Abstract
A survey of a vast range of mycobacterial strains led us to discover a new Pps1 intein allele in Mycobacterium gastri which differs from those of Mycobacterium tuberculosis and Mycobacterium leprae in both its sequence and insertion site. While little is known about Pps1, except that it belongs to the YC24 family of ABC transporters, we show that, unlike the other inteins described so far from Eubacteria, the MgaPps1 intein possesses a specific endonuclease activity. The intein is the first eubacterial intein to be characterised as an endonuclease. Like other intein endonucleases, its minimal sequence for recognition and cleavage is quite large, with 22 bp spanning the Pps1-c site. The fact that an active endonuclease is found among the mycobacterial inteins supports the concept of a cyclical model of invasion by horizontal transfer of these genes, followed by degeneration and loss until a new invasion event, thus explaining their long-term persistence in closely related eubacterial species.
INTRODUCTION
Inteins are protein introns that are embedded in-frame within a precursor protein and are post-translationally excised by a self-catalytic protein splicing process. Since the discovery of inteins, 10 years ago, more than 110 intein genes have been identified, mostly in Archaebacteria (Inbase, the New England Biolabs intein database at http://www.neb.com/neb/inteins/). Interestingly, among the 32 intein sequences that have been found in Eubacteria, 22 are in mycobacterial genes. These inteins are located in four host proteins, i.e. the DNA gyrase GyrA subunit, RecA, the DnaB helicase and Pps1, whose function is unknown, and are present in 15 different mycobacterial species (Inbase). While most of these species harbour only one or two inteins, Mycobacterium leprae, the causative agent of leprosy, harbours an intein in each of the four host proteins. Apart from Mycobacterium tuberculosis, the most serious mycobacterial pathogen responsible for more deaths annually than any other single human pathogen, which also contains three inteins, in RecA, DnaB and Pps1, the other mycobacterial species harbour inteins at precisely the same locations as the M.leprae inteins. Therefore, these inteins are considered as alleles of the M.leprae inteins, as defined by Perler and co-workers (1).
While over 90% of all known inteins contain the conserved LAGLIDADG sequence motifs of the DOD family of homing endonucleases (Inteins-Protein Introns web site at http://bioinfo.weizmann.ac.il/~pietro/inteins/), endonuclease activity has been experimentally demonstrated for only 11 inteins. Ten of these are from Archaea (1–7) and one from the yeast Saccharomyces cerevisiae (8). This later endonuclease activity seems only to be involved in perpetuation and transfer of the intein. Importantly, no intein endonuclease activity has ever been identified from Eubacteria. To investigate the potential activities of eubacterial inteins, we deliberately studied the functions of those invading mycobacterial genes, reasoning that the Mycobacterium genus, which comprises both pathogenic and non-pathogenic species harbouring various inteins in their genes, would represent a wealth of new and important biological activities. While extending the host range of the intein in RecA and confirming its preferential location in the M.leprae locus (9), we have been unable to detect an endonuclease activity for the intein gene product from both M.leprae and M.tuberculosis. In contrast, the new allele that we have identified in the pps1 gene of Mycobacterium gastri, called MgaPps1, exhibits a new insertion site and a specific endonuclease activity.
MATERIALS AND METHODS
Mycobacterial strains and growth conditions
The type strain of M.gastri (W471) and strain HB4389 were used in the present study and were kindly provided by Dr V.Vincent Lévy-Frébault from the Reference Centre for Mycobacteria (Institut Pasteur, Paris). Mycobacteria were grown at 37°C on Löwenstein–Jensen medium for 3–4 weeks.
Genomic DNA isolation and PCR assays
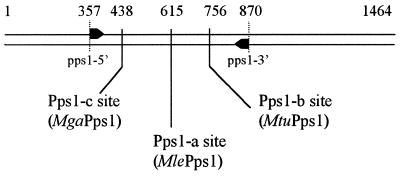
The glass bead disruption method for genomic DNA isolation from Mycobacteria was used as previously described (9). Primer sequences were chosen in the most conserved part of the pps1 gene surrounding both intein insertion sites in M.leprae and M.tuberculosis. The primers pps1-5′ (5′-CATCCGCAACACCTACGACCGG-3′) and pps1-3′ (5′-GTCGTTGTTCGACCAGTTCTGGATGGT-3′) correspond to the M.tuberculosis pps1 gene sequence or its complementary sequence between positions 357–378 and positions 844–870, respectively (Fig. 1).
Figure 1.
Location of intein insertion sites in mycobacterial pps1 genes. Arrows represent the oligonucleotides used to amplify the gene fragment. The positions of the Pps1-a, Pps1-b and Pps1-c sites and of oligonucleotides are indicated in numbers of bp from the translation start site in the M.tuberculosis sequence. The inteins inserted at each site are specified in parentheses.
PCR amplifications were performed using 5 µl of genomic DNA preparation as the DNA matrix. Several PCR assays were performed under different reaction conditions with regard to oligonucleotide concentrations and hybridisation temperature to ensure amplification specificity. In the absence of invading sequence, the amplified fragment consists of 513 bp of the pps1 gene. The presence of an intein can thus be revealed by specific amplification of a longer DNA fragment.
Cloning and sequencing of the M.gastri pps1 gene fragment containing the intein coding sequence
The M.gastri (HB4389) PCR fragment was purified from a 1% agarose gel with a QiaQuick extraction kit (Qiagen) and directly cloned in plasmid PCR2.1-TOPO using a TOPO-TA cloning kit (Invitrogen). The resulting plasmid was double-strand sequenced (MWG Biotech) using universal primers M13Forward and M13Reverse. GenBank accession numbers for the pps1 genes from M.leprae and M.tuberculosis and for the partial pps1 gene from M.gastri are U00013, AL123456 and AJ276128, respectively.
Production and purification of the MgaPps1 intein
In order to express the MgaPps1 intein gene in Escherichia coli, we constructed two different expression vectors. The coding sequence of the MgaPps1 intein was amplified by PCR using Taq DNA polymerase in 10 mM Tris–HCl, pH 8.3, 1.5 mM MgCl2, 50 mM KCl, 0.2 mM dNTP with 10 pmol of each oligonucleotide of either the primer pair Mga-ATG (5′-ATGTGCCTGGCCGGCGACAC-3′) and Mga-3′-TGA (5′-TCAGTTGTGCACCACCAGGCC-3′) or Mga-ATG and Mga-3′ (5′-GTTGTGCACCACCAGGCC-3′) and 5 µl of M.gastri genomic DNA. These 50 µl mixes were incubated for 10 min at 92°C, 29 times for 1 min at 92°C, 1 min at 53°C and 1.5 min at 72°C and finally for 5 min at 72°C. After purification (QiaQuick PCR purification kit), both amplified sequences were inserted in plasmid pCR T7/CT-TOPO according to the manufacturer’s instructions (Invitrogen) and the resulting plasmids were sequenced (Isoprim). In the first construct, pCRT7-MgaPps1, a TGA codon was introduced immediately downstream from the intein coding sequence, while in the second construct, pCRT7-MgaPps1+tag, the intein coding sequence was cloned in-frame with a DNA sequence encoding the C-terminal V5 and 6×His epitopes. Hence, the resulting constructs allowed expression of the intein fused or not to C-terminal V5 and 6×His tags under control of the T7 promoter and T7 RNA polymerase in E.coli.
Escherichia coli BL21(DE3)[pLysS] bacteria transformed with these expression vectors were grown at different temperatures in Luria broth culture medium supplemented with 100 µg/ml ampicillin (Sigma) and 30 µg/ml chloramphenicol. Induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma) was performed for 6 h and cells were lysed in 20 mM sodium phosphate, pH 7.5, by six cycles of freezing–thawing. The lysate was centrifuged at 10 000 g for 30 min to separate the soluble intein from insoluble proteins.
The soluble fraction was dialysed against 10 mM Tris–HCl, pH 7.5, 0.1 mM EDTA, 1 mM DTT, 200 µg/ml BSA and 50 mM NaCl for storage and the protein concentration measured according to the standard Bradford procedure (10).
The non-tagged intein was partially purified by anion exchange chromatography. The soluble extracts were treated with benzonase (Sigma), dialysed against 20 mM sodium phosphate, pH 7.5, loaded in a Q Fast Flow column (Amersham Pharmacia Biotech) and eluted in 2.5 ml fractions with a linear gradient of 0–1 M NaCl at a flow rate of 5 ml/min.
The intein fused to a C-terminal poly(His) was partially purified by Ni2+ affinity chromatography. After digestion of the genomic DNA by benzonase, the soluble proteins were dialysed against binding buffer (20 mM Tris–HCl, pH 7.9, 5 mM imidazole, 500 mM NaCl), loaded on His·Bind resin (Novagen), washed with binding buffer and wash buffer (20 mM Tris–HCl, pH 7.9, 60 mM imidazole, 500 mM NaCl) and finally eluted with buffers containing high concentrations of imidazole (20 mM Tris–HCl, pH 7.9, 0.5 or 1 M imidazole, 500 mM NaCl).
The purified fractions were immediately dialysed against various storage buffers and the protein concentration measured. Samples containing 0.5–5 mg/ml protein were denatured at 95°C for 2 min in an SDS, urea, β-mercaptoethanol buffer and analysed by 10% SDS–PAGE (homogeneous) according to Laemmli (11). Separated proteins were either detected by Coomassie blue (R250) staining or electrically transferred to nitrocellulose membrane (BA45; Schleicher & Schull) in a semi-dry apparatus (12). Membranes were incubated for 30 min in Tris-buffered saline (TBS) (10 mM Tris–HCl, pH 8, 137 mM NaCl) containing 1% Tween-20 and 10% non-fat dry milk, for 1 h with anti-V5 antibodies (Invitrogen) diluted 1:5000 in TBS containing 1% Tween-20 and 1% non-fat dry milk and for 1 h with a peroxidase-labelled anti-mouse IgG conjugate diluted 1:10000 in the same buffer. The detection reaction with chemiluminescent substrate was performed using an ECL detection kit, according to the manufacturer’s instructions (Amersham Pharmacia Biotech).
Endonuclease assay and minimal recognition and cleavage site determination
To assay MgaPps1 endonuclease activity, the 40 bp DNA sequence spanning its homing site was inserted between the XbaI and HindIII restriction sites of plasmid pUC19. Partially complementary oligonucleotides Mga-XbaI (5′-CTAGAGCGTAGCTGCCCAGTATGAGTCAGAGGTGGTGTACCA-CCA-3′) and Mga-HindIII (5′-AGCTTGGTGGTACACCACCTCTGACTCATACTGGGCAGCTACGCT-3′) were annealed by boiling a mix of 1 nmol each oligonucleotide in 10 mM Tris–HCl, pH 7.5, 50 mM NaCl, for 5 min and slow cooling to room temperature. The annealed oligonucleotides were then inserted into pUC19 overdigested with HindIII and XbaI (New England Biolabs). The resulting construct, pMgaSite, was sequenced. In the same way, the 40 bp sequences spanning the Pps1-c site in the M.leprae and M.tuberculosis pps1 genes were inserted between the HindIII and XbaI sites of pUC19 after annealing of oligonucleotide pairs Mle-XbaI (5′-CTAGAGCGTAGCGGCGCAATACGAGTCAGAGGTAGTTTACCACCA-3′) and Mle-HindIII (5′-AGCTTGGTGGTAAACTACCTCTGACTCGTATTGCGCCGCTACGCT-3′) and Mtu-XbaI (5′-CTAGAGAGTAGCCGCACAATACGAAAGTGAAGTTGTATATCACCA-3′) and Mtu-HindIII (5′-AGCTTGGTGATATACAACTTCACTTTCGTATTGTGCGGCT-ACTCT-3′). The resulting constructs, pMleSite and pMtuSite, respectively, were sequenced. These potential substrates were linearized by ScaI, purified from a 1% agarose gel in TBE buffer (90 mM Tris–borate, 2 mM EDTA) and finally this linear DNA was diluted in water to 100 ng/µl for cleavage assays.
Endonuclease activity assays were performed with 100 ng plasmid substrate in a final volume of 10 µl, in various reaction buffers and temperatures ranging from 20 to 50°C. The reaction mixtures were analysed on 1% agarose gels in TBE buffer. The amounts of undigested substrate and products were quantified with the ImageQuant program (Molecular Dynamics).
The endonuclease minimal recognition site for PI-MgaI was determined by a primer extension method (13). The sequencing and digestion procedures, using the T7 polymerase sequencing kit (Pharmacia), universal primers SeqPuc (5′-GTAACGCCAGGGTTTTCC-3′) and M13Rev (5′-GGAAACAGCTATGACCATG-3′) and the plasmid pMgaSite as matrix, were as previously described (6).
RESULTS AND DISCUSSION
The M.gastri pps1 gene harbours an intein gene at a novel Pps1-c site
Pps1, whose function in Mycobacteria is unknown, is an intein host protein in M.leprae and M.tuberculosis but no intein has been found to date in Pps1 of any non-pathogenic Mycobacteria. The same situation prevailed for RecA inteins, which were, until recently, believed to be confined to pathogenic species (14). Through a survey of a vast range of mycobacterial species we recently showed that RecA inteins are widely distributed among Mycobacteria (9). Consequently, we first addressed the question of the distribution of Pps1 inteins in 39 non-tuberculous mycobacterial strains belonging to 32 species previously described (9). PCR amplification of the genomic DNA of these strains was performed with oligonucleotides allowing the detection of a 513 bp sequence of the pps1 gene containing both the Pps1-a (MlePps1; 15) and Pps1-b (MtuPps1; 16) insertion sites (Fig. 1). Among the genomic DNA from the strains examined by PCR, only DNA from two strains of M.gastri yielded identical DNA fragments of ∼1600 bp, revealing the presence in pps1 of an invading sequence of ∼1100 bp in size. Cloning and sequencing of these fragments confirmed the presence of a 1134 bp long intein gene inserted at a site different from the two previously characterised sites, i.e. Pps1-a and Pps1-b. This novel site, Pps1-c, was located 177 bp upstream of the Pps1-a site (Fig. 1) and the corresponding intein was named MgaPps1, according to the current nomenclature. Thus, in contrast to what was observed with the intein invading the recA gene of non-tuberculous Mycobacteria, insertion of the intein in pps1 is strain specific and different from that of M.leprae. In addition, the presence of an intein in pps1 of M.gastri, a non-pathogenic mycobacterial species, rules out a strict relationship between the presence of inteins in the pps1 gene and pathogenicity.
The peptide sequence of the Pps1 extein is highly conserved, with 82–92% identity between the M.gastri, M.leprae and M.tuberculosis proteins. Submitting the Pps1 sequence to a BLAST search in the ProDom database (17) revealed that Pps1 belongs to the YC24 family of ABC transporters, whose sequence is strongly conserved among various bacterial species. While MgaPps1 is located immediately after a conserved glutamic acid residue, corresponding to E146 in the M.tuberculosis protein sequence, the MlePps1 and MtuPps1 inteins are inserted 59 and 106 residues downstream, respectively. The M.leprae intein is located precisely at the boundary of the ABC transporter domain, as defined by the ProDom search, while the M.tuberculosis intein is located in the middle of a sequence that is found in all bacterial ABC transporters of this family, suggesting an important role for this peptide domain.
Although the three Pps1 inteins are of similar size (359, 378 and 386 amino acids for MtuPps1, MgaPps1 and MlePps1, respectively), they differ significantly in their peptide sequences, with only 17–32% identity between the three. Nevertheless, the eight peptide motifs typifying inteins are present in all three sequences (Fig. 2). Within motifs C, D and E, MgaPps1 shares more identity with MlePps1 (70%) than MtuPps1 (only 33%). A lower sequence identity was observed for the other motifs between the three inteins; it is noteworthy that, globally, the MtuPps1 sequence is more divergent from the MgaPps1 sequence than the MlePps1 sequence. Despite the presence in MgaPps1 of the four central motifs (C, D, E and H), forming the DOD endonuclease signature that predicts an endonuclease activity for this new intein (18), it should be remembered that over 90% of all known inteins contain these motifs within their peptide sequences but only 11 inteins, among which none are from Eubacteria, have actually been shown to have endonuclease activity. For instance, although all mycobacterial inteins, except MxeGyrA, have the sequence signature of DOD endonucleases we were unable to find any endonuclease activity for the MtuRecA and MleRecA inteins (unpublished results). This low percentage of active endonucleases among inteins is not surprising in view of the data of Goddard and Burt (19); these authors showed that homing endonuclease genes with no selective advantage to the host are expected to be found mostly as non-functional elements, rather than active endonucleases, due to the length of the cycle of invasion by horizontal transmission.
Figure 2.
Comparison between the intein signatures of MgaPps1, MlePps1 and MtuPps1. Sequences of endonuclease motifs C, D, E and H and splicing motifs A, B, F and G are aligned. The position of the last residue of each block appears in parentheses. Residues present in at least two inteins appear in black boxes.
MgaPps1 intein is a site-specific endonuclease (PI-MgaI)
In order to assay for a putative endonuclease activity of MgaPps1, we constructed a pUC19 plasmid containing the 40 bp sequence spanning the Pps1-c intein insertion site in the pps1 gene of M.gastri. This plasmid (pMgaSite) was then linearized by ScaI to serve as a DNA substrate in the endonuclease assays.
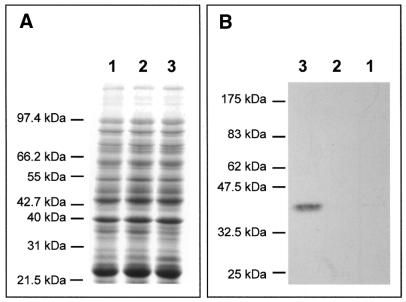
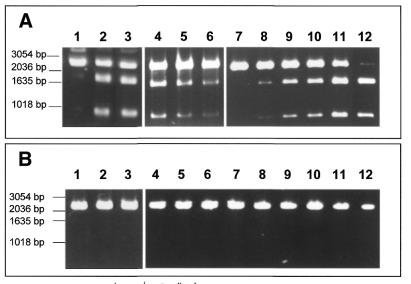
The intein gene was expressed in E.coli. BL21(DE3)[pLysS] bacteria were transformed with the two constructs pCRT7-MgaPps1 and pCRT7-MgaPps1+tag and expression of the intein unfused or fused to C-terminal V5 and 6×His epitopes, respectively, was induced with 0.5 mM IPTG at 37°C. In both cases, the presence of the intein was not detectable in soluble crude extracts (Fig. 3A); a low level of expression was nonetheless detected by western blotting using anti-V5 antibodies (Fig. 3B). Moreover, incubation of the soluble extracts with the potential DNA substrate of the intein showed that MgaPps1 specifically cleaves its target DNA sequence. Indeed, the 2730 linear substrate was cleaved into two products of 940 and 1790 bp only in the presence of the intein, the amount of cleaved DNA depending on both the amount of intein extract and the reaction time (Fig. 4A), while the wild-type pUC19 plasmid was not cleaved (Fig. 4B). This observation implies that sufficient amounts of soluble intein were present in both extracts to detect its activity.
Figure 3.
Expression of MgaPps1 in E.coli. Proteins contained in soluble crude extracts of (lane 1) non-transformed BL21(DE3)[pLysS], (lane 2) untagged MgaPps1 and (lane 3) tagged MgaPps1 were separated by 10% SDS–PAGE. Proteins were detected either by (A) Coomasie blue staining or (B) western blotting using anti-V5 antibodies.
Figure 4.
Cleavage assay for MgaPps1. Aliquots of 100 ng ScaI-linearized pMgaSite substrate (A) or linearized pUC19 (B) were incubated with 250 ng of a crude extract of (lane 1) non-transformed BL21(DE3)[pLysS], (lane 2) untagged MgaPps1 or (lane 3) tagged MgaPps1, or with (lane 4) 100, (lane 5) 50 or (lane 6) 25 ng of a crude extract of untagged MgaPps1 for 10 min at 37°C, or with 25 ng of a crude extract of untagged MgaPps1 for (lanes 7–12) 0, 10, 30, 40, 100 or 150 min, in 10 mM Tris–HCl, pH 8, buffer containing 10 mM MgCl2 and 25 mM KCl.
Before any further study of its endonuclease activity, we attempted to purify the MgaPps1 intein. It was hypothesised that the poor expression of the MgaPps1 intein in E.coli was due to the high GC content of its coding sequence (63.2%) and the different codon usages between E.coli and Mycobacteria. Moreover, we observed that the tagged intein was mainly insoluble when produced at 37°C. The optimisation of growth and induction parameters, particularly a reduction in growth temperature to 30°C, allowed us to increase the amount of soluble intein expressed after a 6 h induction (data not shown).
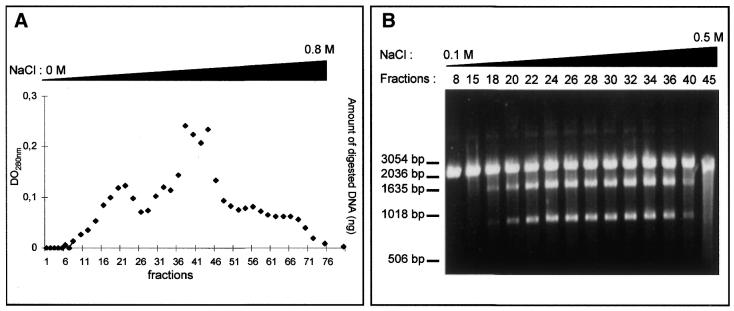
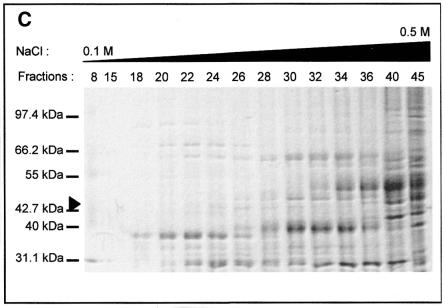
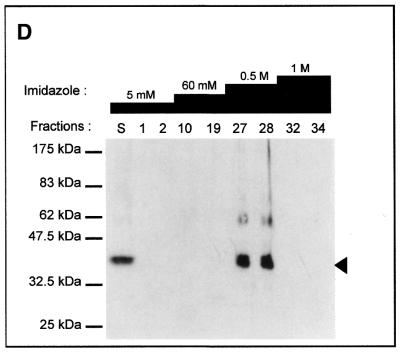
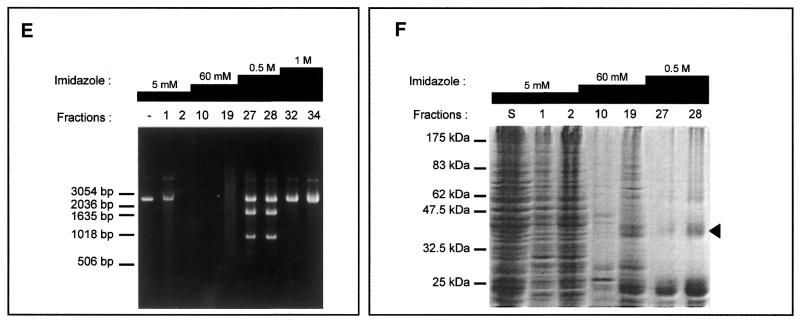
Different strategies were used to purify the intein. For instance, MgaPps1 unfused to C-terminal tags was submitted to anion exchange chromatography. Proteins contained in each fraction were estimated by measuring the absorbance at 280 nm (Fig. 5A) and analysed by SDS–PAGE. In parallel, the endonuclease activity of various fractions was determined. MgaPps1 was eluted with 0.2–0.4 M NaCl according to the endonuclease activity in fractions 18–40 (Fig. 5B). However, the protein profiles of the fractions were heterogeneous and the intein was a minor protein (Fig. 5C). All subsequent dialyses performed for further purification of the intein led to its inactivation. As an alternative, extracts containing the intein fused to a 6×His tag were fractioned, reasoning that presence of the tag at the end of the C-terminal fusion peptide would allow purification of the intein by affinity chromatography with Ni2+. The intein was eluted in 0.5 M imidazole as judged by SDS–PAGE followed by western blotting using anti-V5 antibodies (Fig. 5D) and by the observation of specific endonuclease activity in fractions 27 and 28 (Fig. 5E). Again, the active fractions were heterogeneous in terms of protein profiles and a major contaminant protein co-eluted with the intein (Fig. 5F). It is noteworthy that, once again, the endonuclease activity was rapidly lost whatever the storage buffer, hampering the purification process. Consequently, all the enzymatic assays were done with crude extracts.
Figure 5.
Tentative purification of untagged and tagged MgaPps1. The untagged and tagged inteins were semi-purified by anion exchange chromatography (A–C) and Ni2+ affinity chromatography (D–F), respectively. (A) Proteins eluted from the column with a linear gradient of NaCl were quantified by measuring the absorbance at 280 nm. (B) Aliquots of 100 ng ScaI-linearized MgaSite substrate were incubated with 250 ng of proteins present in the indicated fractions for 10 min at 37°C, in 10 mM Tris–HCl, pH 8, buffer containing 10 mM MgCl2 and 25 mM KCl. (C) Proteins present in the indicated fractions were separated by 10% SDS–PAGE and detected by Coomasie blue staining. The arrowhead indicates the position of the MgaPps1 intein. (D) Proteins eluted at different imidazole concentrations were separated by 10% SDS–PAGE and detected by western blotting using anti-V5 antibodies. (E) Aliquots of 100 ng ScaI-linearized MgaSite substrate were incubated with 100 ng of the corresponding fractions for 10 min at 37°C, in 10 mM Tris–HCl, pH 8, buffer containing 10 mM MgCl2 and 25 mM KCl. (F) Proteins contained in the indicated fractions were separated by 10% SDS–PAGE and detected by Coomasie blue staining. The arrowhead indicates the position of MgaPps1 intein.
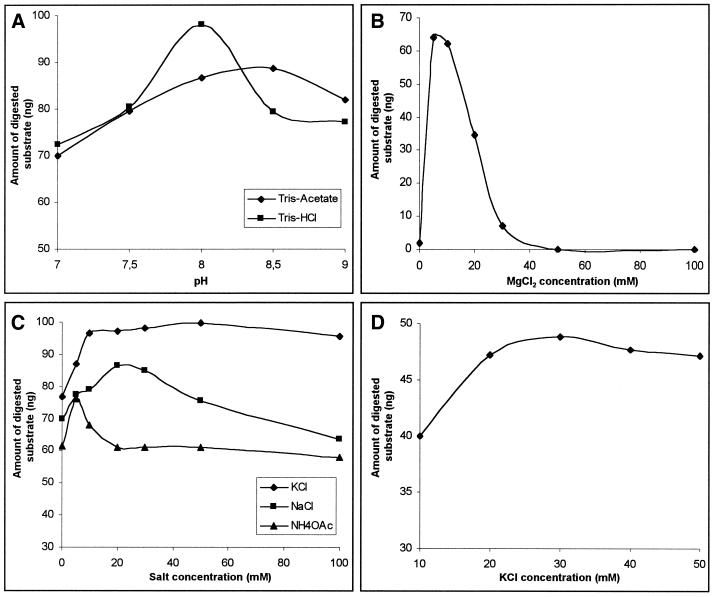
We tested various cleavage conditions and found that MgaPps1 specifically cleaved its linear substrate under a wide range of experimental conditions. The reaction was slightly dependent on the buffer composition and pH (Fig. 6A), maximal activity being observed with 10 mM Tris–HCl, pH 8; the reaction required Mg2+ as a cofactor (Fig. 6B), the enzyme being 10-fold less active with Mn2+ (not shown). The cleavage efficiency was enhanced by monovalent cations, KCl being slightly more efficient than NaCl or NH4OAc (Fig. 6C and D). The endonuclease activity increased between 20 and 50°C but the intein was rapidly inactivated at high temperatures (not shown), while the kinetics of cleavage are linear at 37°C (Fig. 4A). It follows then that the recombinant intein possesses an endonuclease activity that is optimal in 10 mM Tris–HCl, pH 8, buffer containing 10 mM MgCl2 and 25 mM KCl at 37°C. It was named PI-MgaI according to current nomenclature. To the best of our knowledge, this is the first experimental demonstration of an endonuclease activity of a eubacterial intein.
Figure 6.
Optimisation of the PI-MgaI endonuclease activity. Aliquots of 100 ng pMgaSite DNA substrate were incubated for 60 min at 37°C with the soluble crude extract of untagged MgaPps1 under different conditions. These data represent single observations. (A) Effect of buffer composition and pH. The assays were performed at different pH values with 70 ng proteins, in 10 mM Tris–acetate (diamonds) or Tris–HCl (squares) buffers containing 10 mM MgCl2 and 10 mM NaCl. (B) Effect of MgCl2 concentration. The assays were performed with 50 ng proteins, in 10 mM Tris–HCl, pH 8, 10 mM NaCl and increasing concentrations of MgCl2. (C) Effect of monovalent ions. The assays were performed with 60 ng proteins, in 10 mM Tris–HCl, pH 8, 10 mM MgCl2 and increasing concentrations of NaCl (squares), KCl (diamonds) or NH4OAc (triangles). (D) Effect of KCl concentration. The assays were performed in 10 mM Tris–HCl, pH 8, 10 mM MgCl2 and increasing KCl concentrations ranging from 10 to 50 mM, in the presence of only 20 ng of the crude extract of untagged MgaPps1.
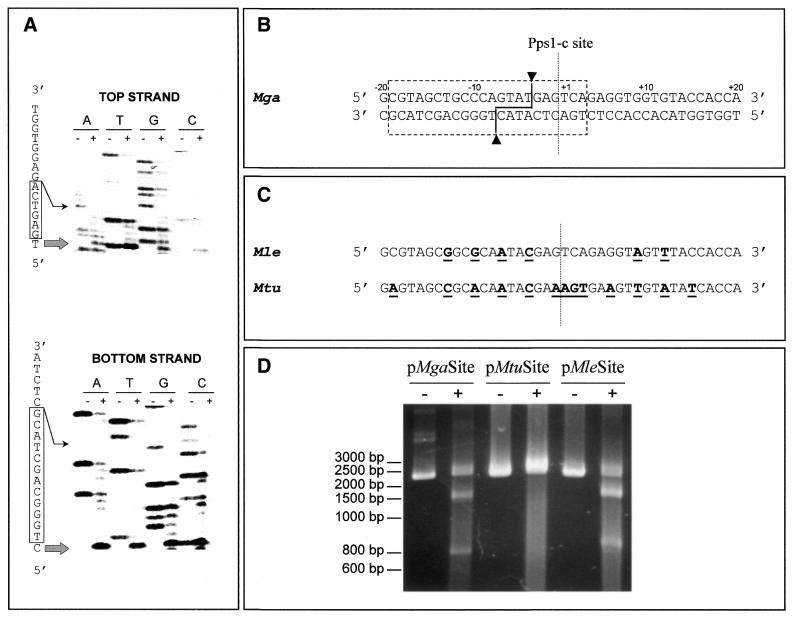
The minimal site for recognition and cleavage by PI-MgaI was determined using the plasmid pMgaSite as DNA matrix for the primer elongation procedure. Although the Wenzlau procedure is not the most accurate way to precisely delineate the recognition site, the results being dependent on DNA conformation and enzyme concentration (6,20), it remains the best way to define the exact cleavage site on both DNA strands. Comparison of PI-MgaI-digested and undigested DNA patterns (Fig. 7A) led to definition of the cleavage site and recognition sequence (Fig. 7B). The results obtained were atypical compared to the sequences cleaved by other endonuclease inteins. Cleavage by PI-MgaI yielded non-identical 3′-overhangs of four bases positioned on the 5′-side of the Pps1-c site; whereas some other known intein cleavage sites are shifted relative to their homing sites, e.g. PI-PkoII, PI-PfuII and PI-PspI, this shift generally occurs towards the 3′-side (3–5). It should be noted that horizontal transmission of homing endonuclease genes requiring double-strand break repair by homologous recombination does not necessitate the DNA cleavage site to strictly overlap the homing site. Moreover, while the 22 bp size of the recognition site is in agreement with the known specificity of inteins, this site is atypically short on the 3′-side of the cleavage site, with only six bases 3′ to this site on the upper DNA strand, compared to 16 bases on the 5′-side. This may imply that most of the specific DNA recognition is on the 5′-side of the cleavage site, in contrast to what was shown for other inteins such as PI-SceI and PI-TfuII (6,21). That the uncommon position of the cleavage site and singular location of the minimal recognition site relative to the cleavage site were not attributable to experimental conditions is a moot point. This hypothesis implies that the presence of contaminant proteins in the crude extract containing dilute intein would perturb the interaction between PI-MgaI and its target DNA and, as a consequence, the endonuclease would adapt its recognition pathway. However, this is unlikely, because of the specificity of the reaction.
Figure 7.
Site specificity of PI-MgaI. (A) Determination of the minimal recognition site using the primer extension procedure. Autoradiograms of the sequencing gels are presented. The sequencing reactions were performed in both the direct and reverse orientations. PI-MgaI-digested (+) and undigested (–) reactions were loaded side-by-side on a 6% denaturing polyacrylamide gel. A large arrow indicates the cleavage site on each DNA strand. Boxes represent bases belonging to the minimal site, in each direction. (B) PI-MgaI recognition sequence. The dashed box delineates the minimal nucleotide sequence necessary for recognition and cleavage by PI-MgaI and arrowheads designate the cleavage points on each DNA strand of the MgaSite plasmid. The dashed line indicates the Pps1-c insertion site of the intein in the pps1 gene. (C) Sequences of pps1 genes from M.leprae and M.tuberculosis around the Pps1-c site. Nucleotides at variance from the M.gastri pps1 sequence appear in bold and are underlined. (D) Cleavage assays using different DNA substrates. Aliquots of 100 ng ScaI-linearized MgaSite, MleSite and MtuSite substrates were incubated with 250 ng of the crude extract of tagged MgaPps1 for 10 min at 37°C in 10 mM Tris–HCl, pH 8, buffer containing 10 mM MgCl2 and 25 mM KCl.
No intein coding sequence has been found to interrupt the pps1 genes of M.leprae and M.tuberculosis at the Pps1-c site. This observation could be explained by the specificity of cleavage of the homing endonuclease, a hypothesis consistent with the sequence divergence between the M.tuberculosis pps1 gene around the Pps1-c site and that of M.gastri. However, the latter sequence is very similar to that of M.leprae (Fig. 7C). Nevertheless, in both cases divergence in the 22 bp sequence corresponding to the PI-MgaI minimal recognition site may prevent cleavage of these sequences. In order to estimate the specificity of PI-MgaI, we constructed the plasmids pMleSite and pMtuSite by cloning the 40 bp spanning the Pps1-c site in the Mle and Mtu pps1 genes in pUC19 and submitted these substrates to cleavage assays. As shown in Figure 7D, pMleSite, but not pMtuSite, is a good DNA substrate for PI-MgaI since it was cleaved as efficiently as the wild-type substrate pMgaSite. The comparison of cleaved and uncleaved sequences highlighted that the nucleotides at positions –1 to +4 of the Pps1-c site are unsubstitutable while the nucleotides at positions –4 and –7, which are engaged in the scissible phosphodiester bonds, as well as the nucleotides at positions –10 and –13, are not crucial for DNA recognition and cleavage. Hence, among the 22 bp constituting the minimal recognition and cleavage sequence, at least 4 nt on the 5′-side of the cleavage site are mutable while at least 4 nt at the 3′-extremity of this sequence are critical for endonuclease activity. These results are not surprising in view of the published data on the specificity of other inteins such as PI-SceI, which tolerates the substitution of 22 nt along the 31 bp minimal site (22), and the isoschizomers belonging to the PI-TliI family, which cleave a consensus sequence of 24 bp of which four are substitutable by any nucleotide and four are substitutable by a pyrimidine (7). Therefore, if the absence of an intein at the Pps1-c site in the M.tuberculosis pps1 gene could be explained by a defective homing event due to the high specificity of the homing endonuclease, the absence of an intein at this insertion site in M.leprae could rather be due to defective genetic exchange between mycobacterial species.
Importantly, and despite sequence divergence, an endonuclease activity was also clearly found when the M.tuberculosis pps1 intein was expressed in E.coli (unpublished data), extending our knowledge on mycobacterial inteins. The purification and characterisation of this latter endonuclease, currently under investigation, and the functional analyses of other mycobacterial inteins should lead to a better understanding of the role of these proteins in bacterial physiology. Finally, the sequence differences between the inteins and the specificity of their insertion sites may help in rapid identification of Mycobacteria at the species level.
DDBJ/EMBL/GenBank accession no. AJ276128
References
- 1.Perler F.B., Olsen,G.J. and Adam,E. (1997) Compilation and analysis of intein sequences. Nucleic Acids Res., 25, 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perler F.B., Comb,D.G., Jack,W.E., Moran,L.S., Qiang,B., Kucera,R.B., Benner,J., Slatko,B.E., Nwankwo,D.O., Hempstead,S.K., Carlow,C.K.S. and Jannash,H. (1992) Intervening sequences in an Archaea DNA polymerase gene. Proc. Natl Acad. Sci. USA, 89, 5577–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu M.Q., Southworth,M.W., Mersha,F.B., Hornstra,L.J. and Perler,F.B. (1993) In vitro protein splicing of purified precursor and the identification of a branched intermediate. Cell, 75, 1371–1377. [DOI] [PubMed] [Google Scholar]
- 4.Nishioka M., Fujiwara,S., Takagi,M. and Imanaka,T. (1998) Characterization of two intein homing endonucleases encoded in the DNA polymerase gene of Pyrococcus kodakaraensis strain KOD1. Nucleic Acids Res., 26, 4409–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komori K., Fujita,N., Ichiyanagi,K., Shinagawa,H., Morikawa,K. and Ishino,Y. (1999) PI-PfuI and PI-PfuI, intein-coded homing endonucleases from Pyrococcus furiosis. I. Purification and identification of the homing-type endonuclease activities. Nucleic Acids Res., 27, 4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saves I., Ozanne,V., Dietrich,J. and Masson,J.-M. (2000) Inteins of Thermococcus fumicolans DNA polymerase are endonucleases with distinct enzymatic behaviors. J. Biol. Chem., 275, 2335–2341. [DOI] [PubMed] [Google Scholar]
- 7.Saves I., Eleaume,H., Dietrich,J. and Masson,J.-M. (2000) The Thy Pol-2 intein of Thermococcus hydrothermalis is an isoschizomer of PI-TliI and PI-TfuII endonucleases. Nucleic Acids Res., 28, 4391–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gimble F.S. and Thorner,J. (1992) Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature, 357, 301–306. [DOI] [PubMed] [Google Scholar]
- 9.Saves I., Lanéelle,M.-A., Daffé,M. and Masson,J.-M. (2000) Inteins invading mycobacterial RecA proteins. FEBS Lett., 480, 221–225. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M.M. (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilising the principle of protein dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 12.Burnette W.N. (1981) “Western blotting”: electrophoresis transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem., 112, 195–203. [DOI] [PubMed] [Google Scholar]
- 13.Wenzlau J.M., Saldanha,R.J., Butow,R.A. and Perlman,P.S. (1989) A latent intron-encoded maturase is also an endonuclease needed for intron mobility. Cell, 56, 421–430. [DOI] [PubMed] [Google Scholar]
- 14.Davis E.O., Thangaraj,H.S., Brooks,P.C. and Colston,M.J. (1994) Evidence of selection for protein introns in the recAs of pathogenic mycobacteria. EMBO J., 13, 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pietrokovski S. (1994) Conserved sequence features of inteins (protein introns) and their use in identifying new inteins and related proteins. Protein Sci., 3, 2340–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole S.T., Brosch,R., Parkhill,J., Garnier,T., Churcher,C., Harris,D., Gordon,S.V., Eiglmeier,K., Gas,S., Barry,C.E. et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- 17.Gouzy J., Corpet,F. and Kahn,D. (1999) Whole genome protein domain analysis using a new method for domain clustering. Comput. Chem., 23, 333–340. [DOI] [PubMed] [Google Scholar]
- 18.Pietrokovski S. (1998) Modular organization of inteins and C-terminal autocatalytic domains. Protein Sci., 7, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goddard M.R. and Burt,A. (1999) Recurrent invasion and extinction of a selfish gene. Proc. Natl Acad. Sci. USA, 96, 13880–13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wende W., Grindl,W., Christ,F., Pingoud,A. and Pingoud,V. (1996) Binding, bending and cleavage of DNA substrates by the homing endonuclease PI-SceI. Nucleic Acids Res., 24, 4123–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimble F.S. and Stephens,B.W. (1995) Substitutions in the conserved dodecapeptide motifs that uncouple the DNA binding and DNA cleavage activities of PI-SceI endonuclease. J. Biol. Chem., 270, 5849–5856. [DOI] [PubMed] [Google Scholar]
- 22.Gimble F.S. and Wang,J. (1996) Substrate recognition and induced DNA distortion by the PI-SceI endonuclease, an enzyme generated by protein splicing. J. Mol. Biol., 263, 163–180. [DOI] [PubMed] [Google Scholar]