Abstract
Findings of immense microbial diversity are at odds with observed functional redundancy, as competitive exclusion should hinder coexistence. Tradeoffs between dispersal and competitive ability could resolve this contradiction, but the extent to which they influence microbial community assembly is unclear. Because fungi influence the biogeochemical cycles upon which life on earth depends, understanding the mechanisms that maintain the richness of their communities is critically important. Here, we focus on ectomycorrhizal fungi, which are microbial plant mutualists that significantly affect global carbon dynamics and the ecology of host plants. Synthesizing theory with a decade of empirical research at our study site, we show that competition–colonization tradeoffs structure diversity in situ and that models calibrated only with empirically derived competition–colonization tradeoffs can accurately predict species–area relationships in this group of key eukaryotic microbes. These findings provide evidence that competition–colonization tradeoffs can sustain the landscape-scale diversity of microbes that compete for a single limiting resource.
Introduction
The importance of microbial communities to global biogeochemical cycles [1], ecosystem productivity [2], macroorganism ecology [3, 4], and human health [5] underscores the critical need to determine the forces creating and maintaining their diversity. However, because many microbial taxa are functionally similar to one another [6], speciose microbial communities often appear to contradict the competitive exclusion principle, which states that species must differ from each other in order to coexist [7]. A primary goal in microbial community ecology is therefore determining how functional redundancy and hyperdiversity may occur simultaneously.
In plant [8, 9] and animal communities [10], numerous species that compete for identical limiting resources can coexist when they experience a tradeoff between their ability to colonize new habitat patches and their ability to competitively displace species from existing habitat patches. Because organisms are more likely to interact with others in their immediate vicinity than those that are far away, inferior competitors that can escape from their competitive superiors by way of dispersal persist at landscape scales [11]. A sufficient quantity of habitat patches at which such dynamics can occur allows the coexistence of a theoretically unlimited number of species [9].
Competition–colonization tradeoffs may facilitate coexistence in microbes as well [12], but this subject remains understudied, likely due to perceptions of microbial dispersal, a central trait in the competition–colonization framework, as a random and unlimited process largely decoupled from species identity [13]. Thus, although the existence of competition–colonization tradeoffs has been established in some microbial taxa [14, 15], the extent to which they contribute to diversity in nature is unclear. Because theory suggests that competition–colonization tradeoffs could be central to microbial community assembly, we measured whether they structured communities of a keystone microbial plant symbiont in situ.
Our study organisms, ectomycorrhizal (EcM) fungi, are ubiquitous associates of many tree species that dominate boreal and temperate forests, and thus have a strong influence on ecosystem carbon cycling [16] and soil carbon sequestration [17, 18]. EcM fungi mobilize soil nutrients and provide them to plant hosts in return for photosynthetically fixed carbon [19]. Although most of them produce macroscopic fruiting structures, they interact with other soil microbes and their environment via a microscopic filamentous growth form similar to that of actinomycetous bacteria [20]. Many EcM host plants cannot complete their life cycles without EcM fungal mutualists; they therefore also strongly affect plant ecology because EcM dispersal and distribution can effectively control that of associated plants [21, 22].
EcM fungi are not only pivotally important in terrestrial ecosystems, but also useful model organisms for studying microbial dispersal and competition. They compete with one another for a single source of biotrophic carbon, host roots [23]. EcM fungi produce microscopic spores as their primary dispersal stage, are molecularly identifiable, and benefit from well-defined species concepts relative to other microbial taxa [24]. Finally, they associate only with specific tree species, allowing researchers to precisely delineate patches of habitat and thereby dispersal sources within a landscape.
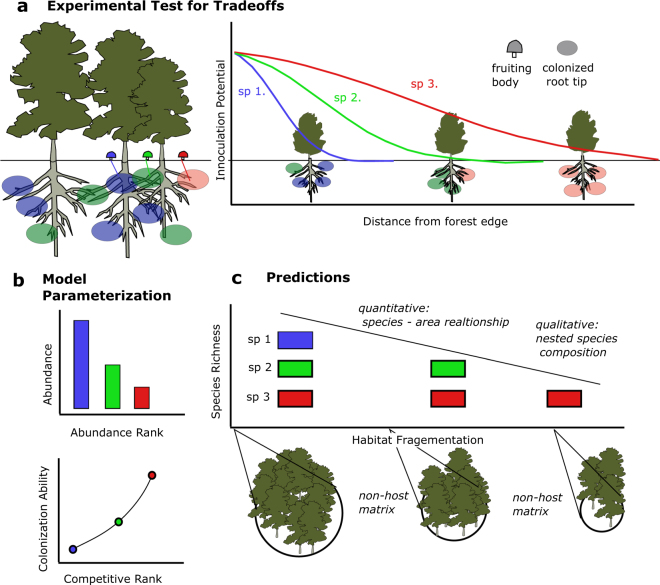
We made use of a ‘forest island’ system at Point Reyes National Seashore, California, USA (PRNS), which offers an ideal environment for studying the dispersal of EcM fungi due to its heterogeneous mixture of EcM and non-EcM plants. Prior research at this location has provided insight into the community assembly of EcM fungi, demonstrating dispersal limitation [25] and adherence to the theory of island biogeography: larger forest islands show greater species richness than smaller [26], and islands close to source populations show greater species richness than those farther away [27]. Here, we pair a laboratory experiment with a theoretical model and synthesize over a decade of prior research at PRNS to assess whether competition–colonization tradeoffs can give rise to observed EcM diversity and community structure (Fig. 1).
Fig. 1.
We paired a, an experimental test for competition–colonization tradeoffs performed at the edges of EcM host-dominated forests (Point Reyes National Seashore, CA, USA), with a simple theoretical model [38]. We parameterized the model using prior research at our study site b, and used it to make species richness predictions for forest islands of varying size c.We then compared the model’s predictions with empirical data from these forest islands [26] to determine whether competition–colonization tradeoffs could give rise to observed diversity patterns
Materials and methods
Field site
Our study area, PRNS in Marin County, California, USA (38°04’ N, 122°50’ W), has a Mediterranean climate, with warm dry summers and cool wet winters. The mean annual temperature is 11 °C, whereas mean annual precipitation is approximately 43 cm. PRNS coastal forests consist primarily of Pinus muricata D. Don, an EcM host species that tends to form monodominant stands. Between these stands are non-EcM grasses and shrubs, such as Baccharis pilularis, Toxicodendron diversiloba, and Rubus ursinus; the forests are therefore the only local sources of inoculum for EcM fungi, and their only habitat.
Seventeen sites were established at increasing distances from patches of contiguous P. muricata forest, as in Peay et al. [25] (Fig. S1). Distances were initially estimated using park GIS vegetation maps, and were verified in the field as ranging from 0.5 m to approximately 5.5 km. In 2010, soil samples were taken from each site using a shovel sterilized with ethanol. A total of 17 separate soil samples were transported back to the University of California, Berkeley and stored at 4 °C. Separate soil samples taken at each site were sent to A&L Western Laboratories (Modesto, CA, USA) for analysis of pH, organic matter content, nitrogen, and phosphorus.
Cultivation of seedlings
P. muricata seeds collected from PRNS were surface sterilized and soaked for 48 h in water prior to planting. Ten replicate seedlings for each of the 17 sites were planted in 50 ml Cone-tainers (Steuwe & Sons, Corvallis, OR, USA) containing a 1:1 mixture of the collected field soil inoculum and autoclaved sand, for a total of 170 seedlings. The seedlings received water every second day, and were harvested following 6 months in a growth chamber as in other soil bioassay studies (e.g. Glassman et al. [28]). Negative control seedlings, which received a 1:1 mixture of locally collected autoclaved inoculum (Tomales Point, PRNS, CA, USA) and sterile sand, were also kept to verify that no contamination took place (none did).
Each seedling was visually evaluated for mycorrhizal colonization. Colonized EcM root tips were quantified by visually differentiable operational category, or morphotype. Two root tips per seedling were selected to represent each observed EcM morphotype. Using the Extract N’ Amp protocol (Sigma Aldrich, St. Louis, MO, USA), DNA was extracted and the internal transcribed spacer (ITS) region sequenced as by Peay et al. [25]. Clustering of operational taxonomic units (OTU) clustering was carried out using a 97% similarity cut-off with Geneious v5.3.6 [29]. Results were taxonomically assigned using the US National Center for Biotechnology Information (NCBI) GenBank database.
Statistical analysis
Statistical analyses and modeling were carried out using R [30]. All measures of distance and area were log10 or log10 + 1 transformed, as in previous studies in this system [25–27, 31]. Figures were generated using R packages ggplot2 [32], cowplot [33], and GGally [34]. Permutational multivariate analyses of variance (PERMANOVA) analysis and non-metric multidimensional scaling (NMDS) ordination as implemented in R package vegan [35] were used to determine the influence that distance from forest had on community composition. Generalized additive models as implemented in R package mgcv [36], in which the occurrence of each EcM species on each replicate seedling was treated as a binomial trial, were used to analyze the abundance of species as a function of distance excluding species too rare to model (occurring only at two consecutive sites or fewer). Smoothed fixed effects for distance were included and knots were held at the minimum to avoid overfitting. Total proportion of replicate seedlings colonized by any EcM species at each site (the global model) was modeled using logistic regression.
To quantitatively assess whether species interactions influenced observed colonization patterns, we first generated a null model for each species representing its expected abundance across the gradient in the absence of competitors. Because seedlings were all grown under identical laboratory conditions and our work in this and past studies has demonstrated that EcM inoculum loads for all studied species decline monotonically with distance from forest source [25, 31], the maximal distance at which a species colonizes seedlings is an accurate representation of its dispersal ability. We assumed therefore that declines in species abundance with distance were attributable to dispersal limitation, but that absence near the forest edge was due to competitive exclusion. As such, the null model for each species consisted of a logistic function predicting colonization of at least one root tip on all replicate seedlings at the forest edge, and of none shortly past the greatest distance at which the target species was observed. These logistic functions were of the same form as the global model,
| 1 |
where x is distance, and a and b are intercept and slope, respectively, of the relationship between seedling replicate colonization by a given fungus and distance. Values for a and b were assigned by setting x equal to the greatest distance at which the species occurred and solving the following equation:
| 2 |
The value 0.06 is the EcM abundance, as measured by proportion of replicate seedlings colonized by any species, predicted by the global model at the greatest distance measured, 5457 m.
For each species, differences between predicted and observed abundances were calculated and compared with the observed abundances of every other species using Spearman’s ρ as implemented in R package Hmisc [37]. The dispersal ability of each species, which was used to rank species, was calculated as the average distance from the forest edge of root tips colonized by the species. To look for competitive hierarchies among the species in our data set, for each pair of species ij, we measured the competitive influence of i on j as the correlation (Spearman’s ρ) between the observed abundance of i and the difference between the null (dispersal only) and observed (dispersal and competition) abundances of j.
Theoretical model
To see if competition–colonization tradeoffs are capable of generating realistic landscape-scale patterns of diversity in EcM fungi, we used a deterministic spatial competition model to predict species richness as a function of area [38] and then compared our results with empirical data on EcM richness in forest islands of differing size [26]. In the model, species compete with one another for discrete single-occupancy sites (host root tips), where they must arrive and establish. Species also exhibit a strict competition–colonization tradeoff. Here, the proportion of sites occupied by species i () is modeled deterministically as a function of competitive rank (i), colonization (c), mortality (m), and the proportion of habitat destroyed (D) [38]:
| 3 |
Considering each host root tip to be a site sensu Tilman et al. [38], we acquired data from a prior study [27] on the abundances of all 25 EcM species observed in a large contiguous forest at PRNS and determined the proportion of root tips colonized by each species. A separate data set was used because the empirical findings of Peay et al. [26], to which we compare our model’s output, included observations of fruiting bodies, which are not directly equivalent to colonized root tips [39].
As the model required some sites to be empty, an estimate of total root tip colonization in the forest was necessary. As data on total root tip colonization from the present study were unavailable, we chose to use 0.74, the mean proportion of root tips colonized on P. muricata seedlings grown in the first meter of the forest edge in a prior study at this site [25], but the model was sensitive to changes in this parameter, with higher values resulting in flatter species–area curves (Fig. S2). We assigned a common mortality rate of 0.5 to all species, representing turnover of host root tips independent of colonizing species identity as the main factor resulting in emptying of a site; changing this parameter did not affect our results. Because we used a data set consisting only of root tip observations, we were able to standardize species abundances. This allowed us to derive a competitive hierarchy by solving the following equation to find the c values for each of the 25 species, representing as the proportion of root tips occupied by species i [9]:
| 4 |
We determined D, the proportion of habitat considered inaccessible to species in the model, for the 12 forest islands of Peay et al. [26] by first assigning the largest island a value of 0. We then calculated D for each smaller island as proportional to the log10 transformed area of the largest island. For each D value, we solved Eq. 3 for each of the 25 species, arriving at species richness predictions for each island by counting the number of non-zero results. For comparison, species counts were log10 transformed as before [26] after adding one to each in order to account for any zeroes. We compared the species–area curves of the model and of Peay et al. [26] using analysis of covariance analysis predicting species richness as a function of area, including an interaction term for the source of the prediction.
Results
Laboratory experiment
A total of 18 species of EcM fungi from 12 genera colonized the root tips of bait seedlings harvested after 6 months (Table S1). EcM community on seedlings exhibited a nested community structure with a few common species present at nearly all sites and many rare taxa only present at sites most proximal to the forest edge (Fig. 2). There was significant community change along the isolation gradient as quantified by PERMANOVA analysis (Fig. 3; F = 6.86, R2 = 0.35, p = 0.001). No significant trends in soil chemistry with distance from forest edge were found (Fig. S3).
Fig. 2.
EcM community nestedness plots. In each figure, rows represent sites and columns species of EcM fungi. Filled squares indicate colonization of a site by that EcM fungus. a Our empirical study, with distance from forest edge along the Y axis. b From Peay et al. [26] and shows communities in forest islands of varying size, with island size along the Y axis. cShows the output of our theoretical model, which predicts species richness based on forest island size, with island size along the Y axis
Fig. 3.

EcM community dissimilarity at each location along the isolation gradient at Point Reyes National Seashore. Points represent sites, excluding two at which no species were observed (n = 15). There is a significant effect of distance from forest on community composition (p = 0.001, R2 = 0.345, F = 6.8). Points are jittered for visibility
Of the 18 EcM species, 7 occurred with sufficient frequency to allow further analysis. Generalized additive modeling revealed that species abundances, as measured by the proportion of replicate seedlings colonized at each location, were significantly affected by distance from forest edge, and that the effects differed by species (Fig. 4). As P. muricata forests are the closest source of EcM inoculum where the study was conducted and EcM inoculum loads decline with distance from forest source [25, 31], an increase in species abundance with an increase in distance, observed in five of seven species, is most consistent with competitive displacement by an inferior disperser closer to the forest edge.
Fig. 4.
Abundance of species across the isolation gradient. Solid lines are species-specific logistic generalized additive models (GAMs) containing a smoothed term for distance. Shared letters indicate a lack of significant difference between GAMs at α = 0.05; * indicates a p-value of 0.053 for the comparison. +++ indicates a significant interaction between distance and abundance at α = 0.005, ++ at 0.05, and + at 0.1. Dashed lines are logistic null models representing the expected abundance of each species in the absence of all competitors. Null models are based on the global model, which includes all species. Points represent sites (n = 17)
Theoretical models of competition–colonization tradeoffs predict that species with different dispersal abilities can coexist when inferior dispersers are able to competitively displace superior dispersers. When superior dispersers instead outcompete inferior dispersers, there is no tradeoff in functional traits and species with different dispersal abilities cannot coexist [9]. To assess the nature of EcM species interactions at PRNS, we quantified the competitive impact of EcM species with differing dispersal abilities on one another in both directions: from inferior to superior dispersers (in agreement with theory) and from superior to inferior dispersers (indicative of a different functional tradeoff). We observed that inferior dispersers affected superior dispersers in most cases of competitive displacement (Fig. 5), supporting the assumptions of the competition–colonization model we used to derive the EcM species–area relationship in forest islands at PRNS.
Fig. 5.
Directionality and strength of species interactions at Point Reyes National Seashore, as measured by the correlation (Spearman’s ρ) of the difference between observed and expected abundance of the focal species with the observed abundance of each other species; interactions significant at α = 0.05 are shown. Impacts of inferior dispersers on superior dispersers, which are consistent with competition–colonization tradeoffs, are displayed in the left panel. Impacts of superior dispersers on inferior dispersers, which are not consistent with competition–colonization tradeoffs, are displayed in the right panel. Arrow size and darkness reflect the strength of the interaction, whereas species numbers represent dispersal rank from worst (1) to best (7) disperser. Darkness of species nodes represents the strength of the total impact on the species
Theoretical model
We applied a spatial competition model developed by Tilman et al. [38] to predict species richness in communities exhibiting competition–colonization tradeoffs. In this model, fragmentation of habitat has the effect of reducing the number of single-occupancy sites for which species can compete, resulting in a loss of species richness. After calibrating the model using empirical data, we predicted species richness as a function of area, comparing the output with empirical data on the EcM richness of 12 forest islands of different size at PRNS [26]. There was no significant difference between the species–area curve produced by the model and that found by Peay et al. [26] (Fig. 6; intercept F = 0.0142, p = 0.907; slope F = 0.3086, p = 0.585). The model also produced a similar nested output to that observed with gradients of forest island size and seedling colonization with distance, although the resulting communities from the theoretical model were more tightly packed and exhibited less turnover than in empirical studies (Fig. 2). We find thus that species richness and patterns of community composition in this system of islands are consistent with patterns generated by a model in which identical sets of species varying only in dispersal ability and competitive strength compete for discrete single-occupancy sites.
Fig. 6.

Species–area curve found by Peay et al. [26] and that produced by our spatial competition model. There is no significant difference between the intercepts (F = 0.0142, p = 0.9065) nor the slopes (F = 0.3086, p = 0.5847) of the line
Discussion
Our experimental and theoretical results both show that competition–colonization tradeoffs structure diversity in EcM communities. Across the isolation gradient, ranging from 0.5 m to approximately 5.5 km from the forest edge, tradeoffs allowed EcM species to coexist with one another by habitat partitioning. The observed species distributions were not attributable to variation in soil chemistry, which did not differ significantly across collection sites (Fig. S3), nor to differences in other abiotic factors, as all host seedlings were grown under controlled laboratory conditions. Thus, we conclude that diversity along the gradient was enabled by competition–colonization tradeoffs, largely consistent with those observed in laboratory studies of EcM fungi [14] and analogous to those maintaining diversity in other taxa [8–10, 15]. This resulted in a characteristic pattern in which the abundance of each species peaked at the nearest location within its dispersal range at which it could escape superior competitors, rather than at the immediate forest edge, where inoculum potential is highest (Fig. 4). If there were no tradeoff between competition and colonization, only the species that disperse best would have been present along the isolation gradient. Similarly, if dispersal were unlimited and did not differ between species, only the strongest competitor would be present.
We found that interspecies EcM competition at the root tip scale can further allow competition–colonization tradeoffs to facilitate species coexistence, as evidenced by the accurate species–area relationship generated by our theoretical model (Fig. 6). Other aspects of the model output necessarily differed from prior empirical findings due to several key simplifications. Because it was calibrated using EcM abundance data from a large, mature forest at PRNS [27], its species pool differed from that of the 12 forest islands of Peay et al. [26], leading to differences in species composition between its predictions and empirical findings. In keeping with our hypothesis of competition–colonization tradeoffs as primary drivers of EcM diversity, species richness in forest islands was also assumed to be a function purely of interspecies competition for host root tips, omitting other potential drivers of nested species distributions in island systems. Finally, the deterministic nature of the model prevented substantial departure from perfect nestedness, resulting in a more tightly packed species matrix in comparison with empirical studies at PRNS (Fig. 2).
Despite these significant simplifications, our model nevertheless accurately predicted the EcM species–area relationship among forest islands as a function of the quantity of available host root tips without the inclusion of resource niche differentiation among species, variation in the number of different resource niches available, or intrinsic differences in immigration rates as in MacArthur and Wilson [40] (Fig. 6). Niche differentiation, although certainly present in EcM communities [41], is therefore not necessary to explain observed diversity. Our finding that competition–colonization tradeoffs at the root tip scale (mm2) can generate the species–area relationships visible at the landscape scale (km2) suggests that they may also contribute to larger biogeographical patterns. This is supported by the similarity between the nested patterns of species abundance observed along the isolation gradient, in the 12 forest islands of Peay et al. [26] (Fig. 2b), and in the output of the theoretical model (Fig. 2c), which is consistent with a shared ecological mechanism.
Although further research is necessary to determine conclusively whether our findings are broadly generalizable to other microbial systems, we suggest that dispersal and competitive strength are likely to be crucial co-varying trait axes in other microbial communities, facilitating the observed coexistence of numerous functionally equivalent species [6]. The environmental conditions necessary for competition–colonization tradeoffs to create diversity (that habitat be divisible into patches between which interactions are rare) are common to microbial habitats [42, 43], and are not taxonomically restricted. Bacteria vary in traits influencing motility and competitive success [44], show evidence of competitive hierarchies [45], and have species–area relationships in island systems [46], just as EcM fungi do [26]. Similarly, aquatic microbes exhibit competition–colonization tradeoffs both in the laboratory [47] and in the field [15], and implementation in microcosms of competition–colonization tradeoffs fosters coexistence among strains of Pseudomonas bacteria [12].
Although the magnitude of microbial diversity, with the apparent coexistence of many functionally similar species [6], appears counterintuitive [7], we find here that tradeoffs in the ability to colonize and compete for a single limiting resource are able and sufficient to explain observed structure and diversity in communities of plant-associated eukaryotic microbes. Theories of spatial ecology, heretofore demonstrated largely in communities of plants and animals, thus may offer an underappreciated and useful lens through which to examine microbial communities in nature, and could help to facilitate a needed synthesis of ecological theory and microbial ecology [48]. Moreover, because the competition–colonization tradeoffs we find in EcM communities are identical to mechanisms that can foster coexistence in plants [8, 9], animals [10], and aquatic bacteria [15], our work indicates that they are likely to be among the most general drivers of biodiversity.
Data availability
All data are available upon request. Molecular data have been submitted to GenBank and are available under accession numbers MF186820–MF186837.
Code availability
All computer code used for analyses is available upon request.
Electronic supplementary material
Acknowledgements
This research was supported by NSF grant DBI-1249341 to KGP, NSF grants DBI-1046115 and DEB-0742868 to TDB, and DOE grant DE-SC0016097 to KGP. The work of GRS is supported by an NSF Graduate Research Fellowship. We thank EG Mordecai for feedback on an earlier draft of this manuscript, and J Tan and L Kaplan for mathematical assistance.
Author contributions
KGP and TDB designed and carried out the field experiment; GRS, BSS, and KGP analyzed and interpreted the data; GRS, BSS, TDB, and KGP wrote the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Gabriel R. Smith, Phone: (+1) 650.723.0552, Email: grsmith@stanford.edu
Kabir G. Peay, Email: kpeay@stanford.edu
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0086-0) contains supplementary material, which is available to authorized users.
References
- 1.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–9. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 2.Laforest-Lapointe I, Paquette A, Messier C, Kembel SW. Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature. 2017;546:145–7. doi: 10.1038/nature22399. [DOI] [PubMed] [Google Scholar]
- 3.Bennett JA, Maherali H, Reinhart KO, Lekberg Y, Hart MM, Klironomos J. Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science. 2017;355:181–4. doi: 10.1126/science.aai8212. [DOI] [PubMed] [Google Scholar]
- 4.Teste FP, Kardol P, Turner BL, Wardle DA, Zemunik G, Renton M, et al. Plant-soil feedback and the maintenance of diversity in Mediterranean-climate shrublands. Science. 2017;355:173–6. doi: 10.1126/science.aai8291. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schimel JP, Schaeffer SM. Microbial control over carbon cycling in soil. Front Microbiol. 2012;3:1–11. doi: 10.3389/fmicb.2012.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardin G. The competitive exclusion principle. Science. 1960;131:1292–7. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- 8.Marino PC. Coexistence on divided habitats: mosses in the family Splachnaceae. Ann Zool Fenn. 1988;25:89–98. [Google Scholar]
- 9.Tilman D. Competition and biodiversity in spatially structured habitats. Ecology. 1994;75:2–16. doi: 10.2307/1939377. [DOI] [Google Scholar]
- 10.Stanton ML, Palmer TM, Young TP. Competition-colonization trade-offs in a guild of African acacia-ants. Ecol Monogr. 2002;72:347–63. [Google Scholar]
- 11.Horn HS, MacArthur RH. Competition among fugitive species in a harlequin environment. Ecology. 1972;53:749–52. doi: 10.2307/1934797. [DOI] [Google Scholar]
- 12.Livingston G, Matias M, Calcagno V, Barbera C, Combe M, Leibold MA, et al. Competition–colonization dynamics in experimental bacterial metacommunities. Nat Commun. 2012;3:1234. doi: 10.1038/ncomms2239. [DOI] [PubMed] [Google Scholar]
- 13.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–63. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy PG, Higgins LM, Rogers RH, Weber MG. Colonization-competition tradeoffs as a mechanism driving successional dynamics in ectomycorrhizal fungal communities. PLoS ONE. 2011;6:e25126. doi: 10.1371/journal.pone.0025126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yawata Y, Cordero OX, Menolascina F, Hehemann JH, Polz MF, Stocker R. Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci USA. 2014;111:5622–7. doi: 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Högberg MN, Högberg P. Extramatrical ectomycorrhizal mycelium contributes one-third of microbial biomass and produces, together with associated roots, half the dissolved organic carbon in a forest soil. New Phytol. 2002;154:791–5. doi: 10.1046/j.1469-8137.2002.00417.x. [DOI] [PubMed] [Google Scholar]
- 17.Averill C, Turner BL, Finzi AC. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature. 2014;505:543–5. doi: 10.1038/nature12901. [DOI] [PubMed] [Google Scholar]
- 18.Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H, et al. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science. 2013;339:1615–8. doi: 10.1126/science.1231923. [DOI] [PubMed] [Google Scholar]
- 19.Smith S, Read D. Mycorrhizal symbiosis. 3rd ed. Burlington, MA: Elsevier; 2008. [Google Scholar]
- 20.Griffin DM. A comparison of the roles of bacteria and Fungi. In: Edward R. Leadbetter and Jeanne S. Poindexter, editor. Vol. 1. Boston, MA, USA: Springer Bacteria in nature. 1985. p. 221–55.
- 21.Nuñez MA, Horton TR, Simberloff D. Lack of belowground mutualisms hinders Pinaceae invasions. Ecology. 2009;90:2352–9. doi: 10.1890/08-2139.1. [DOI] [PubMed] [Google Scholar]
- 22.Dickie IA, Bolstridge N, Cooper JA, Peltzer DA. Co-invasion by Pinus and its mycorrhizal fungi. New Phytol. 2010;187:475–84. doi: 10.1111/j.1469-8137.2010.03277.x. [DOI] [PubMed] [Google Scholar]
- 23.Deacon JW, Fleming L V. Interactions of ectomycorrhizal fungi. In: Michael Allen, editor. Mycorrhizal functioning: an integrative plant fungal process. New York, NY, USA: Chapman & Hall 1992. p. 249–300.
- 24.Peay KG, Kennedy PG, Bruns TD. Fungal community ecology: a hybrid beast with a molecular master. Bioscience. 2008;58:799. doi: 10.1641/B580907. [DOI] [Google Scholar]
- 25.Peay KG, Schubert MG, Nguyen NH, Bruns TD. Measuring ectomycorrhizal fungal dispersal: macroecological patterns driven by microscopic propagules. Mol Ecol. 2012;21:4122–36. doi: 10.1111/j.1365-294X.2012.05666.x. [DOI] [PubMed] [Google Scholar]
- 26.Peay KG, Bruns TD, Kennedy PG, Bergemann SE, Garbelotto M. A strong species-area relationship for eukaryotic soil microbes: island size matters for ectomycorrhizal fungi. Ecol Lett. 2007;10:470–80. doi: 10.1111/j.1461-0248.2007.01035.x. [DOI] [PubMed] [Google Scholar]
- 27.Peay KG, Garbelotto M, Bruns TD. Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology. 2010;91:3631–40. doi: 10.1890/09-2237.1. [DOI] [PubMed] [Google Scholar]
- 28.Glassman SI, Peay KG, Talbot JM, Smith DP, Chung JA, Taylor JW, et al. A continental view of pine-associated ectomycorrhizal fungal spore banks: a quiescent functional guild with a strong biogeographic pattern. New Phytol. 2015;205:1619–31. doi: 10.1111/nph.13240. [DOI] [PubMed] [Google Scholar]
- 29.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R Core Team. R: a language and environment for statistical computing. https://www.r-project.org/. 2016.
- 31.Peay KG, Bruns TD. Spore dispersal of basidiomycete fungi at the landscape scale is driven by stochastic and deterministic processes and generates variability in plant-fungal interactions. New Phytol. 2014;204:180–91. doi: 10.1111/nph.12906. [DOI] [PubMed] [Google Scholar]
- 32.Wickham H. Ggplot2: elegant graphics for data analysis. New York, NY, USA: Springer; 2009.
- 33.Wilke C. cowplot: streamlined plot theme and plot annotations for ‘ggplot2’. https://cran.r-project.org/package=cowplot. 2016.
- 34.Schloerke B, Crowley J, Cook D, Briatte F, Marbach M, Thoen E, et al. GGally: extension to ‘ggplot2’. https://cran.r-project.org/package=GGally. 2016.
- 35.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: community ecology package. https://cran.r-project.org/package=vegan. 2016.
- 36.Wood SN. Generalized additive models: an introduction with R. Boca Raton, FL, USA: Chapman & Hall/CRC. 2006.
- 37.Harrell Jr FE, with contributions from Charles Dupont, many others. Hmisc: Harrell miscellaneous. https://cran.r-project.org/package=Hmisc. 2016.
- 38.Tilman D, May RM, Lehman CL, Nowak MA. Habitat destruction and the extinction debt. Nature. 1994;371:65–66. doi: 10.1038/371065a0. [DOI] [Google Scholar]
- 39.Gardes M, Bruns TD. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: above- and below-ground views. Can J Bot. 1996;74:1572–83. doi: 10.1139/b96-190. [DOI] [Google Scholar]
- 40.MacArthur RH, Wilson EO. The theory of island biogeography. Princeton, New Jersey: Princeton University Press; 1967. [Google Scholar]
- 41.Bogar LM, Peay KG. Processes maintaining the coexistence of ectomycorrhizal fungi at a fine spatial scale. In: Tedersoo L, editor. Vol. 230. Biogeography of mycorrhizal symbiosis. Cham, Switzerland: Springer. 2017. p. 79–105.
- 42.Rillig MC, Muller LA, Lehmann A. Soil aggregates as massively concurrent evolutionary incubators. ISME J. 2017;11:1943–8. doi: 10.1038/ismej.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Geng J, Tang X, Fan H, Xu J, Wen X, et al. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J. 2014;8:881–93. doi: 10.1038/ismej.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higgins LM, Friedman J, Shen H, Gore J. Co-occurring soil bacteria exhibit a robust competitive hierarchy and lack of non-transitive interactions. bioRxiv. 2017; 1–14. 10.1101/175737.
- 46.Bell T, Ager D, Song JI, Newman JA, Thompson IP, Lilley AK, et al. Larger islands house more bacterial taxa. Science. 2005;308:1884. doi: 10.1126/science.1111318. [DOI] [PubMed] [Google Scholar]
- 47.Cadotte MW, Mai DV, Jantz S, Collins MD, Keele M, Drake JA. On testing the competition-colonization trade-off in a multispecies assemblage. Am Nat. 2006;168:704–9. doi: 10.1086/508296. [DOI] [PubMed] [Google Scholar]
- 48.Buchkowski RW, Bradford MA, Grandy AS, Schmitz OJ, Wieder WR. Applying population and community ecology theory to advance understanding of belowground biogeochemistry. Ecol Lett. 2017;20:231–45. doi: 10.1111/ele.12712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request. Molecular data have been submitted to GenBank and are available under accession numbers MF186820–MF186837.