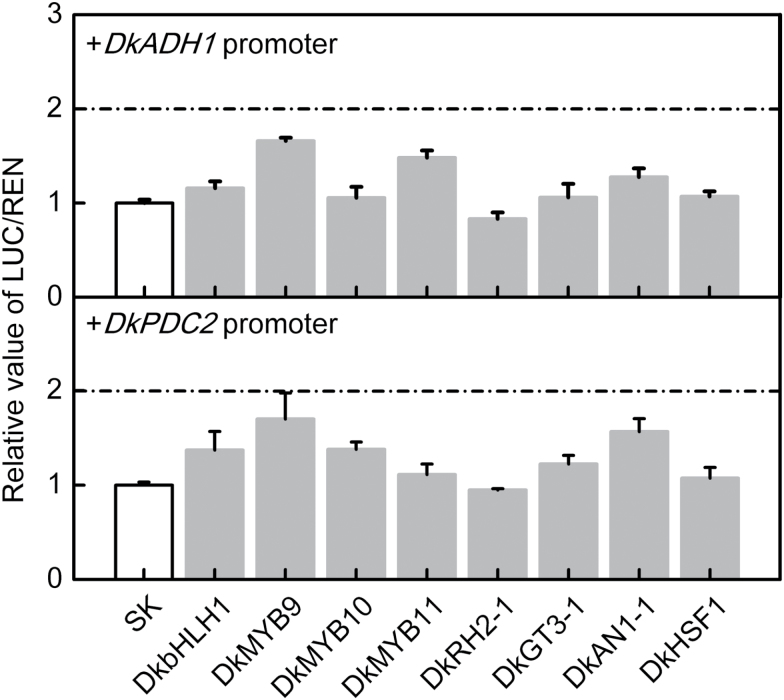
The persimmon genes DkADH1 and DkPDC2 were previously shown to be involved in fruit deastringency and to be induced by AHCA treatment (Min et al., 2012; Mo et al., 2016), and DkERF9 and DkERF10 were shown to have direct interactions with the DkADH1 and DkPDC2 promoters, respectively. In order to investigate the possible roles of other hypoxia-responsive TFs, the promoters of DkADH1 and DkPDC2 were used for dual-luciferase trans-activation assays. The eight AHCA-responsive TFs had either limited or no effects on the DkADH1 and DkPDC2 promoters (less than 2-fold increase) (Fig. 2), suggesting that there is no direct regulation by any of these TFs on the promoters of DkADH1 and DkPDC2.
Fig. 2.
Regulatory effects of artificial high-CO2 atmosphere (AHCA)-responsive TFs on the promoters of DkADH1 and DkPDC2 determined using the dual-luciferase assay. The ratio of LUC/REN of the empty vector (SK) plus promoter was used as the calibrator (set as 1). Values are means (+SE) from five biological replicates.
Relationship between high-CO2/hypoxia-responsive TFs
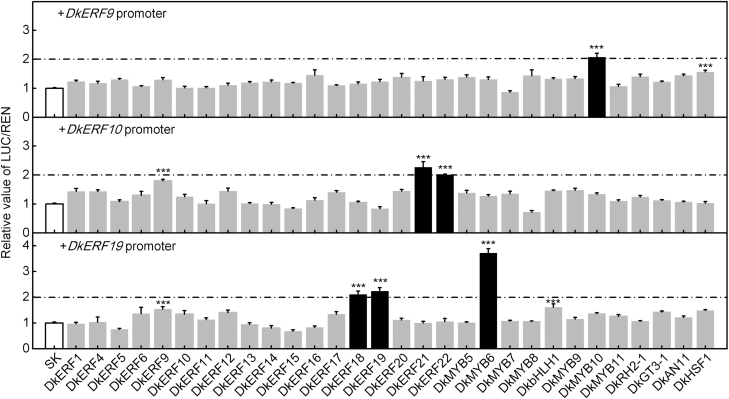
Four DkERF genes, DkERF9,10,19, and 22, were characterized previously as regulators of post-harvest deastringency in persimmon (Min et al., 2012, 2014). A further investigation was conducted to test the possible interaction between hypoxia-responsive TFs, including 18 additional DkERFs and four DkMYBs reported previously (Min et al., 2012, 2014; Fang et al., 2016), and promoters of DkERF9,10, and 19. Dual-luciferase assays indicated various trans-activation reactions, for example between DkMYB10 and the DkERF9 promoter (approximately 2.1-fold response), DkERF21 and 22 and the DkERF10 promoter (approximately 2.3- and 2.0-fold, respectively), and DkERF18 and 19 and DkMYB6 and the DkERF19 promoter (approximately 2.1-, 2.2-, and 3.7-fold, respectively) (Fig. 3). The synergistic effects of DkERF21 and DkERF22 on the promoter of DkERF10, and DkERF18, DkERF19, and DkMYB6 on the promoter of DkERF19 were also investigated, but there were no additive effects of these TFs on their corresponding target promoters (see Supplementary Figs S5 and S6).
Fig. 3.
Regulatory effects of artificial high-CO2 atmosphere (AHCA)-responsive TFs on the promoters of deastringency-related DkERF9, DkERF10, and DkERF19 determined using the dual-luciferase assay. AHCA-responsive DkERF and DkMYB5-8 were isolated by Min et al. (2012) and Fang et al. (2016). The ratio of LUC/REN of the empty vector (SK) plus promoter was used as the calibrator (set as 1). Black columns highlight inductions of at least 2-fold. Values are means (+SE) from five biological replicates (***P<0.001).
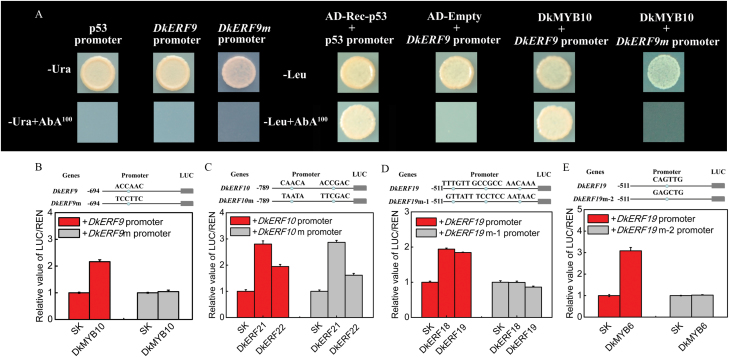
Using the yeast one-hybrid assay, it was found that DkMYB10 could physically bind to the DkERF9 promoter (Fig. 4A). Furthermore, the MBSII (ACCAAC; Grotewold et al., 1994) mutation in the DkERF9 promoter abolished the effects of DkMYB10 (Fig. 4A, B). Since the DkERF10 and DkERF19 promoters auto-activated in yeast (see Supplementary Fig. S7), a combination of cis-element mutations and dual-luciferase assays was used as an alternative way to test the specificity of this interaction. For the DkERF10 promoter, two motifs (CAACA, Kagaya et al., 1999; ACCGAC, DRE element, Stockinger et al., 1997) were mutated to TAATA and TTCGAC, respectively (Fig. 4C). Subsequent dual-luciferase assays indicated that DkERF21 and DkERF22 had similar activation on the DkERF10 promoter or the mutated DkERF10 promoter (DkERF10 m-promoter), suggesting either the absence of a direct interaction or that the interaction occurs with other unknown cis-elements (Fig. 4C). Two different mutations were designed to test the interaction between three transcription factors (DkERF18, 19, and DkMYB6) and the promoter of DkERF19. Three motifs (TTTGTT/AACAAA, TTTGTT, Dinh et al., 2012; GCCGCC, GCC box, Ohme-Takagi and Shinshi, 1995) were mutated to GTTATT/AATAAC and TCCTCC, and designated as DkERF19m-1 (Fig. 4D). To test the DkMYB6 interaction, the CAGTTG motif (MBSI; Solano et al., 1997) in the DkERF19 promoter was mutated to GAGCTG, designated as DkERF19m-2 (Fig. 4E). Dual-luciferase assays indicated that these motif mutations abolished trans-activation of the DkERF19 promoter by DkERF18, 19, and DkMYB6 (Fig. 4D, E).
Fig. 4.
Analysis of interactions between TFs and promoters determined by (A) yeast one-hybrid and (B–E) motif mutation assays. (A) Yeast one-hybrid analysis. Auto-activation of promoters was tested on synthetically defined (SD) medium lacking Ura in the presence of aureobasidin A (–Ura+AbA100). Interactions were determined on SD medium lacking Leu in the presence of aureobasidin A (–Leu+AbA100). Positive control: AD-Rec-p53+p53 promoter, provided with the kit; negative control: AD-Empty+DkERF9 promoter. (B–E) Schematic diagrams of motif mutations for the DkERF9/10/19 promoters and results of dual-luciferase assays performed with original and mutated promoters. The ratio of LUC/REN of the empty vector (SK) plus promoter was used as the calibrator (set as 1). Values are means (+SE) from five replicates.
Transient overexpression analysis in persimmon fruit discs
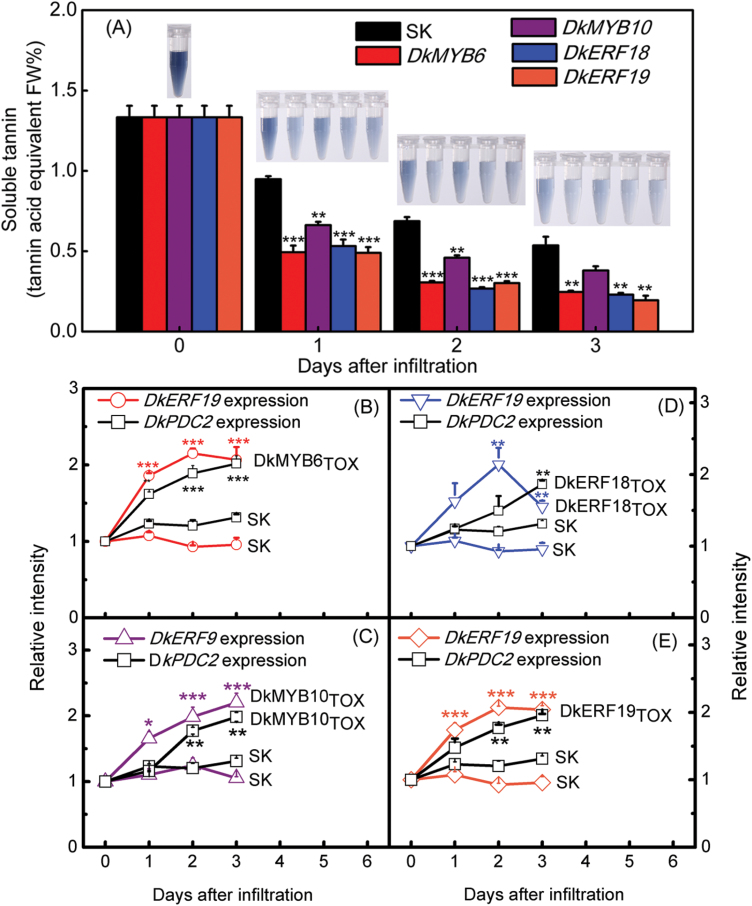
Due to the difficulty of stable transformation of perennial fruit such as persimmon, transient overexpression (TOX) analyses were performed with fruit discs. DkMYB6, DkMYB10, DkERF18, and DkERF19 were selected for analysis in view of their direct trans-activation of the DkERF9 and DkERF19 promoters (Figs 3 and 4), using tannin removal as a reporter for activity. The content of soluble tannins in the discs treated with transcription factors and the empty vector (SK, control) all declined during the incubation, which may have been due to the experimental manipulation (Fig. 5A). All four transcription factors, however, accelerated insolubilization of tannins from 1 d to 3 d, resulting in significantly lower content of soluble tannins than the controls (Fig. 5A). Interactions between transcription factors were also analysed and the results indicated that TOX of DkMYB6 and 10 and DkERF18 and 19 could significantly up-regulate the endogenous DkERF9 or DkERF19 transcripts in persimmon fruit discs, which further supported the interactions of AHCA-responsive transcription factors with the ERF promoters (Fig.5B–E). The expressions of downstream structure genes related deastringency were also analysed, and their expressions were also significantly up-regulated in the fruit discs, indicating that the transcriptional regulatory cascade would ultimately result in the regulation of structural genes (such as DkADH1 and DkPDC2) and hence in regulation of fruit deastringency.
Fig. 5.
Transient over-expression of TFs in persimmon fruit discs. The transient over-expression experiments were conducted with the empty vector pGreen II 002962-SK (SK) and DkMYB6/10 and DkERF18/19. Tissues from each of the infiltrated discs were taken to measure the content of soluble tannins (A) and the relative gene expression levels of related downstream genes compared with the SK control (B–E) during the 3 d of infiltration. Soluble tannin contents were measured using Folin–Ciocalteu reagent and were quantitated as tannin acid equivalents. The images above the bars in (A) show the reaction liquids used for measuring the soluble tannin content: the darker the colour of the test solution the higher the content of soluble tannin. Values are means (±SE) from three biological replicates. (*P<0.05, **P<0.01, ***P<0.001).
Discussion
Multiple TFs associated with the high-CO2/hypoxia response that leads to deastringency
AHCA treatment is the most effective commercial method for reducing persimmon fruit astringency. It functions by stimulating the accumulation of anaerobic metabolites, such as acetaldehyde, which precipitate soluble tannins (Supplementary Fig. S1; Pesis and Ben-Arie, 1984; Taira et al., 1992; Arnal and Del Río, 2004), the loss of which acts as a unique reporter of the anaerobic response. Previous work has highlighted the role of DkERF9,10,19, and 22 in this process (Min et al., 2012, 2014). In this present study, eight new TFs belonging to different families, including MYB, bHLH, Zinc finger, HSF, and IAA, were characterized and their transcripts shown to increase in abundance in response to AHCA treatment in three different cultivars, suggesting that multiple TFs may contribute to the deastringency process. These results are similar to those from omics-based analyses in Arabidopsis, where, in different organs and under different conditions, Liu et al. (2005) found 64 differentially expressed TFs in Arabidopsis seeds under hypoxic conditions, and Licausi et al. (2011b) identified over 180 TF genes, most of which belonged to the ERF, bHLH, MYB, HSF, and Zinc finger families, that were up- or down-regulated in roots under hypoxic conditions.
The ERFs are the best-characterized transcription factor gene family involved in plant hypoxia responses, and members belonging to Group VII play a key role (Hinz et al., 2010; Licausi et al., 2010, 2011a; Gibbs et al., 2011; Yang et al., 2011; Min et al., 2012; Gasch et al., 2016). The ERFs detected in this study and earlier research (Min et al., 2014) in persimmon belong to Group VII (DkERF10), Group IV (the DREB family) (DkERF9), Group IX (DkERF18, 19), and Group X (DkERF21, 22). The DkERF10 protein, the only persimmon Group-VII ERF detected, is assumed to be stabilized due to the MC domain (MCGGAII), which contributes to the stability of hypoxia-responsive ERFs (Gibbs et al., 2011; Licausi et al., 2011a), but there is also a major increase in DkERF10 mRNA on day 1 of anoxia (Min et al., 2012). The other DkERF genes involved in the response lack this MC domain, but our results indicate that they nevertheless participate directly in the regulatory cascade. In Arabidopsis and other plants, multiple groups of ERFs have also been shown to be associated with the responses to hypoxic treatments (Licausi et al., 2011b; Cukrov et al., 2016). Moreover, all these eight AHCA-responsive TFs were not homologous to the known ‘core 49’ hypoxia-responsive genes as identified by Mustroph et al. (2009). Although the expression of MYBs and many other transcription factors has been correlated with hypoxia tolerance (Hoeren et al., 1998; Abe et al., 2003; Mattana et al., 2007; Mustroph et al., 2009; Fang et al., 2016), it is worth emphasizing that among the eight AHCA-responsive TFs, only five genes were characterized as hypoxia-responsive with AHNA treatment, and the other three (DkGT3-1, DkMYB10, and DkAN1-1) were only responsive to high-CO2, and are thus not hypoxia-responsive (Supplementary Fig. S4). These findings indicated the similarities and also the differences between AHCA treatment in persimmon and hypoxia responses in model plants.
Transcriptional regulatory cascade of AHCA-responsive TFs
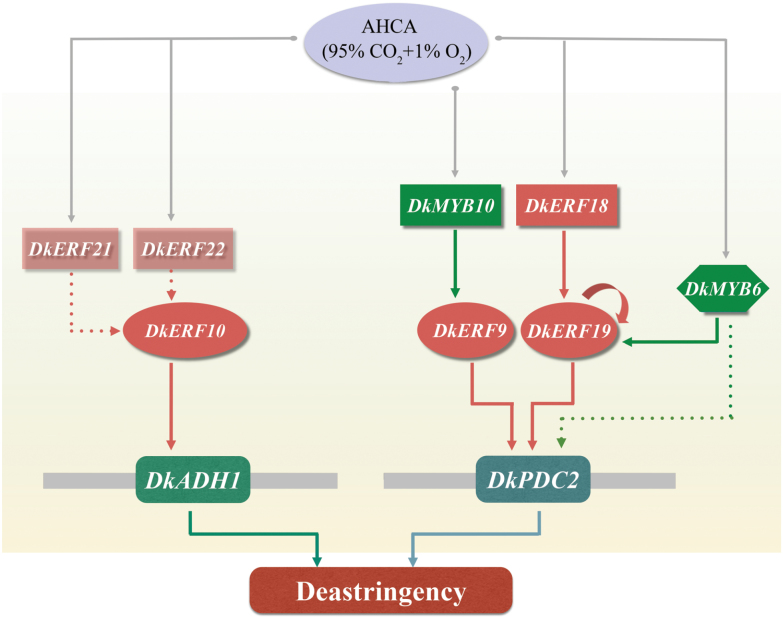
Although the mRNAs for DkbHLH1, DkMYB9,10,11, DkRH2-1, DkGT3-1, DkAN1-1, and DkHSF1 increased in abundance in response to AHCA, the corresponding proteins did not have a significant direct trans-activation effect on the DkADH1 and DkPDC2 promoters (all responses being significantly below 2-fold) (Fig. 2). This suggested that the newly identified factors might function indirectly in stimulating deastringency, and a further investigation was conducted to test possible interactions between the AHCA-responsive TFs and DkERF9,10, and 19, which recognize and trans-activate the DkADH1 or DkPDC2 promoters. The results indicated that there are at least two main types of transcriptional interactions between TFs: MYB–ERF and ERF–ERF interactions (Fig. 6). At least two MYBs, DkMYB6 and DkMYB10, physically bound to, and were putative activators of, the DkERF19 and DkERF9 promoters, respectively. ERF–ERF interactions included an indirect effect of DkERF21 and DkERF22 on the DkERF10 promoter and a direct regulation by DkERF18 and DkERF19 of the DkERF19 promoter (Fig. 6). DkERF19 showed auto-activation in dual-luciferase assays, indicating that its own protein can bind and trans-activate its promoter, and as our knowledge of the TF cascade expands it will be important to test for similar interactions and auto-regulations between specific TFs (e.g. MYB and ERF) that may contribute to regulation of the high-CO2/hypoxia response (Fig. 6). It also worth highlighting DkbHLH1, which was significantly up-regulated by deastringency treatments in all three examined cultivars and had a limited (less than 2-fold, but nonetheless significant) effect on the DkERF19 promoter. Compared to the TFs considered above, the regulatory mechanisms of DkbHLH1 (as well as the other responsive TFs) in the response of fruit to hypoxia require further investigation.
Fig. 6.
Model of TF interactions in response to artificial high-CO2 atmosphere (AHCA). AHCA treatment triggers the expression of various TFs, but only those with confirmed interactions are shown here. DkERF9, 10, and19 bind directly to, and activate promoters of, the anoxia-related genes DkADH1 and DkPDC2, which subsequently accelerate deastringency. DkERF18, 19, and DkMYB6, 10 physically bind and trans-activate DkERF promoters; DkERF21 and 22 are indirect regulators of DkERF10. DkERF19 exhibits auto-activation and binds to its own promoter. Solid arrows indicate direct interactions, while dashed arrows indicate indirect regulation.
Regulatory cascades between TFs have been widely reported in various plants; for example, AtSND1, a NAC transcription factor, is involved in the regulation of secondary wall biosynthesis in Arabidopsis through trans-activation of AtMYB46 (Zhong et al., 2007), and the MdMYB10 protein can bind and transactivate MdMYB10, which is involved in anthocyanin production in red-fleshed apples (Espley et al., 2009). For hypoxia responses, some TFs have been characterized at the transcript level and correlated with the expression of hypoxia-responsive genes (e.g. ADH and PDC, Abe et al., 2003; Licausi et al., 2011b). Our TF–promoter interaction results showed that, although Group-VII ERFs may play a leading role in sensing anaerobic conditions, there is a transcriptional cascade that leads to the up-regulation of the fermentation genes DkADH1 and DkPDC2 that involves the Group-VII DkERF10, and DkERFs from other Groups (DkERF9,18,19,21,22) and DkMYB6 and 10 (Fig. 6). The model presented here is supported by physical-binding and trans-activation studies (Figs 3 and 4; Min et al., 2012, 2014; Zhu et al., 2016) that provide insight into a hierarchy of interactions between the components of a regulatory cascade, leading to anaerobic responses. It enhances our understanding of the mechanism of the fruit hypoxia response, and may also apply to similar responses in other plant organs. One mechanism for the action of DkERF10 is that the protein may be stabilized by the effect of low O2 on the MC domain. No TFs were found that could directly regulate the DkERF10 promoter (Figs 3 and 4), although there was an indirect enhancement by DkERF21 and DkERF22. The possibility that there may be another unknown TF that can directly regulate the DkERF10 promoter and/or that an unknown cis-element exists in the DkERF10 promoter to which ERFs can bind requires further investigation.
In vivo interactions between high-CO2/hypoxia-responsive TFs and their roles in insolublization of tannins in persimmon fruit
TOX analyses showed that DkMYB6TOX and 10TOX and DkERF18TOX and 19TOX could significantly accelerate insolubilization of tannins in persimmon fruit discs, indicating that they participate in causing deastringency, which results directly from anaerobiosis (Taira et al., 2001; Salvador et al., 2007). The advantage of the TOX system is that it allows the analysis of the regulation of the endogenous genes and of the role of their transcriptional regulators and tannin content. Examples of the successful use of TOX include overexpression of DkMYB4 in kiwifruit calluses, which significantly enhanced tannin biosynthesis (Akagi et al., 2009), and expression of DkPDC2 in persimmon leaves, which decreased soluble tannin content (Min et al., 2012). The expression of DkERF9, DkERF19, and DkPDC2 was up-regulated by DkMYB6TOX and 10TOX and DkERF18TOX and 19TOX in discs over a 1–3 d period, indicating the rapid and continuous responses of endogenous genes to these TFs, which occurred concomitantly with the decrease in soluble tannins in fruit discs. These in vitro results (Fig. 5) confirm the potential interactions and roles for ERF and MYB TFs in the response of persimmon fruit to AHCA treatment (Fig. 6).
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Effects of AHCA treatment on post-harvest deastringency in fruit of persimmon ‘Mopanshi’ at 20 °C.
Fig. S2. Expression of transcription factors that were relatively less responsive to AHCA treatment.
Fig. S3. Comparison of tannin printing assays for control and AHNA- and AHCA-treated ‘Gong cheng-shui shi’ fruit at 1 d.
Fig. S4. Expression of transcription factors in response to AHCA and AHNA treatment in ‘Gong cheng-shui shi’ fruit.
Fig. S5. Synergistic trans-activation effects of combinations of DkERF21 and DkERF22 on the DkERF10 promoter.
Fig. S6. Synergistic trans-activation effects of combinations of DkMYB6 and DkERF18/19 on the DkERF19 promoter.
Fig. S7. Auto-activation test for the DkERF10/19 promoters.
Table S1. Primer sequences for 3′-RACE analysis.
Table S2. Primer sequences for 5′-RACE analysis.
Table S3. Primer sequences for full-length TFs.
Table S4. Primer sequences for real-time PCR analysis.
Table S5. Primer sequences for the dual-luciferase assays.
Table S6. Primer sequences for the yeast one-hybrid assay.
Table S7. Primer sequences for site-directed mutagenesis of the DkERF9/10/19 promoters.
Table S8. Summary on hypoxia-responsive transcription factors from persimmon fruit.
Acknowledgements
This research was supported by the National Key Research and Development Program (2016YFD0400102), the National Natural Science Foundation of China (31722042; 31672204), the Natural Science Foundation of Zhejiang Province, China (LR16C150001), and the 111 Project (B17039). The authors have no conflicts of interest to declare.
Abbreviations
- ADH
alcohol dehydrogenase
- AHCA
artificial high-CO2 atmosphere (1% O2 and 95% CO2)
- AHNA
artificial high-N2 atmosphere (99% N2 and 1% O2)
- PDC
pyruvate decarboxylase
- ERF
ethylene-response factor
- HRE
hypoxia-response ERF
- TOX
transient overexpression.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, Yonemori K. 2009. DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiology 151, 2028–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Khan AS, Malik AU, Shahid M. 2016. Effect of controlled atmosphere storage on pericarp browning, bioactive compounds and antioxidant enzymes of litchi fruits. Food Chemistry 206, 18–29. [DOI] [PubMed] [Google Scholar]
- Arnal L, Del Río MA. 2004. Effect of cold storage and removal astringency on quality of persimmon fruit (Diospyros kaki, L.) cv. Rojo Brillante. Food Science & Technology International 10, 179–185. [Google Scholar]
- Banti V, Mafessoni F, Loreti E, Alpi A, Perata P. 2010. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiology 152, 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekele EA, Alis ARR, Hertog MLATM, Nicolai BM, Geeraerd AH. 2016. Dynamics of metabolic adaptation during initiation of controlled atmosphere storage of ‘Jonagold’ apple: effects of storage gas concentrations and conditioning. Postharvest Biology and Technology 117, 9–20. [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. 2005. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Annals of Botany 96, 647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui LT, Giuntoli B, Kosmacz M, Parlanti S, Licausi F. 2015. Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Science 236, 37–43. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter 11, 113–116. [Google Scholar]
- Cukrov D, Zermiani M, Brizzolara S et al. 2016. Extreme hypoxic conditions induce selective molecular responses and metabolic reset in detached apple fruit. Frontiers in Plant Science 7, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TT, Girke T, Liu X, Yant L, Schmid M, Chen X. 2012. The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139, 1978–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagné D et al. 2009. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. The Plant Cell 21, 168–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Wang MM, Zhu QG, Min T, Grierson D, Yin XR, Chen KS. 2016. DkMYB6 is involved in persimmon fruit deastringency, via transcriptional activation on both DkPDC and DkERF. Postharvest Biology and Technology 111, 161–167. [Google Scholar]
- Gasch P, Fundinger M, Müller JT, Lee T, Bailey-Serres J, Mustroph A. 2016. Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in arabidopsis. The Plant Cell 28, 160–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM et al. 2011. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. 1994. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R. 2010. Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiology 153, 757–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. 1998. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami A, Eguchi S, Kitajima A, Inoue K, Yonemori K. 2007. Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit. Plant Science 172, 1037–1047. [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T. 1999. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Research 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RA, Rumpho ME, Fox TC. 1992. Anaerobic metabolism in plants. Plant Physiology 100, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasanthi-Kudahettige R, Magneschi L, Loreti E, Gonzali S, Licausi F, Novi G, Beretta O, Vitulli F, Alpi A, Perata P. 2007. Transcript profiling of the anoxic rice coleoptile. Plant Physiology 144, 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LA, Bailey-Serres J. 2011. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytologist 190, 457–471. [DOI] [PubMed] [Google Scholar]
- Lee TG, Jang CS, Kim JY, Kim DS, Park JH, Kim DY, Seo YW. 2007. A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: roles in response to the oxygen concentration in root environment and abiotic stresses. Physiology Plantarum 129, 375–385. [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LA, Perata P, van Dongen JT. 2011a. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422. [DOI] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P. 2010. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. The Plant Journal 62, 302–315. [DOI] [PubMed] [Google Scholar]
- Licausi F, Weits DA, Pant BD, Scheible WR, Geigenberger P, van Dongen JT. 2011b. Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytologist 190, 442–456. [DOI] [PubMed] [Google Scholar]
- Liu F, Vantoai T, Moy LP, Bock G, Linford LD, Quackenbush J. 2005. Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiology 137, 1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Matityahu I, Marciano P, Holland D, Ben-Arie R, Amir R. 2016. Differential effects of regular and controlled atmosphere storage on the quality of three cultivars of pomegranate (Punica granatum L.). Postharvest Biology and Technology 115, 132–141. [Google Scholar]
- Mattana M, Vannini C, Espen L et al. 2007. The rice Mybleu transcription factor increases tolerance to oxygen deprivation in Arabidopsis plants. Physiologia Plantarum 131, 106–121. [DOI] [PubMed] [Google Scholar]
- Min T, Fang F, Ge H, Shi YN, Luo ZR, Yao YC, Grierson D, Yin XR, Chen KS. 2014. Two novel anoxia-induced ethylene response factors that interact with promoters of deastringency-related genes from persimmon. PLoS ONE 9, e97043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min T, Wang MM, Wang H, Liu X, Fang F, Grierson D, Yin XR, Chen KS. 2015. Isolation and expression of NAC genes during persimmon fruit postharvest astringency removal. International Journal of Molecular Sciences 16, 1894–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min T, Yin XR, Shi YN, Luo ZR, Yao YC, Grierson D, Ferguson IB, Chen KS. 2012. Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon (Diospyros kaki) fruit de-astringency. Journal of Experimental Botany 63, 6393–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo RL, Yang SC, Huang YM, Chen WX, Zhang QL, Luo ZR. 2016. ADH and PDC genes involved in tannins coagulation leading to natural de-astringency in Chinese pollination constant and non-astringency persimmon (Diospyros kaki Thunb.). Tree Genetics & Genomes 12, 17. [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. 2009. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. 1995. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. The Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C, Pérez-Salamó I, Joseph MP, Giuntoli B, Bögre L, Koncz C, Szabados L. 2015. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. The Plant Journal 82, 772–784. [DOI] [PubMed] [Google Scholar]
- Pesis E, Ben-Arie R. 1984. Involvement of acetaldehyde and ethanol accumulation during induced deastringency of persimmon fruits. Journal of Food Science 49, 896–899. [Google Scholar]
- Salvador A, Arnal L, Besada C, Larrea V, Quiles A, Pérez-Munuera I. 2007. Physiological and structural changes during ripening and deastringency treatment of persimmon fruit cv. ‘Rojo Brillante’. Postharvest Biology and Technology 46, 181–188. [Google Scholar]
- Sasidharan R, Mustroph A. 2011. Plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis. The Plant Cell 23, 4173–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Fuertes A, Sánchez-Pulido L, Valencia A, Paz-Ares J. 1997. A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. The Journal of Biological Chemistry 272, 2889–2895. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. 1997. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proceedings of the National Academy of Sciences, USA 3, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira S, Ikeda K, Ohkawa K. 2001. Comparison of insolubility of tannins induced by acetaldehyde vapor in fruit of three types of astringent persimmon. Journal of the Japanese Society for Horticultural Science 48, 684–687. [Google Scholar]
- Taira S, Oba S, Watanabe S. 1992. Removal of astringency from ‘Hiratanenashi’ persimmon fruit with a mixture of ethanol and carbon dioxide. Journal of the Japanese Society for Horticultural Science 61, 437–443. [Google Scholar]
- van Veen H, Akman M, Jamar DC, Vreugdenhil D, Kooiker M, van Tienderen P, Voesenek LA, Schranz ME, Sasidharan R. 2014. Group VII ethylene response factor diversification and regulation in four species from flood-prone environments. Plant, Cell & Environment 37, 2421–2432. [DOI] [PubMed] [Google Scholar]
- Voesenek LA, Bailey-Serres J. 2015. Flood adaptive traits and processes: an overview. New Phytologist 206, 57–73. [DOI] [PubMed] [Google Scholar]
- Xie XL, Yin XR, Chen KS. 2016. Roles of APETALA2/Ethylene Responsive Factors in regulation of fruit quality. Critical Reviews in Plant Sciences 35, 120–130. [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. 2006. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708. [DOI] [PubMed] [Google Scholar]
- Yang CY, Hsu FC, Li JP, Wang NN, Shih MC. 2011. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiology 156, 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XR, Shi YN, Min T, Luo ZR, Yao YC, Xu Q, Ferguson I, Chen KS. 2012. Expression of ethylene response genes during persimmon fruit astringency removal. Planta 235, 895–906. [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. 2007. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. The Plant Cell 19, 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QG, Wang MM, Gong ZY, Fang F, Sun NJ, Li X, Grierson D, Yin XR, Chen KS. 2016. Involvement of DkTGA1 transcription factor in anaerobic response leading to persimmon fruit postharvest de-astringency. PLoS ONE 11, e0155916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.