Abstract
Catalytic activity of four structural variants of the antigenomic delta ribozyme, two cis- and two trans-acting, has been compared in the presence of selected divalent metal ions that effectively support catalysis. The ribozymes differ in regions that are not directly involved in formation of the ribozyme active site: the region immediately preceding the catalytic cleavage site, the P4 stem and a stretch of the viral RNA sequence extending the minimal ribozyme sequence at its 3′-terminus. The variants show high cleavage activity in the presence of Mg2+, Ca2+ and Mn2+, lower with Co2+ and Sr2+ and some variants are also active with Cd2+ and Zn2+ ions. In the presence of a particular metal ion the ribozymes cleave, however with different initial rates, according to pseudo-first or higher order kinetics and to different final cleavage extents. On the other hand, relatively small differences are observed in the reactions induced by various metal ions. The cleavage of trans-acting ribozymes induced by Mg2+ is partially inhibited in the presence of Na+, spermidine and some other divalent metal ions. The inert Co(NH3)63+ complex is unable to support catalysis, as reported earlier for the genomic ribozyme. The results are discussed in terms of the influence of structural elements peripheral to the ribozyme active site on its cleavage rate and efficiency as well as the role of metal ions in the cleavage mechanism. Some implications concerning further studies and possible applications of delta ribozymes are also considered.
INTRODUCTION
Hepatitis delta virus RNA (HDV RNA) is a single-stranded circular RNA which replicates via the double rolling circle mechanism. In the genomic RNA strand as well as in its antigenomic counterpart generated during virus replication there are two sequences with ribozyme activities. These ribozymes, the genomic and antigenomic types, are required for self-cleavage of polymeric RNA transcripts into monomeric units (for selected reviews see 1–4). The spatial structure of the genomic ribozyme determined by X-ray analysis (5,6) and recent propositions on the mechanism of cleavage by delta ribozymes according to a general acid–base catalysis (7–9) has greatly increased our understanding of their functioning. However, one of the most important issues, concerning the role of divalent metal ions in folding or the mechanism of catalysis of delta ribozymes, remains unsolved.
Most evidence has consistently supported a requirement for divalent metal ions in HDV catalysis under physiological pH conditions. The delta ribozymes definitely require divalent ions, while other ribozymes, such as the hammerhead, hairpin and VS ribozymes, are also active in very high concentrations of monovalent ions (10). The presence of a ‘general’ metal ion-binding site in the genomic ribozyme has been suggested based on the results of metal ion-induced cleavage experiments. Specific cleavages are induced in the J4/2 region with Pb2+ (11) and also with Mg2+, Ca2+ and Mn2+ (12). In the trans-acting antigenomic ribozyme, a specific Mg2+-induced cleavage occurs at the bottom of the P2 stem (13). Moreover, although the 3′,5′-phosphodiester linkage at ‘the functional cleavage site’ is cleaved slightly faster in Ca2+ than in Mg2+, the 2′,5′-linkage is cleaved in Mg2+ (or Mn2+) but not Ca2+ (14). This dramatic difference is strongly suggestive of a crucial metal ion interaction at the active site. Recently, it has been shown that imidazole buffer rescues the activity of a mutant ribozyme with a C76→U substitution (7). These data are consistent with imidazole-enhanced cleavage by a general base mechanism, but another possibility is that imidazole could coordinate a catalytic metal ion, presumably replacing a ligand lost with mutation of C76 (7). In another mechanistic proposition (8), C75 acts as the general acid with the pKa of the ring nitrogen N3 shifted upwards to ∼7 as a consequence of interaction with a phosphate residue and the presence of a tight metal ion-binding site in its vicinity. An ionized metal ion hydrate acts as the general base in the proposed mechanism. All the above observations suggest the presence of essential divalent metal ion-binding sites in the delta ribozymes, although the crystal structure of the 3′ cleavage product of a genomic ribozyme (5,6) does not reveal a metal ion in the catalytic pocket.
Catalytic activity of delta ribozymes in the presence of various divalent metal ions has been compared for the genomic variant (15). For the antigenomic ribozyme, several authors describe experiments for testing the activity of different variants with divalent ions (14,16–18). In most cases, however, these data cannot be directly compared since it is not possible to separate the two effects on catalysis: the kind of catalytic divalent metal ion and the structure of a particular ribozyme variant. Precise kinetic data are needed in order to compare the activity of the same ribozyme in the presence of a broader spectrum of divalent metal ions. In order to gain more information on the role of divalent metal ions in catalysis we compared the activity of closely related variants of the antigenomic ribozyme in the presence of various divalent ions. The variants differed in regions that were not directly involved in formation of the ribozyme catalytic core. Thus the role of these peripheral elements in modulating ribozyme activity could also be assessed.
MATERIALS AND METHODS
Materials
The materials used in this study were from the following sources. [γ-32P]ATP (5000 Ci/mmol) was from Amersham and all the chemicals were from Serva or Fluka. Polynucleotide kinase, T7 RNA polymerase, RNase inhibitor and T4 DNA ligase were purchased from MBI Fermentas. AmpliTaq polymerase was from Perkin Elmer. Chemically synthesized oligoribonucleotides S1 (5′-GGGCGGGUCGG-3′), S2 (5′-CUUCGGGUCGG-3′) and S3 (5′-CUUUCCUCUUCGGGUCGGCA-3′) were purchased from Xeragon AG.
DNA template constructs
All oligodeoxyribonucleotides were synthesized at the 0.2 µmol scale, deprotected after synthesis and purified by electrophoresis on denaturing 8% (w/v) polyacrylamide gels. DNA bands were excised, eluted with 0.3 M sodium acetate, pH 5.2, 1 mM EDTA and precipitated with ethanol. The DNA was recovered by centrifugation and dissolved in TE buffer.
The DNA templates for in vitro transcription of the cis(W) ribozyme and the R oligomer, the ribozyme component of trans(S1) and trans(S2), were prepared as follows (12,19). For cis(W), two DNA oligomers were synthesized, A1 (5′-CTCCCTTAGCCATCCGAGTGGACGTGCGTCCTCCTTCGGATGCCCAGGTCGGACCGCGAGGAGGTGGAGATGCCATGCCGACCC-3′) and B1 (5′-TAATACGACTCACTATAGGGTCCTTCTTTCCTCTTCGGGTCGGCATGGCA-3′) (letters in italic mark the T7 RNA polymerase promoter; complementary sequences are underlined). For the R oligomer, two other oligomers were synthesized, A2 (5′-GAAAAGTGGCTCTCCCTTAGCCATCCGAGTGCTCGGATGCCCAGGTCGGACCGCGAGGAGGTGGAGATGCCC-3′) and B2 (5′-TAATACGACTCACTATAGGGCATCTCCACC-3′). Equimolar amounts of both oligomers (A1, B1 or A2, B2) were annealed and double-stranded DNA templates were generated by PCR. The reaction mixtures contained 1 µM both DNA oligomers, 10 mM Tris–HCl, pH 8.3, 2 mM MgCl2, 50 mM KCl, 200 µM each dNTP and 25 U/ml AmpliTaq polymerase. The reactions were performed on a Biometra UNO II thermocycler for five cycles of 30 s at 94°C, 30 s at 46°C and 2 min at 72°C. The mixtures were extracted with phenol/chloroform (1:1) and the reaction products precipitated with ethanol, dissolved in TE buffer and used in transcription reactions.
RNA preparation
The in vitro transcription reactions contained 0.4 µM DNA template, 40 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 2 mM spermidine, 5 mM DTT, 1 mM each NTP, 750 U/ml RNase inhibitor and 2000 U/ml T7 RNA polymerase (MBI Fermentas). To obtain the R oligomer with a monophosphate group at its 5′-end for subsequent preparation of cis(L), 5 mM 5′-GMP was used in the transcription reaction. Following incubation of the mixtures at 37°C for 4 h, the RNA transcripts were purified on 8% denaturing polyacrylamide gels, localized by UV shadowing, eluted with 0.3 M sodium acetate, pH 5.5, 1 mM EDTA, precipitated with ethanol and dissolved in sterile water containing 0.1 mM EDTA.
The cis(W) ribozyme was internally labeled with 32P during transcription by including [γ-32P]ATP in the reaction mixture. To minimize ribozyme self-cleavage the mixture was incubated at 4°C for 48 h, the RNA purified and, following autoradiography, recovered as described above. 5′-32P-labeled cis(L) was prepared by ligation of 5′-32P-labeled oligomer S3 and oligomer R with T4 DNA ligase in the presence of a splint oligodeoxynucleotide spanning 10 nt on each side of the ligation junction (14,20). Equimolar amounts of the three oligomers were heated at 95°C for 2 min, cooled to 25°C for 10 min in 10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl buffer. Subsequently, the ligation reaction (total volume 15 µl) was carried out in 50 mM Tris–HCl, pH 7.5, 16.7 mM NaCl, 11 mM MgCl2, 10 mM DTT, 0.3 mM EDTA, 1 mM ATP buffer with 1000 U/ml T4 DNA ligase for 4 h at room temperature. The ligated RNA was purified on a denaturing 8% polyacrylamide gel. The oligoribonucleotide substrates S1, S2 and S3 were labeled at their 5′-ends using [γ-32P]ATP and T4 polynucleotide kinase under standard conditions.
Cleavage reaction
Prior to self-cleavage the cis-acting ribozymes, internally 32P-labeled cis(W) (100 000 c.p.m., 0.5–1 pmol RNA in 50 µl reaction volume) or 5′-32P-labeled cis(L) (∼50 000 c.p.m., 0.1 pmol RNA in 50 µl reaction volume), were subjected to a denaturation–renaturation procedure in the standard reaction buffer, 50 mM Tris–HCl, pH 7.5, 0.1 mM EDTA by incubating at 100°C for 2 min, 0°C for 10 min and finally at 37°C for 10 min. The reactions of cis(L) were performed in the presence of 100 mM NaCl which was added during the denaturation–renaturation procedure, after incubation of the samples at 0°C (17). The trans-acting ribozymes, trans(S1) or trans(S2), were prepared by mixing the 5′-32P-labeled substrates S1 or S2 (∼50 000 c.p.m., 0.1 pmol RNA in 50 µl reaction volume) with the R oligomer in the standard reaction buffer to obtain the final RNA concentrations of <2 nM and 700 nM for the substrate and ribozyme components, respectively. The mixtures were subjected to the same denaturation–renaturation procedure as described above and under these conditions all the substrate molecules were bound in substrate–ribozyme complexes (16,21). The cleavage reactions were initiated by adding an appropriate divalent metal chloride solution and the reactions proceeded at 37°C. In some experiments (see Figs 5 and 6) additional cofactors (NaCl, spermidine or other divalent metal ions) were added during the denaturation–renaturation procedure after incubation at 0°C and the samples were incubated at 37°C for 10 min prior to initiation of the reaction. Aliquots of the reaction mixtures (5 µl) were removed at specified time points and quenched with equal volumes of 20 mM EDTA, 7 M urea mixture. The digestion products were analyzed by electrophoresis on 12% polyacrylamide, 0.75% bis-acrylamide, 8 M urea gels. Autoradiography was performed at –70°C with an intensifying screen. For quantitative analysis radioactive bands were cut off and counted in a Beckman LS5000TA counter. Some gels were quantified using phosphorimaging screens, a Typhoon 8600 Imager with ImageQuant software (Molecular Dynamics). The fraction of substrate cleaved was plotted versus time and fitted to single or double exponential equations. First order rate constants (kobs) were calculated from data fitted to a single exponential equation: [P]t = [EP](1 – e–kobs × t), where kobs is the first order rate constant and [P]t and [EP] are the fractions cleaved at time t and the reaction end point, respectively. The double exponential equation was [P]t = [ep′](1 – e–k′ × t) + [ep″](1 – e–k″ × t), where ep′ and ep″ represent the amplitudes of the biphasic time course and k′ and k″ the corresponding rate constants. These parameters were estimated by non-linear regression analysis using the Levenberg–Marquardt algorithm and Microcal Origin 5.0 software. Reactions of the same ribozyme variant in the presence of various divalent metal ions were usually performed on a gel side by side. At least two independent data sets were collected and variation between experiments was not greater than 20%.
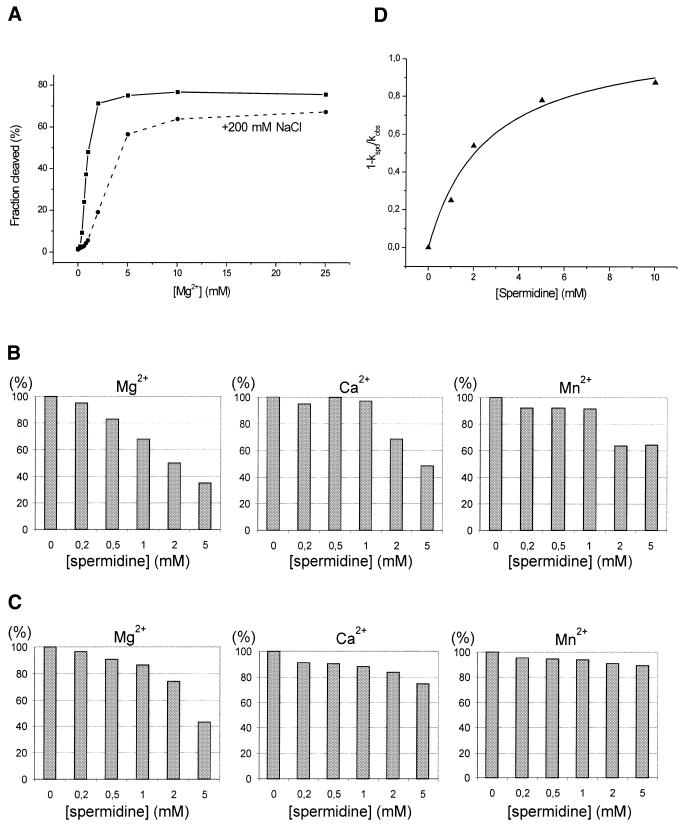
Figure 5.
Effect of sodium ions and spermidine on cleavage of trans-acting antigenomic ribozymes induced by Mg2+, Ca2+ and Mn2+ ions. (A) Cleavage reaction of the trans(S1) ribozyme as a function of Mg2+ ion concentration. The reaction was carried out in the presence or absence of 200 mM NaCl at 37°C for 5 min. (B) Dependence of the extent of cleavage of the trans(S1) ribozyme induced by 1 mM Mg2+, Ca2+ and Mn2+ ions at 37°C for 10 min on the presence of spermidine at concentrations in the range 0–5 mM. The extents of cleavage were normalized to those obtained in control reactions with no spermidine added and the relative values are expressed as percentages. (C) Dependence of the extent of cleavage of the trans(S2) ribozyme induced by 10 mM Mg2+, Ca2+ and Mn2+ ions at 37°C for 10 min on the presence of spermidine at concentrations in the range 0–5 mM. As above, the relative extents of cleavage are shown as percentages. (D) Concentration dependence of spermidine inhibition of the cleavage reaction of the trans(S2) ribozyme induced by 10 mM Mg2+. The data were fitted to a hyperbolic, bimolecular binding equation to give Ki of 2.6 ± 0.7 mM. The symbols kspd and kobs are first order cleavage rate constants determined in the presence and absence of spermidine, respectively.
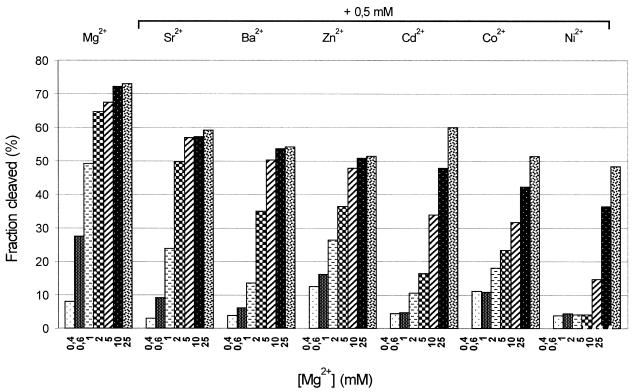
Figure 6.
Inhibition of Mg2+-induced cleavage of the trans(S1) ribozyme caused by a 0.5 mM concentration of selected divalent metal ions. The reactions were carried out in 50 mM Tris–HCl, pH 7.5, 0.1 mM EDTA buffer at 37°C for 5 min (for details see Materials and Methods).
RESULTS
Cleavage in the presence of Mg2+, Ca2+ and Mn2+ ions
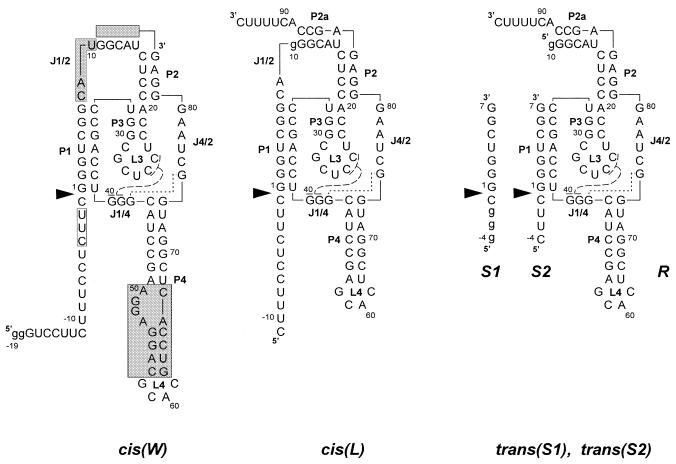
We synthesized four variants of the antigenomic delta ribozyme, two cis- and two trans-acting, which are shown in Figure 1. The sequence of variant cis(W) matches the minimal ribozyme sequence of the antigenomic RNA strand of the HDV virus. The transcript contains 19 nt upstream of the catalytic cleavage site, 17 nt of the wild-type sequence and two extra G residues at its 5′-end, facilitating in vitro transcription by T7 RNA polymerase. In the second variant, cis(L), the 5′-leader sequence has 11 nt, a single mutation U10→G is present in the J1/2 region, the P4 stem is shortened and an additional 11 nt extend the minimal ribozyme sequence at its 3′-end. Both the trans-acting ribozymes, trans(S1) and trans(S2), are derivatives of cis(L) in which the substrate and ribozyme strands are separated in the J1/2 region between A9 and G10 and two nucleotides, C8 and A9, are deleted from the oligonucleotide substrates. The substrates differ in the regions preceding the cleavage site; the wild-type sequence –1CUUC–4 in S2 is replaced by –1CGGG–4 in the S1 oligomer.
Figure 1.
Secondary structure models of cis- and trans-acting antigenomic delta ribozymes. Numbering of nucleotides corresponds to the wild-type ribozyme sequence and nucleotide changes are shown in lower case letters. Base paired segments are denoted P1–P4 and single-stranded regions as J1/2, J1/4 and J4/2. Nucleotides connected with broken and dotted lines indicate the 2 bp helix P1.1 and non-standard G-G interactions analogous to those found in the crystal structure of the genomic variant. Catalytic cleavage sites are marked by filled triangles. The shaded segments in cis(W) denote regions changed in the ribozyme variants.
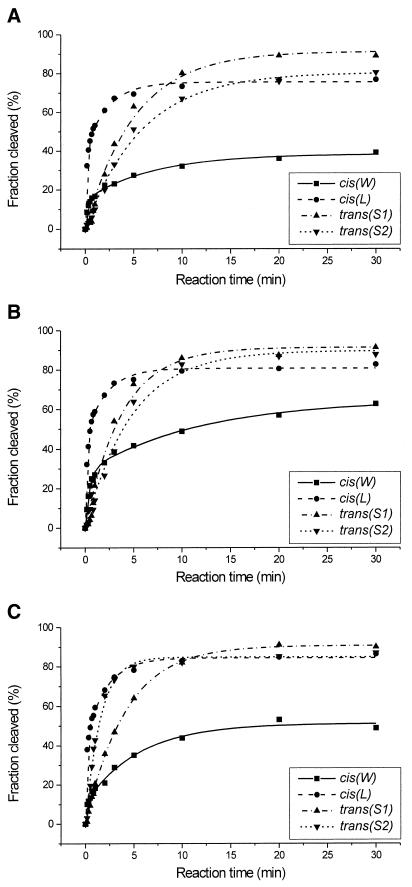
Catalytic activity of the four ribozyme variants was determined at 1 mM concentrations of Mg2+, Ca2+ and Mn2+ ions (Fig. 2 and Table 1). The cis-acting ribozymes, cis(W) and cis(L), are cleaved in the presence of all three metal ions in a multi-phasic mode and the data fit well to curves expressed by the sum of two exponentials. It suggests that each of these ribozymes contains a mixture of fast and slow cleaving molecules (17,22). In the case of cis(W) the percentages of fast cleaving fractions are low, ∼10–30%, depending on the metal ion used in catalysis, and the corresponding rate constants k′ range between 2 and 9 min–1. The rate constants k″ for slow cleaving fractions are lower by a factor of 30–50 and, finally, after 30 min only 40–60% of the ribozyme molecules are cleaved. The cis(L) ribozyme self-cleaves with the k′ constant ranging between 6 and 10 min–1, has an almost identical k″ value of 0.4–0.5 min–1 and the final cleavage extent reached 70% for Mg2+- and 80% for Ca2+- and Mn2+-induced reactions. The trans-acting ribozymes, with either the S1 or S2 substrates, are cleaved under single turnover conditions much slower than the cis variants. The reactions are, however, well approximated by pseudo-first order kinetics and the cleavage extent reached 80–90% in the presence of all three metal ions. The calculated rate constants kobs for Mg2+-, Ca2+- and Mn2+-induced reactions are 0.19, 0.26 and 0.24 min–1 for trans(S1) and 0.17, 0.20 and 0.57 min–1 for trans(S2), respectively. Cleavage rate constants for the trans-acting ribozymes at 10 mM concentration of the metal ions were also determined (Table 1). The S1 substrate is cleaved at 10 mM Mg2+ with a rate constant kobs of 0.92 min–1. The kobs values with the S2 oligonucleotide are 1.32, 2.31 and 1.87 for Mg2+-, Ca2+- and Mn2+-induced reactions.
Figure 2.
Cleavage kinetics of the cis(W), cis(L), trans(S1) and trans(S2) ribozymes in the presence of Mg2+ (A), Ca2+ (B) and Mn2+ (C) ions at 1 mM ion concentration.
Table 1. Summary of kinetic data.
| Ribozyme | Metal ion | 1 mM |
|
| |
|
k′ (ep′); (min–1) (%) |
k″ (ep″); (min–1) (%) |
| cis(W) | Mg | 3.95 ± 0.75 (15) | 0.14 ± 0.02 (23) |
| Ca | 2.16 ± 0.25 (29) | 0.08 ± 0.02 (36) | |
| Mn | 8.8 ± 3.9 (11) | 0.18 ± 0.02 (40) | |
| cis(L) | Mg | 7.65 ± 0.92 (40) | 0.44 ± 0.05 (36) |
| Ca | 5.61 ± 0.69 (45) | 0.48 ± 0.07 (36) | |
| |
Mn |
10.36 ± 1.61 (41) |
0.49 ± 0.05 (44) |
|
kobs (EP); (min–1) (%) |
|
||
| |
|
1 mM |
10 mM |
| trans(S1) | Mg | 0.19 ± 0.01 (92) | 0.92 ± 0.07 (87) |
| Ca | 0.26 ± 0.02 (92) | ||
| Mn | 0.24 ± 0.02 (92) | ||
| trans(S2) | Mg | 0.17 ± 0.01 (81) | 1.32 ± 0.16 (86) |
| Ca | 0.20 ± 0.02 (90) | 2.31 ± 0.32 (86) | |
| Mn | 0.57 ± 0.04 (86) | 1.87 ± 0.25 (86) |
It has been shown that a trans-acting antigenomic ribozyme with a long L4 stem is cleaved in the presence of 10 mM Mg2+ with a kobs of 0.91 min–1 (21), which is essentially identical to the value found for the trans(S1) variant. Another ribozyme, with stem L4 shortened but stem P2 extended by two additional GC base pairs, was cleaved with a lower, but comparable to our data, rate constant, ranging between 0.22 and 0.32 min–1 (16,23). A more rigid structure of stem P2 could explain the lower activity in that case (24). On the other hand, there are only very limited data on cleavage rates of antigenomic delta ribozymes in the presence of Ca2+ and Mn2+. For two trans-acting ribozymes that differed in stems P4 and P2, in the presence of 10 mM Ca2+ the kobs values ranged from 0.4 to 0.9 min–1 (13,14,16). The reported rate constant of 0.06 min–1 in 10 mM Mn2+ (13), much lower than in Mg2+ and Ca2+, seems to be unexpectedly low. Comparable cleavage extents in the presence of Mg2+, Ca2+ and Mn2+ were shown on an autoradiogram by other authors (14). However, for the Mn2+-induced reaction the cleavage rate constant has not been determined.
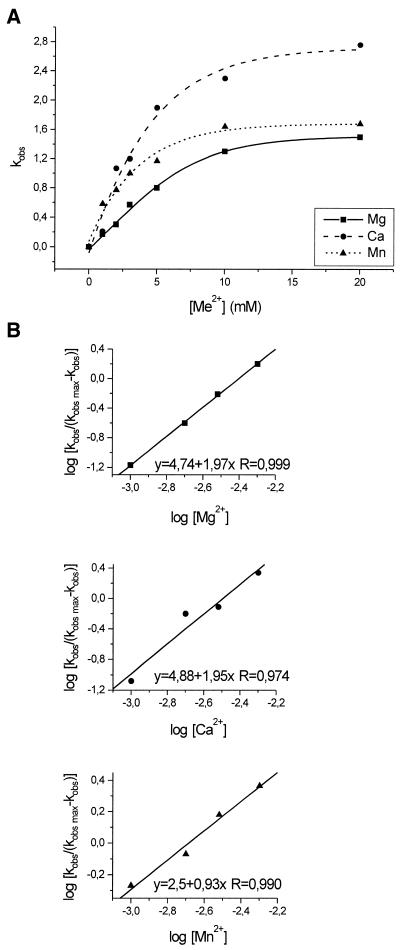
The dependency of the observed kobs values for trans(S1) on concentration of Mg2+, Ca2+ and Mn2+ is shown in Figure 3A. The rates increase almost linearly up to ∼5 mM concentration of divalent ions and reaches a plateau at ∼10 mM. At that ‘saturating concentration’ the cleavage reaction is fastest with Ca2+, then with Mn2+, and slowest with Mg2+. It should be noted that this order of relative activity is essentially observed over the entire range of metal ion concentrations. The concentrations of divalent ions at half maximal cleavage rates, K, are ∼4.5 for Mg2+, 3.5 for Ca2+ and 2.5 for Mn2+. The Hill coefficients calculated from the linear parts of curves are 1.97, 1.95 and 0.93 for the Mg2+-, Ca2+- and Mn2+-induced reactions. These values suggest the involvement of two active Mg2+ or Ca2+ or one Mn2+ ion in catalysis.
Figure 3.
Dependence of the cleavage rate constants kobs for the trans(S2) ribozyme on the concentration of Mg2+, Ca2+ and Mn2+ ions (A) and Hill analysis of the data (B). The correlation coefficients R were calculated during the least squares regression analysis using the Microcal Origin 5.0 computer program.
Other divalent metal ions active in catalysis
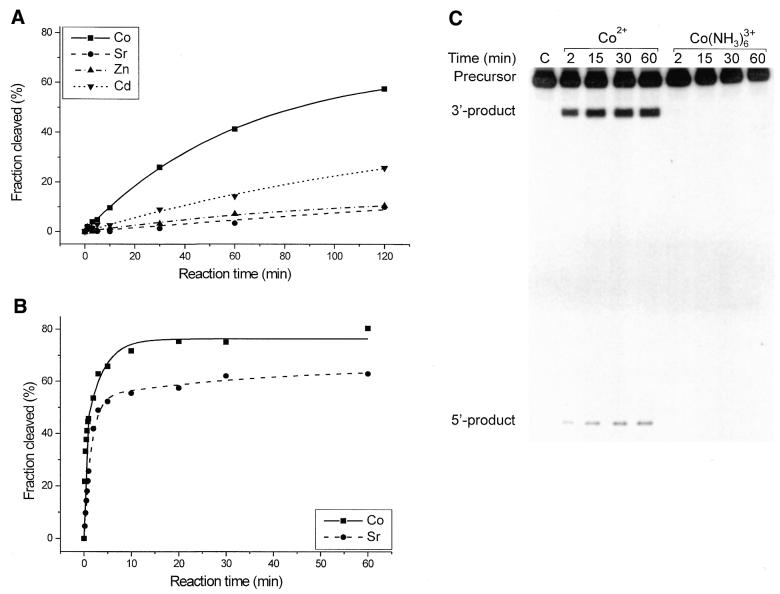
Some other divalent metal ions are also active in catalysis (Fig. 4). The trans(S2) ribozyme is cleaved in the presence of 1 mM Co2+, Sr2+, Zn2+ and Cd2+ with the following first order rate constants and reaction end points: 0.016 min–1 and 67%, 0.003 min–1 and 25%, 0.009 min–1 and 16% and 0.006 min–1 and 49%. These values were, however, determined at 1 mM, likely a sub-saturating concentration of these ions. Since the ions might differ in metal ion–RNA binding affinities the rate constants do not necessarily reflect the relative effectiveness of these metals in catalysis. This effect may be responsible for differences in the cleavage of trans(S1); after 60 min the cleavage extents at 1 mM divalent ions were Co2+, 45% and Sr2+, 5% (Fig. 4), but at 5 mM Co2+ it was 45%, whereas at 10 mM Sr2+ it was 25% (data not shown), suggesting a higher binding affinity of Co2+ than Sr2+ for the RNA. On the other hand, it is known that at higher concentration some of these metal ions may form polyhydroxy species, insoluble precipitates, perturb the RNA structure or even cause its degradation. To avoid misleading interpretations we therefore consider cleavage data obtained at low, ‘safe’ concentrations.
Figure 4.
Cleavage reaction of the antigenomic ribozyme variants in the presence of Co2+, Sr2+ Zn2+, Cd2+ and Co(NH3)63+. (A) Cleavage reaction of the trans(S2) ribozyme induced by Co2+, Sr2+, Zn2+ and Cd2+ at 1 mM ion concentration. The corresponding pseudo-first order rate constants kobs and the reaction end points were 0.016 min–1 and 67%, 0.003 min–1 and 25%, 0.009 min–1 and 16% and 0.006 min–1 and 49%, respectively. (B) Cleavage reaction of the cis(L) ribozyme in the presence of 1 mM Co2+ or Sr2+. The data were fitted to an expression describing the sum of two exponentials (see Materials and Methods) and the following rate constants k′ and k″ were obtained: 4.67 and 0.29 for Co2+ and 0.65 and 0.02 min–1 for the Sr2+-induced reaction. The reaction end points were 76 and 66% for Co2+- and Sr2+-induced cleavage. (C) Autoradiogram of cleavage of the cis(W) ribozyme in the presence of 5 mM Co2+ and 5 mM Co(NH3)63+.
An interesting observation concerns a high cleavage activity of cis(L) in the presence of 1 mM Co2+ and Sr2+, with corresponding rate constants k′ of 4.67 and 0.65 min–1 and reaction end points of 76 and 66% for the Co2+- and Sr2+-induced reactions, respectively (Fig. 4B). These values are comparable to those obtained with the ions most active in catalysis, i.e. Mg2+, Ca2+ and Mn2+ (Table 1). Clearly, cleavage rates depend strongly on the structure of the particular ribozyme variant and cis(L) turns out to be exceptionally active. Figure 4C shows that cis(W) is also active in the presence of 5 mM Co2+, although the cleavage extent does not exceed ∼10%. On the other hand, the inert complex Co(NH3)63+ at the same 5 mM concentration does not induce the reaction. The other ribozyme variants, cis(L) and trans(S2), also remained inactive under these conditions (data not shown). This suggests either direct coordination of the active metal ion to the RNA chain or that ionization of a ligand coordinated to the ion is required in catalysis (discussed later).
Influence of Na+, spermidine and other divalent metal ions on the Mg2+-induced cleavage reaction
The cleavage of trans(S1) induced by Mg2+ is inhibited by sodium ions present at 200 mM concentration (Fig. 5A). This inhibition is almost complete at concentrations of Mg2+ <1 mM, at 2 mM the effect becomes weaker and it almost disappears at >10 mM. This influence of Na+ on the cleavage of cis-acting ribozyme variants has also been observed by other authors (17,22). In that case, the effect depended strongly on ribozyme structure, on the ability of the 3′-terminus extension of the minimal ribozyme sequence to pair with the J1/2 region, forming an additional helix P2a. With a variant unable to form P2a the cleavage rate at 10 mM Mg2+ decreased in 100 mM NaCl from 0.61 to 0.38 min–1, while the presence of P2a stimulated self-cleavage by a factor of 1500 in 100 mM NaCl compared to low Na+/Mg2+ conditions (17,22).
The influence of spermidine on the activity of trans-acting ribozymes in the presence of Ca2+, Mg2+ and Mn2+ has also been tested (Fig. 5B and C). Two concentrations of metal ions were used, 1 and 10 mM. Spermidine was at concentrations ranging between 0 and 5 mM, the cleavage extents were determined after 10 min reaction and the values were normalized to those obtained in control reactions with no spermidine added. The first concentration of Mg2+ is close to its physiological concentration (25), while the second minimizes simple competition for binding to the RNA chain between the positively charged polyamine and metal ions due to electrostatic interactions. It turns out that spermidine inhibits the cleavage of trans(S1) and trans(S2) under all the conditions tested. The effect is stronger at 1 mM concentration of divalent ions and the Mg2+-induced reaction seems to be the most affected. We determined the concentration of spermidine that inhibited by 50% (Ki) the cleavage of trans(S2) induced in the presence of 10 mM Mg2+ (Fig. 5D). The fraction inhibition was plotted as a function of spermidine concentration and the data gave a Ki of 2.6 mM. A similar experiment performed in the presence of 200 mM NaCl gave a higher Ki of ∼16 mM (data not shown). The inhibitory effect of spermidine is, therefore, relatively weak compared to, for example, that observed for the delta ribozymes and some antibiotics (Ki values in the micromolar range) (11).
Catalytic activity of the trans(S1) ribozyme induced by Mg2+ is inhibited in the presence of a 0.5 mM concentration of some other divalent metal ions (Fig. 6). This effect may result from binding of inactive metal ions or ones showing low catalytic activity to the same sites that bind Mg2+. Alternatively, these ions may interact with the ribozyme interfering with formation of its active structure. The trans(S1) ribozyme is active in the presence of various metal ions, including those that are not considered to be particularly efficient in promoting the folding of functional RNA molecules (see the preceding section). This suggests that the observed inhibition of the Mg2+-induced reaction by other metals is rather a consequence of competition with the catalytically active ion than a result of interfering with folding of the global ribozyme structure. Since the observed concentration of Mg2+ at half-maximal cleavage rate (K) is ∼4.5 mM (Fig. 3A), such competition by other divalent ions should be particularly evident in reactions occurring at 2 and 5 mM Mg2+ (Fig. 6). The effect is more pronounced with Cd2+, Co2+ and Ni2+ than with Sr2+, Ba2+ and Zn2+. Such a classification of these ions into two groups, according to their higher and lower ability to compete with Mg2+, is consistent with the observation that at high concentrations of Mg2+ (5, 10 and 25 mM) the extents of cleavage are very similar in the presence of Sr2+, Ba2+ and Zn2+, but differentiated in the case of Cd2+, Co2+ and Ni2+.
DISCUSSION
Structural elements peripheral to the ribozyme active site influence cleavage rate and efficiency
Four variants of the antigenomic ribozyme (Fig. 1) show high cleavage activity in the presence of 1 mM Mg2+, Ca2+ and Mn2+ (Fig. 2 and Table 1). The reactions induced by a particular metal ion occur, however, according to pseudo-first or higher order kinetics, with different rates, and result in different final cleavage extents. These differences are a consequence of structural changes introduced in the ribozyme regions not directly involved in formation of its catalytic core.
The P4 stem is present in its wild-type, long form only in the cis(W) variant. The variant undergoes fast self-cleavage in the presence of all three metal ions and the experimental data fit well to biphasic kinetics. However, after 30 min a relatively high percentage of molecules remains uncleaved. Several ribozymes closely related to the wild-type sequence behave similarly, showing a high tendency for conformational heterogeneity (2,4). Such behavior is not as evident in variants in which the P4 stem is shortened. We show that although cleavage of cis(L) also deviates from first order kinetics, the final cleavage extent reaches on average 80% in the presence of all three metals. This observation is consistent with some earlier data showing that other cis-acting ribozymes with short P4 stems were cleaved to a high percentage (26). On the other hand, some variants with the native or a slightly shortened P4 were cleaved well, but they were acting in trans (14,21,27). Therefore, we conclude that the native P4 stem contributes to incomplete cleavage of antigenomic ribozymes but only in ribozyme variants acting in cis. Possibly, in misfolded molecules the polypurine stretch 51GGAG54 of P4 interacts with region 19CCUC16, as we suggested earlier on the basis of structural studies of a 3′-truncated ribozyme (19).
It has been shown that separation of cis-acting delta ribozymes in the J1/2 region into the substrate and ribozyme strands results in a several-fold decrease in catalytic activity of trans-acting variants (4,21). In the case of cis(L), on the one hand, and trans(S1) or trans(S2), on the other, the corresponding rate constants k′ and kobs differ by a factor of 10–40 depending on the ribozyme variant and catalytic metal ion (Fig. 2 and Table 1). What is responsible for these differences is not clear. Interestingly, the catalytic activity of hammerhead ribozymes acting in cis or in trans does not differ substantially unless the substrate–ribozyme association step limits the reaction rate (28). The substrate–ribozyme association step is definitely not responsible for slower cleavage of trans-acting delta ribozymes since reactions are usually performed under single turnover conditions, ensuring binding of all substrate molecules in substrate–ribozyme complexes (16,21). It has been suggested that the active site in trans-acting ribozymes is less rigidly structured relative to that of cis-acting ribozymes, thus slowing the reaction (21). If this is the case, the observed rate constants might reflect an additional conformational change preceding the chemistry step of the cleavage reaction. One may speculate that conformational changes would be more difficult in the presence of Mg2+ than Ca2+ or Mn2+, taking into account the different abilities of these ions to stabilize folded RNA structures. Indeed, the kobs constants for the trans(S1) and trans(S2) ribozymes are lowest in the Mg2+-induced reactions (Table 1).
An additional short duplex, in which a 3 nt sequence 11GGC13 of J1/2 pairs with a sequence just outside the 3′-boundary of the ribozyme, may be formed in the cis(L), trans(S1) and trans(S2) ribozymes. It has been shown that an analogous duplex extended by a U10–A89 interaction, P2a, can both inhibit and enhance ribozyme activity depending on cleavage conditions (17,22). The cis(L) variant was tested in 100 mM NaCl, i.e. under conditions known to greatly enhance the activity of an antigenomic ribozyme with the P2a element present. Indeed, this variant shows exceptionally high activity not only with Mg2+, Ca2+ and Mn2+ but also with other divalent ions, Co2+ and Sr2+ (Fig. 4B). At low Na+ concentrations, as expected, cleavage activity was marginal (data not shown). The P2a element can also be formed in the trans-acting ribozymes, trans(S1) and trans(S2). However, the presence of 200 mM NaCl did not stimulate cleavage of trans(S1). On the contrary, it inhibited the reaction and the effect was particularly strong at low Mg2+ concentrations (Fig. 5A). Thus the P2a element plays a modulatory role only in cis-acting ribozyme variants. It has been suggested that P2a and P2 form a coaxial stacked helix, most likely roughly parallel to P1. The main effect of additional stabilization of this arrangement appears to be consistent with stabilization of both the inactive and active conformations and a slowing down of the conversion from misfolded to active forms (22).
It has been shown that sequences immediately upstream of the ribozyme cleavage site influence its catalytic activity. Closer examination (23) of the role of nucleotides –1 to –4 in efficient cleavage with a collection of small substrates that possessed single and multiple mutations in this region showed the optimal sequence to be –1HRHY–4 (H = U, C or A; R = purine; Y = pyrimidine). The best substrate contained the sequence –1AAUC–4 and was cleaved with a rate constant of 0.69 min–1 at 10 mM MgCl2. Moreover, two consecutive pyrimidines at positions –1 and –2 were suggested to be responsible for the poor cleavage of substrates with the sequences –1UCGG–4 and –1UUGG–4 (23). Oligomers S1 and S2 used in our studies have the sequences –1CGGG–4 and –1CUUC–4 (Fig. 1). Thus both sequences differ from the optimal one, S1 at positions –3 and –4 and S2 at position –2, and S2 contains two consecutive pyrimidines immediately adjacent to the cleavage site, which were suggested earlier to be detrimental to cleavage in other substrates. The oligomers are, however, cleaved well with kobs of 0.92 for S1 and 1.32 min–1 for S2, at 10 mM Mg2+ (Table 1). These values are higher than the rate constant of 0.69 min–1 observed by Perreault and co-workers (23) with the optimal substrate. In another trans-acting ribozyme the sequence –1CUU–3, with two pyrimidines at positions –1 and –2 preceding the cleavage site, the rate constant was in the range 0.7–0.9 min–1 (14,21). It seems, therefore, that sequential preferences for nucleotides –1 to –4 depend on the particular ribozyme variant used in the trans-acting system.
The role of divalent metal ions in the cleavage mechanism
It has been shown that the maximum rate constant for RNA cleavage of 0.022 min–1 can be achieved under strong alkaline conditions. Therefore, natural RNA-cleaving enzymes must make use of alternative or additional catalytic strategies that allow them to exceed the maximum rate constant permitted by general base catalysis alone (29). In two recently proposed mechanisms of self-cleavage the ability of the delta ribozymes to carry out a general acid–base catalysis has been assumed (7–9). The authors postulated an important role of a divalent metal ion in the reaction mechanism. The ion may act either directly as an ionized hydrate or indirectly by binding in the vicinity of the cleavage site and modulating the properties of its close environment.
Although the ribozyme variants used in our studies differ in their catalytic activities, with given variants acting either in cis or in trans, relatively small differences in the reactions induced by several metal ions have been observed. Despite the fact that the ions differ substantially in the ability of their hydrates to ionize, the pKa values of metal hydrates are 11.4, 12.7 and 10.6 for Mg2+, Ca2+ and Mn2+, respectively (30). Strikingly, the cleavage rate constants kobs determined for trans(S2) at a 10 mM concentration of divalent ions differ by not more than 2-fold and increase in the order Mg2+ < Mn2+ < Ca2+ (Table 1 and Fig. 3A). In the case of both cis-acting variants the faster rate constants k′ increase as follows: Ca2+ < Mg2+ < Mn2+, consistent with an increasing concentration of ionized metal ion hydrates. The differences in cleavage rates are, however, far below expectation assuming a direct involvement of an ionized hydrate in catalysis. This is in contrast to hammerhead ribozyme cleavage in 10 mM Ca2+, Mg2+ and Mn2+, for which the relative rates were ∼1/16, 1 and 10, supporting the proposed role of the hydrate (31). Thus, in the antigenomic delta ribozymes the availability of an ionized metal ion hydrate, which has been proposed to act as the general base in the reaction mechanism (8), does not seem to be the rate-limiting step in ribozyme self-cleavage. Recently, it has been suggested that the overall rate-limiting step appears to be cleavage of the bond between the phosphorus and the 5′-leaving oxygen (32).
The metal ions used in our studies differ not only in pKa values of their hydrates but also in binding preferences for nucleic acid ligands, phosphate, ribose and nucleic acid base residues. Similar cleavage rates in the presence of Mg2+ and Ca2+ and also Mn2+, and for some ribozyme variants even with Co2+ and Sr2+, suggest rather low specificity interactions in the metal ion-binding pockets. Presumably, the ions bind to phosphate or ribose residues and not to nucleic acid bases. Support for this conclusion comes from a well-characterized metal ion-binding pocket in the D-TΨC region of yeast tRNAPhe in which a Pb2+ ion binds directly to the nucleic acid bases U59 and C60 (33–35). Specific cleavage of the D loop at D16 is induced by an ionized Pb2+ hydrate and a transesterification mechanism. Some other ions, Mg2+, Mn2+ and Eu3+, also induce cleavage in this region. However, the number of cleavages, their relative intensities and positions are substantially changed, most likely because of different coordination properties of these metal ions (36–39).
Finally, we show that the inert Co(NH3)63+ complex is unable to support catalysis in the antigenomic delta ribozymes, acting either in cis or in trans, consistent with earlier reported data concerning a cis-acting genomic variant (8). These observations suggest inner sphere coordination of the active metal ion to the polynucleotide chain or that ionization of a ligand coordinated to the ion is required in catalysis. Plots of cleavage rates of the trans(S2) ribozyme on concentration of Mg2+, Ca2+ and Mn2+ and Hill analysis of the data suggest that there are two active ions in the reactions induced by Mg2+ and Ca2+ but one ion in the case of Mn2+. Similar analysis performed for a cis-acting genomic ribozyme (8) suggested only one Mg2+ ion involved in catalysis, but the significance of this difference is not clear.
Implications for further studies and possible application of hepatitis delta ribozymes
Catalytic activity of antigenomic delta ribozymes is strongly influenced by changes in the regions located outside the ribozyme catalytic core. Thus in detailed studies, particularly on the cleavage mechanism or the role of metal ions in catalysis, only data obtained with the same ribozyme variant can be directly compared. Since several ribozyme variants have been studied in different laboratories this point has to be stressed. On the other hand, certain ribozyme variants may be particularly well suited to solving specific problems. For example, the cis(L) ribozyme, which can be readily synthesized and is very active with a variety of metal ions, is currently being used in our laboratory in NAIM experiments designed to unravel the specific localization of the metal ions active in catalysis.
The delta ribozymes, as the only ribozymes naturally active in human cells, are potentially attractive tools in the strategy of directed RNA degradation. Cleavage of an mRNA in trans by an antigenomic delta ribozyme has been demonstrated, for the first time, with mRNA encoding the only protein of HDV virus, the delta antigen (40). The studies showed, however, that further characterization of the effects influencing ribozyme performance is needed before successful application of the delta ribozymes as therapeutic agents or useful biochemical tools. In the trans-acting mode, their cleavage activity is comparable to that of the hammerhead ribozyme but at low Mg2+ ion concentrations the delta ribozymes show the highest cleavage rates among all known ribozymes. We show, however, that at low concentrations of Mg2+ considerable inhibition of cleavage occurs in the presence of monovalent ions. Since most activity assays are usually performed in low salt conditions, ribozymes designed for practical applications should be tested at concentrations of divalent and monovalent ions relevant to physiological conditions. Moreover, stimulation of ribozyme activity by polyamines cannot be expected, in contrast to that observed with the hammerhead or hairpin ribozymes (41–44). Although the addition of extra 3′-terminal nucleotides may be detrimental for some ribozyme variants the effect occurs only with ribozymes acting in cis. Similarly, the native form of the P4 stem increases the fraction of misfolded ribozyme molecules that are resistant to cleavage only in cis-acting ribozyme variants. In ribozymes acting in trans the P4 stem could be used as an engineered element that would enable modulation of the activity or performance of the ribozyme. For example, in applicable variants it could govern their intracellular transport or interactions with protein cofactors.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Professors Fritz Eckstein and Michael Gait for helpful discussions. This work was supported by the Polish Committee for Scientific Research, grants 6 P04A 001 12 and 6 P04A 006 18.
References
- 1.Been M.D. (1994) Cis- and trans-acting ribozymes from a human pathogen, hepatitis delta virus. Biochem. Sci., 19, 251–256. [DOI] [PubMed] [Google Scholar]
- 2.Lazinski D.W. and Taylor,J.M. (1995) Regulation of the hepatitis delta virus ribozymes: to cleave or not to cleave? RNA, 1, 225–233. [PMC free article] [PubMed] [Google Scholar]
- 3.Tanner N.K. (1995) The catalytic RNAs from hepatitis delta virus: structure, functions and applications. In Dinter-Gottlieb,G. (ed.), The Unique Hepatitis Delta Virus. Springer-Verlag, New York, NY, pp. 11–29.
- 4.Been M.D. and Wickham,G.S. (1997) The self cleaving ribozymes of hepatitis delta virus. Eur. J. Biochem., 247, 741–753. [DOI] [PubMed] [Google Scholar]
- 5.Ferre-D’Amare A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- 6.Ferre-D’Amare A.R. and Doudna,J.A. (2000) Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol., 295, 541–556. [DOI] [PubMed] [Google Scholar]
- 7.Perrotta A.T., Shih,I. and Been,M.D. (1999) Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science, 286, 123–126. [DOI] [PubMed] [Google Scholar]
- 8.Nakano S., Chadalavada,D.M. and Bevilacqua,P.C. (2000) General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science, 287, 1493–1497. [DOI] [PubMed] [Google Scholar]
- 9.Wadkins T.S., Shih,I., Perrotta,A.T. and Been,M.D. (2001) A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J. Mol. Biol., 305, 1045–1055. [DOI] [PubMed] [Google Scholar]
- 10.Murray J.B., Seyhan,A.A., Walter,N.G., Burke,J.M. and Scott,W.G. (1998) The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol., 5, 587–595. [DOI] [PubMed] [Google Scholar]
- 11.Rogers J., Chang,A.H., von Ahsen,U., Schroeder,R. and Davies,J. (1996) Inhibition of the self-cleavage reaction of the human hepatitis delta virus ribozyme by antibiotics. J. Mol. Biol., 259, 916–925. [DOI] [PubMed] [Google Scholar]
- 12.Matysiak M., Wrzesinski,J. and Ciesiolka,J. (1999) Sequential folding of the genomic ribozyme of the hepatitis delta virus: structural analysis of RNA transcription intermediates. J. Mol. Biol., 291, 283–294. [DOI] [PubMed] [Google Scholar]
- 13.Lafontaine D.A., Ananvoranich,S. and Perreault,J. (1999) Presence of a coordinated metal ion in a trans-acting antigenomic delta ribozyme. Nucleic Acids Res., 27, 3236–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih I. and Been,M.D. (1999) Ribozyme cleavage of a 2′,5′-phosphodiester linkage: mechanism and a restricted divalent metal-ion requirement. RNA, 5, 1140–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suh Y., Kumar,P.K., Taira,K. and Nishikawa,S. (1993) Self-cleavage activity of the genomic HDV ribozyme in the presence of various divalent metal ions. Nucleic Acids Res., 21, 3277–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercure S., Lafontaine,D.A., Ananvoranich,S. and Perreault,J. (1998) Kinetic analysis of delta ribozyme cleavage. Biochemistry, 37, 16975–16982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perrotta A.T. and Been,M.D. (1998) A toggle duplex in hepatitis delta virus self-cleaving RNA that stabilizes an inactive and a salt-dependent pro-active ribozyme conformation. J. Mol. Biol., 279, 361–373. [DOI] [PubMed] [Google Scholar]
- 18.Ananvoranich S., Lafontaine,D.A. and Perreault,J. (1999) Mutational analysis of the antigenomic trans-acting delta ribozyme: the alterations of the middle nucleotides located on the P1 stem. Nucleic Acids Res., 27, 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrzesinski J., Legiewicz,M. and Ciesiolka,J. (2000) Mapping of accessible sites for oligonucleotide hybridization on hepatitis delta virus ribozymes. Nucleic Acids Res., 28, 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore M.J. and Sharp,P.A. (1992) Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science, 256, 992–997. [DOI] [PubMed] [Google Scholar]
- 21.Shih I. and Been,M.D. (2000) Kinetic scheme for intermolecular RNA cleavage by a ribozyme derived from hepatitis delta virus RNA. Biochemistry, 39, 9055–9066. [DOI] [PubMed] [Google Scholar]
- 22.Perrotta A.T., Nikiforova,O. and Been,M.D. (1999) A conserved bulged adenosine in a peripheral duplex of the antigenomic HDV self-cleaving RNA reduces kinetic trapping of inactive conformations. Nucleic Acids Res., 27, 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deschenes P., Lafontaine,D.A., Charland,S. and Perreault,J. (2000) Nucleotides –1 to –4 of hepatitis delta ribozyme substrate increase the specificity of ribozyme cleavage. Antisense Nucleic Acid Drug Dev., 9, 105–116. [DOI] [PubMed] [Google Scholar]
- 24.Kawakami J., Yuda,K., Suh,Y., Kumar,K.R., Nishikawa,F., Maeda,H., Taira,K., Ohtsuka,E. and Nishikawa,S. (1996) Constructing an efficient trans-acting genomic HDV ribozyme. FEBS Lett., 394, 132–136. [DOI] [PubMed] [Google Scholar]
- 25.Traut T.W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem., 140, 1–22. [DOI] [PubMed] [Google Scholar]
- 26.Huang Z.S., Ping,Y.H. and Wu,H.N. (1997) An AU at the first base pair of helix 3 elevates the catalytic activity of hepatitis delta virus ribozymes. FEBS Lett., 413, 299–303. [DOI] [PubMed] [Google Scholar]
- 27.Perrotta A.T. and Been,M.D. (1992) Cleavage of oligoribonucleotides by a ribozyme derived from the hepatitis δ virus RNA sequence. Biochemistry, 31, 16–21. [DOI] [PubMed] [Google Scholar]
- 28.Stage-Zimmermann T.K. and Uhlenbeck,O.C. (1998) Hammerhead ribozyme kinetics. RNA, 4, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y. and Breaker,R.R. (1999) Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc., 121, 5364–5372. [Google Scholar]
- 30.Richens D.T. (1997) The Chemistry of Aqua Ions. Wiley, New York, NY.
- 31.Scott W. and Klug,A. (1996) Ribozymes: structure and mechanism in RNA catalysis. Trends Biochem. Sci., 21, 220–224. [PubMed] [Google Scholar]
- 32.Warashina M., Takagi,Y., Stec,W.J. and Taira,K. (2000) Differences among mechanisms of ribozyme-catalyzed reactions. Curr. Opin. Biotechnol., 11, 354–362. [DOI] [PubMed] [Google Scholar]
- 33.Brown R.S., Hingerty,B.E., Dewan,J.C. and Klug,A. (1983) Pb(II)-catalyzed cleavage of the sugar-phosphate backbone of yeast tRNAPhe –implications for lead toxicity and self-splicing RNA. Nature, 303, 543–546. [DOI] [PubMed] [Google Scholar]
- 34.Rubin J.R. and Sundaralingam,M. (1983) Lead ion binding and RNA chain hydrolysis in phenylalanine tRNA. J. Biomol. Struct. Dyn., 1, 639–646. [DOI] [PubMed] [Google Scholar]
- 35.Brown R.S., Dewan,J.C. and Klug,A. (1985) Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry, 24, 4785–4801. [DOI] [PubMed] [Google Scholar]
- 36.Krzyzosiak W.J., Marciniec,T., Wiewiórowski,M., Romby,P., Ebel,J.P. and Giege,R. (1988) Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry, 27, 5771–5777. [DOI] [PubMed] [Google Scholar]
- 37.Marciniec T., Ciesiolka,J., Wrzesinski,J., Wiewiórowski,M. and Krzyzosiak,W.J. (1989) Specificity and mechanism of the cleavages induced in yeast tRNAPhe by magnesium ions. Acta Biochim. Pol., 36, 183–194. [PubMed] [Google Scholar]
- 38.Ciesiolka J., Marciniec,T. and Krzyzosiak,W. (1989) Probing the environment of lanthanide binding sites in yeast tRNAPhe by specific metal-ion-promoted cleavages. Eur. J. Biochem., 182, 445–450. [DOI] [PubMed] [Google Scholar]
- 39.Wrzesinski J., Michalowski.D., Ciesiolka,J. and Krzyzosiak,W. (1995) Specific RNA cleavages induced by manganese ions. FEBS Lett., 374, 62–68. [DOI] [PubMed] [Google Scholar]
- 40.Roy G., Ananvoranich,S. and Perreault,J. (1999) Delta ribozyme has the ability to cleave in trans an mRNA. Nucleic Acids Res., 27, 942–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahm S.C. and Uhlenbeck,O.C. (1991) Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry, 30, 9464–9469. [DOI] [PubMed] [Google Scholar]
- 42.Hammann Ch., Hormes,R., Sczakiel,G. and Tabler M. (1997) A spermidine-induced conformational change of long-armed hammerhead ribozymes: ionic requirements for fast cleavage kinetics. Nucleic Acids Res., 25, 4715–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowrira B.M., Berzal-Herranz,A. and Burke,J.M. (1993) Ionic requirements for RNA binding, cleavage and ligation by the hairpin ribozyme. Biochemistry, 32, 1088–1095. [DOI] [PubMed] [Google Scholar]
- 44.Earnshaw D.J. and Gait,M.J. (1998) Hairpin ribozyme cleavage catalyzed by aminoglycoside antibiotics and the polyamine spermine in the absence of metal ions. Nucleic Acids Res., 26, 5551–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]