Jasmonate-responsive repressors of FtMYBs can be degraded by the 26S proteasome in a COI1-dependent manner in Fagopyrum tataricum and act together with FtSAD2 or FtJAZ1 in the phenylpropanoid pathway.
Keywords: 26S proteasome, buckwheat, Fagopyrum, tataricum, jasmonates, MYB transcription factor, phenylpropanoid pathway
Abstract
Jasmonates are plant hormones that induce the accumulation of many secondary metabolites, such as rutin in buckwheat, via regulation of jasmonate-responsive transcription factors. Here, we report on the identification of a clade of jasmonate-responsive subgroup 4 MYB transcription factors, FtMYB13, FtMYB14, FtMYB15, and FtMYB16, which directly repress rutin biosynthesis in Fagopyrum tataricum. Immunoblot analysis showed that FtMYB13, FtMYB14, and FtMYB15 could be degraded via the 26S proteasome in the COI1-dependent jasmonate signaling pathway, and that this degradation is due to the SID motif in their C-terminus. Yeast two-hybrid and bimolecular fluorescence complementation assays revealed that FtMYB13, FtMYB14, and FtMYB15 interact with the importin protein Sensitive to ABA and Drought 2 (FtSAD2) in stem and inflorescence. Furthermore, the key repressor of jasmonate signaling FtJAZ1 specifically interacts with FtMYB13. Point mutation analysis showed that the conserved Asp residue of the SID domain contributes to mediating protein–protein interaction. Protoplast transient activation assays demonstrated that FtMYB13, FtMYB14, and FtMYB15 directly repress phenylalanine ammonia lyase (FtPAL) gene expression, and FtSAD2 and FtJAZ1 significantly promote the repressing activity of FtMYBs. These findings may ultimately be promising for further engineering of plant secondary metabolism.
Introduction
Buckwheat is a type of pseudocereal of the genus Fagopyrum, within the Polygonaceae family. Three major species, tartary buckwheat (TB; Fagopyrum tataricum), common buckwheat (CB; Fagopyrum esculentum), and golden buckwheat (Fagopyrum cymosum), are widely cultivated in Asia and Europe (Zhang et al., 2012). Buckwheat can grow in harsh climates and nutrient-poor soils, which suggests that it has great ecological adaptability. Recently, TB research interest has increased due to its higher content of bioactive flavonoids (e.g. rutin, orientin, vitexin, quercetin, isovitexin, and isoorientin) compared with CB, and hence its potential to contribute to diverse health benefits. For instance, the concentration of rutin, a major flavonoid, can reach 81 mg g–1 in the groats of TB compared with the concentration of 0.2 mg g–1 reported in CB (Wijngaard and Arendt, 2006). Additionally, TB flour accumulates ~10-fold more total flavonoids than CB (Qin et al., 2010). Therefore, buckwheat was considered to be a model plant for studying flavonoid biosynthesis due toits high accumulation of rutin, especially in the early days of research.
MYB transcription factors (TFs) are key regulators of plant flavonoid biosynthesis (Du et al., 2010; Zhou and Memelink, 2016). MYB TFs are classified into four subgroups (R2R3-, 1R-, 3R-, and 4R-MYBs) on the basis of the number of adjacent repeats in their DNA-binding domains (Dubos et al., 2010). Most of the MYB TFs that are involved in the regulation of flavonoid biosynthesis belong to the large R2R3-MYB family (Du et al., 2010). In Arabidopsis thaliana, PAP1/MYB75, PAP2/MYB90, MYB112, MYB113, and MYB114 act positively in the flavonoid biosynthesis pathway (Zimmermann et al., 2004; Teng et al., 2005; Gonzalez et al., 2008; Lotkowska et al., 2015). However, MYB3, MYB4, MYB7, and MYB32, which are classified into subgroup 4 (S4) of the R2R3-MYB TFs, repress phenylpropanoid biosynthesis owing to the EAR repression motif (pdLHLD/LLxiG/S) and the conserved LLsrGIDPxT/SHRxI/L motif in their C-termini (Jin et al., 2000; Preston et al., 2004; Dubos et al., 2010; Fornalé et al., 2014; Zhou et al., 2015a; Zhou et al., 2017). It has been reported that MYB4, MYB7, and MYB32 interact with the importin β-like protein SAD2 (Sensitive to ABA and Drought 2) via a conserved GY/FDFLGL motif [the SAD2 Interaction Domain (SID) motif], mediating the transport of MYBs into the nucleus (Zhou et al., 2015a). Recently, our group showed that the repression activity of MYB3 on expression of the C4H (cinnamate 4-hydroxylase) gene is directly regulated by the co-repressors NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED 1 (LNK1) and LNK2 in phenylpropanoid biosynthesis (Zhou et al., 2017). In Brassica rapa subsp. rapa cv. Tsuda turnip, BrMYB4, the ortholog of AtMYB4, acts as a negative regulator in the phenylpropanoid biosynthesis pathway because of its C-terminal repression motif (Zhang et al., 2014). In buckwheat, FtMYB1 and FtMYB2 function positively in the accumulation of proanthocyanidins (Bai et al., 2014).
Jasmonates (JAs) are important plant hormones that induce the biosynthesis of various secondary metabolites, such as flavonoids and anthocyanins, via modulation of JA-responsive TFs (Zhou and Memelink, 2016). The key transcriptional repressors, jasmonate ZIM domain (JAZ) proteins, can interact with several TFs with different roles in regulating JA-responsive gene expression (Yan et al., 2007; Chini et al., 2007; Thines et al., 2007). A variety of MYB TFs have been found to interact with members of the JAZ family. These include the R2R3-MYB activators MYB21 and MYB24, which are involved in stamen development and male fertility (Song et al., 2011), and MYB75, which is involved in anthocyanin biosynthesis and trichome initiation (Qi et al., 2011). It has been also reported that JAs could boost phenylpropanoid biosynthesis in buckwheat (Kim et al., 2011). However, the JA-regulated mechanism of the phenylpropanoid biosynthetic pathway is still unknown in buckwheat.
Here, a clade of JA-responsive MYB repressors, including FtMYB13, FtMYB14, FtMYB15, and FtMYB16, are described in relation to their role in phenylpropanoid biosynthesis. We found that these MYB repressors spatially regulate phenylpropanoid biosynthesis at the transcript and the protein level in different tissues. In addition, the repressing activity of FtMYBs is directly enhanced by FtSAD2 or the co-repressor FtJAZ1 via protein–protein interaction.
Materials and methods
Isolation of FtMYB13, FtMYB14, FtMYB15, and FtMYB16 genes and the FtPAL promoter
The publicly available database of F. tataricum genes in the flower and inflorescence and a genome database were collected (Logacheva et al., 2011). Searches for DNA sequences containing a conserved MYB domain were performed to confirm the candidate MYB genes. In addition, SMARTRACE and PCR technology were utilized to clone the full-length FtMYB13, FtMYB14, FtMYB15, and FtMYB16 transcripts with the following primers: 5ʹ-ATGGGAAGAGCTCCTTGTTGC-3ʹ and 5ʹ-TCAGATGAGCAAAGACTCAGC-3ʹ for FtMYB13; 5ʹ-ATGG GTAGATCTCCATGTTGTG-3ʹ and 5ʹ-TCATTTCATCTCCAATG ATC-3ʹ for FtMYB14; 5ʹ -ATGGGTCGATCTCCATGTTGC-3ʹ and 5ʹ-TCATTTCATCTCCAAAGTTCTATAG-3ʹ for FtMYB15; 5ʹ-ATG GGGAGATCACCTTGCTGC-3ʹ and 5ʹ-CTAGCTTGTTGTGGCAT TAGAAG-3ʹ for FtMYB16; 5ʹ-ATGGATCTTCCAAGCCTCGCT-3ʹ and 5ʹ-TCAAGAGAGTTCTTCGAGCAT-3ʹ for FtSAD2; 5ʹ-ATGA ACTTGTTCCCACTGAAAG-3ʹ and 5ʹ-TCAGGGCTGAATCGACG TCCG-3ʹ for FtJAZ1. The FtPAL promoter fragments corresponding to the sequence from the transcription start site to 974 bp upstream of the transcription start site were isolated using the primers FtPALproF: 5ʹ-GTCGTTAAATATCGTTAAAAT-3ʹ and FtPALproR: 5ʹ-CCACCCCAACGGATCCTGCAC-3ʹ. The RNA isolation, cDNA synthesis, amplification conditions and sequence analysis were as described previously (Zhou et al., 2015b).
Plant materials, growth conditions, and chemical treatments
Two-week-old F. tataricum seedlings and 20-day-old hairy root lines were treated with 50 µM MeJA (Sigma-Aldrich, St. Louis, MO, USA) as described previously (Zhou et al., 2015b). For the construction of transgenic F. tataricum hairy root lines constitutively overexpressing FtMYB13-HA, FtMYB13D281N-HA, FtMYB13D283N-HA, FtMYB13D285N-HA, FtMYB14-HA, FtMYB14D266N-HA, FtMYB15-HA, FtMYB15D258N-HA, FtMYB16-HA, FtSAD2-HA, and FtJAZ1-HA, pRT101 plasmids containing these genes were digested with SphI and the genes were subsequently cloned into pCAMBIA1300. The binary vectors pCAMBIA1300-FtMYB13-HA, pCAMBIA1300-FtMYB13D281N-HA, pCAMBIA1300-FtMYB13D283N-HA, pCAMBIA 1300-FtMYB13D285N-HA, pCAMBIA1300-FtMYB14-HA, pCAMBIA1300- FtMYB14D266N-HA, pCAMBIA1300-FtMYB15-HA, pCAMBIA1300-FtMYB15D258N- HA, pCAMBIA1300-FtMYB16-HA, pCAMBIA1300-FtSAD2-HA, and pCAMBIA1300-FtJAZ1-HA were introduced into the Agrobacterium rhizogenes A4 strain (agropine type). Sterile F. tataricum leaves were cut into squares and transfected with A. rhizogenes A4 strains containing the above plasmids. Roots developed at the cut edges 2–3 weeks after co-cultivation. F. tataricum transgenic hairy roots were identified using the methods described by Zhou et al. (2010). All primer sequences are listed in Supplementary Table S1 at JXB online.
Yeast two-hybrid assays
The full-length genes FtMYB13, FtMYB13D281N, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB14D266N, FtMYB15, FtMYB15D258N, FtMYB16, FtSAD2, and FtJAZ1 were cloned into pACT2 or pAS2.1 (James et al., 1996). Point mutations of the genes were generated using the GeneTailorTM Site-Directed Mutagenesis System (Life Technologies; http://www.thermofisher.com). The PCR fragments and constructed plasmids were confirmed by sequencing. Co-transformation with bait and prey plasmids at a ratio of 1:1 was performed in yeast strain PJ69-4A according to a modified yeast transformation protocol (Gietz et al., 1992). All primer sequences are listed in Supplementary Table S1.
Yeast one-hybrid assays
FtMYB13, FtMYB14, FtMYB15, and FtMYB16 were PCR amplified from pACT2-FtMYB13, pACT2-FtMYB14, pACT2-FtMYB15, and pACT2-FtMYB16, digested with BamHI and PstI, and cloned into pAS2.1. The plasmids were introduced into the yeast strain PJ69-4A with the reporter genes His3 and LacZ following the manufacturer’s instructions (Clontech, Palo Alto, CA, USA). The promoter fragment of FtPALpro was digested with NotI and SmaI and fused to a TATA box-HIS3 gene in the plasmid pHIS3NX. HIS3 gene constructs were integrated into the genome of yeast strain Y187. After transformation with the plasmid pACT2-FtMYB13, pACT2-FtMYB14, pACT2-FtMYB15, or pACT2-FtMYB16, yeast cells were grown on minimal synthetic defined (SD)-glucose medium lacking Leu and His (SD/-LH) supplemented with 3-amino-1,2,4-triazole (3-AT; Sigma) at increasing concentrations ranging from 0 to 50 mM. Yeast cells containing the empty pAS2.1 vector were used as a negative control. The colony-lift filter β-galactosidase assay was carried out according to the Yeast Protocols Handbook (Clontech Laboratories, Inc.). All primer sequences are listed in Supplementary Table S1.
Confocal microscopy
To observe the subcellular localization of FtMYB13, FtMYB13D281N, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB14D266N, FtMYB15, FtMYB15D258N, and FtMYB16, the green fluorescent protein (GFP)-fused open reading frames were cloned into pTH2. For bimolecular fluorescence complementation (BiFC) assays, FtMYB13, FtMYB13D281N, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB14D266N, FtMYB15, FtMYB15D258N and FtMYB16, FtSAD2, and FtJAZ1 were cloned into pRTL2-YNEE (nYFP-) or pRTL2-HAYC (-cYFP). All constructs were transiently expressed or co-expressed with all possible combinations of nYFP and cYFP fusion proteins by polyethylene glycol (PEG)-mediated transfection into Arabidopsis protoplasts, as previously described (Schirawski et al., 2000). Microscopic images of transfected protoplasts were acquired with a Leica DM IRBE confocal laser scanning microscope and analyzed with ImageJ (Abràmoff et al., 2004). Primers used in the BiFC assays are listed in Supplementary Table S1.
Arabidopsis protoplast transactivation assays
The promoter fragment of FtPAL was amplified on genomic DNA and cloned into the reporter plasmid pGusXX (Pasquali et al., 1994). The FtMYB13, FtMYB13D281N, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB14D266N, FtMYB15, FtMYB15D258N and FtMYB16, FtSAD2, and FtJAZ1 were PCR amplified and cloned into the effector plasmid pRT101 (Töpfer et al., 1987). Protoplasts isolated from an Arabidopsis cell suspension were co-transformed with a reporter plasmid carrying the FtPAL promoter fused to the β-glucuronidase gene (GUS) and effector plasmids carrying either the FtMYB13, FtMYB13D281N, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB14D266N, FtMYB15, FtMYB15D258N, FtMYB16, FtSAD2, or FtJAZ1 gene fused to the CaMV 35S promoter. As controls, co-transformations of FtPAL-promoter-GUS with the empty vector pRT101 were used. PEG-mediated transfection of protoplasts and GUS activity assays were performed as described previously (van der Fits and Memelink, 1997; Schirawski et al., 2000). All primers used in the protoplast transactivation assays are listed in Supplementary Table S1.
Quantitative RT–PCR
Reverse transcription of total RNA was carried out by using the Revert AidTM First Strand cDNA Synthesis Kit (Fermentas). The primers of FtPAL, FtC4H, Ft4CL, FtCHS, FtCHI, FtF3H, FtF3ʹH, FtFLS, and the reference gene FtH3 for quantitative real-time (qRT)–PCR were as previously described (Li et al., 2010; Zhou et al., 2015b). The qRT–PCR amplification was performed as described in previous reports (Zhou et al., 2010; Zhou et al., 2015b), and the result was visualized as a heat map generated by TreeView 1.1.3.
Protein extraction and western blot
Protein extraction from F. tataricum hairy roots and immunoblot analysis with anti-hemagglutinin (HA) or anti-GFP peroxidase antibodies followed the methods described by Zhou et al. (2015a, b).
Rutin measurement
Different tissues of 60-day-old F. tataricum plants were harvested and freeze-dried. F. tataricum hairy roots (1 g) were harvested, frozen, and ground in liquid nitrogen. Frozen samples were extracted twice in 50 ml methanol for 24 h at 4 °C and extracts were vacuum-dried at 80 °C and dissolved in 10 ml methanol. The solution was then filtered through a polyvinylidene filter (pore size 0.45 μm) and diluted two-fold with methanol. The concentration of rutin was analyzed by high-performance liquid chromatography (HPLC) of triplicate independent extractions for each line, as described elsewhere (Huang et al., 2016).
Statistical analysis
All data were analyzed using Student’s t-test and one-way ANOVA. Values of P<0.05 were considered to be significant.
Results
Characterization of JA-responsive R2R3-MYB S4 family members
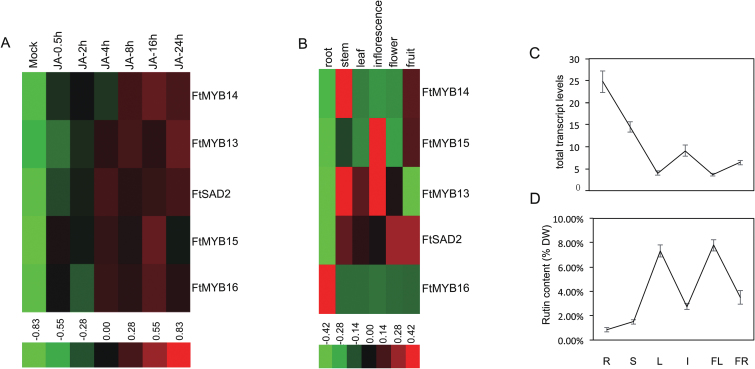
To identify the members of the buckwheat R2R3-MYB S4 family, a BLAST search of the CB genome database (Yasui et al., 2016) and TB transcripts database (Logacheva et al., 2011) was performed, using the EAR domain of MYB4 from Arabidopsis as a query sequence (Zhou et al., 2015a). Of these sequences, FtMYB13 (GenBank accession no. KY290579), FtMYB14 (GenBank accession no. KY290580), FtMYB15 (GenBank accession no. KY290581), and FtMYB16 (GenBank accession no. KY290582) contained an EAR motif (Supplementary Fig. S1) and shared the highest identity with subgroup 4 TFs (Supplementary Fig. S2; Jin et al., 2000; Zhou et al., 2015a). To gain a general perspective on the amount of FtMYBs transcripts response to JAs, qRT–PCR was performed. As shown in Fig. 1A, FtMYB13 mRNA accumulation was highly induced by MeJA treatment and reached its highest level at 24 h, while FtMYB14, FtMYB15 and FtMYB16 were slightly induced. The spatial transcript accumulation analysis showed that the highest level of FtMYB13 was in the stem, leaves, and inflorescence, while the highest level of FtMYB14 was in stem, and that of FtMYB15 was in inflorescence (Fig. 1B). It is noteworthy that the highest level of FtMYB16 was observed in root; levels of this gene were relatively low in the other tissues studied, and none of the other genes showed high levels in root (Fig. 1B). In 60-day-old F. tataricum plants, the mRNA accumulation of four FtMYBs reached its highest level in root and the lowest level in leaf and flower (Fig. 1C), while the largest amount of rutin was found in leaf and flower, and the lowest amount in root (Fig. 1D). The mRNA accumulation of the activators FtMYB1 and FtMYB2 showed similar levels in different tissues of TB (Bai et al., 2014). These results indicate that rutin biosynthesis is probably spatially regulated by a clade of R2R3-MYB repressors, FtMYB13, FtMYB14, FtMYB15, and FtMYB16.
Fig. 1.
Characterization of JA-responsive members of the R2R3-MYB S4 TF family. (A) Expression pattern of FtMYB13, FtMYB14, FtMYB15, and FtMYB16 in 2-week-old F. tataricum seedlings in response to exposure to 50 µM MeJA for various times (in h) as indicated. FtH3 was used as an internal control. Mock indicates exposure to JA for 0 h. (B) Expression patterns of FtMYB13, FtMYB14, FtMYB15, and FtMYB16 in different tissues, as detected by qRT–PCR. Heat maps were generated by hierarchical clustering based on Pearson’s correlation. Different shades indicate higher or lower expression. (C) Measurement of total transcript levels of FtMYB13, FtMYB14, FtMYB15, and FtMYB16 in different tissues by qRT–PCR. (D) Measurement of rutin concentration in different tissues by HPLC. Values are the mean±SD of three biological repeats. DW, Dry weight; FL, flower; FR, fruit; I, inflorescence; L, leaf; R, root; S, stem.
FtMYB13, FtMYB14, and FtMYB15 are JA-responsive nuclear-localized repressors
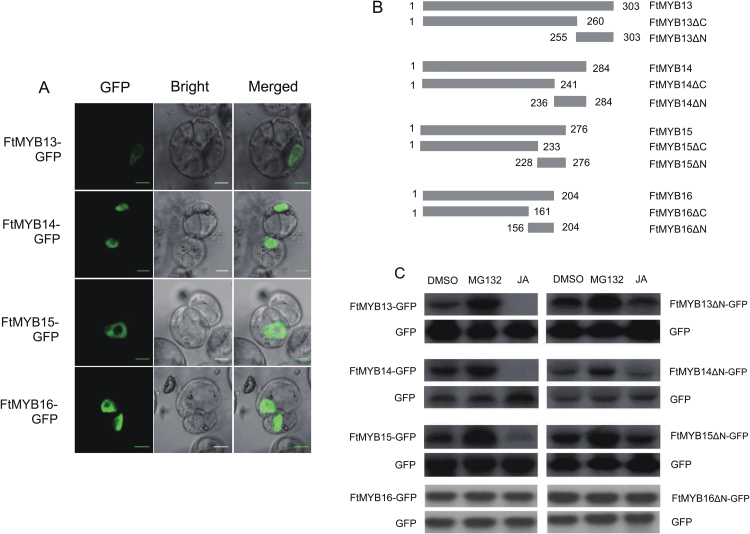
The above data suggest that FtMYB13, FtMYB14, FtMYB15, and FtMYB16 may be nuclear-localized repressors of the rutin biosynthetic pathway (Fig. 1). To examine this hypothesis, the gene encoding GFP was fused to FtMYB13, FtMYB14, FtMYB15, or FtMYB16, positioned so that GFP would be expressed at the C-terminus of the respective protein, with each construct under the control of the CaMV 35S promoter. The resulting plasmid constructs were introduced into Arabidopsis protoplasts. As shown in Fig. 2A, all of these MYB factors were localized in the nucleus, as hypothesized. Transcriptional activity assays in the yeast PJ69-4A strain suggested that only transformants containing pAS2.1 (GAL4-BD: GAL4 DNA binding domain) that were fused with FtMYBs grew well on SD-glucose medium lacking Trp (SD/-W) plates. The β-galactosidase activity assay consistently showed that the β-galactosidase activity of the transformant with pAS2.1-FtMYBs was ~2-fold lower than that of pAS2.1 (Supplementary Fig. S3), demonstrating that these FtMYBs repressed the expression of the reporter gene LacZ in the genome of PJ69-4A.
Fig. 2.
FtMYB13, FtMYB14, and FtMYB15 are JA-responsive, nuclear-localized repressors. (A) A FtMYB13-GFP, FtMYB14-GFP, FtMYB15-GFP, or FtMYB16-GFP construct was transformed into protoplasts isolated from an Arabidopsis cell suspension and examined by confocal laser scanning microscopy. The confocal microscopic image is shown at the left (GFP), the corresponding differential interference contrast (Bright) image is in the middle, and the merged image is at the right. Scale bar=20 µm. (B) Schematic diagram of the N- and C-terminus of FtMYB13, FtMYB14, FtMYB15, and FtMYB16 proteins. (C) Immunoblot analysis with anti-GFP antibodies of total protein extracts from Arabidopsis cell suspension protoplasts transiently co-expressing GFP and FtMYB13-GFP, FtMYB14-GFP, FtMYB15-GFP, FtMYB16-GFP, FtMYB13ΔN-GFP, FtMYB14ΔN-GFP, FtMYB15ΔN-GFP, or FtMYB16ΔN-GFP. Arabidopsis protoplasts were harvested 18 h after transformation of protoplasts treated for 4 h with DMSO at 0.1% (v/v) final concentration, with 50 µM of the 26S proteasome inhibitor MG132, or with 50 µM MeJA.
As shown in Fig. 1, transcripts of FtMYBs were induced by JAs. This finding could not exclude the possibility that JAs also affect the activity of FtMYBs at the protein level. Arabidopsis protoplasts were co-transformed with a GFP-expressing plasmid and a plasmid expressing a FtMYB-GFP fusion, and the transfected protoplasts were treated for 4 h with MeJA or the 26S proteasome inhibitor MG132 or the solvent DMSO. Immunoblot analysis of total cellular protein with anti-GFP antibodies revealed that MG132 increased and MeJA decreased the accumulation of FtMYB13-GFP, FtMYB14-GFP, and FtMYB15-GFP drastically (Fig. 2C), indicating that these TFs are subject to JA-related 26S proteasome-mediated degradation. However, neither MeJA nor MG132 treatment influenced the abundance of GFP and FtMYB16-GFP. Furthermore, we examined the role of COI1, the receptor of the JA signaling pathway (Yan et al., 2009), in JA-induced degradation of FtMYBs. In coi1-1 leaf protoplasts, MG132 and MeJA did not affect the amount of FtMYB13-GFP, FtMYB14-GFP, or FtMYB15-GFP fusion protein, indicating that JA-induced degradation of FtMYBs is COI1 dependent (Supplementary Fig. S4). Our previous study showed that the C-terminal of the R2R3-MYB S4 TFs performs the regulatory activity (Zhou et al., 2015a). To determine whether the C-terminus of FtMYBs is responsible for JA-induced degradation, we constructed plasmids expressing the C-terminus and the N-terminus of FtMYBs fused to GFP (Fig. 2B). The results showed that MG132 and MeJA still affected the levels of FtMYB13ΔN-GFP, FtMYB14ΔN-GFP, and FtMYB15ΔN-GFP protein (Fig. 2C); however, FtMYB13ΔC-GFP, FtMYB14ΔC-GFP, and FtMYB15ΔC-GFP were not subject to MeJA-mediated degradation (Supplementary Fig. S10), demonstrating that the C-terminus of FtMYB13, FtMYB14, and FtMYB15 contain the degron responsive to JAs. Taken together, these results indicate that FtMYB13, FtMYB14, and FtMYB15 are JA-responsive, nuclear-localized transcriptional repressors.
Asp to Asn mutation of the SID domain releases FtMYBs from interaction with FtSAD2
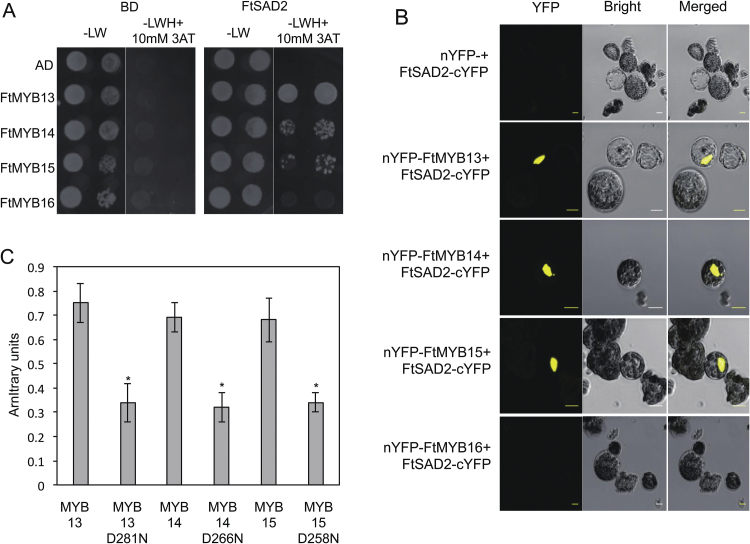
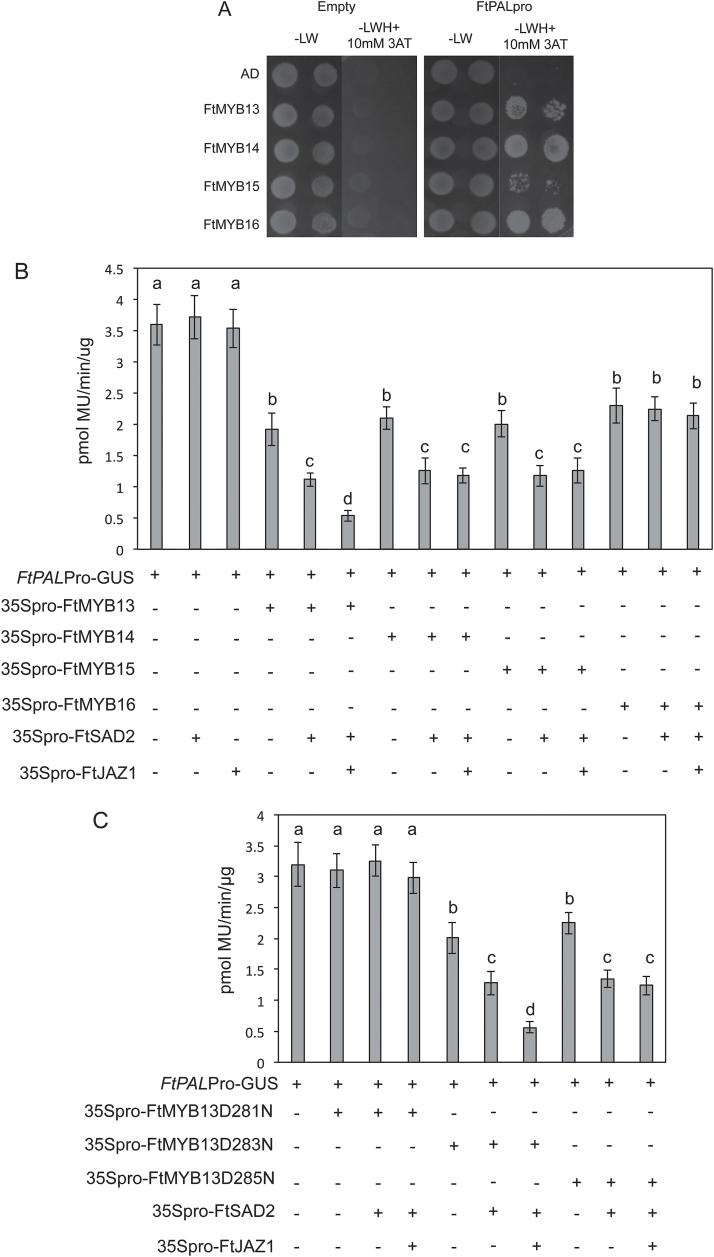
Our previous results showed that R2R3-MYB S4 TFs interact with SAD2 protein via the SID domain in their C-terminus (Zhou et al., 2015a). FtMYB13, FtMYB14, and FtMYB15 possess a conserved SID or SID-like domain (Supplementary Figs S1 and S5); thus, we hypothesized that these three FtMYBs may interact with FtSAD2 as well. Yeast two-hybrid (Y2H) assays were used as a first step to test the interaction between FtMYB13, FtMYB14, FtMYB15, or FtMYB16 and FtSAD2. Results showed that yeast cells co-expressing FtSAD2 and FtMYB13, FtMYB14, or FtMYB15 were able to sustain growth at 3-AT concentrations up to 10 mM on selective medium (Fig. 3A). However, no interaction was detected between FtSAD2 and FtMYB16. To confirm the results of the Y2H assays, BiFC assays were performed. Strong fluorescent signals were observed in the nucleus of Arabidopsis protoplasts upon co-expression of nYFP-FtMYB13 and FtSAD2-cYFP, nYFP-FtMYB14 and FtSAD2-cYFP, or nYFP-FtMYB15 and FtSAD2-cYFP, respectively (Fig. 3B). No or only background YFP fluorescence was detected in negative controls (nYFP co-expressed with FtSAD2-cYFP) and with co-expression of FtSAD2-cYFP and nYFP-FtMYB16. These results indicate that FtSAD2 interacts with FtMYB13, FtMYB14, and FtMYB15 in the nucleus.
Fig. 3.
The Asp to Asn mutation of the SID domain releases FtMYBs from FtSAD2 interaction. (A) FtMYB13, FtMYB14, and FtMYB15 interact with FtSAD2 in yeast. Yeast cells expressing FtMYB13, FtMYB14, FtMYB15, and FtMYB16 proteins fused to the GAL4 AD, and FtSAD2 fused to the GAL4 BD, were spotted on SD/-LW medium to select for the plasmids, and on SD/-LWH medium with 10 mM 3-AT to select for transcriptional activation of the His3 gene. Growth was monitored after 7 days. Yeast cells transformed with the empty plasmids pAS2.1 and pACT2, expressing GAL4 BD and GAL4 AD, respectively, were used as controls. (B) BiFC assays in planta. YFP fluorescence images (alone or merged with bright-field images) of Arabidopsis cell suspension protoplasts co-transformed with constructs encoding the indicated fusion proteins with YFP at the N-terminus (nYFP) or the C-terminus (cYFP). Scale bar=20 µm. (C) Interaction of FtMYB13D281N, FtMYB14D266N, and FtMYB15D258N with FtSAD2 in quantitative Y2H assays. A liquid culture β-galactosidase assay was performed on the transformed yeasts. The activity of β-galactosidase was measured in arbitrary units. Values are the mean±SD of three biological repeats. Asterisks indicate statistically significant differences compared with FtMYB13, FtMYB14, and FtMYB15 (P<0.05, Student’s t-test).
It has been demonstrated that the conserved Asp residue of the SID motif contributes to the protein–protein interaction (Zhou et al., 2015a). Thus, we hypothesized that the conserved Asp residue (FtMYB13Asp281, FtMYB14Asp266, FtMYB15Asp258) could be essential for the interaction of FtMYB13, FtMYB14, or FtMYB15 with FtSAD2. As the first step to test this hypothesis, we generated derivatives of these FtMYBs, including FtMYB13D281N, FtMYB14D266N, and FtMYB15D258N, by introducing point mutations, and we then used these mutants in Y2H assays. As shown in Fig. 3C, in quantitative Y2H assays the interaction of FtMYB13D281N, FtMYB14D266N, and FtMYB15D258N with FtSAD2 was significantly diminished, indicating that these FtMYB derivatives had lost the ability to interact with FtSAD2 protein.
The importin β-like protein SAD2 can mediate the nuclear localization of MYBs in Arabidopsis (Zhou et al., 2015a). Therefore, FtMYB13D281N, FtMYB14D266N, and FtMYB15D258N protein may be absent from the nucleus. To test this hypothesis, subcellular localization analysis was performed. As expected, FtMYB13D281N-GFP, FtMYB14D266N-GFP, and FtMYB15D258N-GFP protein were found only in the cytoplasm (Supplementary Fig. S6). These results suggest that the conserved Asp residue of the SID domains of FtMYB13, FtMYB14, and FtMYB15 are necessary for their transport into the nucleus mediated by FtSAD2. As shown in Fig. 1, FtSAD2 transcription was also induced by JA and transcripts accumulated in stem, leaves and flowers, similar to the pattern of transcript accumulation of FtMYB13, FtMYB14, and FtMYB15. This indicates that FtSAD2 probably directly regulates the transcriptional activity of these FtMYBs.
FtMYB13 interacts with the JA-responsive repressor FtJAZ1
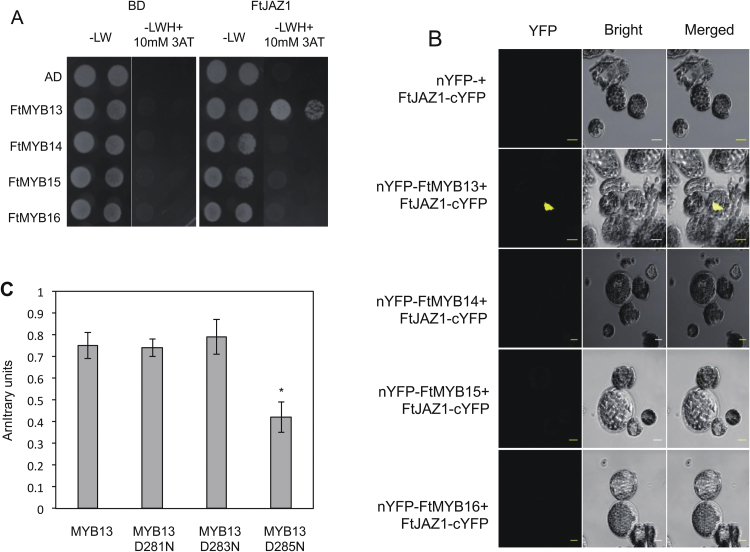
In Arabidopsis, AtJAZ proteins interact with MYBs, modulating the expression of key enzyme genes, such as C4H, in the anthocyanin biosynthesis pathway (Qi et al., 2011). As FtMYB13, FtMYB14, and FtMYB15 are sensitive to JAs at the transcript and protein levels, we assumed that these FtMYBs interact with FtJAZ protein in a similar manner. Interestingly, Y2H assays suggested that FtMYB13 was able to bind FtJAZ1, but FtMYB14, FtMYB15, and FtMYB16 did not show any interaction with FtJAZ1 (Fig. 4A). The same results were observed via BiFC assays (Fig. 4B). Furthermore, none of the FtMYBs interacted with FtJAZ2 (data not shown). The conserved SID domain of FtMYB13 possesses three conserved Asp residues (GLDFDLDL; Asp281, Asp283, and Asp285), among which Asp281 is essential for interaction with SAD2 (Fig. 3). Thus, we assume that Asp283 and Asp285 may be considered more specific for interaction with FtJAZ1. The interaction of FtMYB13D285N with FtJAZ1, tested by quantitative Y2H assays, was significantly diminished (Fig. 4C), while FtMYB13D281N and FtMYB13D283N showed no difference relative to FtMYB13. Moreover, FtMYB13D283N-GFP and FtMYB13D285N-GFP proteins were found in the nucleus (Supplementary Fig. S6). In summary, the Asp285 to Asn285 mutation abolished the interaction between FtMYB13 and FtJAZ1.
Fig. 4.
FtMYB13 interaction with FtJAZ1 depends on a conserved Asp residue. (A) FtMYB13 interacts with FtJAZ1 in yeast. Yeast cells expressing FtMYB13, FtMYB14, FtMYB15, or FtMYB16 proteins fused to the GAL4 AD, and FtJAZ1 fused to the GAL4 BD, were spotted on SD/-LW medium to select for the plasmids, and on SD/-LWH medium with 10 mM 3-AT to select for transcriptional activation of the His3 gene. Growth was monitored after 7 days. Yeast cells transformed with the empty plasmids pAS2.1 and pACT2, expressing GAL4 BD and GAL4 AD, respectively, were used as controls. (B) FtMYB13 interacts with FtJAZ1 in planta. YFP fluorescence images (alone or merged with bright-field images) of Arabidopsis cell suspension protoplasts co-transformed with constructs encoding the indicated fusion proteins with YFP at the N-terminus (nYFP) or the C-terminus (cYFP). Scale bar=20 µm. (C) Interaction of FtMYB13D281N, FtMYB13D283N, and FtMYB13D285N with FtJAZ1 in quantitative Y2H assays. A liquid culture β-galactosidase assay was performed on the transformed yeasts. The activity of β-galactosidase was measured in arbitrary units. Values are the mean±SD of three biological repeats. Asterisks indicate statistically significant differences compared with FtMYB13, FtMYB13D281N, and FtMYB13D283N (P<0.05, Student’s t-test).
The transcriptional repression activity of FtMYBs is regulated by FtSAD2 or FtJAZ1
The first common step of the phenylpropanoid pathway is catalyzed by phenylalanine ammonia lyase (PAL). Analysis of the FtPAL gene promoter revealed a MYB binding element (AATAGTT). Yeast one-hybrid (Y1H) assays were performed to test the direct interaction between the FtMYBs and the FtPAL promoter. A 974 bp fragment of the FtPAL promoter was cloned into the pHIS3NX reporter vector, and its interaction with pACT2-FtMYB13, pACT2-FtMYB14, pACT2-FtMYB15, and pACT2-FtMYB16, harboring FtMYB13, FtMYB14, FtMYB15, and FtMYB16 [GAL4 activation domain (AD) fused], respectively, was assessed. As shown in Fig. 5A, these four MYBs could bind to the promoter of FtPAL.
Fig. 5.
The repression activity of FtMYB13, FtMYB14, and FtMYB15 is modulated by FtSAD2 or FtJAZ1. (A) Y1H analysis of FtMYB13, FtMYB14, FtMYB15, and FtMYB16 binding to the FtPALpro fragment. Transformed yeasts (SD/-L) were grown on SD/-LH medium supplemented with 10 mM 3-AT. Yeast cells were allowed to grow for 7 days at 30 °C. (B, C) Protoplast transactivation assays. Arabidopsis protoplasts were co-transformed with 2 µg of a reporter construct of FtPALpro-GUS and 2 µg of effector plasmids. The effector constructs consisted of an expression vector carrying the CaMV 35S promoter with or without the FtMYB13, FtMYB13D281N, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB15, FtMYB16, FtSAD2, or FtJAZ1 gene. Values represent the mean±SE of triplicate experiments. Significant differences between values are indicated with different letters (P<0.05, one-way ANOVA).
To elucidate the functional significance of the interaction between the four FtMYBs and FtSAD2 or FtJAZ1, we co-transformed an Arabidopsis cell suspension with identical amounts of effector plasmids carrying FtMYB13, FtMYB13D281N, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB14D266N, FtMYB15, FtMYB15D258N, or FtMYB16 with FtSAD2 or FtJAZ1, or in combination with the FtPALpro-GUS reporter constructs. As shown in Fig. 5B and C, the repressing activity of FtMYB13, FtMYB13D283N, and FtMYB13D285N on FtPALpro-GUS was significantly enhanced by FtSAD2 and FtJAZ1. In addition, FtSAD2 also significantly increased the repression activity of both FtMYB14 and FtMYB15. However, neither FtSAD2 nor FtJAZ1 affected the transcriptional activity of FtMYB13D281N, FtMYB14D266N, FtMYB15D258N, or FtMYB16 (Fig. 5B, C; Supplementary Fig. S7), indicating that they act dependently via protein–protein interaction. These results demonstrated that FtMYBs directly repress FtPAL expression, which is modulated by their interacting partners FtSAD2 and FtJAZ1.
Overexpression of FtMYB13, FtMYB14, FtMYB15, or FtMYB16 represses rutin biosynthesis in F. tataricum hairy roots
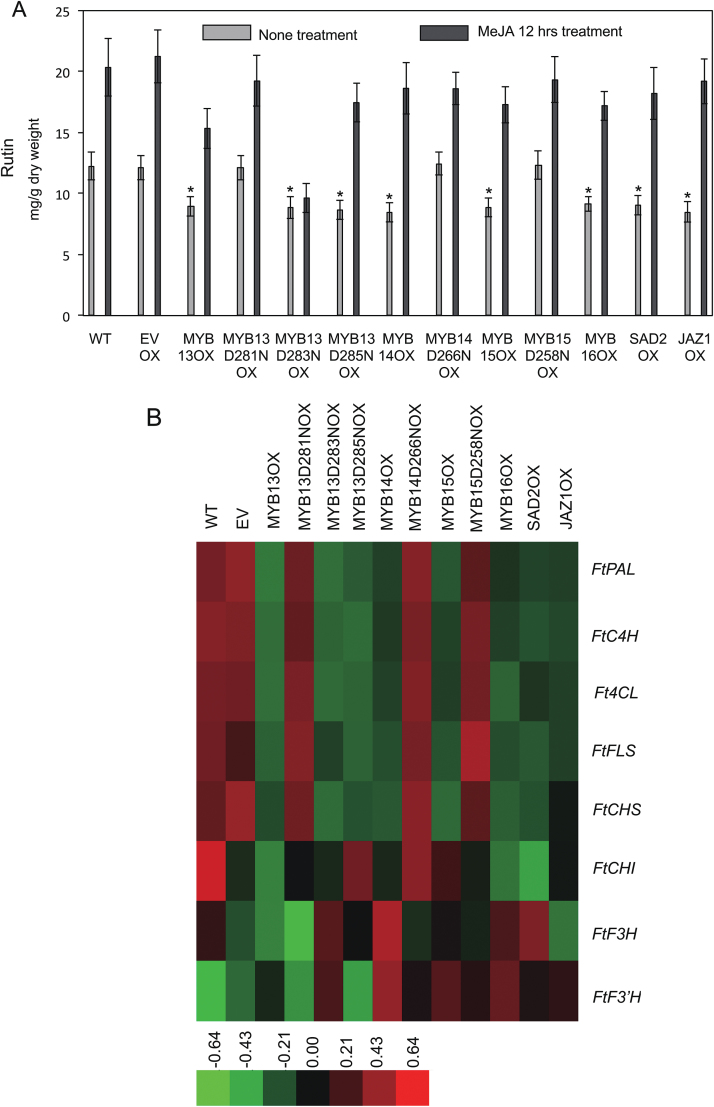
The above data show that FtMYB13, FtMYB14, FtMYB15, and FtMYB16 repress the expression of FtPAL, suggesting that these factors probably regulate rutin biosynthesis. To functionally test the roles of these MYB TFs and their derivatives in planta, we investigated the accumulation of rutin in F. tataricum hairy roots overexpressing FtMYB13-HA, FtMYB13D281N-HA, FtMYB13D283N-HA, FtMYB13D285N-HA, FtMYB14-HA, FtMYB14D266N-HA, FtMYB15-HA, FtMYB15D258N-HA, FtMYB16-HA, FtSAD2-HA, or FtJAZ1-HA. Three independent transgenic hairy root lines of each genotype were identified by western blot (Supplementary Fig. S8) and used for further functional analysis. The hairy root growth occurred primarily during the first 5 days of cultivation, and the maximum content of rutin peaked at 12.15 mg g–1 dry weight at day 20 of cultivation. The 20-day-old hairy root lines were chosen to study the function of these FtMYBs in rutin accumulation. As shown in Fig. 6A, the rutin levels in the FtMYB13, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB15, and FtMYB16 overexpressing lines were significantly lower than those of the wild-type (WT), empty vector (EV), and FtMYB13D281N, FtMYB14D266N, and FtMYB15D258N overexpressing lines, ranging from 8.45 to 9.16 mg g–1 dry weight. MeJA treatment drastically increased the accumulation of rutin in all the lines except the FtMYB13D283N overexpressing line (Fig. 6A), indicating that FtMYB13D283N protein is stable in response to MeJA. Immunoblot analysis of total hairy root protein with anti-HA antibodies revealed that MeJA caused a decrease in the amount of FtMYB13, FtMYB13D281N, and FtMYB13D285N, but not FtMYB13D283N (Supplementary Fig. S9), indicating that the Asp 283 residue is responsible for JA-induced degradation.
Fig. 6.
FtMYB13, FtMYB14, FtMYB15, and FtMYB16 represses rutin biosynthesis in F. tataricum hairy roots. (A) Rutin content in different genotypes of F. tataricum hairy roots with and without treatment for 12 h with 50 µM JA. WT, wild-type hairy roots. The transgenic hairy root lines were generated by transformation with strains of A. rhizogenes A4 harboring T-DNA containing no open reading frame (empty vector; EV), FtMYB13, FtMYB13D281N, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB14D266N, FtMYB15, FtMYB15D258N, FtMYB16-HA, FtSAD2-HA, or FtJAZ1-HA. Values are the mean±SE of three biological repeats of each independent transgenic line. Asterisks indicate statistically significant differences compared with WT and EV (P<0.05). (B) Expression levels of FtPAL, FtC4H, Ft4CL, FtFLS, FtCHS, FtCHI, FtF3H, and FtF3ʹH in different transgenic hairy root lines, as indicated. FtH3 was used as an internal control.
Subsequently, qRT–PCR was performed to investigate the expression patterns of eight rutin biosynthesis genes (FtPAL, FtC4H, FtCHI, FtF3H, Ft4CL, FtCHS, FtF3ʹH, and FtFLS) in different F. tataricum hairy root lines. The transcripts of FtPAL, FtC4H, Ft4CL, FtCHS, and FtFLS were significantly repressed in the FtMYB13, FtMYB13D283N, FtMYB13D285N, FtMYB14, FtMYB15, and FtMYB16 overexpressing lines relative to the transcript levels of WT, EV, and FtMYB13D281N, FtMYB14D266N, and FtMYB15D258N overexpressing lines (Fig. 6B). Additionally, in the FtSAD2 and FtJAZ1 overexpressing lines, six of the eight rutin biosynthesis genes showed lower transcript levels compared with the corresponding levels in WT lines. These results demonstrate that FtMYB13, FtMYB14, FtMYB15, and FtMYB16 regulate rutin biosynthesis by repressing the gene expression of key enzymes, including FtPAL, FtC4H, Ft4CL, FtCHS, and FtFLS, and that this repression is modulated by FtSAD2 and FtJAZ1 via the conserved Asp residues of the SID-like motif.
Discussion
The R2R3-MYB TFs are the main regulators of the phenylpropanoid biosynthetic pathway in plants (Du et al., 2010; Zhou et al., 2015a). It has been demonstrated that the MYB factors in the same subgroup share a similar function (Zhou et al., 2015a). In Arabidopsis, AtMYB4, together with AtMYB7, AtMYB32, and AtMYB3, belong to the S4 MYB TFs, which act as transcriptional repressors owing to the EAR motif that they possess (Jin et al., 2000; Dubos et al., 2010; Fornalé et al., 2014). In this study, we identified a clade of S4 MYB TFs from buckwheat, FtMYB13, FtMYB14, FtMYB15, and FtMYB16, containing the EAR repressing motif. It has been reported that the EAR motif is not essential for mediating protein–protein interaction (Zeng et al., 2015). Our previous findings showed that AtMYB4, AtMYB7, and AtMYB32 possess the conserved GY/FDFLGL motif or SID motif in their C-terminus, which mediates the transport of these MYBs into the nucleus (Zhou et al., 2015a). Deletion of the SID motif of AtMYB4 results in the loss of a loop structure in its C-terminus, which abolishes protein–protein interaction (Zhou et al., 2015a). FtMYB13, FtMYB14, and FtMYB15 possess the conserved SID domain in their C-terminus and can interact with FtSAD2, while FtMYB16 exhibits no interaction with FtSAD2 due to the absence of a SID motif (Fig. 3). Supporting evidence for an additional function of the SID motif was provided by the fact that deletion of the SID motif of BrMYB4 leads to the loss of the capacity to repress the expression of its target gene, BrC4H (Zhang et al., 2014).
As shown in Fig. 1C, the total FtMYBs mRNA exhibits the highest accumulation in root, and the lowest in leaf and flower. Interestingly, the highest content of rutin was found in leaf and flower and the lowest in root Fig. 1D, suggesting that rutin biosynthesis is spatially regulated. Further qRT–PCR analysis showed different transcript accumulation patterns of FtMYB13, FtMYB14, FtMYB15, and FtMYB16 in different tissues. In roots, FtMYB16 showed the highest expression level, while FtSAD2 showed the lowest (Fig. 1). Therefore, the lowest accumulation of FtSAD2 protein in root may well be another explanation for the fact that FtMYB16 does not interact with FtSAD2 (Fig. 3). This strongly suggests the existence of an organ-specific signaling pathway from the lower part of the root to the upper part of the hypocotyl and to the fruit.
BLAST searching on the amino acid sequences of FtMYB13, FtMYB14, FtMYB15, and FtMYB16 reveals that the conserved SID domain has the sequence GXXDFxxxG/DL. It was previously reported that the conserved Asp (D) of the SID domain contributes to protein–protein interaction (Zhou et al., 2015a). Therefore, the presence of two conserved Asp residues in the SID domain of the FtMYBs led us to hypothesize that the FtMYBs probably interact with one or more regulators. Indeed, the first Asp residue in the SID domain contributes to the interaction with FtSAD2. Interestingly, the second Asp residue in the SID domain of FtMYB13 contributes to its interaction with FtJAZ1, which is consistent with previous findings that the conserved Asp residue in the JAZ interacting domain of MYC2 and MYC3 plays an important role for binding to most JAZ proteins in the JA signaling pathway (Goossens et al., 2015). This provides a mechanistic understanding of how the protein–protein interaction could be fine-tuned by the Asp residue of the SID motif.
The transcriptional complexes formed by protein–protein interactions between MYBs and other factors are necessary to control gene expression. For instance, the MYB–basic helix-loop-helix (bHLH)–WD40 complex is responsible for the regulation of expression of genes involved in anthocyanin biosynthesis (Zhou and Memelink, 2016). In Arabidopsis, AtJAZ proteins interact not only with bHLHs but also with MYBs, such as MYB75, which then disrupts the interactions between MYB and bHLH activators, decreasing their transcriptional activity (Qi et al., 2011). On the basis of our findings, FtSAD2, as an interacting partner of FtMYB13, FtMYB14, and FtMYB15, mediates the transport of these MYBs into the nucleus, while FtJAZ1 interacts with only FtMYB13. The repression activity of FtMYB13 is significantly regulated by FtSAD2 or FtJAZ1.
The rutin biosynthesis pathway is induced by MeJA (Fig. 6A), indicating that JAs could regulate the TFs or key enzymes of this pathway at the protein or the transcript level. In this study, JAs could induce FtMYB13, FtMYB14, and FtMYB15 expression (i.e. regulation at the transcript level). Moreover, JAs directly affected FtMYB13, FtMYB14, and FtMYB15 protein stability via the 26S proteasome pathway (Fig. 2), which established these TFs as components of JA signal transduction. These four MYB proteins share relatively high amino acid sequence identity except for the C-terminal SID motif, which is totally absent in FtMYB16. Immunoblot analysis showed that FtMYB16 and FtMYB13D283N are stable after MeJA treatment, indicating that the SID motif is a unique functional domain for degradation. Our results demonstrated that the degradation of FtMYBs in response to JAs is COI1 dependent. The JA receptor COI1 binds bioactive JAs and interacts with JAZ repressors, leading to their degradation (Zhou and Memelink, 2016). The way in which these FtMYBs function as repressors could be similar to the action of JAZ on gene repression. Therefore, further research is needed to demonstrate the role of these MYB factors in JA signaling and the mechanism of signal transduction.
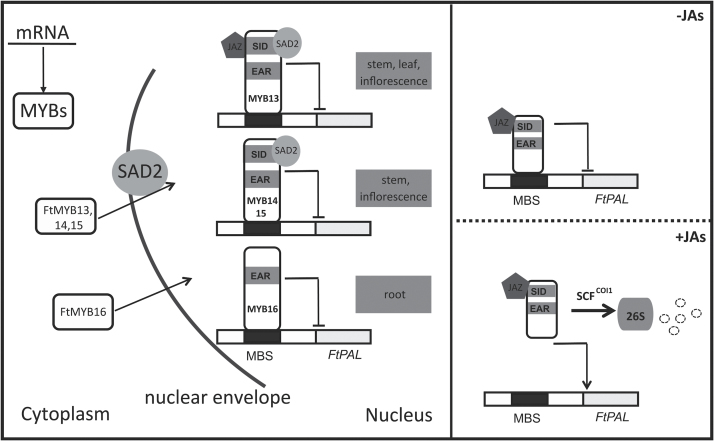
In conclusion, in this study we identified a clade of JA-responsive S4 R2R3-MYB TFs—FtMYB13, FtMYB14, FtMYB15, and FtMYB16—that act as repressors of phenylpropanoid biosynthesis in buckwheat. FtMYB13, FtMYB14, and FtMYB15 can be degraded via the 26S proteasome in the COI1-dependent JA signaling pathway, and this degradation is due to the SID motif of their C-terminus. FtSAD2 directly interacts with FtMYB13, FtMYB14, and FtMYB15, and subsequently mediates the transport of these repressors into the nucleus in stem and inflorescence. Additionally, FtJAZ1 interacts with FtMYB13 and promotes the repression activity of FtMYB13, mainly in stem, leaves, and inflorescence. FtMYB16 specifically acts as a repressor, dependent on the EAR motif, in roots, while the repression activity of FtMYB13, FtMYB14, and FtMYB15 is due to the conserved Asp residues of the SID domain. Based on these data, we propose a coherent model of the spatial repression of rutin biosynthesis by JA-responsive MYB factors in F. tataricum (Fig. 7), which could provide valuable molecular knowledge for further plant metabolic engineering purposes.
Fig. 7.
Model of spatial repression of rutin biosynthesis by JA-responsive MYB factors in F. tataricum. Left panel: FtMYB13, FtMYB14, FtMYB15, and FtMYB16 are synthesized in the cytoplasm. FtMYB13, FtMYB14, and FtMYB15 are recruited by an importin protein, Sensitive to ABA and Drought 2 (SAD2). FtSAD2 recognizes the nuclear pore complex, which then transports the FtSAD2/FtMYB13, FtSAD2/FtMYB14, and FtSAD2/FtMYB15 complexes into the nucleus, increasing the repression activity of FtMYB factors in stem and inflorescence. FtMYB16 specifically acts as a repressor, depending on the EAR motif, in root. In the nucleus, FtJAZ1 interacts with FtMYB13 and acts as a co-repressor to repress expression of the FtMYB13 target genes of phenylpropanoid biosynthesis via binding to MYB binding sites (MBS), mainly in stem, leaves, and inflorescence. Right panels: Simplified transcriptional networks of JA signaling in rutin biosynthesis. (Upper panel) Binding of the JAZ repressor to FtMYB TFs represses the expression of rutin biosynthesis pathway genes. (Lower panel) The presence of bioactive JAs results in COI1-mediated degradation of JAZ and FtMYB repressors via the 26S proteasome pathway, thus inducing rutin accumulation. Arrows indicate activation, and T-shaped lines indicate inhibition. SCF, Skp–Cullin–F-box protein.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Amino acid sequence alignment of a clade of subgroup 4 R2R3-MYB TFs from F. tataricum with subgroup 4 R2R3-MYB TFs from Arabidopsis.
Fig. S2. Phylogeny of R2R3-MYB TFs including two activators, FtMYB1 (JF313345) and FtMYB2 (JF313347), from F. tataricum with subgroup 4 R2R3-MYB TFs from Arabidopsis.
Fig. S3. Transcriptional repression activity assays of FtMYB13, FtMYB14, FtMYB15, and FtMYB16.
Fig. S4. Immunoblot analysis with anti-GFP antibodies of total protein extracts from Arabidopsis coi-1 leaf protoplasts transiently co-expressing GFP and FtMYB13-GFP, FtMYB14-GFP, or FtMYB15-GFP.
Fig. S5. Amino acid sequence alignment of the SID domain of FtMYB13, FtMYB14, and FtMYB15 with AtMYB4, AtMYB7, and AtMYB32.
Fig. S6. Subcellular localization of FtMYB13D281N-GFP, FtMYB13D283N-GFP, FtMYB13D285N-GFP, FtMYB14D2 66N-GFP, and FtMYB15D258N-GFP in Arabidopsis protoplasts.
Fig. S7. The transcriptional activity of FtMYB14D266N or FtMYB15D258N was not affected by FtSAD2 or FtJAZ1.
Fig. S8. Immunoblot analysis with anti-HA antibodies of total protein extracts from FtMYB13-HA, FtMYB13D281N-HA, FtMYB13D283N-HA, FtMYB13D285N-HA, FtMYB14-HA, FtMYB14D266N-HA, FtMYB15-HA, FtMYB15D258N-HA, FtMYB16-HA, FtSAD2-HA, or FtJAZ1-HA overexpressing hairy root lines of F. tataricum.
Fig. S9. Immunoblot analysis with anti-HA antibodies of total protein extracts from F. tataricum hairy roots expressing FtMYB13-HA, FtMYB13D281N-HA, FtMYB13D283N-HA, and FtMYB13D285N-HA.
Fig. S10. Immunoblot analysis with anti-GFP antibodies of total protein extracts from Arabidopsis cell suspension protoplasts transiently co-expressing GFP and FtMYB13ΔC-GFP, FtMYB14ΔC-GFP, FtMYB15ΔC-GFP, or FtMYB16ΔC-GFP.
Table S1. List of primers used in this study.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (grant number 31572457), the earmarked fund for China Agriculture Research System (CARS-08), the Investigation of Forage Germplasms in Central China (grant number 2017FY100604), and the Young Elite Scientists Sponsorship Program by CAST (YESS).
Glossary
Abbreviations:
- CB
Common buckwheat
- GFP
green fluorescent protein
- JA
jasmonate
- JAZ
jasmonate ZIM domain
- PAL
phenylalanine ammonia lyase
- qRT–PCR
quantitative real-time PCR
- SAD2
Sensitive to ABA and Drought 2
- SID
SAD2 Interaction Domain
- TB
Tartary buckwheat
- TF
transcription factor
- Y1H
yeast one-hybrid
- Y2H
yeast two-hybrid
- YFP
yellow fluorescent protein.
References
- Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics International 11, 36–42. [Google Scholar]
- Bai YC, Li CL, Zhang JW, Li SJ, Luo XP, Yao HP, Chen H, Zhao HX, Park SU, Wu Q. 2014. Characterization of two tartary buckwheat R2R3-MYB transcription factors and their regulation of proanthocyanidin biosynthesis. Physiologia Plantarum 152, 431–440. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, et al. 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Du H, Huang Y, Tang Y. 2010. Genetic and metabolic engineering of isoflavonoid biosynthesis. Applied Microbiology and Biotechnology 86, 1293–1312. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornalé S, Lopez E, Salazar-Henao JE, Fernández-Nohales P, Rigau J, Caparros-Ruiz D. 2014. AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant & Cell Physiology 55, 507–516. [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Berger B, Gigolashvili T. 2014. bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiology 166, 349–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. 1992. Improved method for high efficiency transformation of intact yeast cells.Nucleic Acids Research 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Goossens J, Swinnen G, Vanden Bossche R, Pauwels L, Goossens A. 2015. Change of a conserved amino acid in the MYC2 and MYC3 transcription factors leads to release of JAZ repression and increased activity. New Phytologist 206, 1229–1237. [DOI] [PubMed] [Google Scholar]
- Huang X, Yao J, Zhao Y, Xie D, Jiang X, Xu Z. 2016. Efficient rutin and quercetin biosynthesis through flavonoids-related gene expression in Fagopyrum tataricum gaertn. Hairy root cultures with UV-B irradiation. Frontiers in Plant Science 7, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. 2000. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. The EMBO Journal 19, 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Park KJ, Lim JH. 2011. Metabolomic analysis of phenolic compounds in buckwheat (Fagopyrum esculentum M.) sprouts treated with methyl jasmonate. Journal of Agricultural and Food Chemistry 59, 5707–5713. [DOI] [PubMed] [Google Scholar]
- Li X, Park NI, Xu H, Woo SH, Park CH, Park SU. 2010. Differential expression of flavonoid biosynthesis genes and accumulation of phenolic compounds in common buckwheat (Fagopyrum esculentum). Journal of Agricultural and Food Chemistry 58, 12176–12181. [DOI] [PubMed] [Google Scholar]
- Logacheva MD, Kasianov AS, Vinogradov DV, Samigullin TH, Gelfand MS, Makeev VJ, Penin AA. 2011. De novo sequencing and characterization of floral transcriptome in two species of buckwheat (Fagopyrum). BMC Genomics 12, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B. 2015. The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiology 169, 1862–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali G, Ouwerkerk PB, Memelink J. 1994. Versatile transformation vectors to assay the promoter activity of DNA elements in plants. Gene 149, 373–374. [DOI] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. 2004. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. The Plant Journal 40, 979–995. [DOI] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. 2011. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. The Plant Cell 23, 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Wang Q, Shan F, Hou Z, Ren G. 2010. Nutritional composition and flavonoids content of flour from different buckwheat cultivars. International Journal of Food Science & Technology 45, 951–958. [Google Scholar]
- Schirawski J, Planchais S, Haenni AL. 2000. An improved protocol for the preparation of protoplasts from an established Arabidopsis thaliana cell suspension culture and infection with RNA of turnip yellow mosaic tymovirus: a simple and reliable method. Journal of Virological Methods 86, 85–94. [DOI] [PubMed] [Google Scholar]
- Shyu C, Figueroa P, Depew CL, Cooke TF, Sheard LB, Moreno JE, Katsir L, Zheng N, Browse J, Howe GA. 2012. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. The Plant Cell 24, 536–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. 2011. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. The Plant Cell 23, 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal 50, 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. 2005. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiology 139, 1840–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Töpfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH. 1987. A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Research 15, 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. 1997. Comparison of the activities of CaMV 35S and FMV 34S promoter derivatives in Catharanthus roseus cells transiently and stably transformed by particle bombardment. Plant Molecular Biology 33, 943–946. [DOI] [PubMed] [Google Scholar]
- Wijngaard HH, Arendt EK. 2006. Buckwheat. Cereal Chemistry 83, 391–401. [Google Scholar]
- Yan J, Zhang C, Gu M, et al. 2009. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant Cell 21, 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. 2007. A downstream mediator in the growth repression limb of the jasmonate pathway. The Plant Cell 19, 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y, Hirakawa H, Ueno M, Matsui K, Katsube-Tanaka T, Yang SJ, Aii J, Sato S, Mori M. 2016. Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Research 23, 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng JK, Li X, Xu Q, Chen JY, Yin XR, Ferguson IB, Chen KS. 2015. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnology Journal 13, 1325–1334. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang Y, Sun M, Wang J, Kawabata S, Li Y. 2014. BrMYB4, a suppressor of genes for phenylpropanoid and anthocyanin biosynthesis, is down-regulated by UV-B but not by pigment-inducing sunlight in turnip cv. Tsuda. Plant & Cell Physiology 55, 2092–2101. [DOI] [PubMed] [Google Scholar]
- Zhang ZL, Zhou ML, Tang Y, Li FL, Tang YX, Shao JR, Xue WT, Wu YM. 2012. Bioactive compounds in functional buckwheat food. Food Research International 49, 389–395. [Google Scholar]
- Zhou M, Memelink J. 2016. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnology Advances 34, 441–449. [DOI] [PubMed] [Google Scholar]
- Zhou M, Sun Z, Wang C, Zhang X, Tang Y, Zhu X, Shao J, Wu Y. 2015a. Changing a conserved amino acid in R2R3-MYB transcription repressors results in cytoplasmic accumulation and abolishes their repressive activity in Arabidopsis. The Plant Journal 84, 395–403. [DOI] [PubMed] [Google Scholar]
- Zhou M, Tang Y, Zhang K, Li F, Yang P, Tang Y, Wu Y, Shao J. 2013. Identification of TT2 gene from floral transcriptome in Fagopyrum tataricum. Food Research International 54, 1331–1333. [Google Scholar]
- Zhou M, Wang C, Qi L, Yang X, Sun Z, Tang Y, Tang Y, Shao J, Wu Y. 2015b. Ectopic expression of Fagopyrum tataricum FtMYB12 improves cold tolerance in Arabidopsis thaliana. Journal of Plant Growth Regulation 34, 362–371. [Google Scholar]
- Zhou M, Zhang K, Sun Z, Yan M, Chen C, Zhang X, Tang Y, Wu Y. 2017. LNK1 and LNK2 corepressors interact with the MYB3 transcription factor in phenylpropanoid biosynthesis. Plant Physiology 174, 1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ML, Zhu XM, Shao JR, Wu YM, Tang YX. 2010. Transcriptional response of the catharanthine biosynthesis pathway to methyl jasmonate/nitric oxide elicitation in Catharanthus roseus hairy root culture. Applied Microbiology and Biotechnology 88, 737–750. [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. 2004. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. The Plant Journal 40, 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.