Abstract
Taro, Colocasia esculenta, is one of the world’s oldest root crops and is of particular economic and cultural significance in Hawai’i, where historically more than 150 different landraces were grown. We developed a genome-wide set of more than 2400 high-quality single nucleotide polymorphism (SNP) markers from 70 taro accessions of Hawaiian, South Pacific, Palauan, and mainland Asian origins, with several objectives: 1) uncover the phylogenetic relationships between Hawaiian and other Pacific landraces, 2) shed light on the history of taro cultivation in Hawai’i, and 3) develop a tool to discriminate among Hawaiian and other taros. We found that almost all existing Hawaiian landraces fall into 5 monophyletic groups that are largely consistent with the traditional Hawaiian classification based on morphological characters, for example, leaf shape and petiole color. Genetic diversity was low within these clades but considerably higher between them. Population structure analyses further indicated that the diversification of taro in Hawai’i most likely occurred by a combination of frequent somatic mutation and occasional hybridization. Unexpectedly, the South Pacific accessions were found nested within the clades mainly composed of Hawaiian accessions, rather than paraphyletic to them. This suggests that the origin of clades identified here preceded the colonization of Hawai’i and that early Polynesian settlers brought taro landraces from different clades with them. In the absence of a sequenced genome, this marker set provides a valuable resource towards obtaining a genetic linkage map and to study the genetic basis of phenotypic traits of interest to taro breeding such as disease resistance.
Keywords: genotyping-by-sequencing, phylogeny, population structure
Taro, Colocasia esculenta (L.) Schott, has served as a food source for an estimated 28000 years (Loy et al. 1992) and is an ancient crop, cultivated for over 9000 years (Rao et al. 2010). Ranking among the world’s most important root crops, its starchy underground stem or corm serves as a staple food in many tropical and subtropical areas, where it is grown across a large range of environments (Caillon et al. 2006). Typically, taro is grown using vegetative propagules rather than from seed, because most areas of the world where it is cultivated lack the specialized insects that serve as natural pollinators.
Taro is believed to have originated in the Indo-Malayan region, although it is unclear whether domestication occurred once or multiple times. The distribution of natural pollinators, and of species in the genus Colocasia itself, point to northeast India, Southeast Asia, or New Guinea as the origin of taro as a species (Matthews 1995). Widespread distribution of wild taro may have facilitated multiple, independent domestication events in India, Southern China, Melanesia, and northern Australia (Lebot 2009; Chaïr et al. 2016). Asian taros display the greatest genetic diversity, a trend consistently observed across RAPD markers (Irwin et al. 1998), isozymes (Lebot and Aradhya 1991), AFLP markers (Kreike et al. 2004; Lebot et al. 2004), and microsatellites (Chaïr et al. 2016). Within the Pacific Island region, a comprehensive survey of taro germplasm identified the greatest genetic diversity in accessions from New Guinea and the Solomon Islands (Mace et al. 2006). With multiple ploidy levels (2n=2x=28, 2n=3x=42, and 2n=4x=56), mainland Asia is supported also as a center of domestication by cytological evidence, whereas only diploids are found in the Pacific Islands (Coates et al. 1988; Kreike et al. 2004).
Despite the Asiatic origins of taro, it is not a staple crop in mainland Asia. In fact, it is much more important historically in the South and East Pacific (Onwueme 1999; Rao et al. 2010). Humans have formed strong cultural connections to taro, especially in the epistemology of the peoples of the Pacific Islands. For example, taro is considered the “mother of life” in Palau and is an important food to share with people in gatherings on special occasions in American Samoa (Tipton et al. 1993). In many other Pacific Islands, taro is used as part of religious ceremonies, celebrations, and funerals. The dispersal history of taro in the Pacific has not been studied systematically, but there is evidence of taro cultivation during the Lapita era (ca. 3050–2500 years BP) as far east as Fiji (Horrocks and Nunn 2007). Taro was brought to Hawai‘i by Polynesian seafarers, who most likely settled Hawai‘i between 1000 and 1250 AD (Wilmshurst et al. 2011; Kirch 2014) and used it as a staple food during their expansion across the Pacific. Known as “kalo” in Hawaiian, taro is deeply bound to Hawaiian culture, with specific landraces having particular uses as food, medicine, or in religious ceremonies (Handy 1940; Abbott 1992). The significance of taro in Hawai‘i is attested to by its prominent position in the creation chant Kumulipo (Beckwith 1951).
Due to extensive cultivation, more than 150 landraces of taro were estimated to have existed in Hawai‘i prior to European contact. Of these, approximately 70 have been preserved and are grown alongside cultivars and landraces—lineages that have and have not undergone modern breeding, respectively—more recently introduced from the South Pacific and mainland Asia. Drawing on a collection of 84 distinct taro accessions (69 native to Hawai‘i), Whitney et al. (1939) developed a classification system that mostly maintained traditional Hawaiian group names based on origin, use and vegetative morphology. The eight main groups according to this system are 1) Mana (characterized by branched corms, mana meaning ‘branched’ in Hawaiian), 2) Piko (with leaf blades cut to the petiole attachment or piko), 3) Kāī (with pendant leaves), (4) Lauloa (meaning ‘long leaf’, with large leaves), 5) ‘Ele‘ele (‘ele meaning ‘dark’, referring to the petiole), 6) ‘Ula‘ula (‘ula meaning ‘red’, referring to the petiole), 7) Manini (with striped petioles, named after the convict tang Acanthurus triostegus, a striped fish), and 8) Lehua (with reddish-purple corm color, similar to the flower of the ‘Ōhi‘a tree, Metrosideros polymorpha).
Despite the cultural importance of taro in Hawai‘i, production has steadily declined over the 20th century, with the current area under cultivation approximately one third of what it was in 1900 (Cho et al. 2007). However, interest in growing taro is increasing, mostly due to the cultural revitalization efforts of native Hawaiian communities, and the recognition of taro as a medicinal crop (Brown et al. 2005; Reyad-ul-Ferdous et al. 2015). Total production of taro in Hawai‘i for 2015 was valued at $2.4 million, up from $1.9 million in 2014 (USDA National Agricultural Statistics Service 2015), and this market may further increase as taro remains very popular in Hawaiian cuisine.
The use of different taro landraces has been a part of Hawaiian agriculture and society for centuries. Understanding the relationships among the different landraces, modern cultivars, and hybrids is thus essential to creating both breeding and conservation plans. Efforts to preserve the original Hawaiian landraces will benefit from using DNA-based methods for genetic stock identification, a goal that proved previously intractable using unpublished SSR data (Supplementary Material and Supplementary Tables S1 and S2). Moreover, despite the extraordinary variety of landraces and dramatic subsequent losses since the time of first European contact, a genome-wide exploration of genetic relatedness and diversity of Hawaiian taro is still missing. To fill this gap, this study aims to investigate the phylogenetic relationships among the majority of extant Hawaiian landraces, as well as other Pacific and Asian taro lineages. In doing so, we also sought to elucidate the cultivation history of taro in Hawai‘i, evaluate and update the morphology-based classification system proposed by Whitney et al. (1939) using genetic data, and determine if a subset of genetic markers developed in this study could be used to efficiently identify Hawaiian landraces. In addition, the present dataset may prove valuable to advance previous efforts using SNP markers to construct a genetic linkage map of taro (Soulard et al. 2017), and study resistance against diseases threatening taro production around the world (Shintaku et al. 2016).
Materials and Methods
Sample Collection, DNA Extraction and Sequencing
A total of 77 taro accessions—63 of Hawaiian, 6 of South Pacific, 6 of Palauan, and 2 of mainland Asian origin—were sourced from the germplasm collections at Moloka‘i and Hawai‘i Islands and subjected to genotyping-by-sequencing (GBS, Elshire et al. 2011). These “pure-lineage” accessions represent the majority of old Hawaiian landraces in existence today, as well as all extant, morphologically recognized groups of Hawaiian taro (Whitney et al. 1939). In contrast, the South Pacific and Asian landraces and cultivars were introduced to Hawai‘i more recently, after the arrival of European and Asian immigrants.
Concurrently, 2 additional, previously unpublished datasets were produced and used here to improve single nucleotide polymorphism (SNP) calling: 1) GBS data of 113 Hawaiian hybrid cultivars, raising the total number of GBS samples to 190, and 2) restriction-site associated DNA sequencing (RAD-seq, Davey and Blaxter 2010) data obtained from cultivars #230 and #255 (Shintaku et al. 2016), as well as two pools of their progeny which were resistant and non-resistant to Taro Leaf Blight (TLB), respectively.
Genomic DNA was isolated from fresh or freeze-dried leaf tissue using the Qiagen Plant DNeasy Mini kit or Macherey-Nagel Nucleospin Plant II kit according to each manufacturer’s protocol. Libraries for both GBS and RAD-seq were prepared using the restriction enzyme PstI. GBS was conducted at Cornell University’s Genomic Diversity Facility (Ithaca, NY) on 2 flow cell lanes of an Illumina HiSeq 2500 with 100 bp single end reads. Across all 190 GBS samples, approximately 410 million usable reads were obtained. RAD-seq was performed at Floragenex (Portland, OR) on an Illumina Genome Analyzer IIx system. Parent and progeny libraries were sequenced during two separate runs on one flow cell lane each, using 95 bp paired-end and single reads, respectively, and which yielded 95 million reads. The software GBSX v1.1.4 (Herten et al. 2015) was applied to demultiplex and trim barcodes from both GBS and RAD-seq data.
SNP Calling and Filtering
Preliminary SNP calling was conducted using GBS data from all 190 samples with the UNEAK pipeline (Lu et al. 2013) implemented in TASSEL v3.0.160. Parameters employed in this analysis are described in Supplementary Table S3. The dataset was subsequently reduced to the 77 pure-lineage samples, and after filtering sites by minor allele frequency ≥0.1 and minimum mean depth of ≥10 reads using VCFtools v0.1.14 (Danecek et al. 2011), 70 samples with more than 400 sites were retained (Table 1).
Table 1.
List of 70 Hawaiian, Southern Pacific, Palauan, and Asian taro accessions included in the present study
| Accession | No. | Traditional classification | Revised classification | Geographic Origin |
|---|---|---|---|---|
| ‘Iliuaua (“Pake”) | 4 | — | Asian | ? |
| Bun-long | 5 | — | Asian | China |
| ‘Āweu (“wild taro”) | 6 | — | — | Hawai‘i |
| Kakakura | 7 | — | ‘Ula‘ula | South Pacific |
| Mana ‘Ulu | 8 | Mana | Mana | Hawai‘i |
| Mana ‘Ōpelu | 9 | Mana | Mana | Hawai‘i |
| Mana Uliuli | 11 | Mana | Mana | South Pacific |
| Mana ‘Ula‘ula | 12 | Mana | Mana | Hawai‘i |
| Mana Lauloa | 13 | Mana | Mana | Hawai‘i |
| Mana Ke‘oke‘o | 14 | Mana | Mana | Hawai‘i |
| Piko Lehua Apei | 16 | Piko | Mana | Hawai‘i |
| Piko Kea | 18 | Piko | Lehua | Hawai‘i |
| Piko Ke‘oke‘o | 19 | Piko | Lehua | Hawai‘i |
| Piko Uaua | 20 | Piko | Lehua | Hawai‘i |
| Piko Uliuli | 21 | Piko | Lehua | Hawai‘i |
| Piko ‘Ele‘ele | 22 | Piko | Lauloa-Manini | Hawai‘i |
| ‘Elepaio Hā Kea | 23 | — | Lehua | Hawai‘i |
| Uahiapele | 24 | — | Lauloa-Manini | Hawai‘i |
| Manapiko | 25 | — | Mana | Hawai‘i |
| Tahitian | 26 | — | Mana | South Pacific |
| Kāī Uliuli | 27 | Kāī | Kāī | Hawai‘i |
| Kāī Kea | 29 | Kāī | Kāī | Hawai‘i |
| ‘Apuwai | 30 | — | Lehua | Hawai‘i |
| Pi‘i‘ali‘i | 32 | — | Lehua | Hawai‘i |
| Pa‘akai | 33 | — | Lehua | Hawai‘i |
| Moana | 34 | — | Lehua | Hawai‘i |
| Akuugawai | 35 | — | Lauloa-Manini | South Pacific |
| Lauloa ‘Ele‘ele ‘Ōma‘o | 36 | Lauloa | Lauloa-Manini | Hawai‘i |
| Lauloa ‘Ele‘ele ‘Ula | 37 | Lauloa | Lauloa-Manini | Hawai‘i |
| Lauloa Palakea ‘Ele‘ele | 38 | Lauloa | Lauloa-Manini | Hawai‘i |
| Lauloa Palakea ‘Ula | 39 | Lauloa | Lauloa-Manini | Hawai‘i |
| Lauloa Palakea Papamū | 40 | Lauloa | Lauloa-Manini | Hawai‘i |
| Lauloa Palakea Ke‘oke‘o | 41 | Lauloa | Lehua | Hawai‘i |
| Lauloa Ke‘oke‘o | 42 | Lauloa | Lehua | Hawai‘i |
| ‘Ele‘ele Mākoko | 43 | ‘Ele‘ele | Lehua | Hawai‘i |
| ‘Ele‘ele Naioea | 44 | ‘Ele‘ele | Lehua | Hawai‘i |
| Manini ‘Ōwali | 45 | — | Lauloa-Manini | Hawai‘i |
| Kūmū ‘Ele‘ele | 46 | — | Lauloa-Manini | Hawai‘i |
| Nāwao | 47 | — | Lehua | Hawai‘i |
| ‘Ula‘ula Kūmū | 48 | ‘Ula‘ula | ‘Ula‘ula | Hawai‘i |
| ‘Ula‘ula Poni | 49 | ‘Ula‘ula | ‘Ula‘ula | Hawai‘i |
| ‘Ula‘ula Moano | 50 | ‘Ula‘ula | ‘Ula‘ula | Hawai‘i |
| Niue ‘Ula‘ula | 51 | — | ‘Ula‘ula | South Pacific |
| ‘O‘opukai | 52 | — | ‘Ula‘ula | Hawai‘i |
| Manini Uliuli | 53 | Manini | Lauloa-Manini | Hawai‘i |
| Manini Kea | 54 | Manini | Lauloa-Manini | Hawai‘i |
| Pāpākolea Koa‘e | 56 | — | Lehua | Hawai‘i |
| Nihopu‘u | 58 | — | Lauloa-Manini | Hawai‘i |
| Manini ‘Ōpelu | 59 | — | Lauloa-Manini | Hawai‘i |
| Lehua Ke‘oke‘o | 64 | Lehua | Lehua | Hawai‘i |
| Lehua ‘Ele‘ele | 65 | Lehua | Lehua | Hawai‘i |
| Lehua Pala‘i‘i | 66 | Lehua | Lehua | Hawai‘i |
| ‘Apowale | 67 | — | Lehua | Hawai‘i |
| Wehiwa | 68 | — | Lehua | Hawai‘i |
| Papapueo | 69 | — | Lehua | Hawai‘i |
| Kū‘oho | 70 | — | Lehua | Hawai‘i |
| Māea | 72 | — | Lehua | Hawai‘i |
| Haokea | 73 | — | Lehua | Hawai‘i |
| Hāpu‘u | 75 | — | Lehua | Hawai‘i |
| False Lihilihimōlina | 78 | — | — | Hawai‘i |
| Mana ‘Oko‘a | 80 | — | Mana | Hawai‘i |
| Moi | 81 | — | Lehua | Hawai‘i |
| Pololū | 84 | — | Lehua | Hawai‘i |
| Moi ‘Ula‘ula | n/a | n/a | Lehua | Hawai‘i |
| Makalau (Moloka‘i) | n/a | n/a | Mana | Hawai‘i |
| Ngesuas (P-1) | n/a | n/a | Palauan | Palau |
| Ochelochel (P-7) | n/a | n/a | Palauan | Palau |
| Ngeruuch (P-10) | n/a | n/a | Palauan | Palau |
| Merii (P-12) | n/a | n/a | Palauan | Palau |
| Dirratengadik (P-20) | n/a | n/a | Palauan | Palau |
Number, traditional (morphology-based) classification, and presumed geographic origin according to Whitney et al. (1939). The revised classification is based on phylogenetic analyses presented in this study. Dashes denote accessions that could not be assigned unambiguously under each classification; n/a indicates accessions not covered by Whitney et al. 1939. South Pacific and Asian landraces and cultivars were introduced to Hawai‘i in modern times.
In a second step, a de novo reference was assembled from all GBS and RAD-seq data combined using IDBA-UD v1.1.2 (Peng et al. 2012). GBS reads of all samples were aligned to the reference with bowtie2 (Langmead and Salzberg 2012) using a minimum alignment length of 48 nt, followed by SNP calling using TASSEL v5.2.17 (Bradbury et al. 2007, GBS v2 pipeline). This dataset was subset to contain the same 70 samples identified during preliminary SNP calling. Further filtering steps were then performed on the basis of per-site sequencing depth, data missingness, and site-specific heterozygosity. Sequencing depth and data missingness were calculated using VCFtools, while site-specific heterozygosity was calculated with the VariantsToTable tool included in GATK v3.6 (McKenna et al. 2010). Data were visualized as 3D plots using the R package plot3D (Soetaert 2016). Excessive heterozygosity was observed for higher read depths, likely because different forms of repeats map to the same reference contig. Thus, SNPs exceeding 200 per-site sequencing depth were excluded from the final analysis. Per-site sequencing depth minimum was set to 10 reads, and maximum genotype missingness to 20%.
Last, the remaining SNPs were filtered by allele characteristics using VCFtools, applying a range of thresholds regarding minor allele count (mac) and minor allele frequency (maf). A threshold of mac ≥ 2 was finally chosen for providing good balance between the number of SNPs and allelic diversity (Supplementary Table S4), yielding 2447 high-quality SNPs. Additional datasets were obtained using the same filtering parameters as outlined above by constructing alternative references from a subset of GBS data only, and publicly available taro transcriptome data. However, these datasets provided fewer SNPs and lower phylogenetic resolution (see Supplementary Material and Supplementary Tables S5 and S6) than the primary dataset.
Phylogenetic Analyses and Classification
To reconstruct the phylogenetic relationships among accessions, we conducted maximum likelihood analyses with RAxML v8.1.16 (Stamatakis 2014). The data was first converted from VCF to FASTA format using NGSToolsApp.jar v2.1.5 (Duitama et al. 2014), considering only 2088 homozygous positions (heterozygous sites were replaced with unknown values). The best tree of 100 maximum likelihood inferences starting from randomized maximum parsimony trees was outfitted with confidence values derived from 1000 regular bootstrap replicates. Both final maximum likelihood inferences and bootstrapping runs were performed under the GTRCAT model. Trees were drawn with iTOL v3 (Letunic and Bork 2016, http://itol.embl.de). Clade composition inferred by phylogenetic analyses was compared to the classification system developed by Whitney et al. (1939), and the phylogenetic distribution of morphological traits assumed to be diagnostic was evaluated.
To visualize phylogenetic conflict and uncertainty in the data, network-based analyses were conducted with SplitsTree v4.14.2 (Huson and Bryant 2006), using the NeighborNet approach under the Jukes-Cantor model. Bootstrap networks were computed based on 250 bootstrap replicates. Population splits and admixture events between populations were assessed using TreeMix v1.12 (Pickrell and Pritchard 2012). For the latter, input files were prepared using NGSToolsApp.jar v2.1.5, assigning samples to one of seven populations as inferred by the RAxML analyses above. Maximum likelihood trees were built assuming 0, 1, 2, and 3 migration events, respectively.
Genetic Differentiation and Population Structure
Overall genetic similarity among accessions was assessed by principal component analysis (PCA). First, SNP data was converted from VCF to PLINK format using Plink v1.9 (Chang et al. 2015), coding half-calls as missing data. Principal components were then computed and plotted with the packages adegenet (Jombart 2008) and ade4 (Dray and Dufour 2007) in R.
Population structure was analyzed with the program STRUCTURE v2.3.4 (Pritchard et al. 2000). SNP data filtered by maf ≥ 0.1 instead of the default mac ≥ 2 was used here, consisting of 1030 SNPs. VCF files were converted to STRUCTURE format files by PGDSpider v2.1.0.0 (Lischer and Excoffier 2012). Cluster (K) values were set to 2 through 10, and 5 iterations each were run with a burn-in period of 10000 and Markov chain Monte Carlo repetitions set to 50000. Results were uploaded to the Structure Harvester website (Earl et al. 2012, http://taylor0.biology.ucla.edu/structureHarvester/) where an optimal K was calculated using the Evanno method (Evanno et al. 2005), although we also considered the number of clades found by phylogenetic analysis to evaluate the informativeness of K values. STRUCTURE results were also uploaded to the CLUMPAK main pipeline (Kopelman et al. 2015, http://clumpak.tau.ac.il/index.html) for visualization and output. CLUMPAK provided singular membership coefficients for each cluster, calculated over all 5 iterations per K and individual. Assignment to clusters was compared to known membership in phylogenetic clades.
Results
Phylogenetic Analyses and Classification
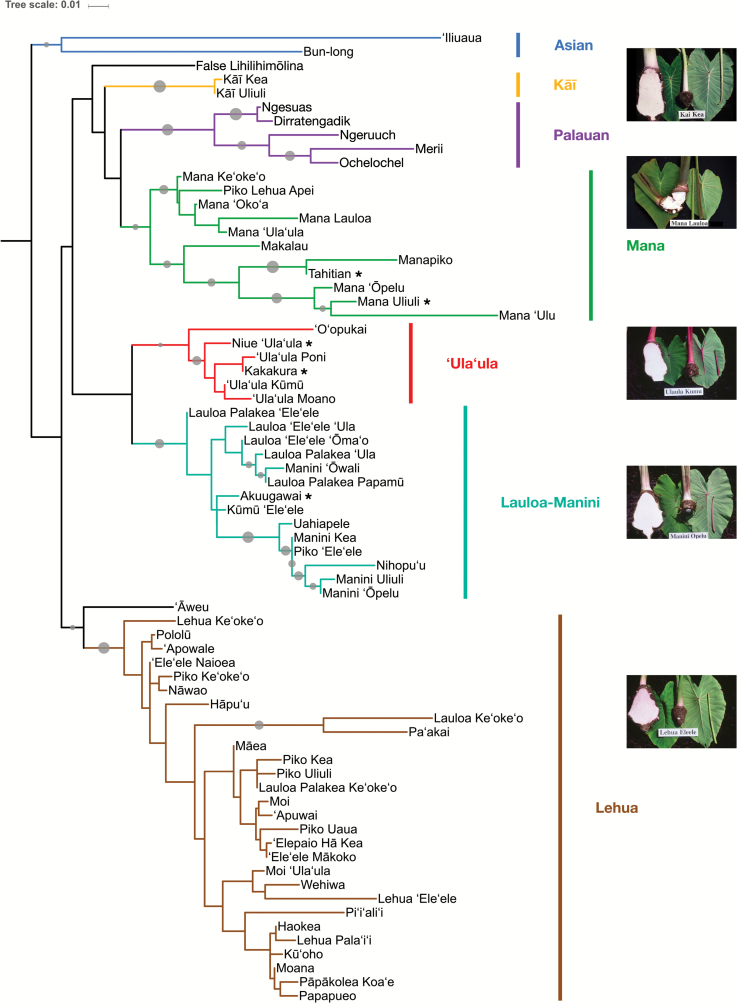
Our phylogenetic analyses recovered five clades among the Hawaiian taro accessions represented in this study, which are referred to here as Lehua, Lauloa-Manini, ‘Ula‘ula, Mana, and Kāī (Figure 1). These names correspond to the traditional Hawaiian classification system based on shared morphological characters (Whitney et al. 1939), which is largely in agreement with our phylogenetic results (Table 1). The Palauan accessions formed an additional clade, while the two Asian accessions were defined as the root. Bootstrap support for clades was generally high, especially with regard to the Palauan, Lauloa-Manini, and Kāī groups. The ‘Ula‘ula and Lehua groups were also recovered consistently, although more divergent accessions (e.g. ‘O‘opukai in ‘Ula‘ula) seemed to suppress the support values for their clades. The Mana group was moderately supported by the primary dataset (Figure 1), but appeared paraphyletic in some analyses using alternative datasets (Supplementary Figure S1). The latter also suggests that the Lauloa-Manini clade may consist of 2 smaller clades, Lauloa and Manini, in a sister-group relationship.
Figure 1.
Phylogenetic relationships of 70 Hawaiian, South Pacific, Palauan and Asian taro accessions inferred from 2088 homozygous, high-quality single nucleotide polymorphisms (SNPs). The maximum likelihood reconstruction was conducted using RAxML under the GTRCAT model, and illustrates that Hawaiian accessions fall into 5 generally well-supported clades. The tree was rooted by 2 Asian accessions. Clade names were given in agreement with the traditional, morphology-based Hawaiian classification system. Grey circles indicate confidence values derived from 1000 bootstrap replicates, and are proportional in size to values from 50–100. Asterisks denote accessions which were introduced to Hawai’i from the South Pacific in modern times. Images of accessions representative for each clade are reproduced with permission by photographers J. Sugano and S. Fukuda, http://www.ctahr.hawaii.edu/site/taro.aspx.
The six accessions of South Pacific Origin were found firmly embedded in clades otherwise consisting of Hawaiian landraces (Figure 1): Tahitian and Mana Uliuli grouped with Mana, Kakakura and Niue ‘Ula‘ula with ‘Ula‘ula, and Akuugawai with Lauloa-Manini. Only 2 accessions, ‘Āweu and False Lihilihimōlina, could not be assigned reliably to any clade. The membership of accessions in phylogenetic clades, along with their traditional, morphology-based classification, is given in Table 1.
In contrast to the high confidence obtained for the clades, the backbone of the tree, i.e., the relationship between clades, remained unresolved as indicated by a lack of bootstrap support values greater than 50 at the deeper nodes. This pattern of well-defined clades and a lack of resolution between clades was also reflected in the NeighborNet analyses that showed distinct subgraphs for each clade emanating from a star-like center (Supplementary Figures S2 and S3). Resolution within clades was found to range from moderate (e.g., Mana, Palauan) to low (Lehua, ‘Ula‘ula, and parts of Lauloa-Manini). Weak resolution in the clades was exacerbated by some accessions displaying nearly identical genotypes in the primary dataset (1–3 allelic differences; 1 pair appeared identical). However, due to the relatively high number of undetermined positions in these accessions, the true number of differences is likely to be higher. Since Manini Kea and Piko ‘Ele‘ele, the only genotypically indistinguishable accessions, are also morphologically very distinct, we assume no identical clones exist in this dataset.
Genetic Differentiation and Population Structure
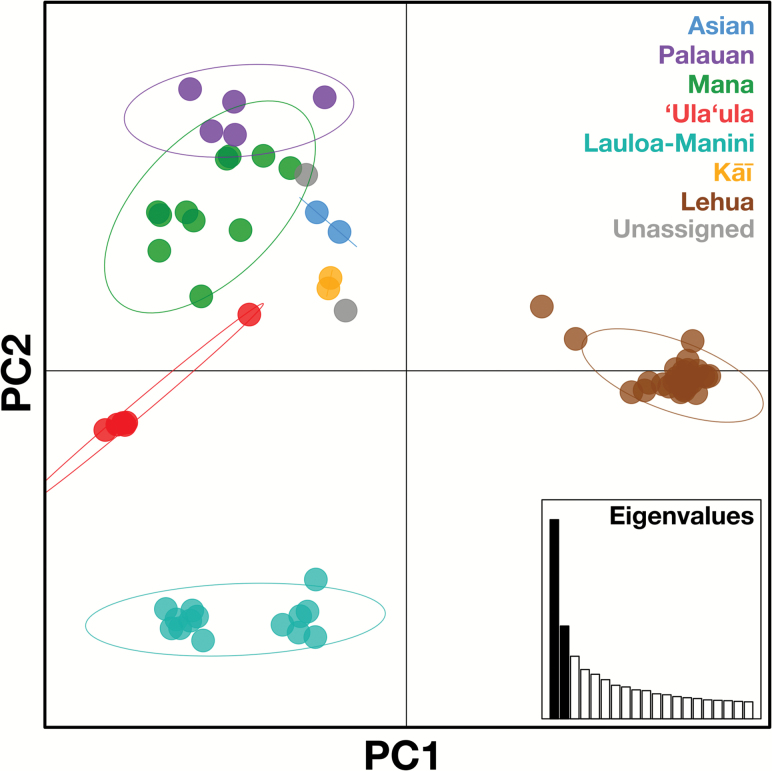
Consistent with the broad phylogenetic pattern, mapping the genetic similarity among samples by principal component analysis (PCA) revealed low genetic diversity in several clades, and clear differentiation among clades (Figure 2). The first principal component, which accounted for 16.3% of the total variation, separated a tightly clustered group composed of the Lehua accessions from all others. The second principal component, accounting for 7.6% of the total variation, isolated Lauloa-Manini from all other accessions. The remaining accessions were separated more gradually along both axes, and thus exhibited higher within-clade diversity, but still formed clusters consistent with phylogenetic clades, displaying no or only moderate overlap between groups. The PCA also supported Lauloa-Manini consisting of 2 distinct sub-groups, Lauloa and Manini (compare Supplementary Figure S1).
Figure 2.
Principal component analysis (PCA) of 70 taro accessions based on 2447 high-quality SNPs. Strong genetic differentiation is apparent between the Lehua group and all other accessions (first component, 16.3% of variation), and between the Lauloa–Manini group and the remaining accessions (second component, 7.6% of variation). Colors highlight clade membership following Figure 1, with oval outlines indicating 95% inertia ellipses. The inset shows the relative eigenvalues of the first 20 principal components, with the represented components in black.
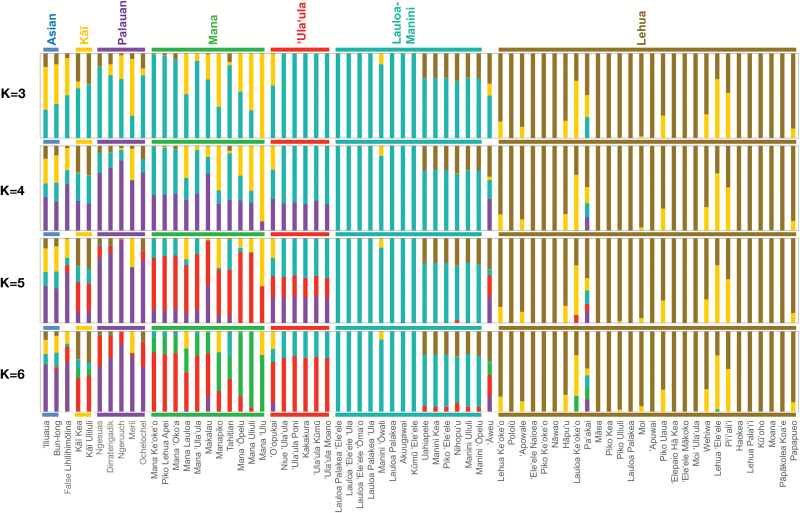
To assess the genetic structure among Hawaiian taros in more detail and identify admixture between clades and individual accessions, we applied the Bayesian inference program STRUCTURE (Pritchard et al. 2000). This method assigns samples to one (or proportionally to several, in the case of admixture) of K clusters based on the allele frequencies across all loci, without relying on prior knowledge of ancestry or origin. We found a primary partition between a cluster of accessions corresponding to clade Lehua, and all remaining accessions at K = 3 (Figure 3). A secondary split between accessions of the Lauloa-Manini clade and all other accessions was becoming notable at this K, which was deemed optimal by the post-hoc Evanno statistic (Evanno et al. 2005) for most runs. However, in line with the phylogenetic results, increasing K resulted in the further separation of the ‘Ula‘ula, Mana and Palauan accessions (K = 5). Additional, unambiguous clusters did not emerge beyond K = 6, which matched our intuition based on the number of clades recovered phylogenetically. While all clades were identifiable by a distinct pattern of group membership coefficients (proportions), only Lehua and Lauloa-Manini (in particular the subgroup Lauloa) accessions were defined by high membership coefficients for a single cluster. Similar results were achieved using datasets generated by alternative SNP calling methods (Supplementary Figure S5). Further support for hybridization between clades was obtained by inferring admixture events using the TreeMix software, most notably between ‘Ula‘ula and the Manini subgroup (Supplementary Figure S6).
Figure 3.
Population structure analysis assuming admixture and K = 3–6 clusters from top to bottom. Taro accessions are represented by vertical bars, which are partitioned into colored segments representing estimated membership fractions for each cluster. Horizontal bars indicate clade membership according to the phylogenetic analysis (Figure 1). Dominant segments were color-matched to clades when possible.
Regarding individual landraces, several accessions stood out by displaying strong signatures of admixture. In line with the phylogenetic results, this admixture signal applied to the unclassifiable accessions False Lihilihimōlina and ‘Āweu, as well as ‘O‘opukai, whose phylogenetic position as part of the ‘Ula‘ula clade was only weakly supported (Figure 1). Other accessions characterized by signatures of admixture and long branches in the phylogenetic tree included Lauloa Ke‘oke‘o, Pa‘akai, Wehiwa, Lehua ‘Ele‘ele, and Pi‘i‘ali‘i, all of which fall into the Lehua clade.
Discussion
Clade Composition and Morphological Traits
We observed that the Hawaiian taro landraces contain sufficient genetic variation across the genome to discriminate among the majority of accessions, and identify several well-demarcated clades using phylogenetic analysis (Figure 1). The composition of these clades largely parallels the traditional classification based on morphological characters such as petiole color, leaf shape, and corm shape and color (Whitney et al. 1939, Table 1). For instance, as defined morphologically, the ‘Ula‘ula group is characterized by red petioles (‘ula meaning red in Hawaiian). Accessions regarded as group members (carrying binomial names beginning with ‘Ula‘ula) indeed proved to be closely related to each other, forming a monophyletic group together with accessions considered loose affiliates due to partially red petioles by Whitney et al. (1939). Similarly, Mana, Kāī, Lauloa, and Manini (the last two joined in a single clade, Lauloa-Manini, but with unclear relations to each other) were also found to be largely natural groups, with diagnostic morphological traits—the division of the parent corm into several branches (Mana), pendant leaf position (Kāī), large leaves (Lauloa), and striped petioles (Manini)—being mostly confined to one clade. This suggests that these traits are under strong genetic control, and were carefully maintained during centuries of cultivation. Compared to other vegetatively propagated crop plants like cassava (Elias et al. 2001; 2000), such broad agreement between genetic and morphological variation is unusual.
However, our phylogenetic analyses also revealed that most Hawaiian taro groups contain more members than accounted for by Whitney et al. (1939), often in the form of accessions previously left unclassified because they failed to unambiguously display diagnostic morphological traits. In addition to those within ‘Ula‘ula discussed above, these accessions include several new members of the Mana clade (e.g., Manapiko, which was considered neither a member of Mana nor Piko, as well as Mana ‘Oko‘a and Makalau). Similarly, the Lauloa-Manini clade contains several traditionally unaffiliated Hawaiian accessions – Manini ‘Ōwali, Kūmū ‘Ele‘ele (both with larger leaves), as well as Piko ‘Ele‘ele, Uahiapele, Nihopu‘u and Manini ‘Ōpelu. The last 3 possess streaked or striped petioles, suggesting that this trait is concentrated in an extended group Manini, which is well supported as a monophyletic group of its own (Figure 1). However, whether Lauloa forms a paraphyletic group with respect to Manini (as shown in the primary maximum likelihood tree, Figure 1), or is its sister group (as indicated by Figure 2 and additional phylogenetic analyses, Supplementary Figures S1–S3) remains ambiguous. We therefore suggest the provisional name Lauloa–Manini for this clade.
The theme of clades including accessions beyond the members of the morphology-based groups was especially pronounced in the Lehua group: only a few landraces with purple corm color were traditionally classified as Lehua (named after the red flower of the ‘Ōhi‘a tree), but phylogenetic analysis showed that this clade encompasses far more accessions, many of which display white to cream-colored corm and were previously left unclassified. Loss of corm color has been observed in hybrid cultivars with originally purple corm flesh after tissue-culturing (personal observation), indicating that this phenotype can revert by somatic mutation. The revised Lehua group also contains several accessions that were traditionally assigned to other groups (e.g., Lauloa Ke‘oke‘o), indicating that the relation between phylogenetic kinship and the traits described above is not always perfect. Indeed, the traditional groups Piko and ‘Ele‘ele are not supported by genetic data as monophyletic, with most of their accessions joining the Lehua group (except Piko Lehua Apei and Piko ‘Ele‘ele, which are found in the Mana and Lauloa-Manini clades, respectively). In traditional Hawaiian nomenclature, the Piko group is characterized by a leaf sinus cut to the point of attachment to the petiole, or a dark-colored spot on the upper surface of the leaf at the point of junction with the petiole (Whitney et al. 1939). ‘Ele‘ele on the other hand is morphologically defined by dark or black petioles (from Hawaiian ‘ele, meaning dark or black). While the observation of a controlled cross suggests that the Piko trait is genetically controlled (unpublished data), the Piko and ‘Ele‘ele phenotypes seem to have originated several times independently. Assuming that mutations at multiple loci may be able to produce these phenotypes, convergent evolution of (and subsequent selection for) these traits could be more likely to occur. In further support, primarily black or dark petioles are also found in accessions not formerly assigned to group ‘Ele‘ele, including accessions in Lauloa-Manini and Lehua. Hybridization, which is discussed further below, may be another mechanism leading to discrepancies between morphological variation and phylogenetic relationships. Weak correlation between morphology and genetic variation also has been reported as widespread in similar crops like cassava (Elias et al. 2001, 2000).
South Pacific and Unaffiliated Accessions
Interestingly, several accessions collected in 1927 by G. P. Wilder in the South Pacific were found nested within Mana, ‘Ula‘ula and Lauloa-Manini (Figure 1, indicated by asterisks). This circumstance has important implications for the cultivation history of Hawaiian taro. If the Hawaiian landraces had diversified from a single genotype, or only a few closely related ones, we would expect the South Pacific accessions to appear paraphyletic with respect to Hawaiian taros (i.e., they would branch off earlier from a single lineage leading to all Hawaiian landraces). Since this is not the case, we hypothesize that early settlers brought several genetically and morphologically diverse landraces to Hawai‘i (at least representatives of Mana, ‘Ula‘ula, and Lauloa-Manini – though future studies including a larger number of South Pacific accessions may turn up members of additional groups as well). In this scenario, both Hawaiian and South Pacific taro landraces are descendants of the same clades that were already cultivated as humans first moved into Polynesia. Similar to other canoe plants, these taro landraces may have been introduced during a single pulse of settlement of Hawai‘i (Wilmshurst et al. 2011), or successively in multiple arrivals (Kirch 2014; Cho et al. 2007) until contact with the rest of East Polynesia ceased for several centuries.
Two accessions that could not be assigned reliably to any clade, ‘Āweu and False Lihilihimōlina, also deserve further mention. Called “wild taro” by Whitney et al. (1939), ‘Āweu was historically widespread in the mountains and forests of Hawai‘i. Despite undesirable characteristics such as high acridity and long rhizomes, it was occasionally collected as food when other sources were scarce. The so-called wild taros in Melanesia were hypothesized by Lebot and Aradhya (1991) to be either clones from actual wild taros or ferals, i.e., escapes from cultivation. Based on this SNP analysis, ‘Āweu does appear to be genetically distinct from cultivated Hawaiian taro, but it seems unlikely that actual wild taro was able to reach Hawai‘i without human help. Instead, it might have escaped from cultivation early during the settlement of Hawai‘i, and its closest relatives were lost. On the other hand, False Lihilihimōlina in all likelihood does not represent an older Hawaiian lineage. The original Hawaiian landrace Lihilihimōlina has been described as having uniquely bi-colored corm flesh, lilac-purple at the center and surrounded by white (Whitney et al. 1939). However, the accessions currently identified as Lihilihimōlina in Hawaiian taro collections (including our sample) do not fit the description of this phenotype (Frank Matsuno, personal communication). Genetically, the accession in our study also did not resemble any of the Hawaiian landraces and was instead found unaffiliated to any major group (Figure 1), or closer to the Asian accessions in our phylogenetic analyses (Supplementary Figures S2 and S3). Moreover, its allelic composition points to a hybrid origin, possibly involving Asian stock (Figure 3). Both anecdotal and experimental evidence thus suggest that the original Lihilihimōlina has been lost and was replaced with a more recent hybrid (hence our designation “false”).
Evidence for Somatic Mutation and Hybridization
In contrast to the pattern of clear differentiation between clades, we found evidence for low genetic diversity within several clades. This is especially apparent in the lack of phylogenetic resolution (Figure 1) and tight clustering by genetic similarity (Figure 2) seen in the ‘Ula‘ula, Lauloa-Manini, and Lehua clades. Since taro is typically grown vegetatively from suckers (‘ohā) or petioles attached to the meristem of the upper corm (huli), clades may often be composed of close genetic relatives derived from somatic mutations. Indeed, according to Whitney et al. (1939) the Lauloa group is prone to somatic mutations, and the landraces Lauloa Palakea ‘Ula and Lauloa Palakea Papamū are considered to be somatic mutations that originated from Lauloa Palakea ‘Ele‘ele. A possible mechanism for this would be selection of desirable sports (phenotypically divergent plant parts resulting from somatic mutation) by farmers rather than crossing genotypes to create new cultivars. In the absence of specialized insect pollinators, such practice may have preserved the identity of clades over centuries. Accessions originating from somatic mutants appear to be old enough to have accumulated additional genetic changes. For instance, we detected 2–10 pairwise allelic differences between the 3 Lauloa landraces mentioned above. Other closely related accessions are distinguished by a similar number of differences, which is likely to be an underestimate due to the relatively high number of undetermined positions in these accessions. However, it remains unclear whether selection of mutants and vegetative propagation became widespread because pollination by hand was less efficient, or because of the ability to fix useful genotypes, making them more uniform and stable.
Lack of phylogenetic resolution also may be caused by hybridization. In particular, hybridization between members of different clades, occurring early in their evolutionary history, may obscure inter-clade relationships despite subsequent diversification within clades. Indeed, we detected compelling signatures of admixture in several clades using STRUCTURE (Figure 3), and found additional evidence in the TreeMix analyses (Supplementary Figure S6). While the majority of Lehua and Lauloa-Manini (especially the putative subgroup Lauloa) accessions were characterized by high membership coefficients for a single population cluster, accessions of other clades could not be assigned unambiguously. This indicates that Lehua and Lauloa-Manini are genetically more distinct (in line with the PCA results, Figure 2), while all other clades—particularly Mana, ‘Ula‘ula, and Kāī—have been subject to hybridization to some degree. For example, ‘Ula‘ula and the Manini subgroup shared a large fraction of alleles according to STRUCTURE (turquoise in Figure 3), suggesting hybridization between members of these clades. This scenario is supported by the inference of admixture events using the TreeMix software (Supplementary Figure S6). Considering this and similar patterns were found very consistently in all members, it seems likely admixture occurred early in the evolutionary history of these accessions, presumably before taro was introduced to Hawai‘i by Polynesian settlers. Note that the 2 Asian taros also showed a strong pattern of allelic diversity consistent with originating closer to the likely geographic center of origin for taro. Interestingly, several accessions within the genetically more uniform clade Lehua possess membership coefficients also pointing to a mixed genetic origin. Some of these have special roles in Hawaiian culture—for instance, Pi‘i‘ali‘i or royal taro was used in religious ceremonies, whereas Lauloa Ke‘oke‘o is known for its medical purposes (Whitney et al. 1939, Cho et al. 2007). As these taros have both a signature of admixture and very specific cultural uses, this might be an indication that these accessions were purposefully bred, possibly by crossing, as opposed to most other Hawaiian taros. In summary, the pattern of low to moderate genetic differentiation within clades, higher levels of differentiation between clades, and lack of phylogenetic resolution between clades was likely shaped by a combination of early hybridization and frequent propagation of somatic mutants. Such patterns of diversification by multiple processes have been observed in other cultivated plants with mixed modes of reproduction (sexual versus clonal) such as sweet potatoes (Roullier et al. 2013).
Conclusions
As a center of taro cultivation both presently and historically, Hawai‘i possesses a wealth of morphologically diverse taro landraces that were bred or selected for different uses. Exploring this diversity based on a genome-wide set of SNP markers, we made several discoveries concerning the phylogenetic relationships, cultivation history, and genetic basis of morphological traits in Hawaiian taros. Phylogenetic analyses, patterns of genetic similarity, and population structure suggest that Polynesian settlers introduced several genetically and morphologically distinct lineages of taro, which then likely further diversified by selection of mutants, genetic drift and occasional (possibly deliberate) hybridization. The underlying genetic diversity, although relatively low compared to cultivation centers closer to the geographic origin of taro, supports a considerable phenotypic diversity, which may have been carefully maintained through clonal propagation over centuries of cultivation.
By largely confirming, and in some cases revising traditional Hawaiian classification based on morphology, the SNP markers developed in this study hopefully will prove valuable for efforts to conserve the diversity of Hawaiian taros. As demonstrated, they can be used to reliably identify and discriminate among the majority of landraces, many of which have already been lost to introduced pests and devastating pathogens such as taro leaf blight. Although conventional breeding has been very successful to improve agronomically important traits in taro, these markers also may facilitate investigations of the genetic basis of relevant phenotypes including quality, taste, disease resistance, or abiotic stress resistance, and the development of effective genetic fingerprinting methods to further study genetic diversity and gene flow among taro landraces and cultivars outside of Hawai‘i.
Supplementary Material
Supplementary data are available at Journal of Heredity online.
Funding
This work was supported by the National Institute of Food and Agriculture at the United States Department of Agriculture (Hatch Project number 08029-H, managed by the College of Tropical Agriculture and Human Resources); the USDA Agriculture and Food Research Initiative project (grant number 2015–05806); and the USDA Cooperative Agreement (grant number 58-5320-4016). In addition, this material is based upon work supported by the National Science Foundation under grant number 1345247. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Conflict of Interest
The authors declare no conflict of interest.
Data Availability
Raw reads generated for this study can be found in the National Center for Biotechnology Information’s Sequence Read Archive (SRA) under BioProject PRJNA381383 (sequencing runs SRR5468958–63). SNP data and alignments used for phylogenetic analyses have been deposited at Dryad, http://dx.doi.org/doi:10.5061/dryad.r35kr.
Supplementary Material
Acknowledgments
We would like to acknowledge Christopher Bernabe, Agricultural Technician, who conducted the conventional breeding of taro to produce hybrids used for GBS analysis, Lukas Kambic and Steven Starnes, Research Specialists at the University of Hawai‘i at Hilo, for performing DNA extraction, purification and quantification, and Noa Kekuewa Lincoln, University of Hawai‘i at Manoa, for helpful comments on the manuscript.
References
- Abbott IA. 1992. La’au Hawai’i: Traditional Hawaiian uses of plants. Honolulu (HI): Bishop Museum Press. [Google Scholar]
- Beckwith MW, editor. 1951. The Kumulipo: A Hawaiian creation chant. HI: University of Hawai’i Press. [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 23:2633–2635. [DOI] [PubMed] [Google Scholar]
- Brown AC, Reitzenstein JE, Liu J, Jadus MR. 2005. The anti-cancer effects of poi (Colocasia esculenta) on colonic adenocarcinoma cells in vitro. Phytother Res. 19:767–771. [DOI] [PubMed] [Google Scholar]
- Caillon S, Quero-Garcîa J, Lescure JP, Lebot V. 2006. Nature of taro (Colocasia esculenta (L) Schott) genetic diversity prevalent in a Pacific Ocean island, Vanua Lava, Vanuatu. Genetic Resources and Crop Evolution. 53:1273–1289. [Google Scholar]
- Chaïr H, Traore RE, Duval MF, Rivallan R, Mukherjee A, Aboagye LM, Van Rensburg WJ, Andrianavalona V, Pinheiro de Carvalho MA, Saborio Fet al. 2016. Genetic Diversification and Dispersal of Taro (Colocasia esculenta (L) Schott). PLoS ONE. 11:e0157712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JJ, and Yamakawa RA. 2007. Hawaiian kalo, past and future. Agriculture Development in the American Pacific Project. 3:35–39. [Google Scholar]
- Coates DJ, Yen DE, Gaffey PM. 1988. Chromosome variation in taro, Colocasia esculenta. Implications for origin in the Pacific. Cytologia. 53:551–560. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST et al. ; 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics. 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Blaxter ML. 2010. RADSeq: next-generation population genetics. Brief Funct Genomics. 9:416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray S, Dufour AB. 2007. The ade4 package: implementing the duality diagram for ecologists. J Stat Soft. 22:1–20. [Google Scholar]
- Duitama J, Quintero JC, Cruz DF, Quintero C, Hubmann G, Foulquié-Moreno MR, Verstrepen KJ, Thevelein JM, Tohme J. 2014. An integrated framework for discovery and genotyping of genomic variants from high-throughput sequencing experiments. Nucleic Acids Res. 42:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, von Holdt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 4:359–61. [Google Scholar]
- Elias M, Panaud O, Robert T. 2000. Assessment of genetic variability in a traditional cassava (Manihot esculenta Crantz) farming system, using AFLP markers. Heredity (Edinb). 85(Pt 3):219–230. [DOI] [PubMed] [Google Scholar]
- Elias M, Penet L, Vindry P, McKey D, Panaud O, Robert T. 2001. Unmanaged sexual reproduction and the dynamics of genetic diversity of a vegetatively propagated crop plant, cassava (Manihot esculenta Crantz), in a traditional farming system. Mol Ecol. 10:1895–1907. [DOI] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 6:e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14(8):2611–2620. [DOI] [PubMed] [Google Scholar]
- Handy ESC. 1940. The Hawaiian planter. Honolulu (HI): Bishop Museum Press. [Google Scholar]
- Herten K, Hestand MS, Vermeesch JR, Van Houdt JK. 2015. GBSX: a toolkit for experimental design and demultiplexing genotyping by sequencing experiments. BMC Bioinformatics. 16:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks M, and Nunn PD. 2007. Evidence for introduced taro (Colocasia esculenta) and lesser yam (Dioscorea esculenta) in Lapita-era (c. 3050–2500 cal. yr BP) deposits from Bourewa, southwest Viti Levu Island, Fiji. J Archaeol Sci. 34:739–748. [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23(2):254–267. [DOI] [PubMed] [Google Scholar]
- Irwin SV, Kaufusi P, Banks K, de la Pena R, Cho JJ. 1998. Molecular characterization of taro (Colocasia esculenta) using RAPD markers. Euphytica. 99:183–189. [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 24(11):1403–1405. [DOI] [PubMed] [Google Scholar]
- Kirch PV. 2014. The Prehistory of Hawai’i. In: Cochrane E, Hunt T, editors. The Oxford handbook of prehistoric Oceania. Oxford (UK): Oxford University Press. [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 15(5):1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreike CM, Van Eck HJ, Lebot V. 2004. Genetic diversity of taro, Colocasia esculenta (L.) Schott, in Southeast Asia and the Pacific. Theor Appl Genet. 109(4):761–768. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebot V, Aradhya KM. 1991. Isozyme variation in taro (Colocasia esculenta (L) Schott) from Asia and Oceania. Euphytica. 56:55–66. [Google Scholar]
- Lebot V, Prana MS, Kreike N, Van Heck H, Pardales J, Okpul T, Gendua T, Thongjiem M, Hue H, Viet N et al. 2004. Characterisation of taro (Colocasia esculenta (L) Schott) genetic resources in Southeast Asia and Oceania. Genetic Resources and Crop Evolution. 51:381–392. [Google Scholar]
- Lebot V. 2009. Tropical root and tuber crops: cassava, sweet potato, yams, aroids. CAB ebooks, www.cabi.org. [Google Scholar]
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischer HE, Excoffier L. 2012. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics. 28(2):298–299. [DOI] [PubMed] [Google Scholar]
- Loy T, Spriggs M, and Wickler S. 1992. Direct evidence for human use of plants 28,000 years ago – starch residues on stone artifacts from the northern Solomon-Islands. Antiquity 66:898–912. [Google Scholar]
- Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD, Buckler ES, Costich DE. 2013. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet. 9(1):e1003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace ES, Mathur PN, Izquierdo L, Hunter D, Taylor MB, Singh D, DeLacy IH, Jackson GVH, and Godwin ID. 2006. Rationalization of taro germplasm collections in the Pacific Island region using simple sequence repeat (SSR) markers. Plant Genetic Resources. 4:210–2210. [Google Scholar]
- Matthews PJ. 1995. Aroids and the Austronesians. Tropics. 4:105–126. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwueme I. 1999. Taro cultivation in Asia and the Pacific. Rap Publication. 16:1–9. [Google Scholar]
- Peng Y, Leung HC, Yiu SM, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155(2):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VR, Hunter D, Eyzaguirre PB, and Matthews PJ. 2010. Ethnobotany and global diversity of taro. The Global Diversity of Taro. In: Rao RV, Matthews PJ, Eyzaguirre PB, Hunter D, editors. The Global Diversity of Taro: Ethnobotany and Conservation. Rome (Italy): Biodiversity International; p. 1–5. [Google Scholar]
- Reyad-ul-Ferdous M, Arman MSI, Tanvir MMI, Sumi S, Siddique KMM, Billah MM, and Islam MS. 2015. Biologically potential for pharmacologicals and phytochemicals of medicinal plants of Colocasia esculenta: a comprehensive review. Am J Clin Exp Med. 3:7–11. [Google Scholar]
- Roullier C, Benoit L, McKey DB, Lebot V. 2013. Historical collections reveal patterns of diffusion of sweet potato in Oceania obscured by modern plant movements and recombination. Proc Nat Acad Sci. 110:2205–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintaku MH, Kimball HL, Miyasaka SC, Sim SB, Geib SM. 2016. Using genotyping by sequencing (GBS) to identify loci in Colocasia esculenta linked with resistance to Phytophthora colocasiae. In: Schultheis JR, editor. XXIX IHC – Proc. Int. Symp. On Root and Tuber Crops: Sustaining lives and livelihoods into the future. Brisbane (Australia): Acta Hortic 1118; p. 131–137. [Google Scholar]
- Soetaert K. 2016. plot3D: Plotting Multi-Dimensional Data. R package version 1.1. [cited 2017 August 31]. Available from http://www.rforscience.com/rpackages/visualisation/plot3d/ [Google Scholar]
- Soulard L, Mournet P, Guitton B, Chair H. 2017. Construction of two genetic linkage maps of taro using single nucleotide polymorphism and microsatellite markers. Molecular Breeding. 37:37. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton TV, Brown JW, Leung P. 1993. Taro Trade and Cost of Production in Selected Areas of the American Affiliated Pacific. In: Ferentinos L, editor. Proceedings of the Sustainable Taro Culture for the Pacific Conference. Sustainable Taro Culture for the Pacific Conference; 1992 Sept 24–25; Honolulu, Hawaii Honolulu (HI): University of Hawaii; p. 112–124. [Google Scholar]
- USDA National Agricultural Statistics Service 2015. State agriculture overview. [cited 2016 October 14]. Available from https://www.nass.usda.gov/Quick_Stats/Ag_Overview/stateOverview.php?state=HAWAII [Google Scholar]
- Wilmshurst JM, Hunt TL, Lipo CP, Anderson AJ. 2011. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proc Nat Acad Sci. 108(5):1815–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney L, Bowers F, Takahashi M. 1939. Taro cultivars in Hawaii. Hawaii Agricultural Experiment Station of the University of Hawaii Bulletin; 84 [cited 2017 August 31]. Available from https://www.ctahr.hawaii.edu/oc/freepubs/pdf/B-084.pdf [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads generated for this study can be found in the National Center for Biotechnology Information’s Sequence Read Archive (SRA) under BioProject PRJNA381383 (sequencing runs SRR5468958–63). SNP data and alignments used for phylogenetic analyses have been deposited at Dryad, http://dx.doi.org/doi:10.5061/dryad.r35kr.