CSLD3 overexpression enhances root and hypocotyl growth by increasing cell elongation; CSLD3-mediated hypocotyl elongation is independent of ethylene signaling.
Keywords: AtCSLD3, GhCSLD3, cellulose synthesis, cell elongation, ethylene response, phosphate starvation, root growth
Abstract
CSLD3, a gene of the cellulose synthase-like D family, affects root hair elongation, but its interactions with ethylene signaling and phosphate-starvation are poorly understood. Here, we aim to understand the role of CSLD3 in the context of the ethylene signaling and phosphate starvation pathways in Arabidopsis plant growth. Therefore, we performed a comparative analysis of the csld3-1 mutant, CSLD3-overexpressing lines, and ethylene-response mutants, such as the constitutive ethylene-response mutant i-ctr1. We found that CSLD3 overexpression enhanced root and hypocotyl growth by increasing cell elongation, and that the root growth was highly sensitive to ethylene treatment (1 µM ACC), in particular under phosphate starvation. However, the CSLD3-mediated hypocotyl elongation occurred independently of the ethylene signaling pathway. Notably, the typical induction of root hair and root elongation by ethylene and phosphate-starvation was completely abolished in the csld3-1 mutant. Furthermore, i-ctr1 csld3-1 double-mutants were hairless like the csld3-1 parent, confirming that CSLD3 acts downstream of the ethylene signaling pathway during root growth. Moreover, the CSLD3 levels positively correlated with cellulose levels, indicating a role of CSLD3 in cellulose synthesis, which may explain the observed growth effects. Our results establish how CSLD3 works in the context of the ethylene signaling and phosphate-starvation pathways during root hair growth, cell elongation, and cell wall biosynthesis.
Introduction
Root hairs are filamentous cells that grow in a defined direction by depositing flexible cell wall material at one site of the cell and exploiting turgor pressure as the driving force for elongation; a process also referred to as tip growth. The root hairs develop and emerge from root epidermal cells, termed trichoblasts (Dolan and Davies, 2004). In Arabidopsis, a root hair develops in a stepwise manner divided into three phases: cell specification, initiation, and elongation (tip growth) (Cho and Cosgrove, 2002). Root hairs are thus important for studies of cell differentiation, development, and elongation (Kacprzyk et al., 2014; Li et al., 2014).
Root hair development is influenced by a plethora of internal and external factors (Grierson and Schiefelbein, 2002; Wada et al., 2015). In particular, soil nutrients [including phosphorus (P), nitrogen (N), iron (Fe), potassium (K), manganese (Mn), and magnesium (Mg)] impact on root hair growth (Müller and Schmidt, 2004; Shin et al., 2005; Yang et al., 2008; Jung et al., 2009; Bloch et al., 2011). For example, plants assimilate P in the form of phosphate (Pi) that typically promotes primary root growth in plants, including Arabidopsis, maize, rice, and tomato. In contrast, Pi starvation increases the production of root hairs and lateral root growth (López-Arredondo et al., 2014). This response increases the ability of the root to access the surface layers of the soil where Pi availability is typically higher.
Plant hormones, in particular ethylene and auxin, can work as positive regulators of root hair development in tandem with environmental signals (Pitts et al., 1998; Martín-Rejano et al., 2011; Lee and Cho, 2013). Ethylene stimulates root hair development as it influences both hair density (specification and initiation) and length (elongation) (Masucci and Schiefelbein, 1994; Tanimoto et al., 1995; Pitts et al., 1998). In Arabidopsis, the ethylene response pathway is well characterized; ethylene is perceived by five receptors associated with the endoplasmic reticulum (Ji and Guo, 2013). In the absence of ethylene, the receptors activate CTR1 (CONSTITUTIVE TRIPLE1 RESPONSE1), which suppresses the function of its downstream target EIN2 (ETHYLENE INSENSTIVE2). When ethylene binds to the receptors, it inactivates CTR1, which causes the translocation of a C-terminal fragment of EIN2 (EIN2-C′) into the nucleus. EIN2-C′ then activates two key transcription factors EIN3 (ETHYLENE INSENSITIVE3) and EIL1 (ETHYLENE INSENSITIVE3-LIKE1), which then turn on the transcription of a chain of downstream target genes, including plant antimicrobial peptide DEFENSIN (PDF1.2; Penninckx et al., 1998), which initiates a plethora of plant responses (Guo and Ecker, 2004; Li et al., 2015; Song et al., 2016). In addition, ethylene can affect primary root growth by regulating cell division and expansion as it induces cell division in the root meristem zone (MZ; Ortega-Martínez et al., 2007), and inhibits cell expansion in the elongation zone (EZ; Le et al., 2001). Dicotyledonous etiolated seedlings display a characteristic ‘triple response’ when treated with ethylene, manifested by the inhibition of root and hypocotyl growth, radial swelling of the hypocotyl, and exaggerated curvature of the apical hook (Guzmán and Ecker, 1990). In the past two to three decades, more than a dozen unique mutants have been identified with typical phenotypes; for example the ethylene-insensitive mutants etr1-3 and ein2-1 exhibit increased cell elongation, and the constitutive ethylene-response mutant ctr1-1 has reduced cell elongation and increased root hair growth compared with wild-type plants (Stepanova and Ecker, 2000; Le et al., 2001; Song et al., 2016).
Plant cell walls are essential in determining the form of cells during plant growth (Keegstra, 2010). The root epidermis is an excellent tissue to study cell wall structure and function (Foreman and Dolan, 2001). Many genes impact on root epidermal cell specification and initiation, for example TRANSPARENT TESTA GLABRA (TTG), GLABRA2 (GL2), ROOT HAIR DEFECTIVE 6 (RHD6) (Galway et al., 1994; Di Cristina et al., 1996; Wada et al., 1997; Lee and Schiefelbein, 1999; Bernhardt et al., 2003; Menand et al., 2007), but only a few genes are known to affect root hair tip growth (Won et al., 2009). For example, the two root hair-specific expansin genes AtEXPA7 and AtEXPA18 promote hair initiation and elongation, indicating that expansins may affect the interactions between cellulose and xyloglucans for cell wall flexibility in root hair growth (Cho and Cosgrove, 2002; Lin et al., 2011).
In the non-growing tubular portion of root hairs, organized cellulose microfibrils form thick and sturdy cell walls that prevent unwanted outgrowth, whereas at the growing tips, unordered cellulose microfibrils support thin and dynamic cell walls and provide the major strength to expanding cell walls (Sassen et al., 1985; Somerville et al., 2004; Galway, 2006; Taylor, 2008; Park et al., 2011). Cellulose is synthesized by cellulose synthase (CesA) complexes, and CesA1, CesA3, and a CesA6-like protein are required for primary wall cellulose synthesis (Li et al., 2013; Schneider et al., 2016). Although mutations in CesA1 (rsw1-1) or CesA6 (prc1-1) affect root-hair development, tip growth is not abolished (Arioli et al., 1998; Fagard et al., 2000; Singh et al., 2008). Accordingly, fluorescently labelled CesA proteins are not found at the apical plasma membrane region of growing root hairs (Park et al., 2011). Instead, a fluorescently labelled cellulose synthase-like D3 (CSLD3) protein has been observed at this region, supporting a role of CSLD3 in root hair growth. Indeed, mutations in CSLD3, e.g. kjk, csld3-1, rhd7-1, and rhd7-4, abolish root-hair tip growth (Favery et al., 2001; Wang et al., 2001; Bernal et al., 2008; Galway et al., 2011). However, the function of CSLD3 in cell wall synthesis remains unclear and it has been variously suggested to be involved in the synthesis of cellulose, xylan, mannan, or homogalacturonan polysaccharides (Bernal et al., 2007; Wang et al., 2010; Park et al., 2011; Verhertbruggen et al., 2011; Yin et al., 2011; Guo et al., 2014).
Despite detailed genetic analyses of AtCSLD3-null mutants, much remains unknown about the functional context of CSLD3 during root-hair tip growth and plant growth, especially with regards to hormones and nutrients. In this study, we demonstrate that CSLD3 acts downstream of the ethylene response and phosphate pathways in the control of root and root-hair elongation, and provide compelling evidence for how CSLD3 regulates general cell expansion via its impact on cellulose synthesis.
Materials and methods
Amplification of the GhCSLD3 gene from cotton
To isolate the CSLD3 gene in cotton (Gossypium hirsutum L.), the Arabidopsis thaliana CSLD3 cDNA sequence (At3g03050) was compared with cotton databases (https://www.cottongen.org/) using BLASTN, and contig21384 was found to be the characteristic root of the target gene (Supplementary Fig. S1A at JXB online). To obtain the full-length gene, a segmented amplification method was used and primers were designed for contig21384 (see Supplementary Fig. S1B and Table S1). First, the middle fragment was amplified by the sense primer GhD33GSPL and the antisense primer GhD33GSPR. Second, the 5′- and 3′-end fragments were amplified by RACE-PCR. More specifically, we amplified the 5′-end fragment using outer primers (5′ RACE outer and HD35P1-out) and inner primers (5′ RACE inner and HD35P2-in) using a TaKaRa 5′- Full RACE Kit (Code: D315) and we used nested PCR to amplify the 3′-end fragment through double-PCR using outer primers (HD33P1-out and GhBC1-out) and inner primers (HD33P2-in and GhBC1-in). Third, the overlap-extension PCR method was applied to get the full-length sequence using the sense primer D3-FW2 and the antisense primer D3-RV combined with GhD33GSPR and GhD33GSPL, respectively. Finally, a 3815-bp fragment was amplified from young roots in cotton. The fragment consisted of a single 3366-bp ORF and was predicted to encode a protein of 1121 amino acid residues.
Phylogenetic analysis
A phylogenetic tree was developed using the CLUSTALW program (http://www.ebi.ac.uk/clustalw/), choosing the Construct/Test Neighbor-Joining Tree (NJ) algorithm, and was visualized using the TREEVIEW software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) for six AtCSLD proteins, five OsCSLD proteins, and the target protein GhCSLDx.
Plant material and growth conditions
All Arabidopsis samples used in this study were of the Columbia ecotype (Col-0) background. Homozygous mutants of csld3-1 (AtCSLD3) have been described previously (Wang et al., 2001). The SALK T-DNA insertional lines (SALK_059754 and SALK_063966) were obtained from the Arabidopsis Biological Resource Center (ABRC). Identification of T-DNA homozygous lines (t-ein2 and t-pdf1.2) was done by using the three primers (LBb1.3+LP+RP) designed in T-DNA Primer Design (http://signal.salk.edu/tdnaprimers.2.html). For generation of RNAi (RNA interference) constructs of AtCTR1 and AtPDF1.2, the RNAi sequences were generated based on the GeneSil website (http://www.genesil.com/siRNAdesign.asp). The corresponding 626- and 227-bp fragments were cloned into the middle vector PHANNIBAL and then connected to vector PART27. For generation of overexpression constructs, complete AtCSLD3 and GhCSLD3 coding regions driven by the D35S promoter were cloned into the binary vector pD1301s to generate the binary plasmids. Transgenic plants were generated by introduction of the plant expression constructs into Agrobacterium tumefaciens strain GV3101 and transformation was done by floral dipping (Zhang et al., 2006). The overexpression constructs were transformed into the wild-type (WT; for overexpression), csld3-1 (for complementation testing), and the i-ctr1, t-ein2, and t-pdf1.2 mutants. Transformed plants were selected on plates containing kanamycin (50 mg ml–1) for RNAi lines and plates containing hygromycin (50 mg ml–1) for overexpression lines. More than 20 independent transformants were selected. All the primers are listed in Supplementary Table S3. The i-ctr1 csld3-1 and csld3-1 i-ctr1 double-mutants were identified from the F2 offspring of hybrids between the i-ctr1 and csld3-1 mutants by kanamycin screening, and we obtained homozygous double-mutants from the F3 offspring.
Arabidopsis seeds were surface-sterilized using 75% ethanol for 4 min and 10% sodium hypochloride with 0.01% Triton X-100 for 3 min, and were then washed in sterile water several times. Seeds were then imbibed at 4 °C in the dark in sterile water containing 0.1% agar for 3 d, and germinated on plates containing half Murashige and Skoog (MS) media (1% sucrose; pH 5.8) in 1% agar. Plates were incubated in a near-vertical position at 22 °C under 16 h light/8 h dark conditions (light-grown) for photomorphogenesis, or under 24 h dark conditions (dark-grown) for skotomorphogenesis. The seedlings were transplanted into soil after the second real leaf was visible.
Ethylene precursor and phosphate starvation treatments
For the ethylene precursor treatment, seeds were sown directly on half-MS medium containing either 1 μM or 5 μM ACC (1-aminocyclopropane-1-carboxylic acid). After the seeds had been stratified at 4 °C for 3 d, they were placed horizontally in a growth room with a 16-h light/8-h dark cycle at 22 °C for 6 d. For the triple-response test, the growth conditions were 24-h dark at 22 °C for 4 d. For the phosphate starvation (P–) treatment, seeds were sown directly either on P– medium or P– medium containing 1 μM ACC: in the P– medium, the 1.25 mM KH2PO4 in the half-MS medium was replaced with 0.65 mM K2SO4, as described previously (Song et al., 2016).
Root hair, root, and hypocotyl phenotype analysis
Root-hair morphology was examined using 6-d-old light-grown (L6) seedlings. To measure the root-hair length, the differentiation zones (DZs) with root hairs were viewed under light microscopy using a Leica stereomicroscope (Leica S6 D, with Leica DFC295 digital camera). The length of root hairs in these images was measured using the ImageJ 1.32j software (https://imagej.nih.gov/ij/).
To observe root and hypocotyl growth, Arabidopsis seedlings were scanned using a HP Scanjet 8300 scanner at 600 dpi and then analysed using ImageJ 1.32j. Measurements were made of the L6 root length from the root tip to hypocotyl base, and of the 4-d-old dark-grown (D4) hypocotyl length of vertically grown seedlings from the hypocotyl base to the apical hook.
For images of epidermal cell patterns, D4 hypocotyls (2nd to 6th cells in the basal part) and L6 roots (the fully expanded cells in the EZ) were mounted in chloral hydrate and images were viewed using differential interference contrast (80i; Nikon, Japan). The EZ of the root is between the MZ (indicated by small and closely aligned cortical cells) and the DZ (indicated by the formation of root hairs). Epidermal cell lengths in recorded images (D4 hypocotyls and L6 roots) were quantified using Image J 1.32j, and epidermal cells of hypocotyls were visualized under confocal laser scanning microscopy (p58; Leica, Leica Microsystems, Nussloch, Germany) using D4 hypocotyls incubated in the dark for 10 min in a fresh solution of 15 mM (10 mg ml–1) propidium iodide (PI) (Naseer et al., 2012). PI was excited at 488 nm, and fluorescence was detected at 600–700 nm.
Root meristem size (i.e. MZ) was determined as the distance between the quiescent center (QC) and the transition zone (TZ; the position of the first elongating cortical cell), and the number of cortical cells were counted in a file extending from the QC to the TZ (Beemster and Baskin, 1998). To count the number of cortical cells, L6 root MZs were mounted in chloral hydrate, and images were taken using differential interference contrast (80i; Nikon, Japan).
All the experiments were done using at least three biological replications and at least 20 seedlings were measured in each replicate. Least-significant difference (LSD) tests (P<0.01) were used for multiple comparisons.
mRNA expression analyses
RNA extraction and RT-PCR analyses of cotton tissues were done using the method described by Wu and Liu (2004). GhUBQ7 was used as the internal control. L6 Arabidopsis seedlings were germinated and grown on half-MS medium and were placed in liquid nitrogen at harvest. Total RNA was isolated from the collected tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA). The RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) and about 10 μg RNA of each sample was transcribed into cDNA. First-strand cDNA was obtained using OligodT and M-MLV reverse transcriptase (Promega, Madison, WI, USA). Q-PCR amplification was carried out on a Bio-Rad MyCycler thermal cycler with SYBER Premix ExTaq (TakaRa, Tokyo, Japan) according to the manufacturer’s instructions, and AtGAPDH was used as the internal control. The PCR thermal cycle conditions were as follows: one cycle of 95 °C for 2 min, followed by 45 cycles of 95 °C for 15 s, 58 °C for 15 s, and 72 °C for 25 s. The expression value of AtGAPDH was defined as 100, and the expression levels of genes were normalized accordingly. All of the primers used in these assays are listed in Supplementary Table S2, and the assays were carried out for three biological replicates. LSD tests (P<0.01) were used for multiple comparisons.
Immunolocalization of glycan epitopes
L6 roots and the inflorescence 1st internode from 7-week-old plants were embedded with 4% agar and then cut into 60-μm sections using a microtome (VT1000S, Leica). For immunolabelling, transverse sections were incubated in 3% (w/v) milk protein in 1× PBS (MP/PBS) for 1 h to block non-specific binding. Sections were then incubated for 1 h with the following monoclonal antibodies diluted by 1:5 in MP/PBS: CCRC-M93, CCRC-M38, CCRC-M35, and CCRC-M149 (http://glycomics.ccrc.uga.edu/wall2/antibodies/antibodyHome.html), which bind to xyloglucan, de-esterified homogalacturonan, rhamnogalacturonan I, and xylan, respectively (Pattathil et al., 2010; DeMartini et al., 2011). After washing with PBS, sections were incubated with anti-mouse-IgG linked to fluorescein isothiocyanate (FITC; Sigma), and diluted 1:1000 in 1× PBS for 1 h. Counterstaining was performed with Calcofluor White M2R fluorochrome (fluorescent brightener 28; Sigma; 0.25 μg ml–1 in dH2O). Immunofluorescence was observed with an epifluorescence microscope (Olympus BX-61, with Retiga-4000DC digital camera) equipped with the following filter sets: 330~385/450 nm (excitation/emission; ex/em) for visualizing cell walls stained with Calcofluor White (Haigler et al., 1980), and 460~490/520 nm (ex/em) for green emission of the FITC fluorochrome.
Crystalline cellulose extraction
L6 roots were detached, freeze-dried, and ground into powder. Powder samples (40 mesh, 0.1~1.0 g) from both L6 roots and stems from 7-week-old plants were suspended in 5.0 ml acetic acid–nitric acid–water (8:1:2, v/v/v) and heated for 1 h in a boiling water bath with stirring every 10 min. After centrifugation, the pellets were washed several times with 5.0 ml water, and the resulting pellets were defined as the crystalline cellulose samples. The extractions were carried out with three biological replicates. LSD tests (P<0.01) were used for multiple comparisons.
Determination of neutral sugars in the total wall polysaccharides by GC-MS
Powder (40 mesh, 0.1~1.0 g) samples from both L6 roots and stems of 7-week-old plants were washed twice with 5.0 ml phosphate buffer and twice with 5.0 ml distilled water. The remaining pellets were stirred with 5.0 ml chloroform–methanol (1:1, v/v) for 1 h at 40 °C and washed twice with 5.0 ml methanol, followed by 5.0 ml acetone. The pellets were then washed once with 5.0 ml distilled water. A 5.0-ml aliquot of DMSO-water (9:1, v/v) was added to the remaining pellets and they were vortexed for 3 min and then rocked gently on a shaker overnight. After centrifugation, the pellets were washed twice with 5.0 ml DMSO-water, and then with 5.0 ml distilled water three times. The resulting pellets were defined as the total wall polysaccharides. Then, the pellets were dissolved in 2 M trifluoroacetic acid (TFA) and the neutral sugars were determined by GC-MS as described previously (Xu et al., 2012). Three biological replications were performed.
Results
Functional characterization of a cotton CSLD3 gene in Arabidopsis
Since there has been little research on CSLD genes in cotton, we first isolated a CSLD-related gene from young cotton roots through RACE-PCR amplification (see Supplementary Fig. S1). The putative GhCSLD3 gene consisted of a 3366-bp ORF encoding a protein of 1121 amino acid residues, showing 78% and 84% identity with the Arabidopsis AtCSLD3 gene and protein, respectively (Supplementary Fig. S1C). Like other CSLDs, the GhCSLD3 protein also contained a ‘D_D_D_QxxRW’ motif, and appeared in close proximity to AtCSLD2 and AtCSLD3 in a phylogenetic tree (Supplementary Fig. S1D).
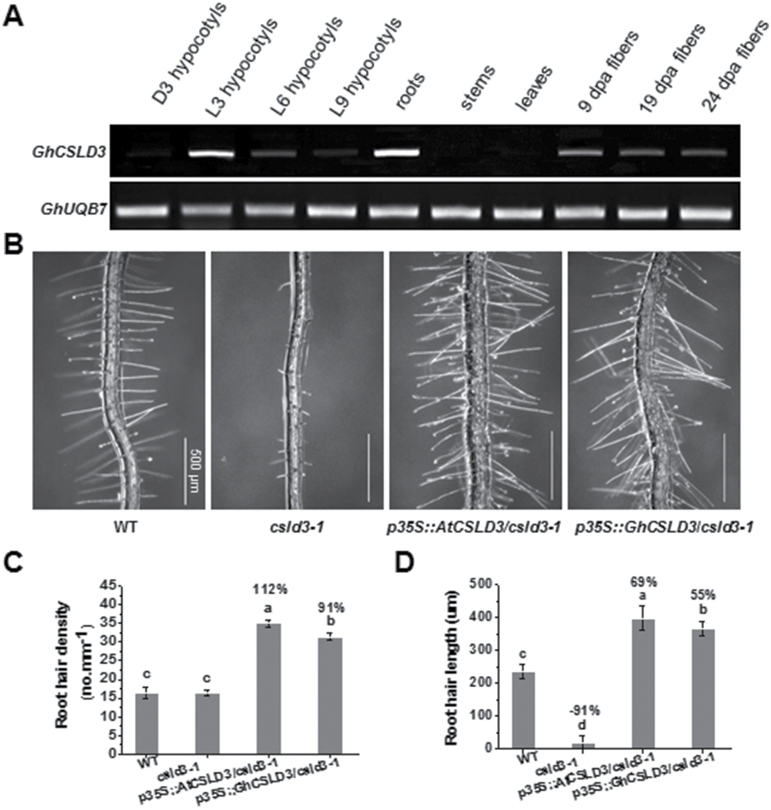
Based on RT-PCR analyses, we found that GhCSLD3 was mainly expressed in cotton roots, in hypocotyls of 3-d-old light-grown (L3) plants, and in fibers at 9 d post-anthesis (dpa) (Fig. 1A). By comparison, AtCSLD3 is also highly expressed in roots, and mutations in the Arabidopsis gene cause defects in root-hair tip growth (Favery et al., 2001; Wang et al., 2001; Galway et al., 2011). Over-expression of GhCSLD3 or AtCSLD3 under the control of a 35S promoter in the Arabidopsis csld3-1 mutant fully restored root-hair (Fig. 1B) and root growth (Supplementary Fig. S2), indicating that GhCSLD3 could functionally rescue the csld3-1 mutant defects. Notably, both the csld3-1 lines overexpressing GhCSLD3 and AtCSLD3 exhibited significantly increased root-hair length and density as compared to the wild-type (WT) (Fig. 1C, D). These data indicate that CSLD3 promotes root hair growth.
Fig. 1.
Genetic complementation test of the Arabidopsis root hairless csld3-1 mutant by AtCSLD3 and GhCSLD3. (A) Expression of GhCSLD3 in different organs in Gossypium hirsutum assessed by RT-PCR. GhUBQ7 was used as the internal control. D indicates seeds germinated and grown on half-MS media under dark (24 h dark) conditions, L indicates light (16 h light:8 h dark) conditions; the number of days of growth is indicated. dpa, days post-anthesis. (B) Root hair patterns of Arabidopsis wild-type (WT), csld3-1, and complementation lines for 6-d-old light-grown (L6) seedlings on half-MS media. (C, D) Quantitative analyses of root-hair density (C) and length (D) of seedlings as shown in (B). Data are means ±SD (three biological replicates), n≥20 seedlings were measured in each replicate. LSD (least-significant difference) tests were used for multiple comparisons. Different letters above bars indicate that the means differ according to ANOVA and LSD tests (P<0.01). The percentage values (%) were calculated by subtraction from the WT value and divided by the WT. Scale bars indicate 500 μm.
Overexpression of AtCSLD3 or GhCSLD3 leads to enhanced cell elongation and root-hair growth
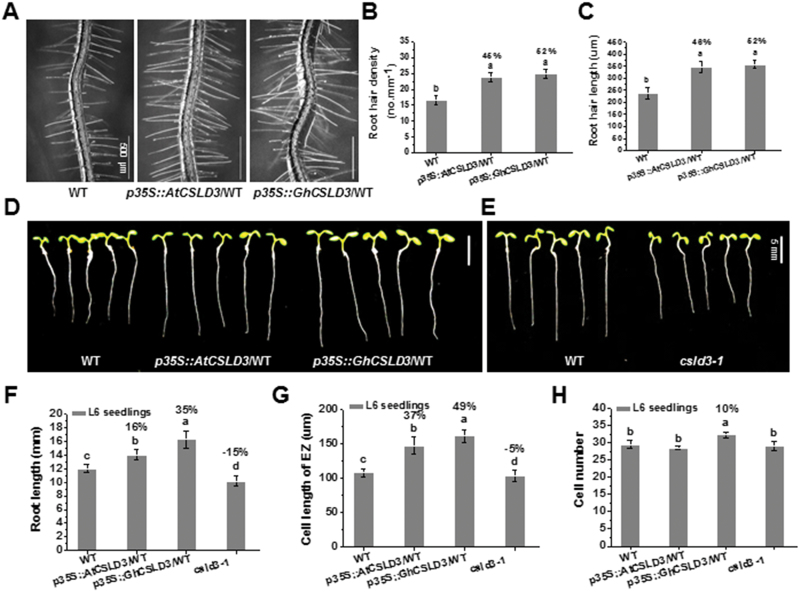
To corroborate the effects of AtCSLD3 and GhCSLD3 on root-hair growth, we also transformed the WT with the AtCSLD3- and GhCSLD3-overexpression constructs (Fig. 2A; Supplementary Fig. S4A, B and Table S2). Compared with the WT, both the p35S::AtCSLD3/WT and p35S::GhCSLD3/WT transgenic seedlings exhibited increased root-hair length and density of up to 50% (Fig. 2A–C). Interestingly, the transgenic lines also displayed longer roots, whereas the clsd3-1 mutant exhibited shorter roots (Fig. 2D–F). Arabidopsis primary roots contain three distinct regions, namely the meristematic zone (MZ), the elongation zone (EZ), and the differentiation zone (DZ) (Somssich et al., 2016). To assess in what region of the root the improved growth of the transgenic lines occurred, we measured the cell number of the MZ and the cell length of the EZ (for the fully expanded cells) in the different lines. Both the p35S::AtCSLD3/WT and p35S::GhCSLD3/WT transgenic lines exhibited much increased cell lengths (+37% and +49%, respectively) compared to the WT (Fig. 2G). By contrast, csld3-1 displayed decreased cell lengths (–5%) compared to the WT. Interestingly, p35S::GhCSLD3/WT seedlings also showed significantly increased cell numbers in the MZ (Fig. 2H). Taken together, these data indicate that CSLD3 positively affects root-hair growth, cell elongation, and cell division during root growth.
Fig. 2.
Phenotypes of the wild-type (WT), csld3-1, and CSLD3-overexpressing seedlings. (A) Root hair patterns of the WT, AtCSLD3-, and GhCSLD3-overexpressing seedlings on half-MS media. Seedlings were grown under light conditions and were 6-d old (L6). (B, C) Quantitative analyses of root-hair density (B) and length (C) for seedlings shown in (A). (D, E) WT, AtCSLD3-, and GhCSLD3-overexpressing L6 seedlings grown on half-MS media. (F) Measurements of root lengths of seedlings shown in (D, E). (G, H) Quantitative analyses of (G) cell lengths (the fully expanded cells) in the root elongation zone (EZ) and (H) cell numbers in the root meristem zone (MZ) of seedlings shown in (D, E). Data are means ±SD (three biological replicates); the number of seedlings measured in each replicate were: n≥20 in (B, C), n≥50 in (F), n≥30 in (G, H). Least-significant difference (LSD) tests were used for multiple comparisons. Different letters above bars indicate that the means differ according to ANOVA and LSD tests (P<0.01). The percentage values (%) were calculated by subtraction from the WT value and divided by the WT. Scale bars indicate 500 μm in (A), 5 mm in (D, E). (This figure is available in colour at JXB online.)
Ethylene-response mutants mimic CSLD3-related root growth phenotypes
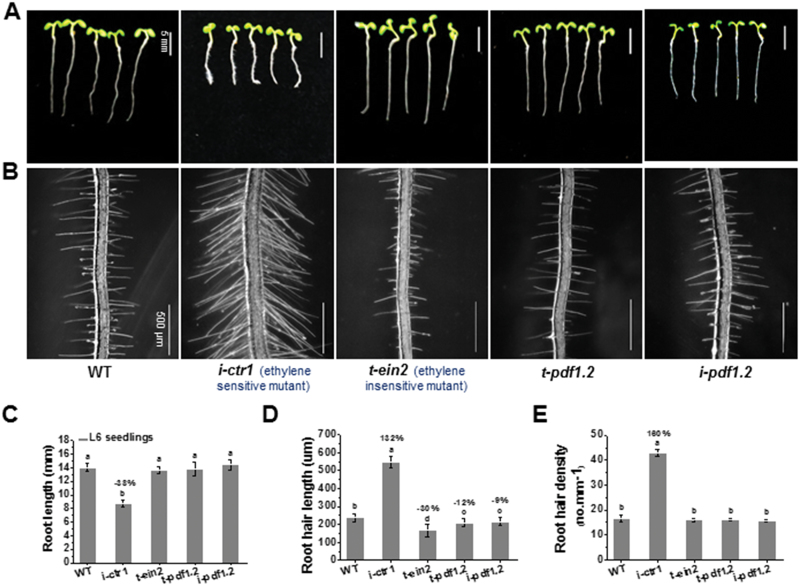
Because ethylene induces root-hair growth and inhibits primary root elongation (Le et al., 2001; Ortega-Martínez et al., 2007), we aimed to investigate potential links between CSLD3 function and ethylene in root development. We therefore obtained T-DNA insertion mutants and generated RNAi lines of some representative genes in the ethylene response pathway (AtCTR1, AtEIN2, AtPDF1.2; T-DNA insertion mutants t-ein2, t-pdf1.2 and RNAi lines i-ctr1, i-pdf1.2; see Supplementary Fig. S3 and Table S3). Mutants and RNAi lines were confirmed by Q-PCR analyses (Supplementary Fig. S4C and Table S2). As compared to the WT, the i-ctr1 lines exhibited remarkably reduced primary root growth and increases in root-hair length and density; by contrast, the other mutants/lines (t-ein2, t-pdf1.2, and i-pdf1.2) showed a reduction in root-hair length (Fig. 3), consistent with previous reports on ethylene-sensitive and -insensitive mutants (Kieber et al., 1993; Alonso et al., 1999).
Fig. 3.
Analyses of growth characteristic of Arabidopsis mutants related to ethylene-response pathways. (A, B) Growth characteristics of L6 seedlings (i.e. 6-d-old, grown under light conditions) in the wild-type (WT), i-ctr1, t-ein2, t-pdf1.2, and i-pdf1.2. (C–E) Quantitative analyses of the seedlings shown in (A, B). Data are means ±SD (three biological replicates), n≥30 seedlings were measured in each replicate. Least-significant difference (LSD) tests were used for multiple comparisons. Different letters above bars indicate that the means differ according to ANOVA and LSD tests (P<0.01). The percentage values (%) were calculated by subtraction from the WT value and divided by the WT. Scale bars indicate 5 mm in (A), 500 μm in (B). (This figure is available in colour at JXB online.)
Lesions in CSLD3 abolish ethylene-induced root-hair tip growth
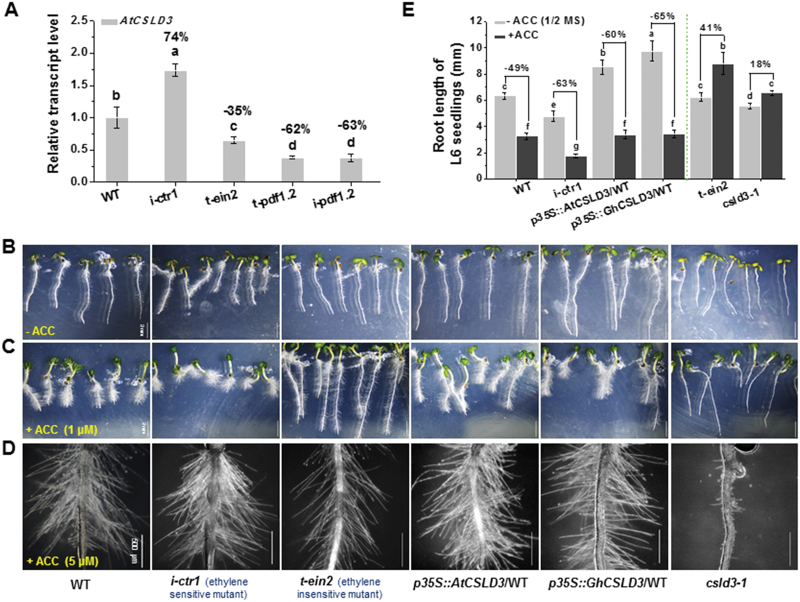
To understand the potential relationship between CSLD3 and ethylene, we assessed the AtCSLD3 expression levels in the ethylene response-pathway mutants. Interestingly, expression of AtCSLD3 was significantly increased in i-ctr1 and decreased in t-ein2, t-pdf1.2, and i-pdf1.2 (Fig. 4A). These data suggested that the expression of AtCSLD3 may be regulated by the ethylene-response pathway. To first assess how the CSLD3 transgenic and mutant lines were affected by ethylene, we grew all the lines on ACC, an ethylene precursor (Masucci and Schiefelbein, 1994, 1996). When grown on media supplemented with 1 μM or 5μM ACC, the CSLD3 overexpression and csld3-1 complemented lines, the ethylene-response mutants, and the WT all exhibited a dramatic increase in root-hair density and length (Fig. 4B–D; Supplementary Fig. S5A–C). Notably, neither the 1 μM nor the 5 μM ACC treatments induced root-hair tip growth in the csld3-1 mutant (Fig. 4B, C). Indeed, the csld3-1 mutants grown on 5 μM ACC showed a maximum root-hair length of 35 μm (WT: 236.58 ± 23.27 μm on half-MS; Fig. 4D; Supplementary Fig. S6B).
Fig. 4.
Comparison of ethylene responses among the wild-type (WT), CSLD3-overexpressing lines, csld3, i-ctr1, and t-ein2 mutants. (A) Gene expression analysis of AtCSLD3 in the WT and ethylene-response mutant seedlings grown on half-MS media. Seedlings were grown under light conditions and were 6-d old (L6). Data are means ±SD (three biological replicates); the results of least-significant difference (LSD) tests are indicated (P<0.01). The percentage values (%) were calculated by subtraction from the WT value and divided by WT. (B–D) Morphological phenotypes of L6 seedlings grown in the absence or presence of ACC. Seedlings were grown on half-MS media alone (B), with addition of 1 μM ACC (C), or with addition of 5 μM ACC (D). (E) Quantitative analyses of root lengths of seedlings as shown in (B, C). Data are means ±SD (three biological replicates), n≥30 seedlings were measured in each replicate. LSD tests were used for multiple comparisons. Different letters above bars indicate that the means differ according to ANOVA and LSD tests (P<0.01). Scale bars indicate 2 mm in (B, C); 500 μm in (D). (This figure is available in colour at JXB online.)
We next looked at primary root growth and, interestingly, root length displayed different responses depending on genotype. Compared with the WT and the two CSLD3 complementary lines (decreased by 49%, 46%, and 51%, respectively, on 1 μM ACC), the CSLD3 overexpressing roots displayed increased sensitivity to ACC (decreased by 60% and 65%), which was also the case for the i-ctr1 mutant seedlings (decreased by 63%; Fig. 4E; Supplementary Fig. S5G). However, the csld3-1 mutant was insensitive to ACC, similar to the ethylene-insensitive and non-responsive mutants t-ein2, t-pdf1.2, and i-pdf1.2 (Fig. 4E; Supplementary Figs. S5G, S6A). Hence, while the root elongation of the CSLD3-overexpressing plants was more sensitive to ethylene, plants lacking CSLD3 were insensitive to ethylene; both in terms of root-hair tip growth and root elongation, which correlated well with ethylene-related signaling processes.
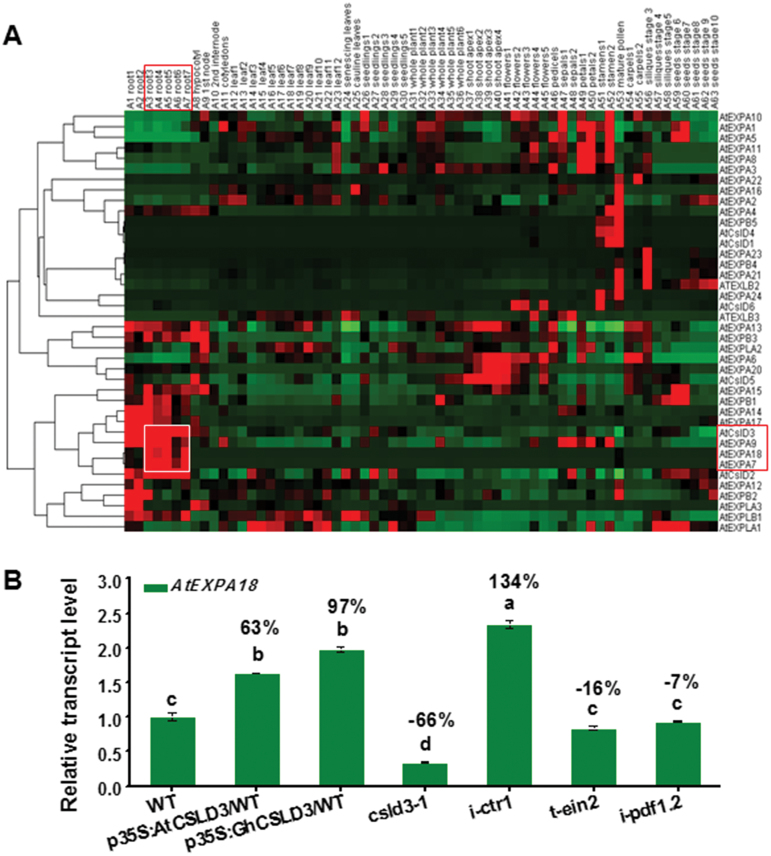
In addition, it has been reported that two root hair-specific expansin genes (encoding cell wall proteins), AtEXPA7 and AtEXPA18, may also act downstream of hormones (e.g. ethylene and auxin) to affect root hair initiation and elongation (Cho and Cosgrove, 2002; Lin et al., 2011). Here, we found AtEXPA18, and its family genes AtEXPA7 and AtEXPA9, were highly co-expressed with AtCSLD3 in the root tissues (Fig. 5A). Indeed, the expression of AtEXPA18 increased in the CSLD3-overexpressing lines and decreased in the csld3-1 mutant (Fig. 5B). However, while AtEXPA18 increased in the i-ctr1 mutant, it showed no significant changes in the t-ein2 and i-pdf1.2 mutants (Fig. 5B), which was somewhat differ to that of the expression of AtCSLD3 in young seedlings (Fig. 4A), suggesting that AtEXPA18 and AtCSLD3 may somehow work together in root-hair growth but perhaps through different pathways.
Fig. 5.
Expression patterns of AtCSLD3 and AtEXPA18. (A) Expression profile of co-expression of expansins and the CSLD gene family in Arabidopsis. (B) AtEXPA18 expression levels in the wild-type (WT), AtCSLD3-, and GhCSLD3-overexpressing lines, and the mutants csld3-1, i-ctr1, t-ein2, t-pdf1.2, and i-pdf1.2 seedlings grown on half-MS. Seedlings were grown under light conditions and were 6-d old (L6). Data are means ±SD (three biological replicates). Least-significant difference (LSD) tests wwereas used for multiple comparisons. Different letters above bars indicated that the means differ according to ANOVA and LSD tests (P<0.01). The percentage values (%) were calculated by subtraction from the WT value and divided by the WT.
CSLD3 acts downstream of the phosphate starvation and enhanced-ethylene signaling pathways in root-hair and root growth
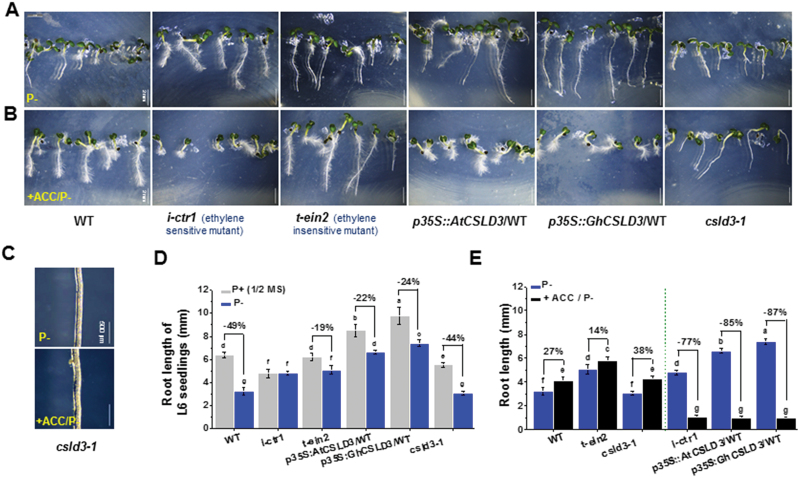
Phosphate starvation (P–) inhibits primary root growth but increases root-hair elongation (López-Arredondo et al., 2014), and we therefore hypothesized that CSLD3 might also work in the context of phosphate perception and signaling. To test this, we grew Arabidopsis seedlings from the different genotypes on P– media. This treatment led to increased root-hair tip growth of the CSLD3-overexpressing and complemented lines, all the ethylene-response mutants, and the WT, but it failed to induce root-hair tip growth in the csld3-1 mutant ((Fig. 6A, C; Supplementary Fig. S5D, E). Because ethylene plays an important role in P starvation-induced root-hair development (Song et al., 2016), we grew Arabidopsis seedlings on P– media containing 1 μM ACC and found that, indeed, the P– treatment did cause a further enhancement of root-hair growth in all the genotypes with the exception of csld3-1 (Fig. 6B, C; Supplementary Fig. S5F). These data suggest that CSLD3 works downstream of the ethylene and phosphate signaling pathways during root-hair growth in Arabidopsis.
Fig. 6.
Increased ethylene sensitivity of root elongation under phosphate starvation (P–). (A, B) Morphological phenotypes of 6-d-old seedlings grown under light conditions (L6). Seedlings grown on P– media (A) or P– co-supplied with 1 μM ACC (B). (C) A close-up view of the root hair patterns in the csld3-1 mutant. (D, E) Quantitative analyses of root lengths of seedlings as shown in (A, B). Data are means ±SD (three biological replicates), n≥30 seedlings were measured in each replicate. Least-significant difference (LSD) tests were used for multiple comparisons. Different letters above bars indicate that the means differ according to ANOVA and LSD tests (P<0.01). Scale bars indicate 2 mm in (A, B); 500 μm in (C). (This figure is available in colour at JXB online.)
We next investigated how phosphate interacted with ethylene and CSLD3 during root growth. Similar to the root-hair growth estimates, we first assessed the root growth on P– media. Here, the CSLD3-overexpressing lines and the ethylene-response mutants showed similar growth responses, which were less affected compared to the WT, the CSLD3 complemented lines, and the csld3-1 mutant plants (Fig. 6D; Supplementary Fig. S5H). We also grew the lines on P– media supplemented with ACC and measured the effects on root growth. Interestingly, here the WT, CSLD3 complemented, t-ein2 and the csld3-1 mutant lines behaved very similarly to each other, with very minor growth differences as compared to roots grown with the P– treatment (Fig. 6E; Supplementary Fig. S5I). In contrast, the root growth of the CSLD3-overexpressing lines and i-ctr1, t-pdf1.2, and i-pdf1.2 were strongly inhibited by the addition of 1 μM ACC. These data suggest that although CSLD3 is important for how roots grow in response to ethylene and phosphate, clearly PDF1.2 contributes another intersection point in this path.
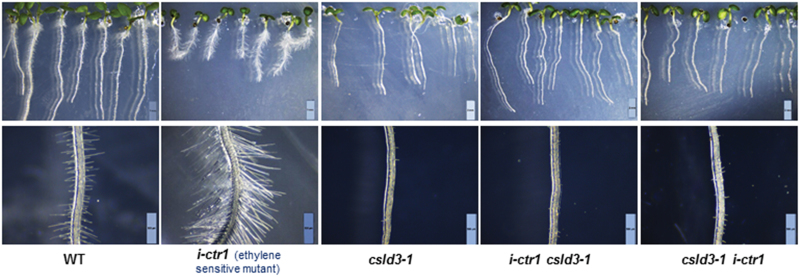
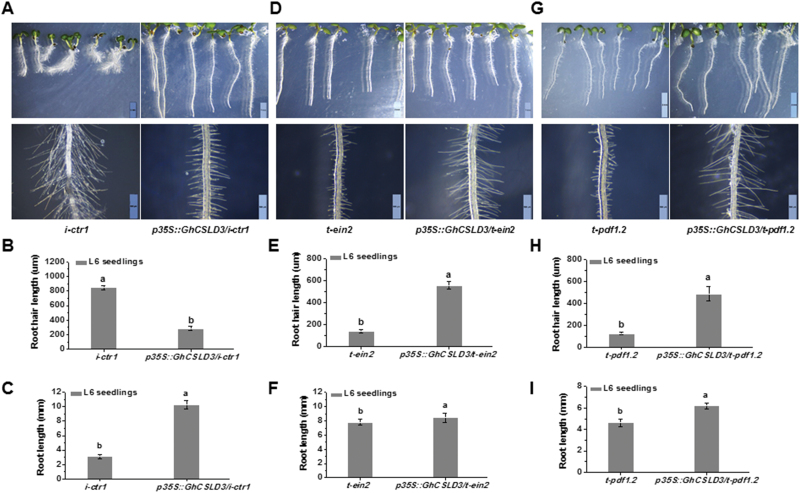
To further explore the association between the ethylene-response pathway genes and CSLD3, we constructed i-ctr1 csld3-1 and csld3-1 i-ctr1 double-mutants. Like csld3-1, the double-mutants did not produce root hairs (Fig. 7), suggesting that CSLD3 acts downstream of CTR1 in the control of root-hair tip growth. Consistent with this, overexpression of CSLD3 in the i-ctr1 mutant displayed markedly shorter root-hair length compared to the i-ctr1 mutant, leading to a similar phenotype to the WT (Fig. 8A, B). By contrast, overexpression of GhCSLD3 in the t-ein2 and t-pdf1.2 mutants led to markedly longer root-hair length (Fig. 8D, E, G, H). Moreover, all the transgenic lines showed longer root length (Fig. 8C, F, I), similar to that of the CSLD3-overexpressing lines (Fig. 2D, F). These data further support the notion that CTR1, EIN2, PDF1.2, and CSLD3 act in the same genetic pathway in the control of root and root hair elongation.
Fig. 7.
The homozygous i-ctr1 csld3-1 and csld3-1 i-ctr1 double-mutants are hairless like the csld3-1 parent. Morphological phenotypes of 9-d-old light-grown (L9) seedlings grown on half-MS media. Scale bars indicate 2 mm (top); 500 μm (bottom). (This figure is available in colour at JXB online.)
Fig. 8.
Overexpression of GhCSLD3 in the i-ctr1, t-ein2, and t-pdf1.2 mutants. (A, D, G) L6 seedlings (i.e. 6-d-old grown under light conditions) grown on half-MS media. Scale bars indicate 2 mm (top) and 500 μm (bottom). (B–I) Quantitative analyses of root-hair length (B, E, H) and root length (C, F, I) for the seedlings shown in (A, D, G). Data are means ±SD (three biological replicates); the number of seedlings measured in each replicate were: n≥20 in (B, E, H) and n≥50 in (C, F, I). Least-significant difference (LSD) tests were used for multiple comparisons. Different letters above bars indicate that the means differ according to ANOVA and LSD tests (P<0.01). (This figure is available in colour at JXB online.)
CSLD3 has positive effects on hypocotyl elongation independently of the ethylene response pathway
To investigate whether CSLD3 affected cell elongation, we also examined etiolated hypocotyl growth. Compared with the WT, CSLD3-overexpressing seedlings exhibited increased hypocotyl lengths, whereas the csld3-1 mutant had reduced lengths (Fig. 9A). Based on measurements of cells at the base of the hypocotyls (2nd to 6th cells), the CSLD3-overexpressing seedlings had much longer cells than the WT, and the csld3-1 seedlings showed significantly shorter cells (Fig. 9B, C). Because the number of epidermal cells in a single, vertical cell file (parallel to the direction of growth) is genetically fixed at approximately 20 in hypocotyl tissues of Arabidopsis (Gendreau et al., 1997), the data indicate that CSLD3 positively affects cell elongation in Arabidopsis hypocotyls.
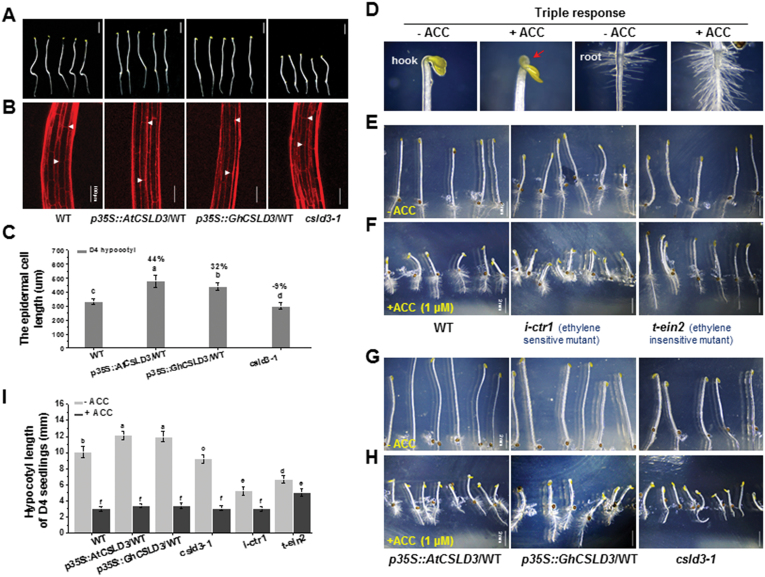
Fig. 9.
Triple response to ethylene among the wild-type (WT), CSLD3-overexpressing lines, and the csld3, i-ctr1, and t-ein2 mutants. (A) Dark-grown 4-d-old (D4) seedlings grown on half-MS media. (B) Confocal laser scanning microscopy images of the basal region of the hypocotyl (2nd to 6th cells) showing the longest epidermal cells (indicated by arrowheads) of the seedlings shown in (A). (C) Quantitative analyses of the longest epidermal cell lengths in the hypocotyls shown in (B). The percentage values (%) were calculated by subtraction from the WT value and divided by the WT. (D) Sample images of the growth characteristics of the ‘tripe response’ to ethylene. (E–H) Morphological phenotypes of D4 seedlings in the absence or presence of 1 μM ACC For the WT, i-ctr1, and t-ein2 mutants (E, (F), and the AtCSLD3- and GhCSLD3-overexpressing lines, and the csld3-1 mutant (G, H). (I) Quantitative analyses of the hypocotyl lengths in the seedlings shown in (E–H). Data are means ±SD (three biological replicates), n≥30 seedlings were measured in each replicate. Least-significant difference (LSD) tests were used for multiple comparisons. Different letters above bars indicate that the means differ according to ANOVA and LSD tests (P<0.01). Scale bars indicate 5 mm in (A); 100 μm in (B); 2 mm in (E–H). (This figure is available in colour at JXB online.)
To again put this in the context of ethylene signaling, we grew etiolated Arabidopsis seedlings on media supplemented with 1 μM ACC. All seedlings exhibited the characteristic ‘triple response’ with exaggerated apical hook curvature, and reduced hypocotyl growth (Fig. 9D–H; Guzmán and Ecker, 1990; Song et al., 2016), with the notable exception of the t-ein2 mutants. Here, the csld3-1 mutant exhibited clear hypocotyl growth inhibition in response to ACC, similar to what we observed in the CSLD3-overexpressing transgenic lines, the WT, and the i-ctr1 mutants (Fig. 9I). These data suggest that, in contrast to root-hair tip and primary root growth, the positive impact of CSLD3 on hypocotyl elongation is probably independent of the ethylene-response pathway, or at least works in parallel with it.
Changes in CSLD3 expression affect cellulose synthesis
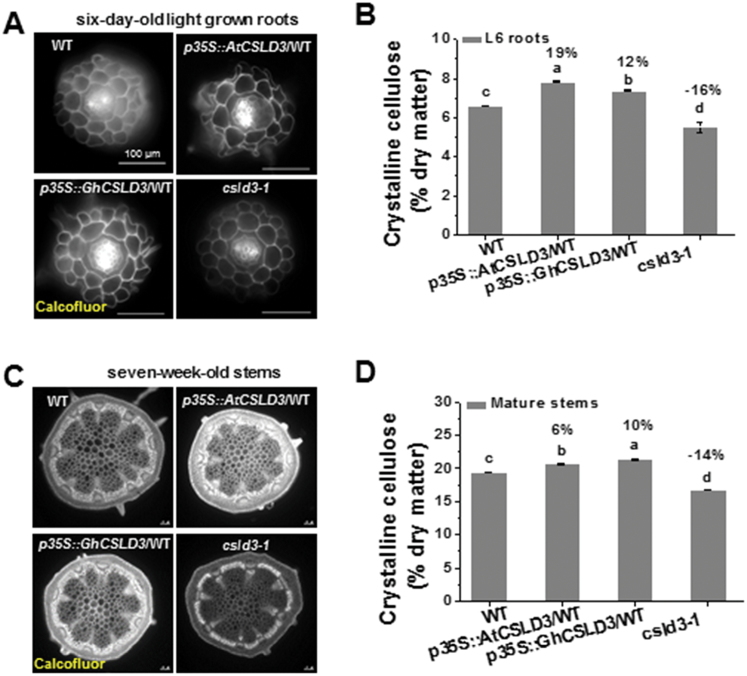
As CSLDs are reportedly associated with the biosynthesis of wall polysaccharides (xylan, pectin, and cellulose) (Bernal et al., 2007; Park et al., 2011; Verhertbruggen et al., 2011; Yin et al., 2011; Qi et al., 2013), we examined how CSLD3 impacted on the cell wall composition of L6 roots and 7-week-old stems. Using Calcofluor staining for β-glucans, we observed stronger fluorescence signals in the CSLD3-overexpressing lines than in the WT in the young roots (Fig. 10A). Chemical analyses revealed that the CSLD3-overexpressing lines had significantly increased crystalline cellulose (12–19% higher than the WT) (Fig. 10B). In contrast, the csld3-1 mutant exhibited both relatively weaker Calcofluor staining, and reduced crystalline cellulose content (16% lower than the WT) (Fig. 10A, B). Using glycan antibodies for immunolabelling wall polymers in situ, we observed similar patterns and intensities for antibodies against xyloglucan, de-esterified homogalacturonan, and rhamnogalacturonan I in the WT, the CSLD3-overexpressing lines, and the csld3-1 mutant (Supplementary Fig. S7A). Moreover, based on GC-MS analyses of neutral sugars within the total wall polysaccharides, both the CSLD3-overexpressing lines and the csld3-1 mutant showed a significant increase in rhamnose and decreases in arabinose and mannose compared to the WT (Supplementary Fig. S7B); however, these changes were more difficult to discern than the changes in cellulose.
Fig. 10.
Analyses of cellulose production in roots of 6-d-old light-grown (L6) seedlings and 7-week-old stems. (A, C) Calcofluor staining for β-glucan. Transverse section of roots (A) and stems (C). (B, D) Detection of crystalline cellulose contents for roots (B) and stems (D). WT, wild-type. Data are means ±SD (three biological replicates). Least-significant difference (LSD) tests were used for multiple comparisons. Different letters above bars indicate that the means differ according to ANOVA and LSD tests (P<0.01). The percentage values (%) were calculated by subtraction from the WT value and divided by the WT. Scale bars indicate 100 μm in (A, C).
In addition, we found that the CSLD3-overexpressing lines also had higher crystalline cellulose levels (6–10%) than the WT in 7-week-old stems, whereas the csld3-1 mutant showed reduced crystalline cellulose (14% lower than the WT) (Fig. 10C, D). Similar to seedling tissues, we did not find any differences in immunolabeling patterns in stems using xylan and de-esterified homogalacturonan antibodies (Supplementary Fig. S7C), and only slight changes in the neutral sugars within the total wall polysaccharides (Supplementary Fig. S7D). These data indicate that CSLD3 positively affects cellulose synthesis, with some moderate effects on other cell wall polymers.
Discussion
Root-hair development is influenced by many factors, such as developmental regulators, hormones, and the environment (Grierson and Schiefelbein, 2002). Ethylene has positive effects on both root-hair initiation and elongation (Tanimoto et al., 1995; Pitts et al., 1998). Although many mutants of key genes in the ethylene-signaling pathway (e.g. etr1 and ein2) affect root-hair growth (Masucci and Schiefelbein, 1996), none of them display complete inhibition of root-hair tip growth, indicating the possible existence of some predominant downstream genes that control tip growth. Moreover, little has been reported on how genes associated with cell wall synthesis and re-modelling function in root-hair development during ethylene signaling, which is surprising as root-hair growth is dependent on rapid alteration of the cell wall. Nevertheless, two root hair-specific expansin genes (AtEXPA7 and AtEXPA18; Cho and Cosgrove, 2002; Lin et al., 2011) are known to act downstream of the ethylene-signaling pathway and to impact on root-hair elongation. In this study, through use of the i-ctr1 csld3-1 double-mutant, we report that CSLD3 acts downstream of the ethylene-response pathway to control root and root-hair elongation (Fig. 7). Overexpression of GhCSLD3 restored the root-hair defects in csld3-1 (Fig. 1), which is consistent with overexpression of PdCSLD5 and PdCSLD6 (poplar orthologs to AtCSLD3) in the atcsld3 mutant reported previously (Qi et al., 2013). In addition, overexpression of either AtCSLD3 or GhCSLD3 increased the sensitivity of root and root-hair elongation to ethylene (Figs 4, 6), suggesting a role of CSLD3 in the ethylene pathway. Notably, we found similar, but also distinct, phenotypes between root hair-specific expansin and CSLD3 mutants, suggesting that those two genes may work together in root-hair growth, but perhaps via involvement in different pathways, as shown in the proposed model presented in Fig. 11. Hence, our results provide insights into root-hair growth that should inform future studies.
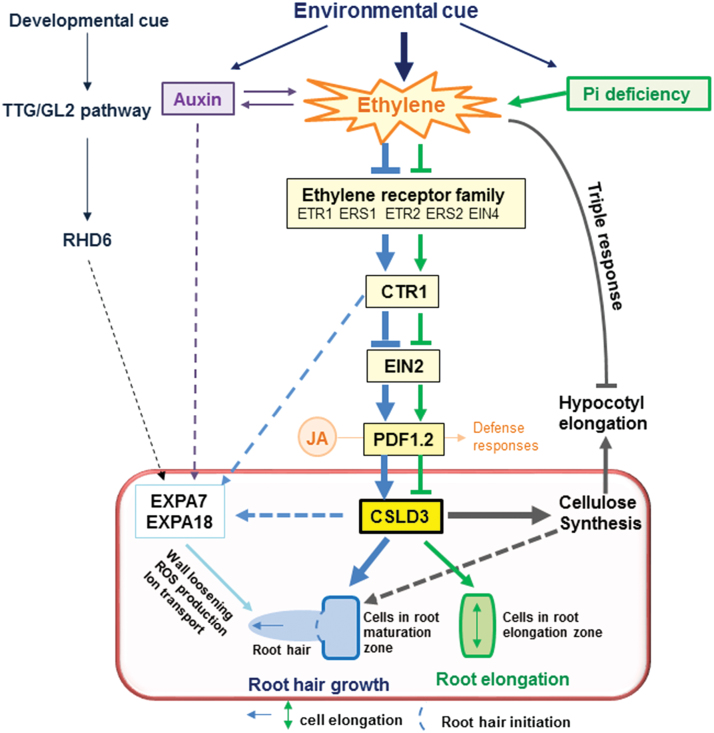
Fig. 11.
A model that illustrates how CSLD3 acts downstream of the ethylene-response pathway in root hairs and during cell elongation in Arabidopsis. CSLD3 controls root-hair growth and root elongation, probably through cellulose biosynthesis and cell wall formation. The model highlights that CSLD3-mediated root elongation is sensitive to ethylene treatment, in particular under phosphate starvation, but that CSLD3-mediated hypocotyl elongation proceeds largely independently of ethylene signaling. The model also assumes a co-ordinated effort between CSLD3 and root hair-specific expansin genes. Arrows indicate positive regulation; lines ending in bars indicate negative action; solid lines indicate that the effects are experimentally supported; dashed lines indicate hypothetical links. (This figure is available in colour at JXB online.)
Previous studies have revealed ethylene-mediated root-hair development under phosphate starvation (Song et al., 2016), but little has been reported on root elongation. In this study, we showed that cell wall changes via CSLD3 influenced how ethylene and phosphate starvation affected root elongation. In particular, when treated with ethylene, the CSLD3-overexpressing lines and ethylene-sensitive mutant i-ctr1 exhibited reduced root lengths under phosphate starvation (+ACC/P–), which was in contrast to the WT, csld3-1, and t-ein2 mutants that were insensitive, or less sensitive, to ethylene (Figs 4E, 6E). Here, the t-pdf1.2 and i-pdf1.2 mutants acted in accordance with the i-ctr1 rather than the t-ein2 mutant (Supplementary Fig. S5I), perhaps because AtPDF1.2 is also involved in other signaling pathways (e.g. jasmonate; Penninckx et al., 1998).
Mutant alleles (kjk/csld3-1/rhd) of AtCSLD3 exhibit inhibited root-hair tip growth (Favery et al., 2001; Wang et al., 2001; Galway et al., 2011), but CSLD3 is also involved in other processes such as female gametophyte development and vegetative growth (Yin et al., 2011; Yoo et al., 2012). In particular, CSLD2, CSLD3, and CSLD5 play important roles in various vegetative tissues during plant growth and development in Arabidopsis (Bernal et al., 2008; Yin et al., 2011; Yoo et al., 2012). For instance, the csld2csld5 and csld3csld5 double-mutants show disturbed cell division and cell elongation in many tissues and organs. In addition, other studies have reported that AtCSLD5 and its orthologous genes in rice and maize play a role in cell division during leaf development (Hunter et al., 2012; Yoshikawa et al., 2013; Gu et al., 2016). In this study, we provided further evidence for how CSLD3 functions in root and hypocotyl elongation. CSLD3 promoted hypocotyl elongation in an ethylene-independent manner (Fig. 9), indicating that the role of CSLD3 in hypocotyl elongation is different from that in the roots. Yin et al. (2011) reported that the csld3 mutant showed a 31% reduction in L7 root length, whereas the triple-mutant csld2/csld3/csld5 had a 70% reduction in L7 root length, and they suggested that disruption of CSLD3 may lead to a lower rate of cell division whereas both cell division and cell elongation may be disrupted in the triple-mutant. We only detected a 15% reduction in L6 root length, probably due to different plant culture conditions. In the present study, the csld3 mutant showed reduced root-cell length by 5% (Fig. 2), but the transgenic line overexpressing CSLD3 showed significantly increased cell number and length, confirming that CSLD3 could affect both cell elongation and cell division. In addition, numerous studies have demonstrated that cellulose affects cell elongation and that mutations in primary wall cellulose synthase (CesAs) typically lead to reduced cell elongation and loss of anisotropic growth (Fagard et al., 2000; Cano-Delgado et al., 2003; Chen et al., 2010, 2016; Hu et al., 2017; Li et al., 2017). Here, both AtCSLD3- and GhCSLD3-overexpressing lines exhibited significantly enhanced cellulose production and the csld3-1 mutant had decreased cellulose production in both seedlings and mature plants, which might explain the increase in cell elongation. Despite the low levels of GhCSLD3 expression in etiolated hypocotyls of cotton (Fig. 1A), the increased length of hypocotyls, roots, and root hairs in Arabidopsis that overexpressed GhCSLD3 suggests that this gene may also affect cell elongation in cotton.
The products of the CSLD enzyme remain largely controversial. Previous studies have shown that mutations of CSLD genes lead to reduced xylan, mannan, or homogalacturonan polysaccharides (Bernal et al., 2007; Li et al., 2009; Verhertbruggen et al., 2011; Yin et al., 2011). However, several studies support a role of CSLD in cellulose synthesis (Doblin et al., 2010; Park et al., 2011). Here, we analysed the cell wall composition of both CSLD3-overexpressing lines and the csld3-1 mutant in primary (young root) and secondary (mature stem) cell walls. Notably, only the cellulose levels were significantly increased in the CSLD3-overexpressing lines and, conversely, levels were reduced in the the csld3-1 mutant (Fig. 10). By comparison, other wall polysaccharides showed more complex changes in both the overexpressing transgenic lines and the mutant (Supplementary Fig. S7), suggesting that CSLD3 may be mainly involved in cellulose biosynthesis with indirect impacts on other wall polysaccharides. In addition, our findings indicate that CSLD3 might not only provide β-1,4-linked glucan synthase activity during root-hair development as previously reported (Park et al., 2011), but it may also promote cellulose synthesis in other tissues.
In conclusion, we propose a hypothetical model to show that the function of CSLD3 is essential for root-hair and root growth even if ethylene and phosphate levels are altered. However, in contrast, CSLD3-mediated hypocotyl elongation occurs largely independently of ethylene signaling (Fig. 11). Our data also provide potential connections between CSLD3 and the two root hair-specific expansin genes AtEXPA7 and AtEXPA18 in the control of root-hair elongation induced by environmental and developmental cues, as reported previously (Cho and Cosgrove, 2002; Guo and Ecker, 2004; Grierson et al., 2014; Hwang et al., 2016; Song et al., 2016).
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used for GhCSLD3 full-length cDNA cloning.
Table S2. Primers used for Q-PCR.
Table S3. Primers used for overexpression vector construction.
Fig. S1. Identification of the GhCSLD3 gene.
Fig. S2. Phenotypes of the WT, AtCSLD3, and GhCSLD3 complemented seedlings.
Fig. S3. Schematic diagram of RNAi screening (i-ctr1 and i-pdf1.2) and the T-DNA insertion mutants (t-ein2 and t-pdf1.2).
Fig. S4. Q-PCR analyses of gene expression levels using L6 seedlings grown on half-MS media.
Fig. S5. Comparison of ethylene responses among pdf1.2 mutants and CSLD3 complemented lines.
Fig. S6. Quantitative analyses of the csld3-1 mutant treated with or without 5 μM ACC.
Fig. S7. Analysis of wall polysaccharides in 6-d-old light-grown roots and 7-week-old stems.
Acknowledgements
This work was supported in part by grants from the National Natural Science Foundation of China (31571721, 31670296) and the National 111 Project (B08032). SP was funded by a R@MAP Professorship at University of Melbourne and an ARC Future Fellowship grant (FT160100218). We would like to thank Dr Mieke Van Lijsebettens (Universiteit Ghent, Belgium) for kindly providing the csld3-1 mutant. We also thank Kexing Xin, Yuyang Li, Shiguang Zhou, and Jiangfeng Huang for technical assistance with the laser scanning confocal microscope, immunofluorescent labeling experiments, and cell wall composition analysis.
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. 1999. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. [DOI] [PubMed] [Google Scholar]
- Arioli T, Peng LC, Betzner AS et al. 1998. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Beemster GT, Baskin TI. 1998. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiology 116, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal AJ, Jensen JK, Harholt J et al. 2007. Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. The Plant Journal 52, 791–802. [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Yoo CM, Mutwil M et al. 2008. Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiology 148, 1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. 2003. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439. [DOI] [PubMed] [Google Scholar]
- Bloch D, Monshausen G, Singer M, Gilroy S, Yalovsky S. 2011. Nitrogen source interacts with ROP signalling in root hair tip-growth. Plant, Cell & Environment 34, 76–88. [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A, Penfield S, Smith C, Catley M, Bevan M. 2003. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. The Plant Journal 34, 351–362. [DOI] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR. 2010. Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proceedings of the National Academy of Sciences, USA 107, 17188–17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Jia H, Zhao H, Liu D, Liu Y, Liu B, Bauer S, Somerville CR. 2016. Anisotropic cell expansion is affected through the bidirectional mobility of cellulose synthase complexes and phosphorylation at two critical residues on CESA3. Plant Physiology 171, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. 2002. Regulation of root hair initiation and expansin gene expression in Arabidopsis. The Plant Cell 14, 3237–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartini JD, Pattathil S, Avci U, Szekalski K, Mazumder K, Hahn MG, Wyman CE. 2011. Application of monoclonal antibodies to investigate plant cell wall deconstruction for biofuels production. Energy and Environment Science 4, 4332–4339. [Google Scholar]
- Doblin M, Pettolino F, Bacic A. 2010. Plant cell walls: the skeleton of the plant world. Functional Plant Biology 37, 357–381. [Google Scholar]
- Dolan L, Davies J. 2004. Cell expansion in roots. Current Opinion in Plant Biology 7, 33–39. [DOI] [PubMed] [Google Scholar]
- Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. 1996. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant Journal 10, 393–402. [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T et al. 2000. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. The Plant Cell 12, 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L. 2001. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes & Development 15, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Dolan L. 2001. Root hairs as a model system for studying plant cell growth. Annals of Botany 88, 1–7. [Google Scholar]
- Galway ME. 2006. Root hair cell walls: filling in the framework. Canadian Journal of Botany 84, 613–621. [Google Scholar]
- Galway ME, Eng RC, Schiefelbein JW, Wasteneys GO. 2011. Root hair-specific disruption of cellulose and xyloglucan in AtCSLD3 mutants, and factors affecting the post-rupture resumption of mutant root hair growth. Planta 233, 985–999. [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. 1994. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Developmental Biology 166, 740–754. [DOI] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H. 1997. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiology 114, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Nielsen E, Ketelaarc T, Schiefelbein J. 2014. Root hairs. The Arabidopsis Book 12, e0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Schiefelbein J. 2002. Root hairs. The Arabidopsis Book 1, e0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Bringmann M, Combs JR, Yang J, Bergmann DC, Nielsen E. 2016. Arabidopsis CSLD5 functions in cell plate formation in a cell cycle-dependent manner. The Plant cell 28, 1722–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR. 2004. The ethylene signaling pathway: new insights. Current Opinion in Plant Biology 7, 40–49. [DOI] [PubMed] [Google Scholar]
- Guo K, Zou W, Feng Y et al. 2014. An integrated genomic and metabolomic framework for cell wall biology in rice. BMC Genomics 15, 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. 1990. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. The Plant Cell 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Brown RM Jr, Benziman M. 1980. Calcofluor white ST alters the in vivo assembly of cellulose microfibrils. Science 210, 903–906. [DOI] [PubMed] [Google Scholar]
- Hu HH, Zhang R, Feng SQ et al. 2017. Three AtCesA6-like members enhance biomass production by distinctively promoting cell growth in Arabidopsis. Plant Biotechnology Journal. In press, doi:10.1111/pbi.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CT, Kirienko DH, Sylvester AW, Peter GF, McCarty DR, Koch KE. 2012. Cellulose Synthase-Like D1 is integral to normal cell division, expansion, and leaf development in maize. Plant Physiology 158, 708–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Lee H, Lee YS, Cho HT. 2016. Cell wall-associated ROOT HAIR SPECIFIC 10, a proline-rich receptor-like kinase, is a negative modulator of Arabidopsis root hair growth. Journal of Experimental Botany 67, 2007–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji YS, Guo HW. 2013. From endoplasmic reticulum (ER) to nucleus: EIN2 bridges the gap in ethylene signaling. Molecular Plant 6, 11–14. [DOI] [PubMed] [Google Scholar]
- Jung JY, Shin R, Schachtman DP. 2009. Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. The Plant Cell 21, 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzyk J, Devine A, McCabe PF. 2014. The root hair assay facilitates the use of genetic and pharmacological tools in order to dissect multiple signalling pathways that lead to programmed cell death. PLoS ONE 9, e94898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K. 2010. Plant cell walls. Plant Physiology 154, 483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. 1993. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Le J, Vandenbussche F, Van Der Straeten D, Verbelen JP. 2001. In the early response of Arabidopsis roots to ethylene, cell elongation is up- and down-regulated and uncoupled from differentiation. Plant Physiology 125, 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. 1999. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Lee RD, Cho HT. 2013. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Frontiers in Plant Science 4, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Xia T, Xu W et al. 2013. An integrative analysis of four CESA isoforms specific for fiber cellulose production between Gossypium hirsutum and Gossypium barbadense. Planta 237, 1585–1597. [DOI] [PubMed] [Google Scholar]
- Li FC, Xie GS, Huang JF et al. 2017. OsCESA9 conserved-site mutation leads to largely enhanced plant lodging resistance and biomass enzymatic saccharification by reducing cellulose DP and crystallinity in rice. Plant Biotechnology Journal 15, 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xiong GY, Li R, Cui J, Tang D, Zhang BC, Pauly M, Cheng ZK, Zhou YH. 2009. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. The Plant Journal 60, 1055–1069. [DOI] [PubMed] [Google Scholar]
- Li T, Lin G, Zhang X, Chen Y, Zhang S, Chen B. 2014. Relative importance of an arbuscular mycorrhizal fungus (Rhizophagus intraradices) and root hairs in plant drought tolerance. Mycorrhiza 24, 595–602. [DOI] [PubMed] [Google Scholar]
- Li W, Ma M, Feng Y, Li H, Wang Y, Ma Y, Li M, An F, Guo H. 2015. EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163, 670–683. [DOI] [PubMed] [Google Scholar]
- Lin C, Choi HS, Cho HT. 2011. Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Molecular Cell 31, 393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L. 2014. Phosphate nutrition: improving low-phosphate tolerance in crops. Annual Review of Plant Biology 65, 95–123. [DOI] [PubMed] [Google Scholar]
- Martín-Rejano EM, Camacho-Cristobal JJ, Herrera-Rodriguez MB, Rexach J, Navarro-Gochicoa MT, Gonzalez-Fontes A. 2011. Auxin and ethylene are involved in the responses of root system architecture to low boron supply in Arabidopsis seedlings. Physiologia Plantarum 142, 170–178. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. 1994. The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiology 106, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. 1996. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. The Plant Cell 8, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. 2007. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316, 1477–1480. [DOI] [PubMed] [Google Scholar]
- Müller M, Schmidt W. 2004. Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiology 134, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N. 2012. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proceedings of the National Academy of Sciences, USA 109, 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Martínez O, Pernas M, Carol RJ, Dolan L. 2007. Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317, 507–510. [DOI] [PubMed] [Google Scholar]
- Park S, Szumlanski AL, Gu F, Guo F, Nielsen E. 2011. A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nature Cell Biology 13, 973–980. [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D et al. 2010. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiology 153, 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF. 1998. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. The Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. 1998. Auxin and ethylene promote root hair elongation in Arabidopsis. The Plant Journal 16, 553–560. [DOI] [PubMed] [Google Scholar]
- Qi GQ, Hu RB, Yu L, Chai GH, Cao YP, Zuo R, Kong YZ, Zhou GK. 2013. Two poplar cellulose synthase-like D genes, PdCSLD5 and PdCSLD6, are functionally conserved with Arabidopsis CSLD3. Journal of Plant Physiology 170, 1267–1276. [DOI] [PubMed] [Google Scholar]
- Sassen MMA, Traas JA, Wolters-Arts AMC. 1985. Deposition of cellulose microfibrils in cell walls of root hairs. European Journal of Cell Biology 37, 21–26. [Google Scholar]
- Schneider R, Hanak T, Persson S, Voigt CA. 2016. Cellulose and callose synthesis and organization in focus, what’s new?Current Opinion in Plant Biology 34, 9–16. [DOI] [PubMed] [Google Scholar]
- Shin R, Berg RH, Schachtman DP. 2005. Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant & Cell Physiology 46, 1350–1357. [DOI] [PubMed] [Google Scholar]
- Singh SK, Fischer U, Singh M, Grebe M, Marchant A. 2008. Insight into the early steps of root hair formation revealed by the procuste1 cellulose synthase mutant of Arabidopsis thaliana. BMC Plant Biology 8, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G et al. 2004. Toward a systems approach to understanding plant-cell walls. Science 306, 2206–2211. [DOI] [PubMed] [Google Scholar]
- Somssich M, Khan GA, Persson S. 2016. Cell wall heterogeneity in root development of Arabidopsis. Frontiers in Plant Science 7, 1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Yu H, Dong J, Che X, Jiao Y, Liu D. 2016. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genetics 12, e1006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Ecker JR. 2000. Ethylene signaling: from mutants to molecules. Current Opinion in Plant Biology 3, 353–360. [DOI] [PubMed] [Google Scholar]
- Tanimoto M, Roberts K, Dolan L. 1995. Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. The Plant Journal 8, 943–948. [DOI] [PubMed] [Google Scholar]
- Taylor NG. 2008. Cellulose biosynthesis and deposition in higher plants. New Phytologist 178, 239–252. [DOI] [PubMed] [Google Scholar]
- Verhertbruggen Y, Yin L, Oikawa A, Scheller HV. 2011. Mannan synthase activity in the CSLD family. Plant Signaling & Behavior 6, 1620–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. 1997. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Wada Y, Kusano H, Tsuge T, Aoyama T. 2015. Phosphatidylinositol phosphate 5-kinase genes respond to phosphate deficiency for root hair elongation in Arabidopsis thaliana. The Plant Journal 81, 426–437. [DOI] [PubMed] [Google Scholar]
- Wang LQ, Guo K, Li Y, Tu YY, Hu HZ, Wang BR, Cui XC, Peng LC. 2010. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biology 10, 282–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M. 2001. AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiology 126, 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho HT. 2009. cis-Element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiology 150, 1459–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YT, Liu JY. 2004. A modified hot borate method for efficient isolation of total RNA from different cotton tissues. Cotton Science 16, 67–71. [Google Scholar]
- Xu N, Zhang W, Ren S et al. 2012. Hemicelluloses negatively affect lignocellulose crystallinity for high biomass digestibility under NaOH and H2SO4 pretreatments in Miscanthus. Biotechnology for Biofuels 5, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJ, Perry PJ, Ciani S, Pandian S, Schmidt W. 2008. Manganese deficiency alters the patterning and development of root hairs in Arabidopsis. Journal of Experimental Botany 59, 3453–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Verhertbruggen Y, Oikawa A et al. 2011. The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Molecular Plant 4, 1024–1037. [DOI] [PubMed] [Google Scholar]
- Yoo CM, Quan L, Blancaflor EB. 2012. Divergence and redundancy in CSLD2 and CSLD3 function during Arabidopsis thaliana root hair and female gametophyte development. Frontiers in Plant Science 3, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Eiguchi M, Hibara K, Ito J, Nagato Y. 2013. Rice SLENDER LEAF 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. Journal of Experimental Botany 64, 2049–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols 1, 641–646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.