Abstract
Exposure to dietary sources of methylmercury (MeHg) is the focus of public health concerns with environmental mercury (Hg) contamination. MeHg is formed in anoxic environments by anaerobic microorganisms. This process has been studied mostly with single-species culture incubations, although the relevance of such studies to Hg(II)-methylation in situ is limited because microbial activities in the environment are critically modulated by interactions among microbial functional groups. Here we describe experiments in which Hg(II)-methylation was examined within the context of various microbial syntrophies. We show enhanced Hg(II)-methylation under conditions that established syntrophy by interspecies hydrogen and acetate transfer. Relative to activity of monocultures, interactions of Hg(II) methylating sulfate-reducing bacteria with a methanogen stimulated potential Hg(II)-methylation rates 2-fold to 9-fold, and with Syntrophobacter sp. 1.7-fold to 1.8-fold; those of a Hg(II) methylating Syntrophobacter sp. with a methanogen increased Hg(II)-methylation 2-fold. Under sulfate-depleted conditions, higher Hg(II)-methylation rates in the syntrophic incubations corresponded to higher free energy yields (ΔG°′) than in the monocultures. Based on energetic considerations, we therefore propose that syntrophic microbial interactions are likely a major source of MeHg in sulfate- and iron-limited anoxic environments while in sulfate-replete environments, MeHg formation via sulfate reduction dominates.
Introduction
Because inorganic Hg is the form most commonly deposited to the environment [1], in situ Hg(II)-methylation is a critical process determining bioavailability and toxicity of Hg [2]. Anaerobic microorganisms, e.g., sulfate and iron reducing bacteria and methanogens, synthesize MeHg by a mechanism that is not fully understood. The identification of genes that specify a Hg methylase (hgcA) and an associated reductase (hgcB) [3] has led to the discovery that fermenters and dehalogenating microbes may methylate as well [4]. To date, methylation has been studied either by incubating environmental samples and deciphering the contribution of individual functional groups to MeHg production by the addition of metabolic inhibitors and stimulators [5], or by comparing the activities of pure cultures [6]. However, microbial activities in anaerobic environments that can be readily depleted of strong electron acceptors are mostly governed by syntrophic interactions among different microorganisms [7, 8] that may lead to biogeochemical cycling under seemingly unfavorable conditions [9]. Yet, the effects of microbial syntrophy on MeHg production, while documented early on [10], has not been evaluated within the currently known complexity of methylating microbes and their involvement in syntrophic interactions in diverse environments.
Syntrophy by interspecies electron transfer, whereby a thermodynamically endergonic reaction may lead to energy conservation and microbial metabolism when coupled to hydrogen consumption [9], is perhaps the best understood such interaction. Modes of syntrophy in natural habitats vary widely, ranging from the classic interspecies electron transfer from sulfate-reducing prokaryotes (SRP) or Syntrophobacteraceae to methanogens [11, 12], to more recently described interactions such as SRP with Syntrophobacteraceae [13], and direct interspecies electron transfer (DIET) between iron reducers [14]. Syntrophobacteraceae are central to propionate degradation, a crucial intermediate step in organic matter degradation [15]. It has been estimated that 30% of methanogenesis in lake sediments and rice paddy soils is driven by the activity of syntrophic guilds [16, 17]. Consistent with this observation, Syntrophobacter-related species have been recently shown to be widespread in wetlands and freshwater ecosystems, and act as the major propionate-dependent sulfate reducers and syntrophs in rice paddy soil [18]. Interestingly, the dominant hgcA gene homologs, specifying Hg(II)-methylation, obtained from a wetland microbial community in the Florida Everglades were most similar to those of the Syntrophobacteraceae [19]. However, Hg(II)-methylation rates of Syntrophobacteraceae species in mono- or co-culture have not been reported.
We assessed the effect of syntrophy on Hg(II)-methylation by three types of co-cultures of Hg(II)-methylating microorganisms with appropriate metabolic partners: (1) Hg(II) methylating sulfate-reducing bacteria (SRB) grown as fermenters with a methanogen; (2) a methylating Syntrophobacter sp. grown as a fermenter with a methanogen; and (3) SRB grown as hydrogenotrophs with a Syntrophobacter sp. The SRB-methanogen and Syntrophobacter–methanogen syntrophies represent microbial interactions likely to occur in environments where sulfate is limited (e.g., freshwater lakes and wetlands), while the Syntrophobacter–SRB syntrophy exemplifies an association likely to be encountered in environments with variable levels of sulfate (brackish marshes) and where the supply of energy is limited either by low levels of organic substrates (e.g., groundwater aquifers) or the recalcitrant nature of these substrates (e.g., Sphagnum-dominated wetlands).
Materials and methods
Cultures and growth conditions
Strains tested in this study included Syntrophobacter wolinii DSM 2805, Syntrophobacter sulfatireducens DSM 16706, Syntrophobacter fumaroxidans DSM 10017, Methanospirillum hungatei DSM 864, Desulfovibrio africanus DSM 2603, and Desulfovibrio desulfuricans ND132. All cultures except strain ND132 (received from Cynthia Gilmour; henceforth referred to as D. desulfuricans) were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ) and were grown in their recommended media at 37 °C. The exceptions were D. africanus and D. desulfuricans, which were grown at 28 °C and in the medium described by Widdel and Bak [20]. Syntrophy was tested between a methanogen and Desulfovibrio spp. in a medium modified from Pak and Bartha [10] (Table S1), between a methanogen and Syntrophobacter spp. in a modified DSMZ Medium 307 (Table S2) or Medium 684 (Table S3), and between S. sulfatireducens with Desulfovibrio spp. in a modified DSMZ Medium 307 (Table S2).
All culture manipulations were performed under strictly anaerobic conditions using O2-free gases obtained by passage through a reduced, hot copper column for bench handling, or in an anaerobic glovebox (Coy Laboratory Products Inc., Grass Lake, MI) with a gas mixture of 95% nitrogen (N2) and 5% hydrogen (H2). All media (except where noted) were reduced with 0.25 mM titanium(III)-nitrilotriacetic acid (TiNTA) (1 mM) in order to minimize the influence of sulfide as a reductant on Hg speciation and methylation [21].
Mercury(II)-methylation experiments and chemical analysis
For Hg(II)-methylation assays by each strain alone, 7 ml (10%) of pre-grown exponential phase culture was added to 63 ml of fresh medium in a 130 ml serum bottles in triplicate. For methylation assays by co-cultures, pre-grown cells were washed twice with the co-culturing medium under strictly anaerobic conditions and used to inoculate at roughly 1–10 ratio of inoculum to fresh medium as described before [22]. Resting cells in co-culture incubations refer to non-growing, but metabolically active cells. Triplicate cultures were employed in each experiment. Abiotic heat-killed controls (80 °C for 1 h) [23] and medium blanks were included in all methylation experiments. All glassware used in methylation experiments was acid-cleaned.
Potential Hg(II)-methylation rates were analyzed by a radiotracer approach [5, 24], which consisted of spiking 203HgCl2 (Eckert and Ziegler Isotope Products, Valencia, CA) at the initiation of the experiment and obtaining toluene extracts of newly synthesized 203Hg-MeHg in 10 ml aliquots of cultures after incubation (see details in the Supplemental Information). MeHg concentrations in the culture media were measured by cold vapor atomic fluorescence spectrometry (CVAFS; a Tekran model 2500 spectrophotometer) as described previously [22]. Propionate and acetate concentrations were analyzed on a Beckman Coulter System Gold HPLC equipped with a Bio-Rad® Aminex HPX-87H organic acid analysis column. Sulfate concentrations in cultures were measured using a Dionex ICS-1000 Ion Chromatography system (see details in the supplementary information).
Protein assay
Cell growth was quantified as total protein concentrations using the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA) as described previously [22].
Cell counting by fluorescence staining and flow cytometry (FC)
To estimate the growth of individual strains in co-cultures, 1 ml of cell cultures taken at each time point were preserved by adding 143 μl 48% betaine stock [25], and stored at −20 °C until analysis. Upon thawing, preserved cells were stained with SYBR Gold concentrated stock (Molecular Probes, Invitrogen, Eugene, OR) at a final dilution of 1 × 10−4 for 20 min in the dark at room temperature. A sterile phosphate-buffered saline solution served as the sheath fluid during FC. Individual strains in co-cultures were distinguished based on differences in light diffraction due to varied cell properties (Supplementary Figure S1), and counted separately by a BD Influx Mariner 209s Flow Cytometer and High Speed Cell sorter (BD Biosciences, Rockville, MD). All signals were collected with logarithmic amplification and were triggered on the fluorescence channel. Triplicate cultures were analyzed. The ability of the FC protocol to distinguish the cells of each individual strain in co-cultures was confirmed by FC analysis of pure cultures of these strains (Supplementary Figure S1), and the counting accuracy was confirmed with an internal bead standard (Polysciences, Warrington, PA).
Statistical analysis
Results were analyzed by repeated two-way ANOVA. Overall effects of the two dimensions of treatments (incubation time and different strains or cultures) were shown by results of type 3 tests. Specific comparisons among different treatments were performed by Tukey’s honest significant difference (HSD) or Tukey–Kramer test using SAS software (SAS Institute, Cary, NC).
Free energy calculations
Free energy yields of reactions catalyzed by SRB, methanogens, and Syntrophobacter spp. alone or in syntrophic associations (Table S4) were estimated using free energies of formation and environmentally relevant concentrations of each reactant and product (Table S5). Standard free energy yields for each reaction (ΔG°reaction) were calculated as:
where νi,j is the stoichiometric coefficient of species i or j, and ΔGf°i-products and ΔGf°j-reactants are the free energies of formation of products and reactants. Free energy yields for each reaction at environmentally relevant reactant and product concentrations (ΔG°'reaction) were calculated as
where 5.7 ≈ 2.3 RT at 25 °C, R = 8.314 × 10−3 kJ mol−1 K−1, and logQ = Σνilog[producti]−Σνjlog[reactantj]. Free energy yields were examined for a range of reactant and product concentrations. However, with the exception of H2 (Supplementary Figure S2), variation in reactant and product concentrations had very little effect on free energy yields [26]. Free energy yields of syntrophic metabolisms were calculated as the sums of free energies of each individual metabolism. Free energies of reactions in saltwater were not corrected for ionic strength as this correction accounts for ≤0.3 kJ mol−1.
Results and discussion
Syntrophy of Desulfovibrio spp. with a methanogen
Pak and Bartha [10] reported enhanced Hg(II)-methylation in syntrophic cultures of D. desulfuricans strains LS or ND132 with Methanococcus maripaludis, a non-Hg methylator which does not contain gene homologs encoding HgcA and HgcB [22]. Thus, we started our examination of the role of syntrophy in Hg(II)-methylation by repeating and extending Pak and Bartha’s results [10] to additional Desulfovibrio spp. and a different methanogen. We then proceeded to study how methylation is affected by other syntrophic interactions. We assessed the impact of syntrophy on Hg(II)-methylation by following MeHg accumulation and cell growth in co-culture incubations of methylating organisms with their metabolic partners. Syntrophy with the methylating methanogen M. hungatei JF1T [22] was tested in a sulfate-free, lactate-bicarbonate medium (Table S1). The SRB we tested were previously examined under sulfate-reducing conditions (sulfate + lactate) with D. desulfuricans defined as a strong methylator [6], and D. africanus subsp. africanus (strain Benghazi) defined as a weak methylator [27]. In this medium, Desulfovibrio, a genus that includes most Hg(II) methylating SRB described so far [4], ferments lactate to acetate, bicarbonate, and H2, conserving energy only when H2 is maintained at low concentrations as a result of its immediate consumption, for example, by a methanogen [12, 13].
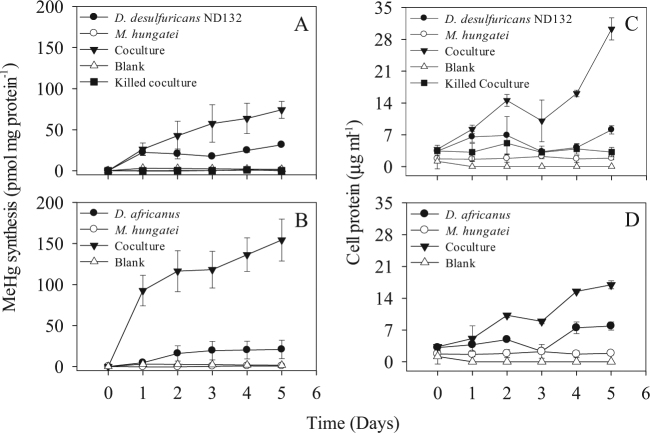
Monocultures of D. desulfuricans and D. africanus methylated Hg(II) in sulfate-free, lactate-bicarbonate medium, while no methylation was observed in blanks or heat-killed controls (p = 0.003 and p = 0.051, respectively) (Fig. 1a, b). The two SRB species grew slowly over 5 days in this medium (Fig. 1c, d). These relatively low levels of Hg(II)-methylation and growth by the SRB monocultures were likely supported by fermentative growth on the yeast extract in the co-culture medium (Table S1) [10].
Fig. 1.
The effects on Hg(II)-methylation of interspecies hydrogen and acetate transfer between a methanogen M. hungatei JF-1 and SRB D. desulfuricans ND132 (a), and D. africanus subsp africanus (b), in a lactate-bicarbonate medium. Samples were withdrawn for determination of MeHg and protein concentrations (c, d) every day for 5 days. Heat-killed (1 h at 80 °C) and medium-only (blank) controls were included. Averages and SD of three replicate cultures are shown
The co-culture of D. desulfuricans with M. hungatei methylated Hg(II) at a specific rate of 29.8 pmol mg initial protein−1 d−1 (all rates were normalized to the initial protein concentrations, hereafter referred to as protein) compared to 22.9 pmol mg protein−1 d−1 in the monoculture of D. desulfuricans and 0.1 pmol mg protein−1 d−1 in the monoculture of M. hungatei (Table 1, Fig. 1a), consistent with the production of more biomass (Fig. 1c) and a higher concentration of MeHg by the co- (334 pM) vs. the monoculture (138 pM) after 5 days of incubation (Table 1). The D. africanus - M. hungatei co-culture catalyzed MeHg at a rate of 78.1 pmol mg protein−1 d−1 compared with 8.0 pmol mg protein−1 d−1 in the D. africanus monoculture (p < 0.0001; Table 1 and Fig. 1b) and had 2-fold higher growth (Fig. 1d) and produced more than 7-fold more MeHg (443 pM) than the SRB monoculture (62.3 pM). Thus, a weak methylator under sulfate-reducing conditions [27], D. africanus, became a much more robust methylator than a strong one, D. desulfuricans, when examined under syntrophy, pointing out the limitation of extrapolating from pure culture studies to environmental processes where interactions among taxa modulate activities. Note that although we cannot rule out that some of the MeHg in these co-cultures was produced by M. hungatei, its contribution was likely small compared with that of the SRB since a monoculture of M. hungatei did not grow (Fig. 1c,d) and barely methylated Hg(II) (Fig. 1a,b) when grown in the co-culture medium used in these experiments. Nonetheless, the discovery of the potential for Hg(II)-methylation by methanogens [3, 22, 28] and their important role in syntrophy by interspecies electron transfer [29] clearly suggests that, methylation by both partners may be stimulated by their interactions.
Table 1.
Cell growth, potential Hg(II)-methylation rates, and MeHg production (yield) in monospecies and syntrophic cultures of mercury methylating sulfate-reducing bacteria (SRB), a methanogen, and a syntroph
| Syntrophic association | Media and growth conditions | Cell growth (fold increase)* | Specific Hg(II)-methylation rates (pmol mg protein−1 d−1)** | MeHg produced (pM, highest levels) |
|---|---|---|---|---|
| SRB/methanogen | Lactate-bicarbonate*** | |||
| D. desulfuricans ND132 | Monoculture by fermentation | 2.4 × | 22.9 ± 3.1 | 137.6 ± 3.7 |
| D. africanus DSM 2603 | Monoculture by fermentation | 2.5× | 8.0 ± 4.6 | 62.3 ± 24.4 |
| M. hungatei DSM 864 | Monoculture, resting | 1.1× | 0.1 ± 0.0 | 1.4 ± 0.2 |
| ND132 with M. hungatei | Co-culture | 7.9× | 29.8 ± 0.1 | 334.4 ± 57.3 |
| DSM 2603 with M. hungatei | Co-culture | 5.1× | 78.1 ± 12.4 | 443.2 ± 28.1 |
| Syntroph/methanogen | Propionate-bicarbonate | |||
| S. wolinii DSM 2805 | Monoculture by fermentation | 1.5 | 4.4 ± 2.2 | 48.7 ± 1.5 |
| M. hungatei DSM 864 | Monoculture, resting | 2.0× | 0.1 ± 0.3 | 16.6 ± 1.1 |
| DSM 2805 with DSM 864 | Co-culture | 2.6× | 9.2 ± 0.2 | 104.0 ± 2.0 |
| Syntroph/SRB | Propionate-sulfate | |||
| S. sulfatireducens DSM 16706 | Monoculture by sulfate reduction | 1.6× | 0.0 ± 0.0 | 13.1 ± 1.9 |
| D. desulfuricans ND132 | Monoculture, resting | 1.1× | 69.1 ± 3.3 | 3798.3 ± 298.6 |
| D. africanus DSM 2603 | Monoculture, resting | 2.1× | 8.9 ± 0.3 | 273.9 ± 28.6 |
| DSM 16706 with ND132 | Co-culture | 2.8× | 119.4 ± 0.9 | 5023.3 ± 322.4 |
| DSM 16706 with DSM 2603 | Co-culture | 3.4× | 16.4 ± 0.9 | 566.7 ± 17.3 |
*Cell growth was calculated by the measured protein content;
**Hg methylation rates were calculated by linear regressions of MeHg production and were then normalized to the initial cell protein levels of each incubation (48 h for most treatments except 24 h for ND132 and its co-culture with M. hungatei). Average ± SD are reported;
***The first compound identifies the electron donor and the second, the electron acceptor; media composition is provided in SM (Tables S1–S2).
A higher specific rate of MeHg synthesis was reported by Pak and Bartha [10] compared to rates reported here, likely due to the 169-fold higher Hg(II) concentration in their incubations (1 mg L−1) compared to the level used in our experiments (5.9 µg L−1). Previous studies showed that there was a strong positive correlation between the spiked Hg(II) concentration and extent of microbial Hg(II)-methylation [30, 31].
Syntrophobacter spp. as Hg(II) methylators
Syntrophobacter spp. are affiliated with a deltaproteobacterial genus that is phylogenetically related to sulfate reducers [32] and that can grow as a monoculture by sulfate reduction [13]. This taxon is widely distributed in natural environments including freshwater wetlands such as Sphagnum moss mats, lake sediments, salt marshes, and rice paddy fields [18, 33, 34]. The association of methanogens with Syntrophobacter spp. is a commonly observed syntrophy in freshwater ecosystems [15].
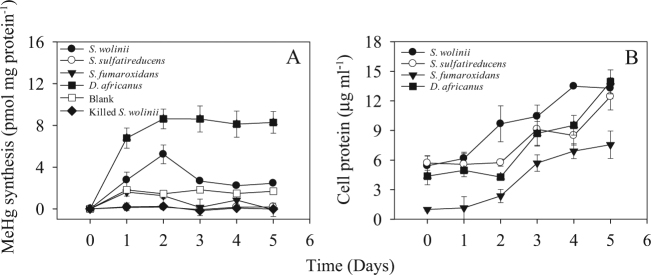
We first tested if monocultures of S. wolinii DB [11], S. sulfatireducens TB8106 [35], and S. fumaroxidans MPOB [36] methylated Hg(II) under sulfate-reducing conditions. After 5 days of incubation, S. wolinii was the only strain to methylate Hg(II) (Fig. 2a) even though all strains grew significantly (p < 0.01; Fig. 2b). Potential methylation rates of S. wolinii were lower (on day 2) than those of D. africanus, 3.1 vs. 8.6 pmol mg protein−1 d−1, respectively (Fig. 2), defining this strain as a weak methylator. Enhanced methylation by S. wolinii was observed when this strain grew alone or was co-cultured with M. hungatei in a propionate-bicarbonate medium (Fig. 3a). We confirmed MeHg production by S. wolinii cultures using column separation of Hg species [22] and CVAFS [37]. After 2 days of monoculture growth with 30 nM Hg(II), S. wolinii formed 27.6 ± 12.0 pM MeHg. Similar analyses with S. sulfatireducens TB8106 and S. fumaroxidans MPOB yielded no MeHg (Figs. 2a and 3b, c), identifying these cultures as non-methylators.
Fig. 2.
Hg(II)-methylation by Syntrophobacter spp. MeHg synthesis (pmol of mg initial protein) (a) and cell protein contents (b) were analyzed in monocultures of S. wolinii DB, S. sulfatireducens TB1806, S. fumaroxidans MPOB, and D. africanus. Cultures were grown in sulfate-reducing media as recommended for each by the DSMZ (Na2S as a reducing agent). Averages and standard deviations of three replicate cultures are shown
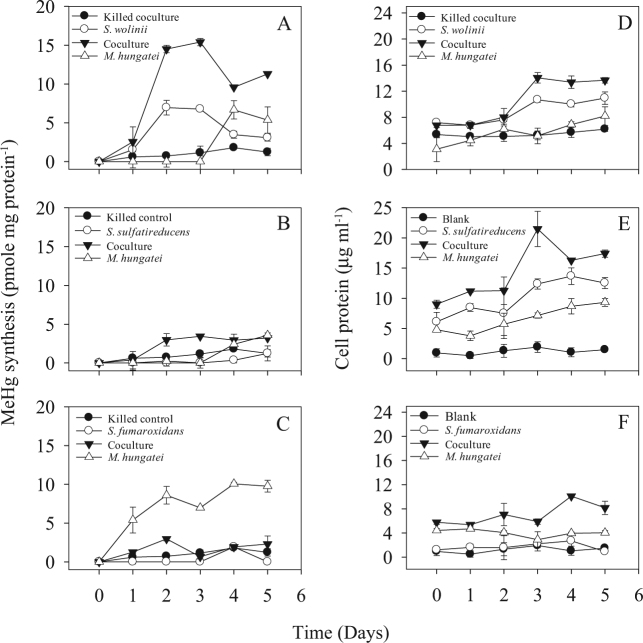
Fig. 3.
The effects on Hg(II)-methylation of interspecies hydrogen and acetate transfer between the syntrophs S. wolinii DSM 2805 (a), S. sulfatireducens DSM 16706 (b), and S. fumaroxidans DSM 10017 (c) and the methanogen M. hungatei JF-1. Samples were withdrawn for determination of MeHg concentrations and protein content (d–f) in a sulfate-free propionate Syntrophobacter medium every day for 5 days. Heat-killed (1 h at 80 °C) controls were included. Averages and standard deviations of three replicate cultures are shown
While this is the first report of Hg(II)-methylation by S. wolinii, Hg(II)-methylation activity was confirmed in another syntroph Syntrophus aciditrophicus SB [4], and Hg(II)-methylation gene hgcA/hgcB homologs were identified in the genomes of Syntrophorhabdus aromaticivorans UI and Syntrophobotulus glycolicus DSM 8271 [3]. Along with S. aciditrophicus, all syntrophic species may associate with methanogens or hydrogenotrophic Desulfovibrio spp. [38–40]. The inability of S. fumaroxidans and S. sulfatireducens to methylate Hg(II) is consistent with a prior report [31] as is the absence of hgcA/hgcB gene homologs in the sequenced genome of S. fumaroxidans (PRJNA13013). Based on these findings, Hg(II)-methylation may be relatively common among syntrophic bacteria, adding to the diversity of Hg(II) methylating anaerobic guilds and potentially explaining observed relationships between their abundance and MeHg production rates in wetland environments [19].
Syntrophy of Syntrophobacter spp. with a methanogen
The importance of environmentally relevant syntrophic interactions of Syntrophobacter spp. to MeHg production was examined in Syntrophobacter-methanogen and Syntrophobacter–Desulfovibrio (SRB, see below) co-cultures. The co-culture of S. wolinii with M. hungatei growing in a sulfate-free propionate-bicarbonate medium increased both the Hg(II)-methylation rate and MeHg production by factors of 2 above those for the fermenting S. wolinii monoculture, and by factors of 92 and 6, respectively, above those for the M. hungatei monoculture (Fig. 3a and Table 1). Both partners in this interaction are Hg(II) methylators and either or both may therefore contribute to the enhancement of methylation in co-cultures. However, the low level and late onset of Hg(II)-methylation by the M. hungatei monoculture in the propionate-bicarbonate medium (Fig. 3a) suggests that the enhancement of methylation by the co-culture resulted from a stimulatory effect of the methanogen on Hg(II)-methylation by the syntroph.
Interestingly, while the co-culture of M. hungatei with the non-methylating S. sulfatireducens resulted in an earlier production of MeHg and more growth relative to M. hungatei monocultures (Fig. 3b, e), the presence of the non-methylating S. fumaroxidans with M. hungatei appeared to have inhibited methylation (Fig. 3c). Syntrophobacter–methanogen interactions may therefore have positive or negative effects on MeHg production in the environment.
After day 2 or 3, MeHg production in the S. wolinii culture declined significantly (p < 0.01) in both monoculture and co-cultures (Figs. 2a and 3a), suggesting that MeHg was likely degraded following its formation as was previously documented for D. desulfuricans ND132 [6]. Compared with the obvious net accumulation of MeHg in the cultures of ND132 and D. africanus (Figs. 1a, b and 2a), the substantial decrease in MeHg concentration in S. wolinii cultures suggests that MeHg degradation might have exceeded methylation by S. wolinii at later incubation times.
Syntrophy of Syntrophobacter spp. with Desulfovibrio spp
In brackish or freshwater environments where microbial activities are limited by organic substrate availability, but not by sulfate, syntrophy between Syntrophobacter spp. and H2-utilizing SRB (e.g., Desulfovibrionaceae) may be established [11, 13], and could play a dominant role in the propionate-dependent sulfate reduction or syntrophic fermentation in the ecosystems [18]. While this mode of syntrophy has been little studied, it may play an important role in MeHg synthesis [19]. To examine how this interaction may affect Hg(II)-methylation, we set up propionate-fermenting S. sulfatireducens–Desulfovibrio spp. co-cultures under sulfate-reducing conditions (initial 3.94 mM sulfate). Propionate is an important recalcitrant intermediate in the mineralization of organic matter [41], and was used here as the electron and carbon donor (Supplementary Table S2). While most Syntrophobacter spp. respire sulfate when cultured alone in propionate-sulfate media, in co-culture with SRB they ferment propionate to produce H2 and acetate even when sulfate is present [42]. In these incubations, the Desulfovibrio spp. were the only Hg(II) methylating partners as S. sulfatireducens does not methylate Hg(II) ([31]; and Fig. 2a).
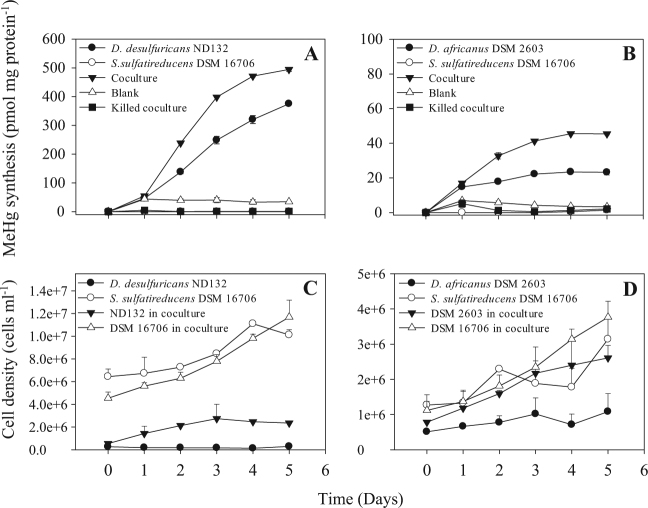
In co-cultures of propionate fermenting S. sulfatireducens with D. desulfuricans or D. africanus, cell densities of the SRB, quantified by flow cytometry (Supplementary Figure S1), increased by factors of 4 and 3, respectively, while monocultures of the two SRB in the propionate-sulfate medium showed little to no changes in growth (Fig. 4c, d), substrate consumption, and metabolite production (Supplementary Table S6 and Figure S3). Specific Hg(II)-methylation rates in co-cultures of each SRB strain with S. sulfatireducens were nearly 1.5-fold to 2-fold higher (p < 0.0001, Fig. 4a, b) than the respective rates of each SRB in monocultures. The co-culture of S. sulfatireducens with D. desulfuricans produced 5023 pM, compared with 3798 pM, MeHg in the D. desulfuricans monoculture, and the maximum MeHg yield doubled from 273 pM in the D. africanus monoculture to 566 pM in the co-culture of D. africanus with S. sulfatireducens (Table 1).
Fig. 4.
The effects on Hg(II)-methylation (a, b) and growth (c, d) of syntrophy between the methylating SRB strains D. desulfuricans ND132 or D. africanus subsp. africanus and the syntroph S. sulfatireducens TB8061 when grown in a propionate-sulfate SRB medium (at an initial sulfate concentration of 3.94 mM). Samples were withdrawn for determination of MeHg concentrations and cell counts by flow cytometry-cell sorting (Figure S1) every day for 5 days. Heat-killed (1 h at 80 °C) and medium-only (blank) controls were included. Averages and standard deviations (positive only for clarity) of three replicate cultures are shown
These results show for the first time that syntrophic co-cultures of S. sulfatireducens with Desulfovibrio spp. significantly stimulated Hg(II)-methylation. We also observed a similar stimulation of MeHg synthesis by a co-culture of S. wolinii with D. desulfuricans (data not shown). Muyzer and Stams [13] proposed that when S. wolinii was co-cultured with Desulfovibrio spp., sulfate reduction by S. wolinii would be suppressed and it would shift to grow as an acetogen by fermentation. Therefore, a likely scenario for the stimulation of Hg(II)-methylation by the syntrophy examined here is that Desulfovibrio spp. outcompete S. sulfatireducens with respect to sulfate reduction, while fermentative propionate oxidation by S. sulfatireducens contributes carbon (acetate) and energy (H2) to Desulfovibrio spp. Desulfovibrio cells would then significantly methylate Hg(II) while respiring sulfate. Although showing little or only modest growth in the monocultures, resting cells of Desulfovibrio alone were likely still metabolically active, resulting in significantly increased synthesis of MeHg (Fig. 4a, b). A previous study showed that non-growing, but metabolically active D. desulfuricans cells could methylate Hg(II) [43]. In the Syntrophobacter–Desulfovibrio associations, acetate provided by the Syntrophobacter strain is apparently incorporated into cell material by the SRB, but does not contribute to the energy budget of the cells. This is also the case for the syntrophy of Desulfovibrio with M. hungatei in which most of the methane produced by the methanogen is from the bicarbonate provided, thus highlighting the importance of hydrogen as an energy source to these syntrophic associations.
Syntrophic associations and mercury(II) methylation in the environment
Our demonstration that three types of syntrophic interactions between fermenting and hydrogenotrophic microbes enhance Hg(II)-methylation may be of particular importance to the microbial production of MeHg in sulfate-limited freshwater, or organic carbon-limited brackish environments. The manner in which syntrophies affect Hg(II)-methylation may vary, enhancing MeHg synthesis of the two partners while both are Hg(II) methylators (Figs. 1 and 3a) or only stimulating the activity of an individual Hg(II) methylator when another partner is a non-Hg(II) methylator (Figs. 3b, c and 4; [10]). In addition, the contribution of syntrophy to MeHg synthesis is not directly proportional to the Hg(II)-methylation ability of each partner when grown in monoculture. We found that Hg(II)-methylation by the syntrophic association of a methanogen with the previously defined “weak” methylating SRB D. africanus (grown under sulfate-reducing conditions) was stimulated to a much greater extent (~10-fold higher than its monoculture, p < 0.0001) than by the syntrophy of the same methanogen with the “strong” methylating SRB strain ND132 (~2-fold, p < 0.001). Thus, a weak methylator under sulfate-reducing conditions may produce the same or even more MeHg than a strong methylator when interacting with other microbes in their native environment.
To examine the broader environmental relevance of microbial syntrophy to Hg(II)-methylation, we plotted free energy yields (molar Gibbs energies of reaction) of methylating guilds that are common in freshwater and saline environments (Fig. 5, Tables S4 and S5). This approach is based on theoretical principles and experimental observations that available metabolic energy determines the growth [26] and activities of microbial organisms in their native habitats [44]. Indeed, we found that Hg(II)-methylation rates for individual organisms or their syntrophies expected in low iron and sulfate freshwater systems, were positively correlated with the free energy yields of their metabolisms (Fig. 5). These methylation rate-free energy yield correspondences are consistent with the current paradigm of methylation as a co-metabolic process [6].
Fig. 5.
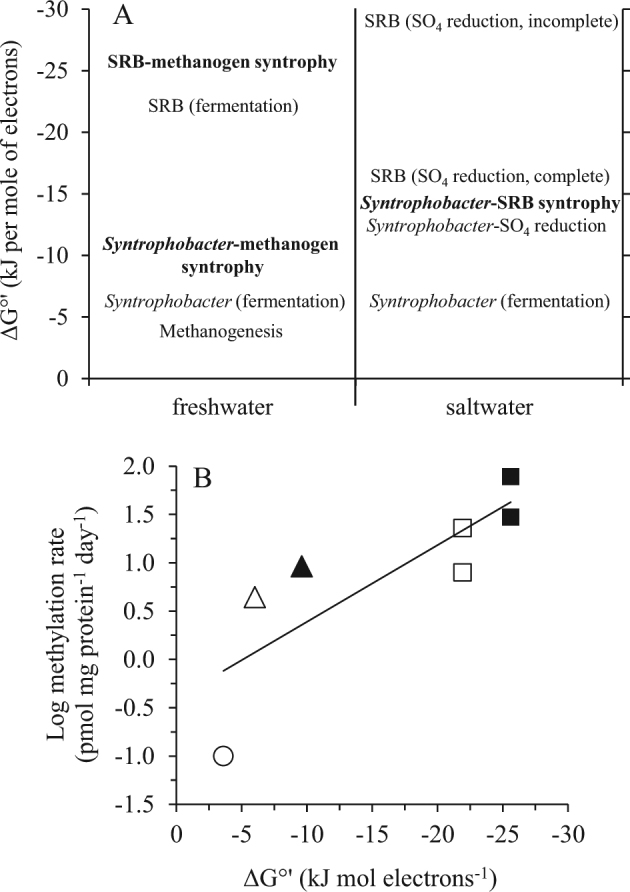
Free energy yields of reaction for individual Hg(II) methylating anaerobes and syntrophies likely to be important in freshwater (low sulfate and iron) and marine systems (low iron). A hypothetical scheme predicting the relative contribution of different anaerobes and their interactions to Hg(II) methylation (a). Entries in bold were tested in this paper. Microbes or syntrophies with higher (more negative) free energy yields (ΔG°') should outcompete those with lower ΔG°' at the specified environmental concentrations of substrates and products (Table S4). Relationship between Hg(II)-methylation rates in low iron and sulfate freshwater systems (Table 1) and the expected free energy yields of their metabolisms (Table S4) (b). Open circle-methanogens; open triangle-syntrophs grown by fermentation; filled triangle-syntrophs grown with methanogens; open squares-SRB grown by fermentation; filled squares-SRB grown with methanogens. The two pairs of symbols depicting SRB potential methylation rates are those observed with strains D. desulfuricans and D. africanus. The relationship between methylation rate and free energy yield is log (methylation rate) = −0.079ΔG°' - 0.4 (R2 = 0.66, p < 0.05). Such a log-linear relationship is in accord with theoretical linear free energy relationships between reaction kinetics and thermodynamics [46]
In freshwater systems where sulfate is scarce, the syntrophy between SRB and methanogens is most energetically favored (ΔG°' = −25.6 kJ mol−1 electrons), highlighting its potential importance to MeHg production (Fig. 5a). While variations in H2 concentrations and electron transfer efficiencies among syntrophic partners are likely critical factors controlling MeHg yield (Supplementary Figure S2), the observation that the SRB-methanogen syntrophy has the highest potential to produce MeHg in freshwater systems (Fig. 5a) is consistent with our findings (Fig. 1), and partially explains how Hg(II)-methylation occurs in ecosystems with little or no sulfate [10] or iron. Likewise, the proposed role of the syntrophy between Syntrophobacter spp. and methanogens in Hg(II)-methylation is in accord with the dominant distribution of syntrophs, characterized by 16S rRNA, hgcA and dsrB genes, in MeHg-producing wetlands [19, 45].
In contrast, Hg(II)-methylation in marine systems is expected to be dominated by sulfate reducers, with syntrophy between propionate oxidizing syntrophs and hydrogenotrophic SRB (ΔG°' = −13.2 kJ mol−1 electrons) less energetically favored than sulfate reduction with complete (ΔG°' = −16.0 kJ mol−1 electrons) or incomplete (ΔG°' = −29.2 kJ mol−1 electrons) oxidation of organic carbon (Fig. 5a, Table S4).
The observed effects of syntrophic interactions on Hg(II)-methylation clearly suggest that our perspectives on the contributions of specific microbial guilds to this process, mostly gleaned from studies of pure cultures, should be revisited. Rather, the numerous and complex interactions among methylating microbes in natural environments where MeHg is produced, need to be taken into account. When assessing methylation activities in the environment, direct prediction based only on laboratory tests of pure cultures may be misleading. The discovery of methylation by sulfate and iron reducing bacteria, methanogens, syntrophs, and fermenters suggests that understanding how MeHg is produced in the environment necessitates the study of methylation within the context of the complexity of microbial interactions in natural habitats.
Electronic supplementary material
Supplemental Information_Syntrophy(DOC 570 kb)
Acknowledgements
We thank Jim Wholt for assistance with obtaining rumen fluid, Peter Strom for assistance with obtaining sewage sludge, Kay Bidle and Frank Natale for help with flow cytometry, and Dwayne Elias for technical suggestions in cell alkaline lysis and protein assay. This research was supported by the US Department of Energy Office of Science (BER) no. DE-SC0007051 grant (to T.B. and J.R.), by the National Science Foundation PLR-1304773 award (to T.B. and M.H.), the New Jersey Water Resources Research Institute (to R.-Q.Y.), and a Hatch/McIntyre-Stennis grant through the New Jersey Agricultural Experiment Station.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0106-0) contains supplementary material, which is available to authorized users.
References
- 1.Lin C-C, Yee N, Barkay T. Microbial transformations in the mercury cycle. In: Liu G, Cai Y, O’Driscoll N (eds). Environmental Chemistry and Toxicology of Mercury. Hoboken, NJ: John Wiley & Sons, 2012. p. 155-191.
- 2.Hsu-Kim H, Kucharzyk KH, Zhang T, Deshusses MA. Mechanisms regulating mercury bioavailability for methylating microorganisms in the aquatic environment: a critical review. Environ Sci Technol. 2013;47:2441–2456. doi: 10.1021/es304370g. [DOI] [PubMed] [Google Scholar]
- 3.Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Jr., Smith SD, et al. The genetic basis for bacterial mercury methylation. Science. 2013;339:1332–1335. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 4.Gilmour CC, Podar M, Bullock AL, Graham AM, Brown SD, Somenahally AC, et al. Mercury methylation by novel microorganisms from new environments. Environ Sci Technol. 2013;47:11810–11820. doi: 10.1021/es403075t. [DOI] [PubMed] [Google Scholar]
- 5.Yu RQ, Flanders JR, Mack EE, Turner R, Mirza MB, Barkay T. Contribution of coexisting sulfate and iron reducing bacteria to methylmercury production in freshwater river sediments. Environ Sci Technol. 2012;46:2684–2691. doi: 10.1021/es2033718. [DOI] [PubMed] [Google Scholar]
- 6.Gilmour CC, Elias DA, Kucken AM, Brown SD, Palumbo AV, Schadt CW, et al. Sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Appl Environ Microbiol. 2011;77:3938–3951. doi: 10.1128/AEM.02993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grosskopf T, Zenobi S, Alston M, Folkes L, Swarbreck D, Soyer OS. A stable genetic polymorphism underpinning microbial syntrophy. ISME J. 2016;10:2844–2853. doi: 10.1038/ismej.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sieber JR, McInerney MJ, Gunsalus RP. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol. 2012;66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 9.Morris BE, Henneberger R, Huber H, Moissl-Eichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 10.Pak K, Bartha R. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl Environ Microbiol. 1998;64:1987–1990. doi: 10.1128/aem.64.6.1987-1990.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boone DR, Bryant MP. Propionate-degrading bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from methanogenic ecosystems. Appl Environ Microbiol. 1980;40:626–632. doi: 10.1128/aem.40.3.626-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant MP, Campbell LL, Reddy CA, Crabill MR. Growth of Desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl Environ Microbiol. 1977;33:1162–1169. doi: 10.1128/aem.33.5.1162-1169.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muyzer G, Stams AJ. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol. 2008;6:441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- 14.Shi L, Dong H, Reguera G, Beyenal H, Lu A, Liu J, et al. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol. 2016;14:651–662. doi: 10.1038/nrmicro.2016.93. [DOI] [PubMed] [Google Scholar]
- 15.McInerney MJ, Sieber JR, Gunsalus RP. Syntrophy in anaerobic global carbon cycles. Curr Opin Biotechnol. 2009;20:623–632. doi: 10.1016/j.copbio.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krylova N, Janssen PH, Conrad R. Turnover of propionate in methanogenic paddy soil. FEMS Microbiol Ecol. 1997;23:107–117. doi: 10.1111/j.1574-6941.1997.tb00395.x. [DOI] [Google Scholar]
- 17.Rothfuss F, Conrad R. Thermodynamics of methanogenic intermediary metabolism in littoral sediment of Lake Constance. FEMS Microbiol Ecol. 1993;12:265–276. doi: 10.1111/j.1574-6941.1993.tb00039.x. [DOI] [Google Scholar]
- 18.Liu P, Conrad R. Syntrophobacteraceae-affiliated species are major propionate-degrading sulfate reducers in paddy soil. Environ Microbiol. 2017;19:1669–1686. doi: 10.1111/1462-2920.13698. [DOI] [PubMed] [Google Scholar]
- 19.Bae HS, Dierberg FE, Ogram A. Syntrophs dominate sequences associated with the mercury methylation-related gene hgcA in the water conservation areas of the Florida Everglades. Appl Environ Microbiol. 2014;80:6517–6526. doi: 10.1128/AEM.01666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH, editors. The Prokaryotes. New York: Springer-Verlag; 1992. pp. 3353–3378. [Google Scholar]
- 21.Jay JA, Murray KJ, Gilmour CC, Mason RP, Morel FM, Roberts AL, et al. Mercury methylation by Desulfovibrio desulfuricans ND132 in the presence of polysulfides. Appl Environ Microbiol. 2002;68:5741–5745. doi: 10.1128/AEM.68.11.5741-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu RQ, Reinfelder JR, Hines ME, Barkay T. Mercury methylation by the methanogen Methanospirillum hungatei. Appl Environ Microbiol. 2013;79:6325–6330. doi: 10.1128/AEM.01556-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiatrowski HA, Ward PM, Barkay T. Novel reduction of mercury(II) by mercury-sensitive dissimilatory metal reducing bacteria. Environ Sci Technol. 2006;40:6690–6696. doi: 10.1021/es061046g. [DOI] [PubMed] [Google Scholar]
- 24.Hines ME, Faganeli J, Adatto I, Horvat M. Microbial mercury transformations in marine, estuarine and freshwater sediment downstream of the Idrija Mercury Mine, Slovenia. Appl Geochem. 2006;21:1924–1939. doi: 10.1016/j.apgeochem.2006.08.008. [DOI] [Google Scholar]
- 25.Cleland D, Krader P, McCree C, Tang J, Emerson D. Glycine betaine as a cryoprotectant for prokaryotes. J Microbiol Methods. 2004;58:31–38. doi: 10.1016/j.mimet.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Roden EE, Jin Q. Thermodynamics of microbial growth coupled to metabolism of glucose, ethanol, short-chain organic acids, and hydrogen. Appl Environ Microbiol. 2011;77:1907–1909. doi: 10.1128/AEM.02425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekstrom EB, Morel FM, Benoit JM. Mercury methylation independent of the acetyl-coenzyme A pathway in sulfate-reducing bacteria. Appl Environ Microbiol. 2003;69:5414–5422. doi: 10.1128/AEM.69.9.5414-5422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamelin S, Amyot M, Barkay T, Wang Y, Planas D. Methanogens: Principal methylators of mercury in lake periphyton. Environ Sci Technol. 2011;45:7693–7700. doi: 10.1021/es2010072. [DOI] [PubMed] [Google Scholar]
- 29.Stams AJ, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol. 2009;7:568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 30.Gilmour CC, Riedel GS. Measurement of Hg methylation in sediments using high specific-activity Hg-203 and ambient incubation. Water Air Soil Poll. 1995;80:747–756. doi: 10.1007/BF01189726. [DOI] [Google Scholar]
- 31.Ranchou-Peyruse M, Monperrus M, Bridou R, Duran R, Amouroux D, Salvado JC, et al. Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: Implications for environmental studies. Geomicrobiol J. 2009;26:1–8. doi: 10.1080/01490450802599227. [DOI] [Google Scholar]
- 32.Harmsen HJ, Wullings B, Akkermans AD, Ludwig W, Stams AJ. Phylogenetic analysis of Syntrophobacter wolinii reveals a relationship with sulfate-reducing bacteria. Arch Microbiol. 1993;160:238–240. doi: 10.1007/BF00249130. [DOI] [PubMed] [Google Scholar]
- 33.Loy A, Kusel K, Lehner A, Drake HL, Wagner M. Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl Environ Microbiol. 2004;70:6998–7009. doi: 10.1128/AEM.70.12.6998-7009.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chauhan A, Ogram A, Reddy KR. Syntrophic-methanogenic associations along a nutrient gradient in the Florida Everglades. Appl Environ Microbiol. 2004;70:3475–3484. doi: 10.1128/AEM.70.6.3475-3484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, Liu X, Dong X. Syntrophobacter sulfatireducens sp. nov., a novel syntrophic, propionate-oxidizing bacterium isolated from UASB reactors. Int J Syst Evol Microbiol. 2005;55:1319–1324. doi: 10.1099/ijs.0.63565-0. [DOI] [PubMed] [Google Scholar]
- 36.Harmsen HJ, Van Kuijk BL, Plugge CM, Akkermans AD, De Vos WM, Stams AJ. Syntrophobacter fumaroxidans sp. nov., a syntrophic propionate-degrading sulfate-reducing bacterium. Int J Syst Bacteriol. 1998;48:1383–1387. doi: 10.1099/00207713-48-4-1383. [DOI] [PubMed] [Google Scholar]
- 37.EPA. Method 1630: Methyl Mercury inWater by Distillation, Aqueous Ethylation, Purge and Trap, and CVAFS (EPA-821-R-01-020, January 2001) Washington, DC: Office of Water, Office of Science and Technology, Engineering and Analysis Division 4303. US Environmental Protection Agency; 2001. [Google Scholar]
- 38.Han C, Mwirichia R, Chertkov O, Held B, Lapidus A, Nolan M, et al. Complete genome sequence of Syntrophobotulus glycolicus type strain (FlGlyR) Stand Genom Sci. 2011;4:371–380. doi: 10.4056/sigs.2004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L, et al. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci USA. 2007;104:7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu YL, Hanada S, Ohashi A, Harada H, Kamagata Y, Sekiguchi Y. Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Appl Environ Microbiol. 2008;74:2051–2058. doi: 10.1128/AEM.02378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bok FA, Plugge CM, Stams AJ. Interspecies electron transfer in methanogenic propionate degrading consortia. Water Res. 2004;38:1368–1375. doi: 10.1016/j.watres.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 42.Wallrabenstein C, Hauschild E, Schink B. Pure culture and cytological properties of Syntrophobacter wolinii. FEMS Microbiol Lett. 1994;123:249–254. doi: 10.1111/j.1574-6968.1994.tb07232.x. [DOI] [Google Scholar]
- 43.Janssen SE, Schaefer JK, Barkay T, Reinfelder JR. Fractionation of mercury stable isotopes during microbial methylmercury production by iron- and sulfate-reducing bacteria. Environ Sci Technol. 2016;50:8077–8083. doi: 10.1021/acs.est.6b00854. [DOI] [PubMed] [Google Scholar]
- 44.Nealson KH. Sediment bacteria: who’s there, what are they doing, and what’s new? Annu Rev Earth Planet Sci. 1997;25:403–434. doi: 10.1146/annurev.earth.25.1.403. [DOI] [PubMed] [Google Scholar]
- 45.Yu RQ, Adatto I, Montesdeoca MR, Driscoll CT, Hines ME, Barkay T. Mercury methylation in Sphagnum moss mats and its association with sulfate-reducing bacteria in an acidic Adirondack forest lake wetland. FEMS Microbiol Ecol. 2010;74:655–668. doi: 10.1111/j.1574-6941.2010.00978.x. [DOI] [PubMed] [Google Scholar]
- 46.Brezonik PL. Principles of linear free-energy and structure-activity relationships and their applications to the fate of chemicals in aquatic systems. In: Stumm W, editor. Aquatic Chemical Kinetics: Reaction Rates of Process in Natural Waters. New York: Wiley-Interscience; 1990. pp. 113–143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information_Syntrophy(DOC 570 kb)